Proteolytic Cleavage of Bioactive Peptides and Protease-Activated Receptors in Acute and Post-Colitis
Abstract
:1. Introduction
2. Results
2.1. Proteolytic Cleavage of Bioactive and PAR-Based Peptides by Purified Enzymes
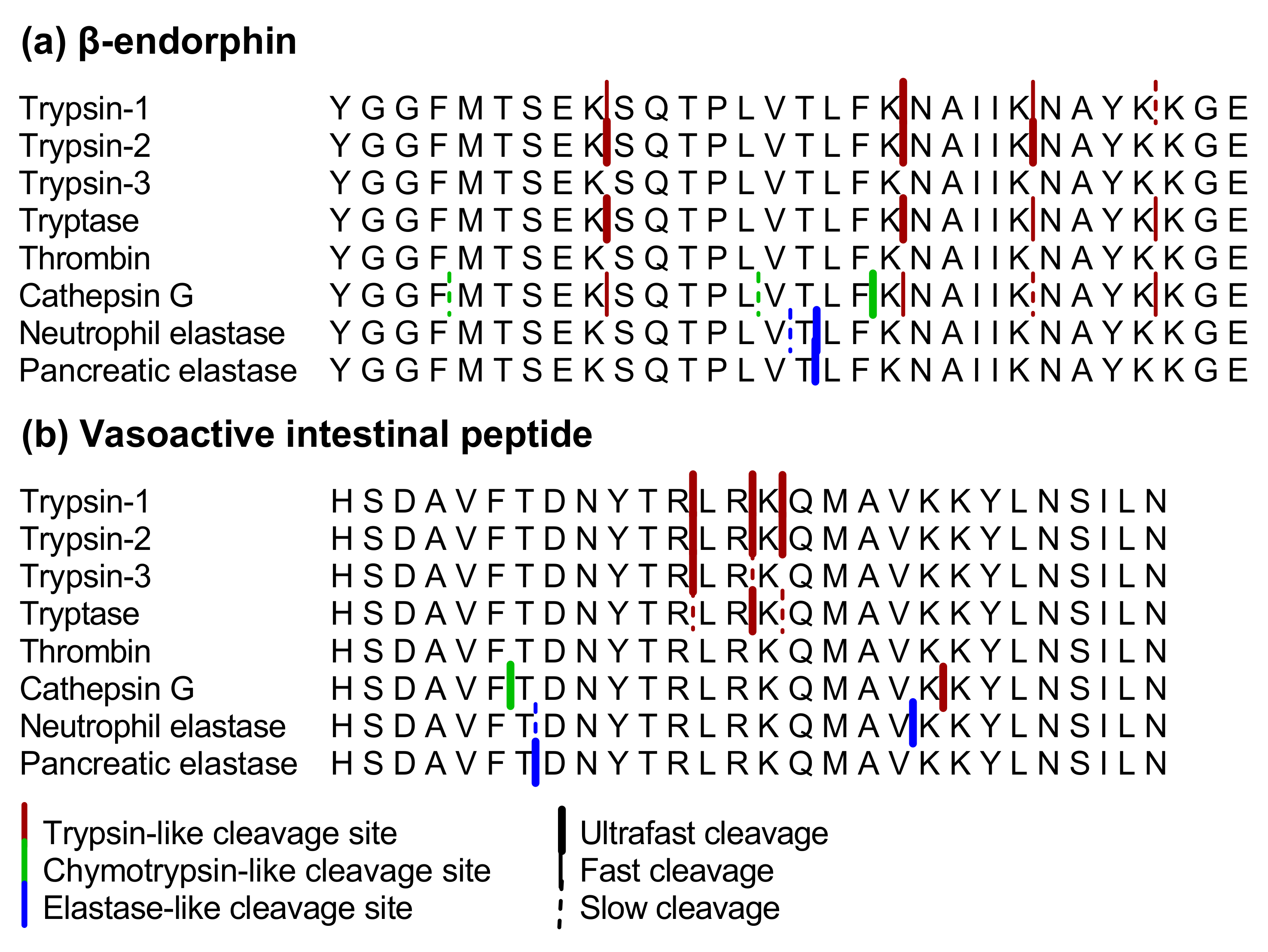
2.1.1. β-endorphin
2.1.2. Vasoactive Intestinal Peptide
2.1.3. Substance P, Bradykinin, Neurotensin, Leu-enkephalin and Met-enkephalin
2.1.4. PAR1-Based Peptide
2.1.5. PAR2-Based Peptide
2.1.6. PAR3-Based Peptide
2.1.7. PAR 4-Based Peptide
2.2. Proteolytic Cleavage of Bioactive and PAR-Based Peptides by Colonic Samples from Acute and Post-Colitis Models
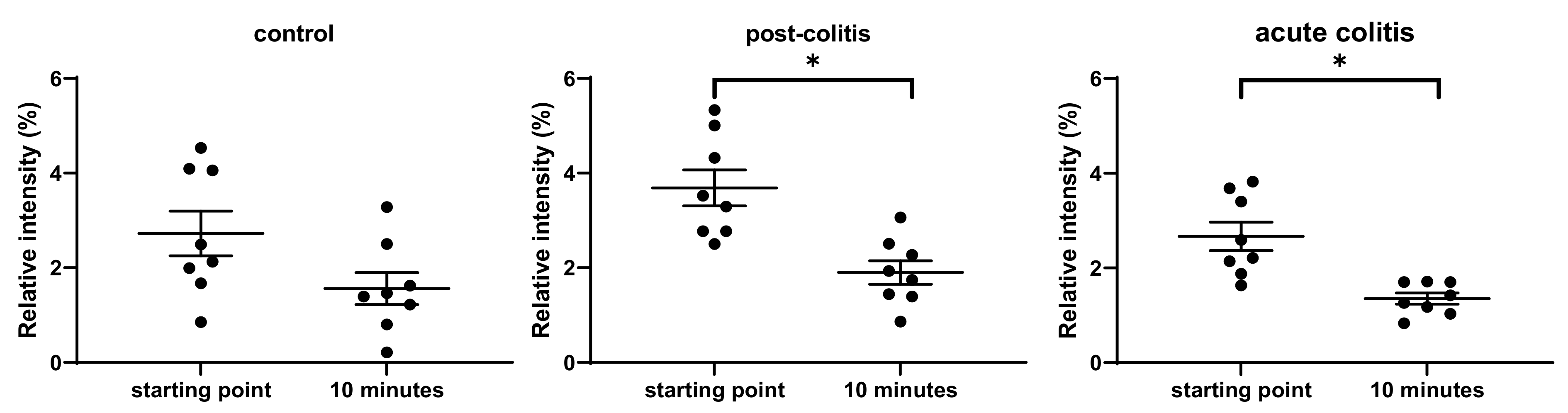
2.2.1. Decreased Processing of Neurotensin by Acute and Post-Colitis Samples
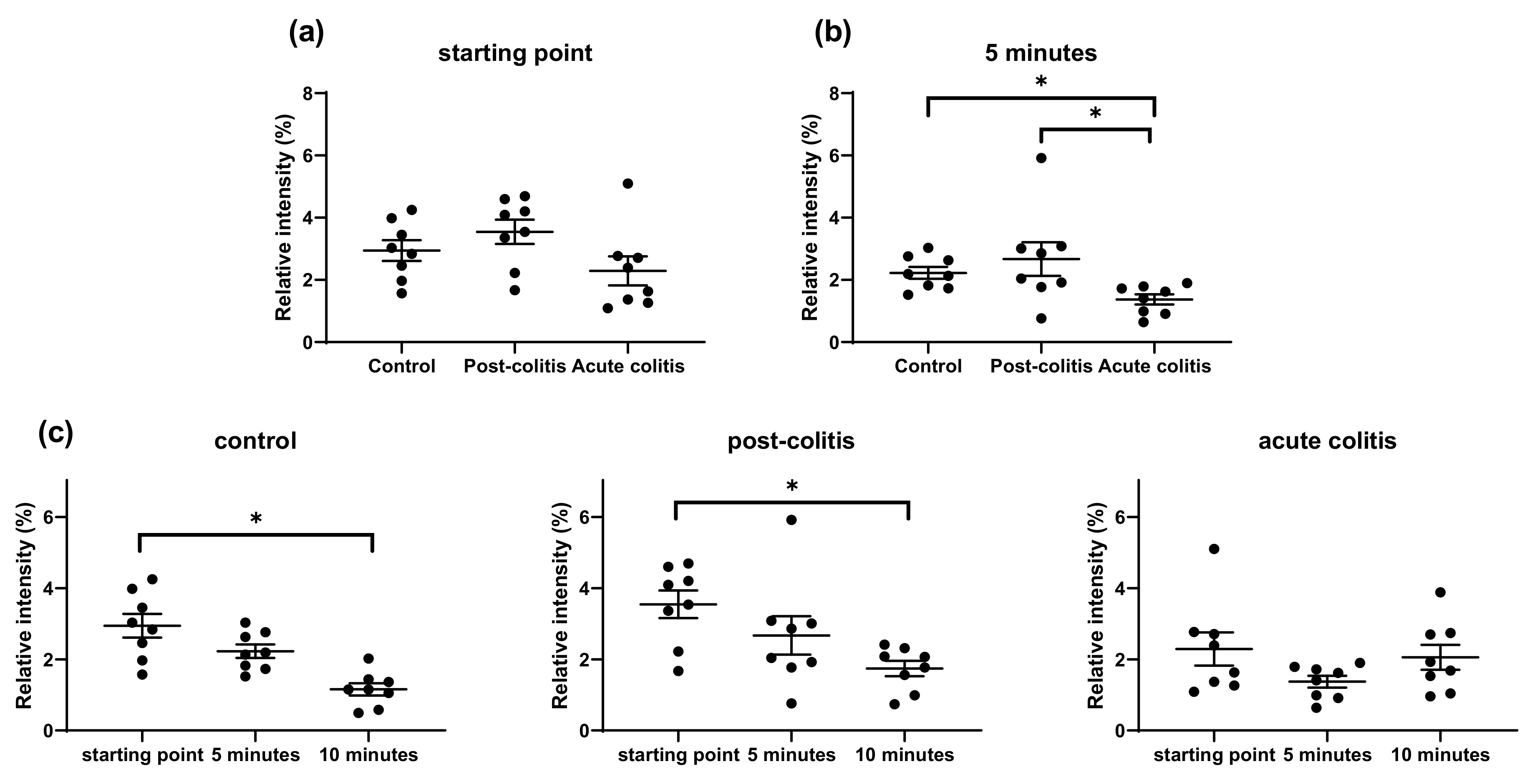
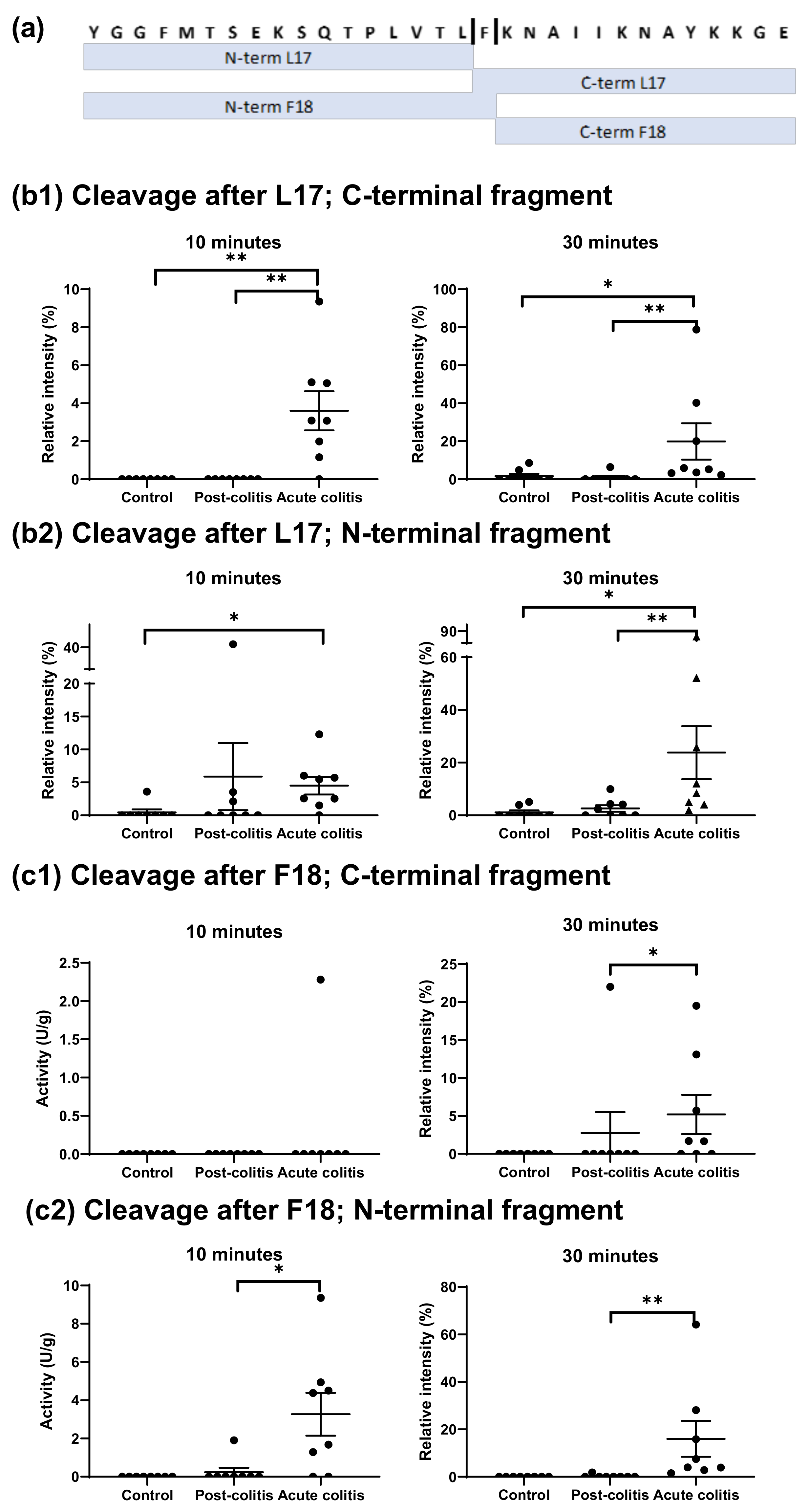
2.2.2. Increased Processing of Leu-enkephalin and β-endorphin by Acute Colitis but Not Post-Colitis Samples
2.2.3. Increased Processing of Met-enkephalin and Bradykinin by Acute Colitis and Post-Colitis Samples
2.2.4. Similar Processing of VIP and Substance P by Control, Post-Colitis and Acute Colitis Samples
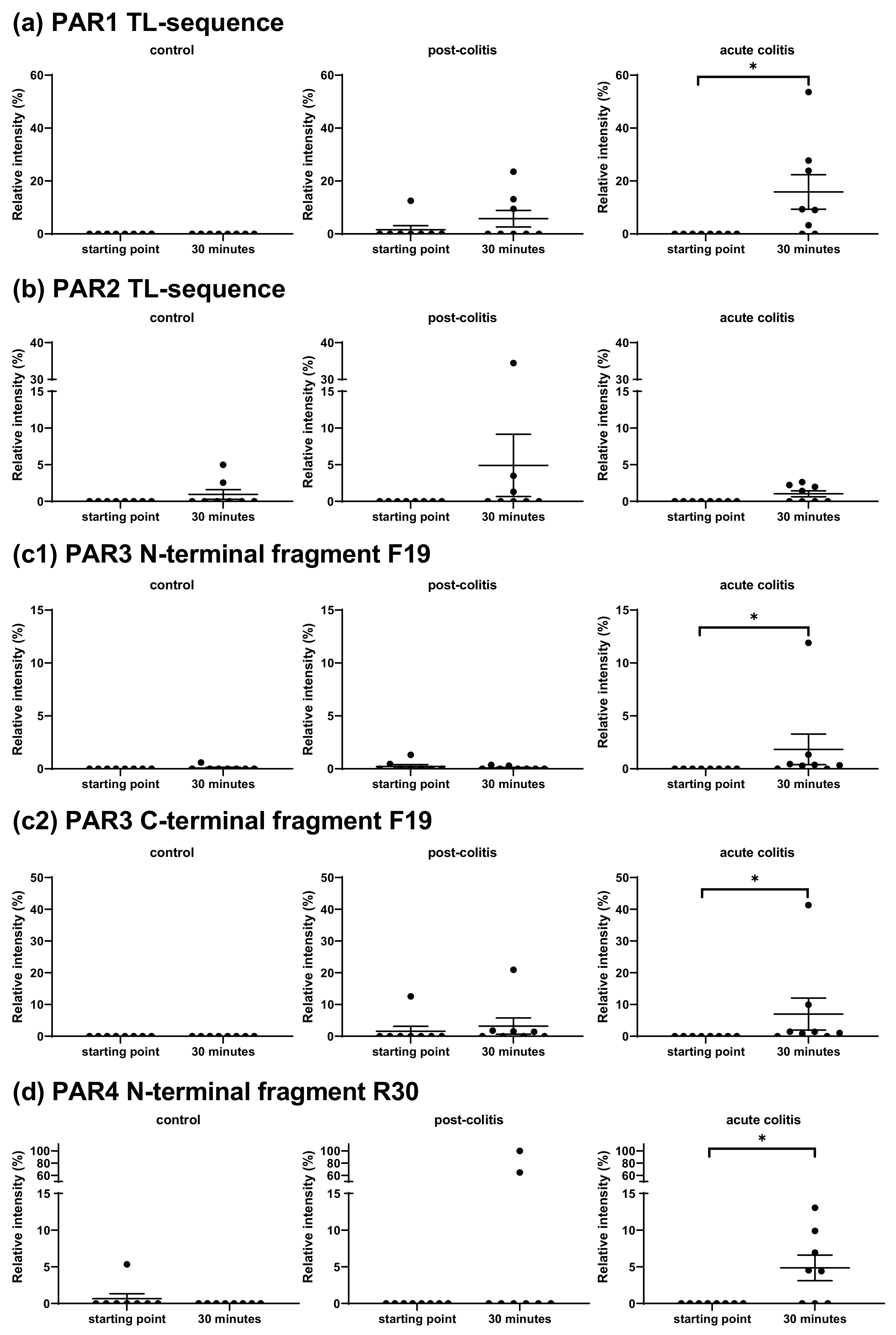
2.2.5. Activation’ or ‘Disarming’ of PAR-Based Peptides
3. Discussion
3.1. Proteolytic Cleavage of Bioactive and PAR-Based Peptides by Purified Enzymes
3.2. Proteolytic Cleavage of Bioactive and PAR-Based Peptides by Colonic Samples from Acute and Post-Colitis Models
3.2.1. Cleavage of Substance P, VIP and the PAR2-Based Peptide Is Not Different in the Acute or Post-Inflammatory Phase of TNBS-Induced Colitis
3.2.2. Cleavage of β-endorphin and Disarming of the TL-Sequence of the PAR3-Based Peptide in Acute Colitis Point to the Involvement of Chymotrypsin-Like Proteases
3.2.3. Unmasking of the TL-Sequence of the PAR1- and PAR4-Based Peptide in Acute Colitis Points to the Involvement of Thrombin
3.2.4. Cleavage of Met-enkephalin, Leu-enkephalin and Neurotensin Point to the Involvement of Proteases with Other Specificities
4. Materials and Methods
4.1. Animal Model
4.2. Preparation of Colonic Lysate
4.3. Proteolytic Activity with Fluorogenic Substrates
4.4. Proteolytic Cleavage of Natural Substrates
4.4.1. Peptides and Proteases of Interest
4.4.2. Reaction Mixtures
4.4.3. MALDI-TOF/TOF
4.4.4. Mass Spectra Data Analysis
4.5. Statistics and Figures
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Annaházi, A.; Gecse, K.; Dabek, M.; Ait-Belgnaoui, A.; Rosztóczy, A.; Róka, R.; Molnár, T.; Theodorou, V.; Wittmann, T.; Bueno, L.; et al. Fecal proteases from diarrheic-IBS and ulcerative colitis patients exert opposite effect on visceral sensitivity in mice. Pain 2009, 144, 209–217. Available online: http://www.ncbi.nlm.nih.gov/pubmed/19450926 (accessed on 27 August 2017). [CrossRef]
- Barbara, G.; Stanghellini, V.; De Giorgio, R.; Cremon, C.; Cottrell, G.S.; Santini, D.; Pasquinelli, G.; Morselli-Labate, A.M.; Grady, E.F.; Bunnett, N.W.; et al. Activated mast cells in proximity to colonic nerves correlate with abdominal pain in irritable bowel syndrome. Gastroenterology 2004, 126, 693–702. Available online: http://linkinghub.elsevier.com/retrieve/pii/S0016508503019966 (accessed on 2 August 2017). [CrossRef] [PubMed] [Green Version]
- Buhner, S.; Li, Q.; Vignali, S.; Barbara, G.; De Giorgio, R.; Stanghellini, V.; Cremon, C.; Zeller, F.; Langer, R.; Daniel, H.; et al. Activation of human enteric neurons by supernatants of colonic biopsy specimens from patients with irritable bowel syndrome. Gastroenterology 2009, 137, 1425–1434. Available online: https://www.sciencedirect.com/science/article/pii/S0016508509011524?via%3Dihub (accessed on 10 December 2018). [CrossRef]
- Cenac, N.; Andrews, C.N.; Holzhausen, M.; Chapman, K.; Cottrell, G.; Andrade-Gordon, P.; Steinhoff, M.; Barbara, G.; Beck, P.; Bunnett, N.W.; et al. Role for protease activity in visceral pain in irritable bowel syndrome. J. Clin. Investig. 2007, 117, 636–647. Available online: http://www.ncbi.nlm.nih.gov/pubmed/17304351 (accessed on 2 August 2017). [CrossRef] [PubMed] [Green Version]
- Gecse, K.; Roka, R.; Ferrier, L.; Leveque, M.; Eutamene, H.; Cartier, C.; Ait-Belgnaoui, A.; Rosztoczy, A.; Izbeki, F.; Fioramonti, J.; et al. Increased faecal serine protease activity in diarrhoeic IBS patients: A colonic lumenal factor impairing colonic permeability and sensitivity. Gut 2008, 57, 591–599. Available online: http://www.ncbi.nlm.nih.gov/pubmed/18194983 (accessed on 16 August 2017). [CrossRef]
- Róka, R.; Rosztóczy, A.; Leveque, M.; Izbéki, F.; Nagy, F.; Molnár, T.; Lonovics, J.; Garcia-Villar, R.; Fioramonti, J.; Wittman, T.; et al. A pilot study of fecal serine-protease activity: A pathophysiologic factor in diarrhea-predominant irritable bowel syndrome. Clin. Gastroenterol. Hepatol. 2017, 5, 550–555. Available online: http://www.ncbi.nlm.nih.gov/pubmed/17336590 (accessed on 16 August 2017). [CrossRef] [PubMed]
- Tooth, D.; Garsed, K.; Singh, G.; Marciani, L.; Lam, C.; Fordham, I.; Fields, A.; Banwait, R.; Lingaya, M.; Layfield, R.; et al. Characterisation of faecal protease activity in irritable bowel syndrome with diarrhoea: Origin and effect of gut transit. Gut 2013, 63, 753–760. Available online: http://www.ncbi.nlm.nih.gov/pubmed/23911555 (accessed on 15 November 2017). [CrossRef]
- Rolland-Fourcade, C.; Denadai-Souza, A.; Cirillo, C.; Lopez, C.; Jaramillo, J.O.; Desormeaux, C.; Cenac, N.; Motta, J.-P.; Larauche, M.; Tache, Y.; et al. Epithelial expression and function of trypsin-3 in irritable bowel syndrome. Gut 2017, 66, 1767–1778. Available online: http://www.ncbi.nlm.nih.gov/pubmed/28096305 (accessed on 17 August 2017). [CrossRef]
- Edogawa, S.; Edwinson, A.; Peters, S.A.; Chikkamenahalli, L.L.; Sundt, W.J.; Graves, S.; Gurunathan, S.V.; Breen-Lyles, M.K.; Johnson, S.; Dyer, R.B.; et al. Serine proteases as luminal mediators of intestinal barrier dysfunction and symptom severity in IBS. Gut 2020, 69, 62–73. [Google Scholar] [CrossRef]
- Motta, J.; Magne, L.; Descamps, D.; Rolland, C.; Squarzoni–Dale, C.; Rousset, P.; Martin, L.; Cenac, N.; Balloy, V.; Huerre, M.; et al. Modifying the protease, antiprotease pattern by elafin overexpression protects mice from colitis. Gastroenterology 2011, 140, 1272–1282. Available online: https://www.sciencedirect.com/science/article/pii/S0016508510018974?via%3Dihub (accessed on 26 July 2018). [CrossRef] [Green Version]
- Motta, J.-P.; Palese, S.; Giorgio, C.; Chapman, K.; Denadai-Souza, A.; Rousset, P.; Sagnat, D.; Guiraud, L.; Edir, A.; Seguy, C.; et al. Increased mucosal thrombin is associated with Crohn’s disease and causes inflammatory damage through protease-activated receptors activation. J. Crohns Coliti 2021, 15, 787–799. Available online: https://academic.oup.com/ecco-jcc/article/15/5/787/5985634 (accessed on 31 May 2021). [CrossRef]
- Ceuleers, H.; Hanning, N.; Heirbaut, J.; Van Remoortel, S.; Joossens, J.; Van der Veken, P.; Francque, S.M.; De Bruyn, M.; Lambeir, A.-M.; De Man, J.G.; et al. Newly developed serine protease inhibitors decrease visceral hypersensitivity in a post-inflammatory rat model for irritable bowel syndrome. Br. J. Pharmacol. 2018, 175, 3516–3533. [Google Scholar] [CrossRef] [Green Version]
- Hanning, N.; De Bruyn, M.; Ceuleers, H.; Boogaerts, T.; Berg, M.; Smet, A.; De Schepper, H.; Joossens, J.; van Nuijs, A.; De Man, J.G.; et al. Local Colonic Administration of a Serine Protease Inhibitor Improves Post-Inflammatory Visceral Hypersensitivity in Rats. Pharmaceutics 2021, 13, 811. Available online: https://www.mdpi.com/1999-4923/13/6/811 (accessed on 31 May 2021). [CrossRef] [PubMed]
- Gelbmann, C.M.; Mestermann, S.; Gross, V.; Köllinger, M.; Schölmerich, J.; Falk, W. Strictures in Crohn’s disease are characterised by an accumulation of mast cells colocalised with laminin but not with fibronectin or vitronectin. Gut 1999, 45, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Motta, J.-P.; Rolland, C.; Edir, A.; Florence, A.-C.; Sagnat, D.; Bonnart, C.; Rousset, P.; Guiraud, L.; Quaranta-Nicaise, M.; Mas, E.; et al. Epithelial production of elastase is increased in inflammatory bowel disease and causes mucosal inflammation. Mucosal Immunol. 2021, 14, 1–12. Available online: http://www.nature.com/articles/s41385-021-00375-w (accessed on 10 March 2021). [CrossRef] [PubMed]
- Jablaoui, A.; Kriaa, A.; Mkaouar, H.; Akermi, N.; Soussou, S.; Wysocka, M.; Wołoszyn, D.; Amouri, A.; Gargouri, A.; Maguin, E.; et al. Fecal serine protease profiling in inflammatory bowel diseases. Front. Cell. Infect. Microbiol. 2020, 10, 21. [Google Scholar] [CrossRef]
- Russo, F.; Chimienti, G.; Riezzo, G.; Linsalata, M.; D’Attoma, B.; Clemente, C.; Orlando, A. Adipose tissue-derived biomarkers of intestinal barrier functions for the characterization of diarrhoea-predominant IBS. Dis. Markers 2018, 2018, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Bugni, J.M.; Pthoulakis, C. Neurotensin. In Handbook of Biologically Active Peptides; Academic Press: Cambridge, MA, USA, 2013; pp. 1265–1270. [Google Scholar]
- Bednarska, O.; Walter, S.A.; Casado-Bedmar, M.; Ström, M.; Salvo-Romero, E.; Vicario, M.; Mayer, E.A.; Keita, Å.V. Vasoactive intestinal polypeptide and mast cells regulate increased passage of colonic bacteria in patients with irritable bowel syndrome. Gastroenterology 2017, 153, 948–960.e3. [Google Scholar] [CrossRef] [PubMed]
- Benarroch, E.E. Endogenous opioid systems: Current concepts and clinical correlations. Neurology 2012, 79, 807–814. [Google Scholar] [CrossRef]
- Sanger, G.J. Neurokinin NK1 and NK3 receptors as targets for drugs to treat gastrointestinal motility disorders and pain. Br. J. Pharmacol. 2004, 141, 1303–1312. [Google Scholar] [CrossRef]
- Fabisiak, A.; Sobocińska, M.; Kamysz, E.; Fichna, J.; Zielińska, M. Antinociceptive potency of enkephalins and enkephalinase inhibitors in the mouse model of colorectal distension-proof-of-concept. Chem. Biol. Drug Des. 2018, 92, 1387–1392. [Google Scholar] [CrossRef] [PubMed]
- Liddle, R. Vasoactive Intestinal Polypeptide-UpToDate. Available online: https://www.uptodate.com/contents/vasoactive-intestinal-polypeptide (accessed on 18 November 2019).
- Dothel, G.; Chang, L.; Shih, W.; Barbaro, M.R.; Cremon, C.; Stanghellini, V.; De Ponti, F.; Mayer, E.A.; Barbara, G.; Sternini, C. µ-opioid receptor, β-endorphin, and cannabinoid receptor-2 are increased in the colonic mucosa of irritable bowel syndrome patients. Neurogastroenterol. Motil. 2019, 31, e13688. [Google Scholar] [CrossRef] [PubMed]
- Szymaszkiewicz, A.; Malkiewicz, A.; Storr, M.; Fichna, J.; Zielinska, M. The place of tachykinin NK2 receptor antagonists in the treatment diarrhea-predominant irritable bowel syndrome. J. Physiol. Pharmacol. 2019, 70, 15–24. [Google Scholar]
- Del Valle-Pinero, A.Y.; Sherwin, L.B.; Henderson, W.A.; Anderson, E.M.; Caudle, R.M. Altered vasoactive intestinal peptides expression in irritable bowel syndrome patients and rats with trinitrobenzene sulfonic acid-induced colitis Basic Study. World J. Gastroenterol. 2015, 21, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Sohn, W.; Lee, O.Y.; Lee, S.P.; Lee, K.N.; Jun, D.W.; Lee, H.L.; Yoon, B.C.; Choi, H.S.; Sim, J.; Jang, K.-S. Mast cell number, substance P and vasoactive intestinal peptide in irritable bowel syndrome with diarrhea. Scand. J. Gastroenterol. 2013, 49, 43–51. [Google Scholar] [CrossRef]
- Holzer, P. Tachykinins. In Handbook of Biologically Active Peptides; Academic Press: Cambridge, MA, USA, 2013; pp. 1330–1337. [Google Scholar]
- Deiteren, A.; de Wit, A.; Van der Linden, L.; de Man, J.G.; Pelckmans, P.A.; de Winter, B.Y. Irritable bowel syndrome and visceral hypersensitivity: Risk factors and patho physiological mechanisms. Acta Gastro Enterol. Belg. 2016, 79, 29–38. [Google Scholar]
- Blackshaw, L.A.; Brierley, S.; Hughes, P. TRP channels: New targets for visceral pain. Gut 2010, 59, 126–135. [Google Scholar] [CrossRef] [Green Version]
- Vergnolle, N. Clinical relevance of proteinase activated receptors (PARs) in the gut. Gut 2005, 54, 867–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ceuleers, H.; Van Spaendonk, H.; Hanning, N.; Heirbaut, J.; Lambeir, A.-M.; Joossens, J.; Augustyns, K.; De Man, J.G.; De Meester, I.; De Winter, B.Y. Visceral hypersensitivity in inflammatory bowel diseases and irritable bowel syndrome: The role of proteases. World J. Gastroenterol. 2016, 22, 10275–10286. Available online: http://www.ncbi.nlm.nih.gov/pubmed/28058009 (accessed on 23 August 2017). [CrossRef]
- Sébert, M.; Sola-Tapias, N.; Mas, E.; Barreau, F.; Ferrand, A. Protease-activated receptors in the intestine: Focus on inflammation and cancer. Front. Endocrinol. 2019, 10, 717. [Google Scholar] [CrossRef]
- Vergnolle, N. Proteinase-activated receptors (PARs) in infection and inflammation in the gut. Int. J. Biochem. Cell Biol. 2008, 40, 1219–1227. [Google Scholar] [CrossRef]
- Zhao, P.; Metcalf, M.; Bunnett, N.W. Biased signaling of protease-activated receptors. Front. Endocrinol. 2015, 5, 67. Available online: http://www.ncbi.nlm.nih.gov/pubmed/24860547 (accessed on 25 January 2018). [CrossRef] [Green Version]
- Thorpe, M.; Fu, Z.; Chahal, G.; Akula, S.; Kervinen, J.; De Garavilla, L.; Hellman, L. Extended cleavage specificity of human neutrophil cathepsin G: A low activity protease with dual chymase and tryptase-type specificities. PLoS ONE 2018, 13, e0195077. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, T.; Minematsu, Y.; Reilly, C.F.; Travis, J.; Powers, J.C. Human leukocyte cathepsin G. Subsite mapping with 4-nitroanilides, chemical modification, and effect of possible cofactors. Biochemistry 1985, 24, 2040–2047. [Google Scholar] [CrossRef]
- Maison, C.M.; Villiers, C.; Colomb, M.G. Proteolysis of C3 on U937 cell plasma membranes. Purification of cathepsin G. J. Immunol. 1991, 147, 921–926. [Google Scholar] [PubMed]
- Polanowska, J.; Krokoszynska, I.; Czapinska, H.; Watorek, W.; Dadlez, M.; Otlewski, J. Specificity of human cathepsin G. Biochim. Biophys. Acta BBA Protein Struct. Mol. Enzym. 1998, 1386, 189–198. Available online: https://pubmed.ncbi.nlm.nih.gov/9675278/ (accessed on 16 June 2021). [CrossRef]
- Herath, H.M.D.R.; Cabot, P.J.; Shaw, P.N.; Hewavitharana, A.K. Study of beta endorphin metabolism in inflamed tissue, serum and trypsin solution by liquid chromatography-tandem mass spectrometric analysis. Anal. Bioanal. Chem. 2012, 402, 2089–2100. [Google Scholar] [CrossRef] [PubMed]
- Schilling, O.; Biniossek, M.L.; Mayer, B.; Elsässer, B.; Brandstetter, H.; Goettig, P.; Stenman, U.-H.; Koistinen, H. Specificity profiling of human trypsin-isoenzymes. Biol. Chem. 2018, 399, 997–1007. [Google Scholar] [CrossRef]
- ENZYME-3.4.21.70 Pancreatic Endopeptidase E. Available online: https://enzyme.expasy.org/EC/3.4.21.70 (accessed on 21 April 2021).
- De Oliveira, E.B.; Salgado, M.C.O. Pancreatic elastases. In Handbook of Proteolytic Enzymes; Academic Press: Cambridge, MA, USA, 2013; Volume 3, pp. 2639–2645. [Google Scholar]
- ENZYME-3.4.21.36 Pancreatic Elastase. Available online: https://enzyme.expasy.org/EC/3.4.21.36 (accessed on 21 April 2021).
- Powers, J.C.; Gupton, B.; Harley, A.; Nishino, N.; Whitley, R.J. Specificity of porcine pancreatic elastase, human leukocyte elastase and cathepsin G Inhibition with peptide chloromethyl ketones. BBA Enzymol. 1977, 485, 156–166. [Google Scholar] [CrossRef]
- ENZYME-3.4.21.71 Pancreatic Elastase II. Available online: https://enzyme.expasy.org/EC/3.4.21.71 (accessed on 21 April 2021).
- Korkmaz, B.; Gauthier, F. Elastase-2/leukocyte elastase. In Handbook of Proteolytic Enzymes; Elsevier: Amsterdam, The Netherlands, 2013; pp. 2653–2661. [Google Scholar]
- ENZYME-3.4.21.37 Leukocyte Elastase. Available online: https://enzyme.expasy.org/EC/3.4.21.37 (accessed on 21 April 2021).
- O’Donoghue, A.J.; Jin, Y.; Knudsen, G.M.; Perera, N.C.; Jenne, D.E.; Murphy, J.E.; Craik, C.S.; Hermiston, T.W. Global substrate profiling of proteases in human neutrophil extracellular traps reveals consensus motif predominantly contributed by elastase. PLoS ONE 2013, 8, e75141. [Google Scholar] [CrossRef] [Green Version]
- Knecht, W.; Cottrell, G.S.; Amadesi, S.; Mohlin, J.; Skåregärde, A.; Gedda, K.; Peterson, A.; Chapman, K.; Hollenberg, M.D.; Vergnolle, N.; et al. Trypsin IV or mesotrypsin and p23 cleave protease-activated receptors 1 and 2 to induce inflammation and hyperalgesia. J. Biol. Chem. 2007, 282, 26089–26100. [Google Scholar] [CrossRef] [Green Version]
- Dabek, M.; Ferrier, L.; Roka, R.; Gecse, K.; Annahazi, A.; Moreau, J.; Escourrou, J.; Cartier, C.; Chaumaz, G.; Leveque, M.; et al. Luminal cathepsin G and protease-activated receptor 4: A duet involved in alterations of the colonic epithelial barrier in ulcerative colitis. Am. J. Pathol. 2009, 175, 207–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sambrano, G.R.; Huang, W.; Faruqi, T.; Mahrus, S.; Craik, C.; Coughlin, S.R. Activates Protease-activated Receptor-4 in Human platelets. J. Biol. Chem. 2000, 275, 6819–6823. [Google Scholar] [CrossRef] [Green Version]
- Bishop, A.E.; Polak, J.M.; Bryant, M.G.; Bloom, S.R.; Hamilton, S. Abnormalities of vasoactive intestinal polypeptide-containing nerves in Crohn’s disease. Gastroenterology 1980, 79, 853–860. [Google Scholar] [CrossRef]
- O’Morain, C.; Bishop, E.A.; McGregor, G.P.; Levi, A.J.; Bloom, S.R.; Polak, J.M.; Peters, T.J. Vasoactive intestinal peptide concentrations and immunocytochemical studies in rectal biopsies from patients with inflammatory bowel disease. Gut 1984, 25, 57–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Surrenti, C.; Renzi, D.; Garcea, M.; Salvadori, G. Colonic vasoactive intestinal polypeptide in ulcerative colitis. J. Physiol. 1993, 87, 307–311. [Google Scholar] [CrossRef]
- Kimura, M.; Masuda, T.; Hiwatashi, N.; Toyota, T.; Nagura, H. Changes in neuropeptide-containing nerves in human colonic mucosa with inflammatory bowel disease. Pathol. Int. 1994, 44, 624–634. [Google Scholar] [CrossRef]
- Bedmar, M.T.C.; Heil, S.D.S.; Myrelid, P.; Söderholm, J.D.; Keita, Å.V. Upregulation of intestinal mucosal mast cells expressing VPAC1 in close proximity to vasoactive intestinal polypeptide in inflammatory bowel disease and murine colitis. Neurogastroenterol. Motil. 2018, 31, e13503. [Google Scholar] [CrossRef]
- Saeed, M.A.; Ng, G.Z.; Däbritz, J.; Wagner, J.; Judd, L.; Han, J.-X.; Dhar, P.; Kirkwood, C.D.; Sutton, P. Protease-activated receptor 1 plays a proinflammatory role in colitis by promoting Th17-related immunity. Inflamm. Bowel Dis. 2017, 23, 593–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bian, Z.X.; Li, Z.; Huang, Z.X.; Zhang, M.; Chen, H.L.; Xu, H.X.; Sung, J.J.Y. Unbalanced expression of protease-activated receptors-1 and -2 in the colon of diarrhea-predominant irritable bowel syndrome patients. J. Gastroenterol. 2009, 44, 666–674. [Google Scholar] [CrossRef]
- Desormeaux, C.; Bautzova, T.; Caraballo, S.C.G.; Rolland, C.; Barbaro, M.R.; Brierley, S.; Barbara, G.; Vergnolle, N.; Cenac, N. Protease-activated receptor 1 is implicated in irritable bowel syndrome mediators–induced signaling to thoracic human sensory neurons. Pain 2018, 159, 1257–1267. [Google Scholar] [CrossRef]
- Cenac, N. Protease-activated receptors as therapeutic targets in visceral pain. Curr. Neuropharmacol. 2013, 11, 598–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schick, J.; Kern, H.; Scheele, G. Hormonal stimulation in the exocrine pancreas results in coordinate and anticoordinate regulation of protein synthesis. J. Cell Biol. 1984, 99, 1569–1574. [Google Scholar] [CrossRef] [Green Version]
- Try4-Trypsin-4 Precursor-Rattus Norvegicus (Rat)-Try4 Gene & Protein. Available online: https://www.uniprot.org/uniprot/P12788 (accessed on 16 August 2021).
- Try3-Cationic Trypsin-3 Precursor-Rattus Norvegicus (Rat)-Try3 Gene & Protein. Available online: https://www.uniprot.org/uniprot/P08426 (accessed on 16 August 2021).
- Prss2-Anionic Trypsin-2 Precursor-Rattus Norvegicus (Rat)-Prss2 Gene & Protein. Available online: https://www.uniprot.org/uniprot/P00763 (accessed on 16 August 2021).
- Prss1-Anionic Trypsin-1 Precursor-Rattus Norvegicus (Rat)-Prss1 Gene & Protein. Available online: https://www.uniprot.org/uniprot/P00762 (accessed on 16 August 2021).
- F2-Prothrombin Precursor-Rattus Norvegicus (Rat)-F2 Gene & Protein. Available online: https://www.uniprot.org/uniprot/P18292 (accessed on 16 August 2021).
- F2-Prothrombin Precursor-Homo Sapiens (Human)-F2 Gene & Protein. Available online: https://www.uniprot.org/uniprot/P00734 (accessed on 16 August 2021).
- Szymaszkiewicz, A.; Storr, M.; Fichna, J.; Zielinska, M. Enkephalinase inhibitors, potential therapeutics for the future treatment of diarrhea predominant functional gastrointestinal disorders. Neurogastroenterol. Motil. 2018, 31, e13526. [Google Scholar] [CrossRef] [PubMed]
- Iwaszkiewicz, K.S.; Schneider, J.J.; Hua, S. Targeting peripheral opioid receptors to promote analgesic and anti-inflammatory actions. Front. Pharmacol. 2013, 4, 132. [Google Scholar] [CrossRef] [Green Version]
- Verma-Gandhu, M.; Verdu, E.F.; Bercik, P.; Blennerhassett, P.A.; Al-Mutawaly, N.; Ghia, J.E.; Collins, S.M. Visceral pain perception is determined by the duration of colitis and associated neuropeptide expression in the mouse. Gut 2007, 56, 358–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, H.; Morise, K.; Kusugami, K.; Furusawa, A.; Konagaya, T.; Nishio, Y.; Kaneko, H.; Uchida, K.; Nagai, H.; Mitsuma, T.; et al. Abnormal neuropeptide concentration in rectal mucosa of patients with inflammatory bowel disease. J. Gastroenterol. 1996, 31, 525–532. [Google Scholar] [CrossRef]
- Safavi, A.; Miller, B.C.; Cottam, L.; Hersh, L.B. Identification of γ-endorphin-generating enzyme as insulin-degrading enzyme. Biochemistry 1996, 35, 14318–14325. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.P.F.; Hong, H.-J.; Enberger, D.A.R.; Wong, W.D.; Buls, J.G. The presence of insulin-degrading enzyme in human ileal and colonic mucosal cells. J. Pharm. Pharmacol. 2011, 48, 1180–1184. [Google Scholar] [CrossRef]
- Kageyama, T.; Ichinose, M.; Yonezawa, S. Processing of the precursors to neurotensin and other bioactive peptides by cathepsin E. J. Biol. Chem. 1995, 270, 19135–19140. [Google Scholar] [CrossRef] [Green Version]
- Hausmann, M.; Obermeier, F.; Schreiter, K.; Spottl, T.; Falk, W.; Schölmerich, J.; Herfarth, H.; Saftig, P.; Rogler, G. Cathepsin D is up-regulated in inflammatory bowel disease macrophages. Clin. Exp. Immunol. 2004, 136, 157–167. [Google Scholar] [CrossRef]
- Menzel, K.; Hausmann, M.; Obermeier, F.; Schreiter, K.; Dunger, N.; Bataille, F.; Falk, W.; Scholmerich, J.; Herfarth, H.; Rogler, G. Cathepsins B, L and D in inflammatory bowel disease macrophages and potential therapeutic effects of cathepsin inhibition in vivo. Clin. Exp. Immunol. 2006, 146, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.-A.; Jiang, M.; Yang, X.; Liu, Y.; Guo, J.; Zheng, J.; Qu, Y.; Song, Y.; Li, R.; Qin, X.; et al. Unconjugated bilirubin ameliorates the inflammation and digestive protease increase in TNBS-induced colitis. Mol. Med. Rep. 2017, 16, 1779–1784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denadai-Souza, A.; Bonnart, C.; Tapias, N.S.; Marcellin, M.; Gilmore, B.; Alric, L.; Bonnet, D.; Schiltz, O.; Hollenberg, M.D.; Vergnolle, N.; et al. Functional proteomic profiling of secreted serine proteases in health and inflammatory bowel disease. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef]
- Van Spaendonk, H.; Ceuleers, H.; Smet, A.; Berg, M.; Joossens, J.; Van der Veken, P.; Francque, S.M.; Lambeir, A.-M.; De Man, J.G.; De Meester, I.; et al. The Effect of a Novel Serine Protease Inhibitor on Inflammation and Intestinal Permeability in a Murine Colitis Transfer Model. Front. Pharmacol. 2021, 12, 1601. [Google Scholar] [CrossRef]
- Buhner, S.; Hahne, H.; Hartwig, K.; Li, Q.; Vignali, S.; Ostertag, D.; Meng, C.; Hörmannsperger, G.; Braak, B.; Pehl, C.; et al. Protease signaling through protease activated receptor 1 mediate nerve activation by mucosal supernatants from irritable bowel syndrome but not from ulcerative colitis patients. PLoS ONE 2018, 13, e0193943. [Google Scholar] [CrossRef]
- Pal, S.; Raghav, N.; Kamboj, R.C.; Singh, H. Dipeptidyl peptidase I from goat brain: Purification, characterization and its action on leu-enkephalin. Neurochem. Int. 1993, 22, 59–68. Available online: https://pubmed.ncbi.nlm.nih.gov/8443565/ (accessed on 26 May 2021). [CrossRef]
- Rose, C.; Voisin, S.; Gros, C.; Schwartz, J.C.; Ouimet, T. Cell-specific activity of neprilysin 2 isoforms and enzymic specificity compared with neprilysin. Biochem. J. 2002, 363, 697–705. [Google Scholar] [CrossRef]
- Tanco, S.; Zhang, X.; Morano, C.; Avilés, F.X.; Lorenzo, J.; Fricker, L.D. Characterization of the substrate specificity of human carboxypeptidase A4 and implications for a role in extracellular peptide processing. J. Biol. Chem. 2010, 285, 18385–18396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyons, P.J.; Fricker, L.D. Substrate specificity of human carboxypeptidase A6. J. Biol. Chem. 2010, 285, 38234–38242. [Google Scholar] [CrossRef] [Green Version]
- Castagliuolo, I.; Wang, C.C.; Valenick, L.; Pasha, A.; Nikulasson, S.; Carraway, R.E.; Pothoulakis, C. Neurotensin is a proinflammatory neuropeptide in colonic inflammation. J. Clin. Investig. 1999, 103, 843–849. [Google Scholar] [CrossRef] [Green Version]
- Brun, P.; Mastrotto, C.; Beggiao, E.; Stefani, A.; Barzon, L.; Sturniolo, G.C.; Palù, G.; Castagliuolo, I. Neuropeptide neurotensin stimulates intestinal wound healing following chronic intestinal inflammation. Am. J. Physiol. Liver Physiol. 2005, 288, G621–G629. [Google Scholar] [CrossRef] [Green Version]
- Akcan, A.; Muhtaroglu, S.; Akgun, H.; Akyildiz, H.; Kucuk, C.; Sozuer, E.; Yurci, A.; Yilmaz, N. Ameliorative effects of bombesin and neurotensin on trinitrobenzene sulphonic acid-induced colitis, oxidative damage and apoptosis in rats. World J. Gastroenterol. 2008, 14, 1222–1230. [Google Scholar] [CrossRef]
- Deiteren, A.; De Man, J.G.; Ruyssers, N.E.; Moreels, T.G.; Pelckmans, P.A.; De Winter, B. Histamine H4 and H1 receptors contribute to postinflammatory visceral hypersensitivity. Gut 2014, 63, 1873–1882. [Google Scholar] [CrossRef]
- Deiteren, A.; van der Linden, L.; de Wit, A.; Ceuleers, H.; Buckinx, R.; Timmermans, J.-P.; Moreels, T.G.; Pelckmans, P.A.; De Man, J.G.; De Winter, B.Y. P2 × 3 receptors mediate visceral hypersensitivity during acute chemically-induced colitis and in the post-inflammatory phase via different mechanisms of sensitization. PLoS ONE 2015, 10, e0123810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, P.; Weisell, J.; Pakkala, M.; Peräkylä, M.; Zhu, L.; Koistinen, R.; Koivunen, E.; Stenman, U.-H.; Närvänen, A.; Koistinen, H. Identification of novel peptide inhibitors for human trypsins. Biol. Chem. 2010, 391, 283–293. [Google Scholar] [CrossRef]
- Koistinen, H.; Koistinen, R.; Zhang, W.-M.; Valmu, L.; Stenman, U.-H. Nexin-1 inhibits the activity of human brain trypsin. Neuroscience 2009, 160, 97–102. [Google Scholar] [CrossRef] [PubMed]
- O’Donoghue, A.J.; Eroy-Reveles, A.A.; Knudsen, G.M.; Ingram, J.; Zhou, M.; Statnekov, J.B.; Greninger, A.L.; Hostetter, D.R.; Qu, G.; Maltby, D.A.; et al. Global identification of peptidase specificity by multiplex substrate profiling. Nat. Methods 2012, 9, 1095–1100. Available online: http://www.ncbi.nlm.nih.gov/pubmed/23023596 (accessed on 28 August 2017). [CrossRef] [PubMed] [Green Version]
- Loew, D.; Perrault, C.; Morales, M.; Moog, S.; Ravanat, C.; Schuhler, S.; Arcone, S.; Pietropaolo, C.; Cazenave, J.-P.; Van Dorsselaer, A.; et al. Proteolysis of the exodomain of recombinant protease-activated receptors: Prediction of receptor activation or inactivation by MALDI mass spectrometry. Biochemistry 2000, 39, 10812–10822. [Google Scholar] [CrossRef] [PubMed]
- Strohalm, M.; Hassman, M.; Košata, B.; Kodíček, M. mMass data miner: An open source alternative for mass spectrometric data analysis. Rapid Commun. Mass Spec. 2008, 22, 905–908. [Google Scholar] [CrossRef]



| Peptide | Trypsin-1 | Trypsin-2 | Trypsin-3 | Tryptase | Thrombin | Cathepsin G | Neutrophil šElastase | Pancreatic Elastase | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β-endorphin | +++ | +++ | − | +++ | − | +++ | +++ | +++ | ||||||||
| VIP | +++ | +++ | +++ | +++ | − | +++ | +++ | +++ | ||||||||
| Substance P | + | + | + | + | + | + | N/A | N/A | ||||||||
| Neurotensin | + | + | + | + | + | + | N/A | N/A | ||||||||
| Bradykinin | + | + | + | + | + | + | N/A | N/A | ||||||||
| L-enkephalin | N/A | N/A | N/A | N/A | N/A | − | − | + | ||||||||
| M-enkephalin | N/A | N/A | N/A | N/A | N/A | − | − | + | ||||||||
| TL Unmasking or Disarming | U | D | U | D | U | D | U | D | U | D | U | D | U | D | U | D |
| PAR1-based | +++ | − | +++ | − | ++ | − | +++ | − | +++ | − | +++ | − | − | − | − | − |
| PAR2-based | +++ | − | +++ | − | − | − | +++ | − | − | − | − | − | − | + | − | − |
| PAR3-based | +++ | +++ | +++ | ++ | − | + | − | +++ | +++ | − | − | +++ | − | +++ | − | +++ |
| PAR4-based | +++ | − | +++ | − | +++ | − | − | − | +++ | − | − | − | − | − | − | − |
| Peptide of Interest | Sequence | # Amino Acids | Molecular Weight (Da) |
|---|---|---|---|
| β-endorphin | YGGFMTSEKSQTPLVTLFKNAIIKNAYKKGE | 31 | 3465.03 |
| VIP | HSDAVFTDNYTRLRKQMAVKKYLNSILN-NH2 | 28 | 3325.84 |
| Substance P | RPKPQQFFGLM-NH2 | 11 | 1347.65 |
| Neurotensin | pELYENKPRRPYIL | 13 | 1672.95 |
| Bradykinin | RPPGFSPFR | 9 | 1060.22 |
| Leu-enkephalin | YGGFL | 5 | 555.63 |
| Met-enkephalin | YGGFM | 5 | 573.67 |
| PAR1-based | ARTRARRPESKATNATLDPRSFLLRNPNDK-NH2 | 30 | 3452.0 |
| PAR2-based | TIQGTNRSSKGRSLIGKVDGTSHV-NH2 | 24 | 2497.9 |
| PAR3-based | GMENDTNNLAKPTLPIKTFRGAPPNSFE-NH2 | 28 | 3059.6 |
| PAR4-based | GGTQTPSVYDESGSTGGGDDSTPSILPAPRGYPGQVCAND-NH2 | 40 | 3911.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De bruyn, M.; Ceuleers, H.; Hanning, N.; Berg, M.; De Man, J.G.; Hulpiau, P.; Hermans, C.; Stenman, U.-H.; Koistinen, H.; Lambeir, A.-M.; et al. Proteolytic Cleavage of Bioactive Peptides and Protease-Activated Receptors in Acute and Post-Colitis. Int. J. Mol. Sci. 2021, 22, 10711. https://doi.org/10.3390/ijms221910711
De bruyn M, Ceuleers H, Hanning N, Berg M, De Man JG, Hulpiau P, Hermans C, Stenman U-H, Koistinen H, Lambeir A-M, et al. Proteolytic Cleavage of Bioactive Peptides and Protease-Activated Receptors in Acute and Post-Colitis. International Journal of Molecular Sciences. 2021; 22(19):10711. https://doi.org/10.3390/ijms221910711
Chicago/Turabian StyleDe bruyn, Michelle, Hannah Ceuleers, Nikita Hanning, Maya Berg, Joris G. De Man, Paco Hulpiau, Cedric Hermans, Ulf-Håkan Stenman, Hannu Koistinen, Anne-Marie Lambeir, and et al. 2021. "Proteolytic Cleavage of Bioactive Peptides and Protease-Activated Receptors in Acute and Post-Colitis" International Journal of Molecular Sciences 22, no. 19: 10711. https://doi.org/10.3390/ijms221910711
APA StyleDe bruyn, M., Ceuleers, H., Hanning, N., Berg, M., De Man, J. G., Hulpiau, P., Hermans, C., Stenman, U.-H., Koistinen, H., Lambeir, A.-M., De Winter, B. Y., & De Meester, I. (2021). Proteolytic Cleavage of Bioactive Peptides and Protease-Activated Receptors in Acute and Post-Colitis. International Journal of Molecular Sciences, 22(19), 10711. https://doi.org/10.3390/ijms221910711






