New Insights into Cancer Targeted Therapy: Nodal and Cripto-1 as Attractive Candidates
Abstract
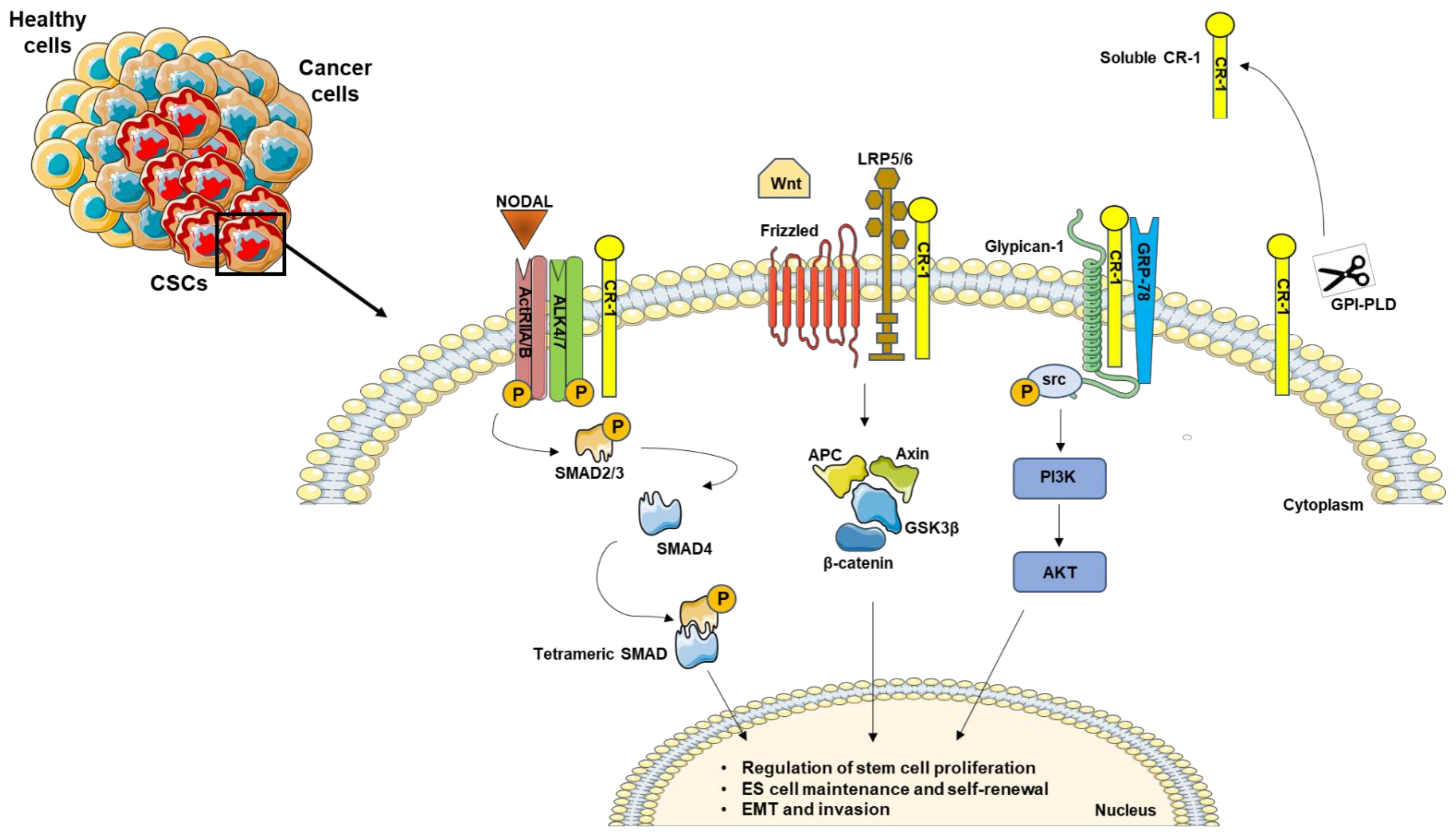
1. Nodal and Cripto-1 Signal Cascade
2. The Role of Nodal and Cripto-1 in Cancer Stem Cells
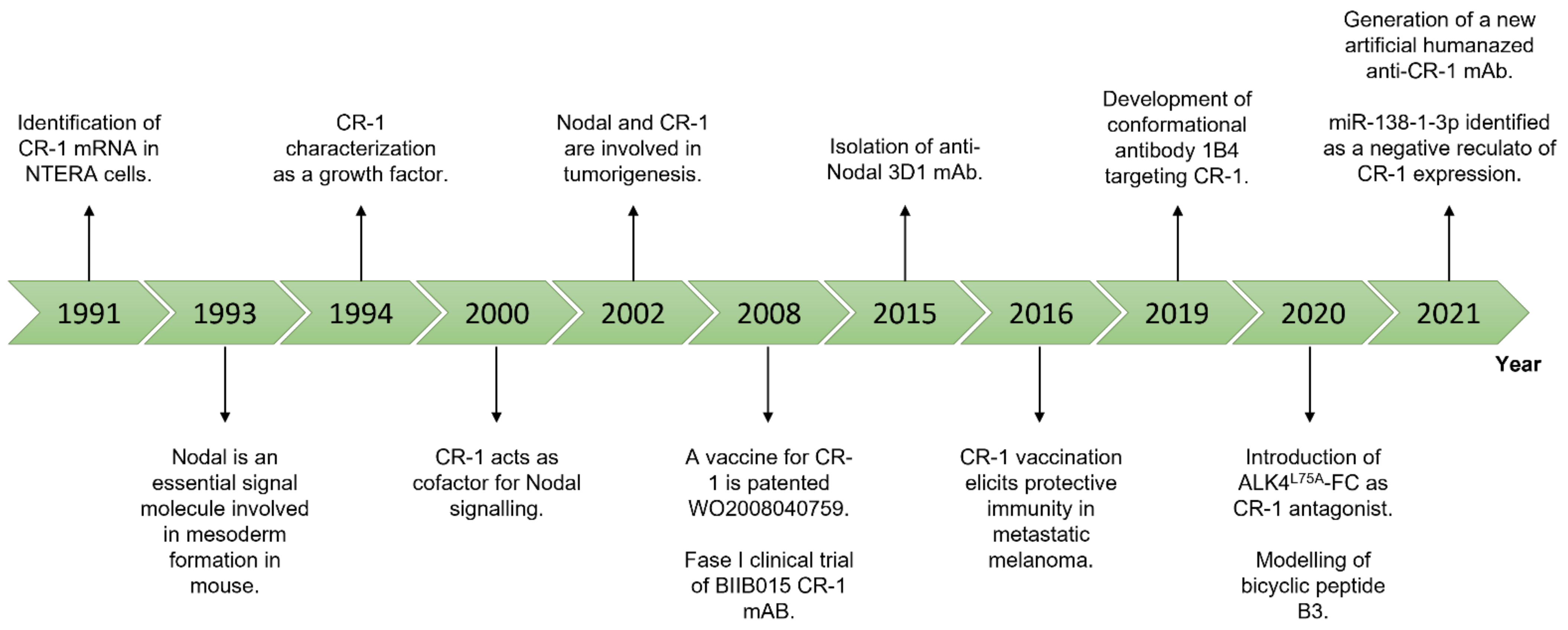
3. Nodal and Cripto-1 as Theranostic Targets
3.1. Monoclonal Antibodies
3.2. Oligonucleotides-Based Therapies: Antisense Oligos, miRNAs, and circRNAs
3.3. Small Molecule Inhibitors
3.4. Cancer Vaccines
4. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Morikawa, M.; Derynck, R.; Miyazono, K. TGF-β and the TGF-β family: Context-dependent roles in cell and tissue physiology. Cold Spring Harb. Perspect. Biol. 2016, 8, a021873. [Google Scholar] [CrossRef] [PubMed]
- Economou, A.D.; Hill, C.S. Temporal Dynamics in the Formation and Interpretation of Nodal and BMP Morphogen Gradients. In Current Topics in Developmental Biology; Academic Press Inc.: Cambridge, MA, USA, 2020; Volume 137, pp. 363–389. ISBN 9780128127902. [Google Scholar]
- Vander Ark, A.; Cao, J.; Li, X. TGF-β receptors: In and beyond TGF-β signaling. Cell. Signal. 2018, 52, 112–120. [Google Scholar] [CrossRef]
- Schier, A.F. Nodal Signaling in Vertebrate Development. Annu. Rev. Cell Dev. Biol. 2003, 19, 589–621. [Google Scholar] [CrossRef]
- Beck, S.; Le Good, J.A.; Guzman, M.; Haim, N.B.; Roy, K.; Beermann, F.; Constam, D.B. Extraembryonic proteases regulate Nodal signalling during gastrulation. Nat. Cell Biol. 2002, 4, 981–985. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.M. Nodal signaling: Development roles and regulation. Development 2007, 134, 1023–1034. [Google Scholar] [CrossRef]
- Wrana, J.L. Signaling by the TGFβ superfamily. Cold Spring Harb. Perspect. Biol. 2013, 5, 11197–11198. [Google Scholar] [CrossRef]
- Massagué, J. TGF-β signal transduction. Annu. Rev. Biochem. 1998, 67, 753–791. [Google Scholar] [CrossRef]
- Attisano, L.; Wrana, J.L. Signal transduction by the TGF-β superfamily. Science 2002, 296, 1646–1647. [Google Scholar] [CrossRef]
- Schmierer, B.; Hill, C.S. TGFβ-SMAD signal transduction: Molecular specificity and functional flexibility. Nat. Rev. Mol. Cell Biol. 2007, 8, 970–982. [Google Scholar] [CrossRef]
- Miyazawa, K.; Miyazono, K. Regulation of TGF-β family signaling by inhibitory smads. Cold Spring Harb. Perspect. Biol. 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Wang, Q. Molecular regulation of Nodal signaling during mesendoderm formation. Acta Biochim. Biophys. Sin. (Shanghai). 2018, 50, 74–81. [Google Scholar] [CrossRef]
- Strizzi, L.; Bianco, C.; Normanno, N.; Salomon, D. Cripto-1: A multifunctional modulator during embryogenesis and oncogenesis. Oncogene 2005, 24, 5731–5741. [Google Scholar] [CrossRef] [PubMed]
- Gritsman, K.; Zhang, J.; Cheng, S.; Heckscher, E.; Talbot, W.S.; Schier, A.F. The EGF-CFC protein one-eyed pinhead is essential for nodal signaling. Cell 1999, 97, 121–132. [Google Scholar] [CrossRef]
- Yan, Y.-T.; Liu, J.-J.; Luo, Y.; E, C.; Haltiwanger, R.S.; Abate-Shen, C.; Shen, M.M. Dual Roles of Cripto as a Ligand and Coreceptor in the Nodal Signaling Pathway. Mol. Cell. Biol. 2002, 22, 4439–4449. [Google Scholar] [CrossRef]
- Aykul, S.; Parenti, A.; Chu, K.Y.; Reske, J.; Floer, M.; Ralston, A.; Martinez-Hackert, E. Biochemical and cellular analysis reveals ligand binding specificities, a molecular basis for ligand recognition, and membrane association-dependent activities of cripto-1 and cryptic. J. Biol. Chem. 2017, 292, 4138–4151. [Google Scholar] [CrossRef] [PubMed]
- Liguori, G.L.; Borges, A.C.; D’Andrea, D.; Liguoro, A.; Gonçalves, L.; Salgueiro, A.M.; Persico, M.G.; Belo, J.A. Cripto-independent Nodal signaling promotes positioning of the A-P axis in the early mouse embryo. Dev. Biol. 2008, 315, 280–289. [Google Scholar] [CrossRef]
- Ciccodicola, A.; Dono, R.; Obici, S.; Simeone, A.; Zollo, M.; Graziella Persico, M. Molecular characterization of a gene of the “EGF family” expressed in undifferentiated human NTERA2 teratocarcinoma cells. EMBO J. 1989, 8, 1987–1991. [Google Scholar] [CrossRef]
- Salomon, D.S.; Bianco, C.; Ebert, A.D.; Khan, N.I.; De Santis, M.; Normanno, N.; Wechselberger, C.; Seno, M.; Williams, K.; Sanicola, M.; et al. The EGF-CFC family: Novel epidermal growth factor-related proteins in development and cancer. Endocr. Relat. Cancer 2000, 7, 199–226. [Google Scholar] [CrossRef]
- Ding, J.; Yang, L.; Yan, Y.T.; Chen, A.; Desai, N.; Wynshaw-Boris, A.; Shen, M.M. Cripto is required for correct orientation of the anterior-posterior axis in the mouse embryo. Nature 1998, 395, 702–707. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.M.; Schier, A.F. The EGF-CFC gene family in vertebrate development. Trends Genet. 2000, 16, 303–309. [Google Scholar] [CrossRef]
- Ravisankar, V.; Singh, T.P.; Manoj, N. Molecular evolution of the EGF-CFC protein family. Gene 2011, 482, 43–50. [Google Scholar] [CrossRef]
- Yeo, C.Y.; Whitman, M. Nodal signals to Smads through Cripto-dependent and Cripto-independent mechanisms. Mol. Cell 2001, 7, 949–957. [Google Scholar] [CrossRef]
- Watanabe, K.; Hamada, S.; Bianco, C.; Mancino, M.; Nagaoka, T.; Gonzales, M.; Bailly, V.; Strizzi, L.; Salomon, D.S. Requirement of glycosylphosphatidylinositol anchor of Cripto-1 for trans activity as a nodal co-receptor. J. Biol. Chem. 2007, 282, 35772–35786. [Google Scholar] [CrossRef]
- Cheng, S.K.; Olale, F.; Bennett, J.T.; Brivanlou, A.H.; Schier, A.F. EGF-CFC proteins are essential coreceptors for the TGF-β signals VG1 and GDF1. Genes Dev. 2003, 17, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Gray, P.C.; Harrison, C.A.; Vale, W. Cripto forms a complex with activin and type II activin receptors and can block activin signaling. Proc. Natl. Acad. Sci. USA 2003, 100, 5193–5198. [Google Scholar] [CrossRef] [PubMed]
- de Julio, M.K.; Alvarez, M.J.; Galli, A.; Chu, J.; Price, S.M.; Califano, A.; Shen, M.M. Regulation of extra-embryonic endoderm stem cell differentiation by Nodal and Cripto signaling. Development 2011, 138, 3885–3895. [Google Scholar] [CrossRef] [PubMed]
- Bianco, C.; Strizzi, L.; Rehman, A.; Normanno, N.; Wechselberger, C.; Sun, Y.; Khan, N.; Hirota, M.; Adkins, H.; Williams, K.; et al. Advances in Brief A Nodal- and ALK4-independent Signaling Pathway Activated by Cripto-1 through. Cancer Res. 2003, 153035, 1192–1197. [Google Scholar]
- Bianco, C.; Adkins, H.B.; Wechselberger, C.; Seno, M.; Normanno, N.; De Luca, A.; Sun, Y.; Khan, N.; Kenney, N.; Ebert, A.; et al. Cripto-1 Activates Nodal- and ALK4-Dependent and -Independent Signaling Pathways in Mammary Epithelial Cells. Mol. Cell. Biol. 2002, 22, 2586–2597. [Google Scholar] [CrossRef] [PubMed]
- Arcaro, A.; Aubert, M.; Espinosa del Hierro, M.E.; Khanzada, U.K.; Angelidou, S.; Tetley, T.D.; Bittermann, A.G.; Frame, M.C.; Seckl, M.J. Critical role for lipid raft-associated Src kinases in activation of PI3K-Akt signalling. Cell. Signal. 2007, 19, 1081–1092. [Google Scholar] [CrossRef] [PubMed]
- Quinones, Q.J.; de Ridder, G.G.; Pizzo, S.V. GRP78: A chaperone with diverse roles beyond the endoplasmic reticulum. Histol. Histopathol. 2008, 23, 1409–1416. [Google Scholar]
- Tsai, Y.L.; Zhang, Y.; Tseng, C.C.; Stanciauskas, R.; Pinaud, F.; Lee, A.S. Characterization and mechanism of stress-induced translocation of 78-kilodalton glucose-regulated protein (GRP78) to the cell surface. J. Biol. Chem. 2015, 290, 8049–8064. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.; Ribaux, P.; Epiney, M.; Irion, O. Role of prostate apoptosis response 4 in translocation of GRP78 from the endoplasmic reticulum to the cell surface of trophoblastic cells. PLoS ONE 2013, 8, e80231. [Google Scholar] [CrossRef]
- Casas, C. GRP78 at the centre of the stage in cancer and neuroprotection. Front. Neurosci. 2017, 11, 177. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Mi, X.; Song, Z.; Li, Y.; Niu, J. Cripto-1 promotes resistance to drug-induced apoptosis by activating the TAK-1/NF-κB/survivin signaling pathway. Biomed. Pharmacother. 2018, 104, 729–737. [Google Scholar] [CrossRef] [PubMed]
- Nagaoka, T.; Karasawa, H.; Castro, N.P.; Rangel, M.C.; Salomon, D.S.; Bianco, C. An evolving web of signaling networks regulated by Cripto-1. Growth Factors 2012, 30, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Lo, R.C.L.; Leung, C.O.N.; Chan, K.K.S.; Ho, D.W.H.; Wong, C.M.; Lee, T.K.W.; Ng, I.O.L. Cripto-1 contributes to stemness in hepatocellular carcinoma by stabilizing Dishevelled-3 and activating Wnt/β-catenin pathway. Cell Death Differ. 2018, 25, 1426–1441. [Google Scholar] [CrossRef] [PubMed]
- Morkel, M.; Huelsken, J.; Wakamiya, M.; Ding, J.; van de Wetering, M.; Clevers, H.; Taketo, M.M.; Behringer, R.R.; Shen, M.M.; Birchmeier, W. β-Catenin regulates Cripto- and Wnt3-dependent gene expression programs in mouse axis and mesoderm formation. Development 2003, 130, 6283–6294. [Google Scholar] [CrossRef]
- Nagaoka, T.; Karasawa, H.; Turbyville, T.; Rangel, M.C.; Castro, N.P.; Gonzales, M.; Baker, A.; Seno, M.; Lockett, S.; Greer, Y.E.; et al. Cripto-1 enhances the canonical Wnt/β-catenin signaling pathway by binding to LRP5 and LRP6 co-receptors. Cell. Signal. 2013, 25, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Pauklin, S.; Vallier, L. Activin/nodal signalling in stem cells. Development 2015, 142, 607–619. [Google Scholar] [CrossRef]
- Bonnet, D.; Dick, E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997, 3, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef]
- Ricci-Vitiani, L.; Lombardi, D.G.; Pilozzi, E.; Biffoni, M.; Todaro, M.; Peschle, C.; De Maria, R. Identification and expansion of human colon-cancer-initiating cells. Nature 2007, 445, 111–115. [Google Scholar] [CrossRef]
- O’Brien, C.A.; Pollett, A.; Gallinger, S.; Dick, J.E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 2007, 445, 106–110. [Google Scholar] [CrossRef]
- Hermann, P.C.; Huber, S.L.; Herrler, T.; Aicher, A.; Ellwart, J.W.; Guba, M.; Bruns, C.J.; Heeschen, C. Distinct Populations of Cancer Stem Cells Determine Tumor Growth and Metastatic Activity in Human Pancreatic Cancer. Cell Stem Cell 2007, 1, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Heidt, D.G.; Dalerba, P.; Burant, C.F.; Zhang, L.; Adsay, V.; Wicha, M.; Clarke, M.F.; Simeone, D.M. Identification of pancreatic cancer stem cells. Cancer Res. 2007, 67, 1030–1037. [Google Scholar] [CrossRef] [PubMed]
- Zito, G.; Richiusa, P.; Bommarito, A.; Carissimi, E.; Russo, L.; Coppola, A.; Zerilli, M.; Rodolico, V.; Criscimanna, A.; Amato, M.; et al. In vitro identification and characterization of CD133(pos)cancer stem-like cells in anaplastic thyroid carcinoma cell lines. PLoS ONE 2008, 3. [Google Scholar] [CrossRef]
- Schatton, T.; Murphy, G.F.; Frank, N.Y.; Yamaura, K.; Waaga-Gasser, A.M.; Gasser, M.; Zhan, Q.; Jordan, S.; Duncan, L.M.; Weishaupt, C.; et al. Identification of cells initiating human melanomas. Nature 2008, 451, 345–349. [Google Scholar] [CrossRef]
- Singh, S.K.; Clarke, I.D.; Terasaki, M.; Bonn, V.E.; Hawkins, C.; Squire, J.; Dirks, P.B. Identification of a Cancer Stem Cell in Human Brain Tumors. Cancer Res. 2003, 63, 5821–5828. [Google Scholar]
- Singh, S.K.; Hawkins, C.; Clarke, I.D.; Squire, J.A.; Bayani, J.; Hide, T.; Henkelman, R.M.; Cusimano, M.D.; Dirks, P.B. Identification of human brain tumour initiating cells. Nature 2004, 432, 396–401. [Google Scholar] [CrossRef]
- Wang, X.; De Julio, M.K.; Economides, K.D.; Walker, D.; Yu, H.; Halili, M.V.; Hu, Y.P.; Price, S.M.; Abate-Shen, C.; Shen, M.M. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature 2009, 461, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Collins, A.T.; Berry, P.A.; Hyde, C.; Stower, M.J.; Maitland, N.J. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005, 65, 10946–10951. [Google Scholar] [CrossRef] [PubMed]
- Prince, M.E.; Sivanandan, R.; Kaczorowski, A.; Wolf, G.T.; Kaplan, M.J.; Dalerba, P.; Weissman, I.L.; Clarke, M.F.; Ailles, L.E. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc. Natl. Acad. Sci. USA 2007, 104, 973–978. [Google Scholar] [CrossRef]
- Plaks, V.; Kong, N.; Werb, Z. The cancer stem cell niche: How essential is the niche in regulating stemness of tumor cells? Cell Stem Cell 2015, 16, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Barbato, L.; Bocchetti, M.; Di Biase, A.; Regad, T. Cancer Stem Cells and Targeting Strategies. Cells 2019, 8, 926. [Google Scholar] [CrossRef]
- Clarke, M.F. Clinical and Therapeutic Implications of Cancer Stem Cells. N. Engl. J. Med. 2019, 380, 2237–2245. [Google Scholar] [CrossRef]
- Dreesen, O.; Brivanlou, A.H. Signaling pathways in cancer and embryonic stem cells. Stem Cell Rev. 2007, 3, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Ning, F.; Wang, H.-F.; Guo, Q.; Liu, Z.-C.; Li, Z.-Q.; Du, J. Expression and significance of Nodal in human cancers: A meta-analysis. Int. J. Clin. Exp. Med. 2015, 8, 20227–20235. [Google Scholar] [PubMed]
- Mori TDGF1 is a novel predictive marker for metachronous metastasis of colorectal cancer. Int. J. Oncol. 2010, 36, 563–568. [CrossRef]
- Park, K.S.; Moon, Y.W.; Raffeld, M.; Lee, D.H.; Wang, Y.; Giaccone, G. High cripto-1 and low miR-205 expression levels as prognostic markers in early stage non-small cell lung cancer. Lung Cancer 2018, 116, 38–45. [Google Scholar] [CrossRef]
- Xu, C.H.; Sheng, Z.H.; Di Hu, H.; Hao, K.K.; Wang, Q.B.; Yu, L.K. Elevated expression of Cripto-1 correlates with poor prognosis in non-small cell lung cancer. Tumor Biol. 2014, 35, 8673–8678. [Google Scholar] [CrossRef]
- Li, P.; Sun, D.; Li, X.; He, Y.; Li, W.; Zhao, J.; Wang, Y.; Wang, H.; Xin, Y. Elevated expression of Nodal and YAP1 is associated with poor prognosis of gastric adenocarcinoma. J. Cancer Res. Clin. Oncol. 2016, 142, 1765–1773. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Meyer, M.J.; Strizzi, L.; Lee, J.M.; Gonzales, M.; Bianco, C.; Nagaoka, T.; Farid, S.S.; Margaryan, N.; Hendrix, M.J.C.; et al. Cripto-1 is a cell surface marker for a tumorigenic, undifferentiated subpopulation in human embryonal carcinoma cells. Stem Cells 2010, 28, 1303–1314. [Google Scholar] [CrossRef] [PubMed]
- Chambers, I.; Colby, D.; Robertson, M.; Nichols, J.; Lee, S.; Tweedie, S.; Smith, A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 2003, 113, 643–655. [Google Scholar] [CrossRef]
- Nichols, J.; Zevnik, B.; Anastassiadis, K.; Niwa, H.; Klewe-Nebenius, D.; Chambers, I.; Schöler, H.; Smith, A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 1998, 95, 379–391. [Google Scholar] [CrossRef]
- Park, S.-W.; Do, H.-J.; Han, M.-H.; Choi, W.; Kim, J.-H. The expression of the embryonic gene Cripto-1 is regulated by OCT4 in human embryonal carcinoma NCCIT cells. FEBS Lett. 2018, 592, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Cui, X.; Yu, X.; Bian, B.-S.-J.; Qian, F.; Hu, X.; Ji, C.; Yang, L.; Ren, Y.; Cui, W.; et al. Cripto-1 acts as a functional marker of cancer stem-like cells and predicts prognosis of the patients in esophageal squamous cell carcinoma. Mol. Cancer 2017, 16, 81. [Google Scholar] [CrossRef]
- Alam, M.J.; Takahashi, R.; Afify, S.M.; Oo, A.K.K.; Kumon, K.; Nawara, H.M.; Khayrani, A.C.; Du, J.; Zahra, M.H.; Seno, A.; et al. Exogenous cripto-1 suppresses self-renewal of cancer stem cell model. Int. J. Mol. Sci. 2018, 19, 3345. [Google Scholar] [CrossRef]
- Francescangeli, F.; Contavalli, P.; De Angelis, M.L.; Baiocchi, M.; Gambara, G.; Pagliuca, A.; Fiorenzano, A.; Prezioso, C.; Boe, A.; Todaro, M.; et al. Dynamic regulation of the cancer stem cell compartment by Cripto-1 in colorectal cancer. Cell Death Differ. 2015, 22, 1700–1713. [Google Scholar] [CrossRef]
- James, D.; Levine, A.J.; Besser, D.; Hemmati-Brivanlou, A. TGFβ/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development 2005, 132, 1273–1282. [Google Scholar] [CrossRef]
- Gordeeva, O. TGFβ Family Signaling Pathways in Pluripotent and Teratocarcinoma Stem Cells’ Fate Decisions: Balancing between Self-Renewal, Differentiation, and Cancer. Cells 2019, 8, 1500. [Google Scholar] [CrossRef]
- Gong, W.; Sun, B.; Sun, H.; Zhao, X.; Zhang, D.; Liu, T.; Zhao, N.; Gu, Q.; Dong, X.; Liu, F. Nodal signaling activates the Smad2/3 pathway to regulate stem cell-like properties in breast cancer cells. Am. J. Cancer Res. 2017, 7, 503–517. [Google Scholar]
- Yang, J.; Jiang, W. The Role of SMAD2/3 in Human Embryonic Stem Cells. Front. Cell Dev. Biol. 2020, 8, 653. [Google Scholar] [CrossRef]
- Vallier, L.; Mendjan, S.; Brown, S.; Ching, Z.; Teo, A.; Smithers, L.E.; Trotter, M.W.B.; Cho, C.H.H.; Martinez, A.; Rugg-Gunn, P.; et al. Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. Development 2009, 136, 1339–1349. [Google Scholar] [CrossRef]
- Harpelunde Poulsen, K.; Nielsen, J.E.; Grønkær Toft, B.; Joensen, U.N.; Rasmussen, L.J.; Blomberg Jensen, M.; Mitchell, R.T.; Juul, A.; Rajpert-De Meyts, E.; Jørgensen, A. Influence of Nodal signalling on pluripotency factor expression, tumour cell proliferation and cisplatin-sensitivity in testicular germ cell tumours. BMC Cancer 2020, 20. [Google Scholar] [CrossRef] [PubMed]
- Lonardo, E.; Hermann, P.C.; Mueller, M.T.; Huber, S.; Balic, A.; Miranda-Lorenzo, I.; Zagorac, S.; Alcala, S.; Rodriguez-Arabaolaza, I.; Ramirez, J.C.; et al. Nodal/activin signaling drives self-renewal and tumorigenicity of pancreatic cancer stem cells and provides a target for combined drug therapy. Cell Stem Cell 2011, 9, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Guo, Y.; Hai, Y.; Yang, H.; Liu, Y.; Yang, S.; Zhang, Z.; Ma, M.; Liu, L.; Li, Z.; et al. Nodal promotes the self-renewal of human colon cancer stem cells via an autocrine manner through Smad2/3 signaling pathway. Biomed Res. Int. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Cave, D.D.; Hernando-Momblona, X.; Sevillano, M.; Minchiotti, G.; Lonardo, E. Nodal-induced L1CAM/CXCR4 subpopulation sustains tumor growth and metastasis in colorectal cancer derived organoids. Theranostics 2021, 11, 5686–5699. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, I.B.; Joe, A. Oncogene addiction. Cancer Res. 2008, 68, 3077–3080. [Google Scholar] [CrossRef]
- Al-Lazikani, B.; Banerji, U.; Workman, P. Combinatorial drug therapy for cancer in the post-genomic era. Nat. Biotechnol. 2012, 30, 679–692. [Google Scholar] [CrossRef]
- Sandomenico, A.; Ruvo, M. Targeting Nodal and Cripto-1: Perspectives Inside Dual Potential Theranostic Cancer Biomarkers. Curr. Med. Chem. 2018, 26, 1994–2050. [Google Scholar] [CrossRef]
- Xu, C.H.; Wang, Y.; Qian, L.H.; Yu, L.K.; Zhang, X.W.; Wang, Q.B. Serum Cripto-1 is a novel biomarker for non-small cell lung cancer diagnosis and prognosis. Clin. Respir. J. 2017, 11, 765–771. [Google Scholar] [CrossRef] [PubMed]
- Spiller, C.M.; Lobo, J.; Boellaard, W.P.A.; Gillis, A.J.M.; Bowles, J.; Looijenga, L.H.J. Cripto and MIR-371A-3P are serum biomarkers of testicular germ cell tumors and are detected in seminal plasma from azoospermic males. Cancers (Basel) 2020, 12, 760. [Google Scholar] [CrossRef]
- Xue, Y.J.; Chen, S.N.; Chen, W.G.; Wu, G.Q.; Liao, Y.F.; Xu, J.B.; Tang, H.; Yang, S.H.; He, S.Y.; Luo, Y.F.; et al. Cripto-1 expression in patients with clear cell renal cell carcinoma is associated with poor disease outcome. J. Exp. Clin. Cancer Res. 2019, 38, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Bianco, C.; Strizzi, L.; Mancino, M.; Rehman, A.; Hamada, S.; Watanabe, K.; De Luca, A.; Jones, B.; Balogh, G.; Russo, J.; et al. Identification of Cripto-1 as a novel serologic marker for breast and colon cancer. Clin. Cancer Res. 2006, 12, 5158–5164. [Google Scholar] [CrossRef]
- De Angelis, E.; Grassi, M.; Gullick, W.J.; Johnson, G.R.; Rossi, G.B.; Tempesta, A.; De Angelis, F.; De Luca, A.; Salomon, D.S.; Normanno, N. Expression of cripto and amphiregulin in colon mucosa from high risk colon cancer families. Int. J. Oncol. 1999, 14, 437–440. [Google Scholar] [CrossRef] [PubMed]
- Adkins, H.B.; Bianco, C.; Schiffer, S.G.; Rayhorn, P.; Zafari, M.; Cheung, A.E.; Orozco, O.; Olson, D.; De Luca, A.; Chen, L.L.; et al. Antibody blockade of the Cripto CFC domain suppresses tumor cell growth in vivo. J. Clin. Investig. 2003, 112, 575–587. [Google Scholar] [CrossRef]
- Focà, G.; Iaccarino, E.; Focà, A.; Sanguigno, L.; Untiveros, G.; Cuevas-Nunez, M.; Strizzi, L.; Leonardi, A.; Ruvo, M.; Sandomenico, A. Development of conformational antibodies targeting Cripto-1 with neutralizing effects in vitro. Biochimie 2019, 158, 246–256. [Google Scholar] [CrossRef]
- Hu, X.F.; Xing, P.X. Cripto monoclonal antibodies. Drug News Perspect. 2005, 18, 293–303. [Google Scholar] [PubMed]
- Iaccarino, E.; Calvanese, L.; Untiveros, G.; Falcigno, L.; D’Auria, G.; Latino, D.; Sivaccumar, J.P.; Strizzi, L.; Ruvo, M.; Sandomenico, A. Structure-based design of small bicyclic peptide inhibitors of Cripto-1 activity. Biochem. J. 2020, 477, 1391–1407. [Google Scholar] [CrossRef]
- Balcioglu, O.; Heinz, R.E.; Freeman, D.W.; Gates, B.L.; Hagos, B.M.; Booker, E.; Mirzaei Mehrabad, E.; Diesen, H.T.; Bhakta, K.; Ranganathan, S.; et al. CRIPTO antagonist ALK4L75A-Fc inhibits breast cancer cell plasticity and adaptation to stress. Breast Cancer Res. 2020, 22, 125. [Google Scholar] [CrossRef]
- Ishii, H.; Zahra, M.H.; Takayanagi, A.; Seno, M. A Novel Artificially Humanized Anti-Cripto-1 Antibody Suppressing Cancer Cell Growth. Int. J. Mol. Sci. 2021, 22, 1709. [Google Scholar] [CrossRef] [PubMed]
- Karches, C.H.; Benmebarek, M.-R.; Schmidbauer, M.L.; Kurzay, M.; Klaus, R.; Geiger, M.; Rataj, F.; Cadilha, B.L.; Lesch, S.; Heise, C.; et al. Bispecific Antibodies Enable Synthetic Agonistic Receptor-Transduced T Cells for Tumor Immunotherapy. Clin. Cancer Res. 2019, 25, 5890–5900. [Google Scholar] [CrossRef]
- Focà, A.; Sanguigno, L.; Focà, G.; Strizzi, L.; Iannitti, R.; Palumbo, R.; Hendrix, M.J.C.; Leonardi, A.; Ruvo, M.; Sandomenico, A. New anti-nodal monoclonal antibodies targeting the nodal pre-helix loop involved in cripto-1 binding. Int. J. Mol. Sci. 2015, 16, 21342–21362. [Google Scholar] [CrossRef]
- Strizzi, L.; Sandomenico, A.; Margaryan, N.V.; Focà, A.; Sanguigno, L.; Bodenstine, T.M.; Chandler, G.S.; Reed, D.W.; Gilgur, A.; Seftor, E.A.; et al. Effects of a novel Nodal-targeting monoclonal antibody in melanoma. Oncotarget 2015, 6, 34071–34086. [Google Scholar] [CrossRef]
- Calvanese, L.; Focà, A.; Sandomenico, A.; Focà, G.; Caporale, A.; Doti, N.; Iaccarino, E.; Leonardi, A.; D’Auria, G.; Ruvo, M.; et al. Structural insights into the interaction of a monoclonal antibody and Nodal peptides by STD-NMR spectroscopy. Bioorganic Med. Chem. 2017, 25, 6589–6596. [Google Scholar] [CrossRef]
- Kelly, R.K.; Olson, D.L.; Sun, Y.; Wen, D.; Wortham, K.A.; Antognetti, G.; Cheung, A.E.; Orozco, O.E.; Yang, L.; Bailly, V.; et al. An antibody–cytotoxic conjugate, BIIB015, is a new targeted therapy for Cripto positive tumours. Eur. J. Cancer 2011, 47, 1736–1746. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, A.F.; ten Dijke, P.; Zhu, H.J. On-Target Anti-TGF-β Therapies Are Not Succeeding in Clinical Cancer Treatments: What Are Remaining Challenges? Front. Cell Dev. Biol. 2020, 8, 605. [Google Scholar] [CrossRef] [PubMed]
- Ciardiello, D.; Elez, E.; Tabernero, J.; Seoane, J. Clinical development of therapies targeting TGFβ: Current knowledge and future perspectives. Ann. Oncol. 2020, 31, 1336–1349. [Google Scholar] [CrossRef] [PubMed]
- Ciardiello, F.; Tortora, G.; Bianco, C.; Selvam, M.P.; Basolo, F.; Fontanini, G.; Pacifico, F.; Normanno, N.; Brandt, R.; Persico, M.G.; et al. Inhibition of CRIPTO expression and tumorigenicity in human colon cancer cells by antisense RNA and oligodeoxynucleotides. Oncogene 1994, 9, 291–298. [Google Scholar] [PubMed]
- De Luca, A.; Selvam, M.P.; Sandomemco, C.; Pepe, S.; Bianco, A.R.; Ciardiello, F.; Salomon, D.S.; Normanno, N. Anti-sense oligonucleotides directed against EGF-related growth factors enhance anti-proliferative effect of conventional anti-tumor drugs in human colon-cancer cells. Int. J. Cancer 1997, 73, 277–282. [Google Scholar] [CrossRef]
- Normanno, N.; Bianco, C.; Damiano, V.; De Angelis, E.; Selvam, M.P.; Grassi, M.; Magliulo, G.; Tortora, G.; Bianco, A.R.; Mendelsohn, J.; et al. Growth inhibition of human colon carcinoma cells by combinations of anti-epidermal growth factor-related growth factor antisense oligonucleotides. Clin. Cancer Res. 1996, 2, 601–609. [Google Scholar]
- De Luca, A.; Arra, C.; D’Antonio, A.; Casamassimi, A.; Losito, S.; Ferraro, P.; Ciardiello, F.; Salomon, D.S.; Normanno, N. Simultaneous blockade of different EGF-like growth factors results in efficient growth inhibition of human colon carcinoma xenografts. Oncogene 2000, 19, 5863–5871. [Google Scholar] [CrossRef]
- Normanno, N.; Tortora, G.; De Luca, A.; Pomatico, G.; Casamassimi, A.; Agrawal, S.; Mendelsohn, J.; Bianco, A.R.; Ciardiello, F. Synergistic growth inhibition and induction of apoptosis by a novel mixed backbone antisense oligonucleotide targeting CRIPTO in combination with C225 anti-EGFR monoclonal antibody and 8-Cl-cAMP in human GEO colon cancer cells. Oncol. Rep. 1999, 6, 1105–1109. [Google Scholar] [CrossRef]
- Casamassimi, A.; De Luca, A.; Agrawal, S.; Stromberg, K.; Salomon, D.S.; Normanno, N. EGF-related antisense oligonucleoltides inhibit the proliferation of human ovarian carcinoma cells. Ann. Oncol. 2000, 11, 319–325. [Google Scholar] [CrossRef]
- Baldassarre, G.; Bianco, C.; Tortora, G.; Ruggiero, A.; Moasser, M.; Dmitrovsky, E.; Bianco, A.R.; Ciardiello, F. Transfection with a CRIPTO anti-sense plasmid suppresses endogenous CRIPTO expression and inhibits transformation in a human embryonal carcinoma cell line. Int. J. Cancer 1996, 66, 538–543. [Google Scholar] [CrossRef]
- Du, T.; Jiang, J.; Chen, Y.; Zhang, N.; Chen, G.; Wang, X.; Long, X.; Feng, X. MiR-138-1-3p alters the stemness and radiosensitivity of tumor cells by targeting CRIPTO and the JAK2/STAT3 pathway in nasopharyngeal carcinoma. Ann. Transl. Med. 2021, 9, 485. [Google Scholar] [CrossRef]
- Yun, S.; Yun, C.W.; Lee, J.H.; Kim, S.; Lee, S.H. Cripto enhances proliferation and survival of mesenchymal stem cells by up-regulating JAK2/STAT3 pathway in a GRP78-dependent manner. Biomol. Ther. 2018, 26, 464–473. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Yan, S.S.; Shi, L.; Wan, Z.Q.; Jiang, N.; Fu, L.S.; Li, M.; Guo, J. MicroRNA-15b suppresses the growth and invasion of glioma cells through targeted inhibition of cripto-1 expression. Mol. Med. Rep. 2016, 13, 4897–4903. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Hou, S.K.; Fan, H.J.; Liu, Y.F. MiR-15a-16 represses Cripto and inhibits NSCLC cell progression. Mol. Cell. Biochem. 2014, 391, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Rengganaten, V.; Huang, C.J.; Tsai, P.H.; Wang, M.L.; Yang, Y.P.; Lan, Y.T.; Fang, W.L.; Soo, S.; Ong, H.T.; Cheong, S.K.; et al. Mapping a circular RNA–microRNA–mRNA-signaling regulatory axis that modulates stemness properties of cancer stem cell populations in colorectal cancer spheroid cells. Int. J. Mol. Sci. 2020, 21, 7864. [Google Scholar] [CrossRef]
- Shi, Y.; Bao, Y.L.; Wu, Y.; Yu, C.L.; Huang, Y.X.; Sun, Y.; Zheng, L.H.; Li, Y.X. Alantolactone inhibits cell proliferation by interrupting the interaction between Cripto-1 and activin receptor type II A in activin signaling pathway. J. Biomol. Screen. 2011, 16, 525–535. [Google Scholar] [CrossRef]
- Inman, G.J.; Nicolás, F.J.; Callahan, J.F.; Harling, J.D.; Gaster, L.M.; Reith, A.D.; Laping, N.J.; Hill, C.S. SB-431542 is a potent and specific inhibitor of transforming growth factor-β superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 2002, 62, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Saxena, M.; van der Burg, S.H.; Melief, C.J.M.; Bhardwaj, N. Therapeutic cancer vaccines. Nat. Rev. Cancer 2021, 21, 360–378. [Google Scholar] [CrossRef] [PubMed]
- Bianco, C.; Salomon, D.S. Human Cripto-1 as a target for a cancer vaccine: WO2008040759. Expert Opin. Ther. Pat. 2009, 19, 141–144. [Google Scholar] [CrossRef] [PubMed]
- Witt, K.; Ligtenberg, M.A.; Conti, L.; Lanzardo, S.; Ruiu, R.; Wallmann, T.; Tufvesson-Stiller, H.; Chambers, B.J.; Rolny, C.; Lladser, A.; et al. Cripto-1 plasmid DNA vaccination targets metastasis and cancer stem cells in murine mammary carcinoma. Cancer Immunol. Res. 2018, 6, 1417–1425. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arboretto, P.; Cillo, M.; Leonardi, A. New Insights into Cancer Targeted Therapy: Nodal and Cripto-1 as Attractive Candidates. Int. J. Mol. Sci. 2021, 22, 7838. https://doi.org/10.3390/ijms22157838
Arboretto P, Cillo M, Leonardi A. New Insights into Cancer Targeted Therapy: Nodal and Cripto-1 as Attractive Candidates. International Journal of Molecular Sciences. 2021; 22(15):7838. https://doi.org/10.3390/ijms22157838
Chicago/Turabian StyleArboretto, Paola, Michele Cillo, and Antonio Leonardi. 2021. "New Insights into Cancer Targeted Therapy: Nodal and Cripto-1 as Attractive Candidates" International Journal of Molecular Sciences 22, no. 15: 7838. https://doi.org/10.3390/ijms22157838
APA StyleArboretto, P., Cillo, M., & Leonardi, A. (2021). New Insights into Cancer Targeted Therapy: Nodal and Cripto-1 as Attractive Candidates. International Journal of Molecular Sciences, 22(15), 7838. https://doi.org/10.3390/ijms22157838





