Genome-Wide Identification, Structure Characterization, and Expression Pattern Profiling of the Aquaporin Gene Family in Betula pendula
Abstract
1. Introduction
2. Results and Discussion
2.1. General Considerations
2.2. Genome-Wide Identification, Diversity and Evolutionary Analysis of Betula Aquaporins
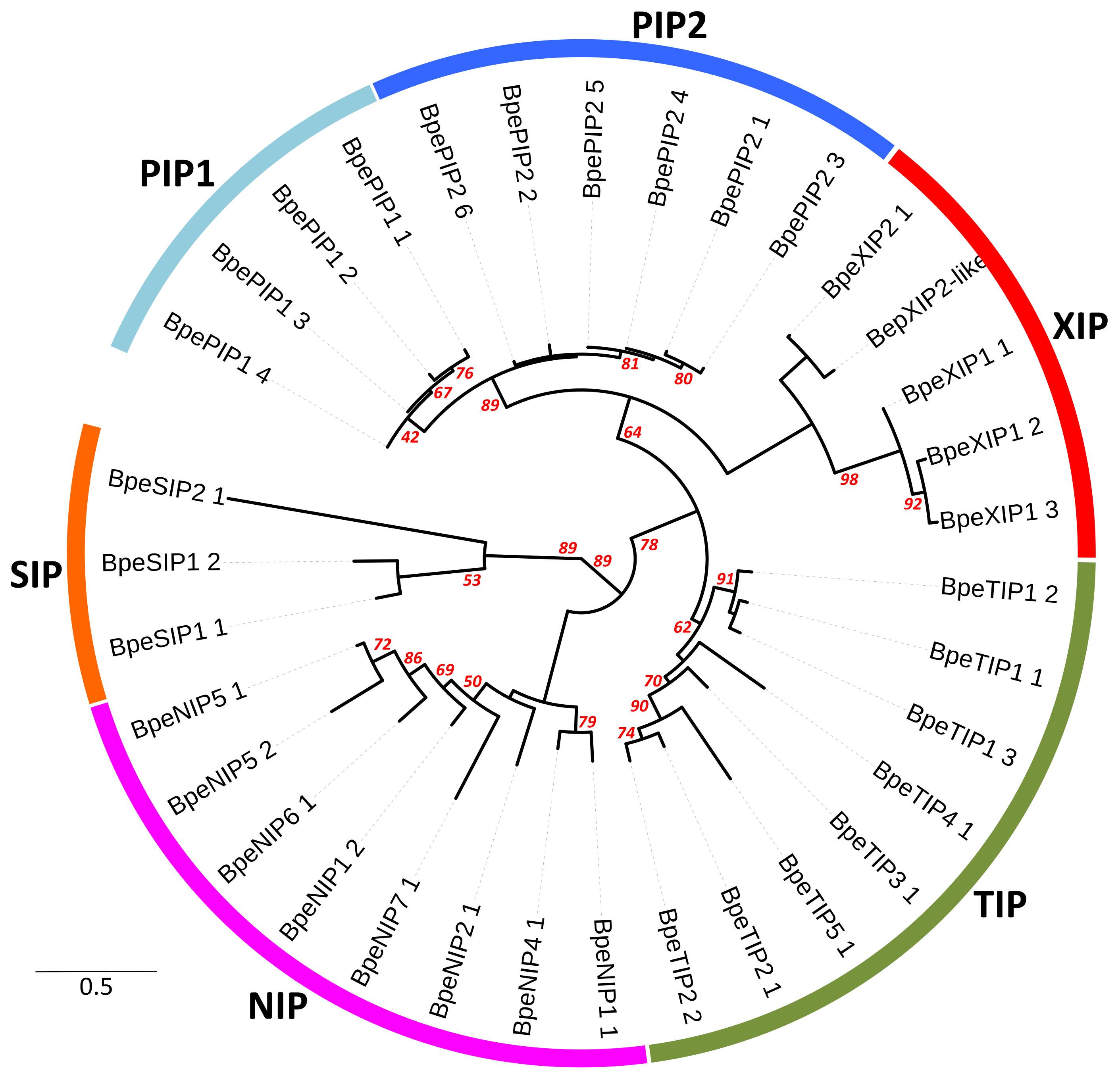
2.2.1. Genetic Structure of BpeAQP Subfamilies
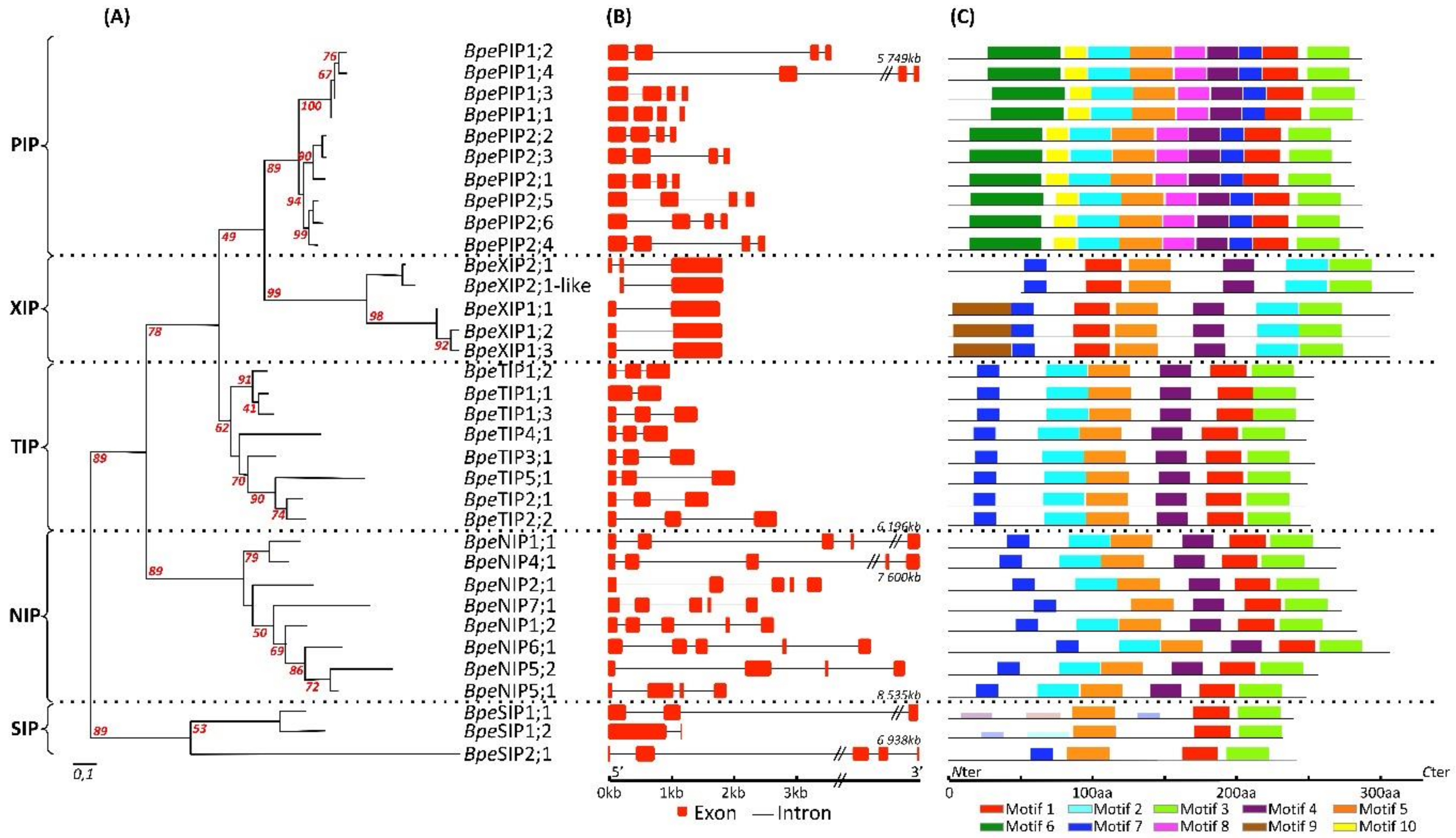
2.2.2. Genome Structure of BpeAQP Genes Models
2.2.3. Expertizing Sequence Discrepancies for Few AQP Candidates
2.3. Protein Transporter Structure Analysis, and Conserved Substrate-Specific Residues of Betula AQP
2.3.1. Protein and Pore 2D and 3D Basic Structure Organizations of BpeAQPs
2.3.2. Sequence Structure and Protein Function Relationship of BpeAQPs
2.3.3. Conserved Substrate-Specific Residue, and Solute Permeability of BpeAQPs per Subfamily
PIP Subfamily
TIP Subfamily
XIP Subfamily
NIP Subfamily
SIP Subfamily
2.3.4. Subcellular Localization Prediction of BpeAQPs
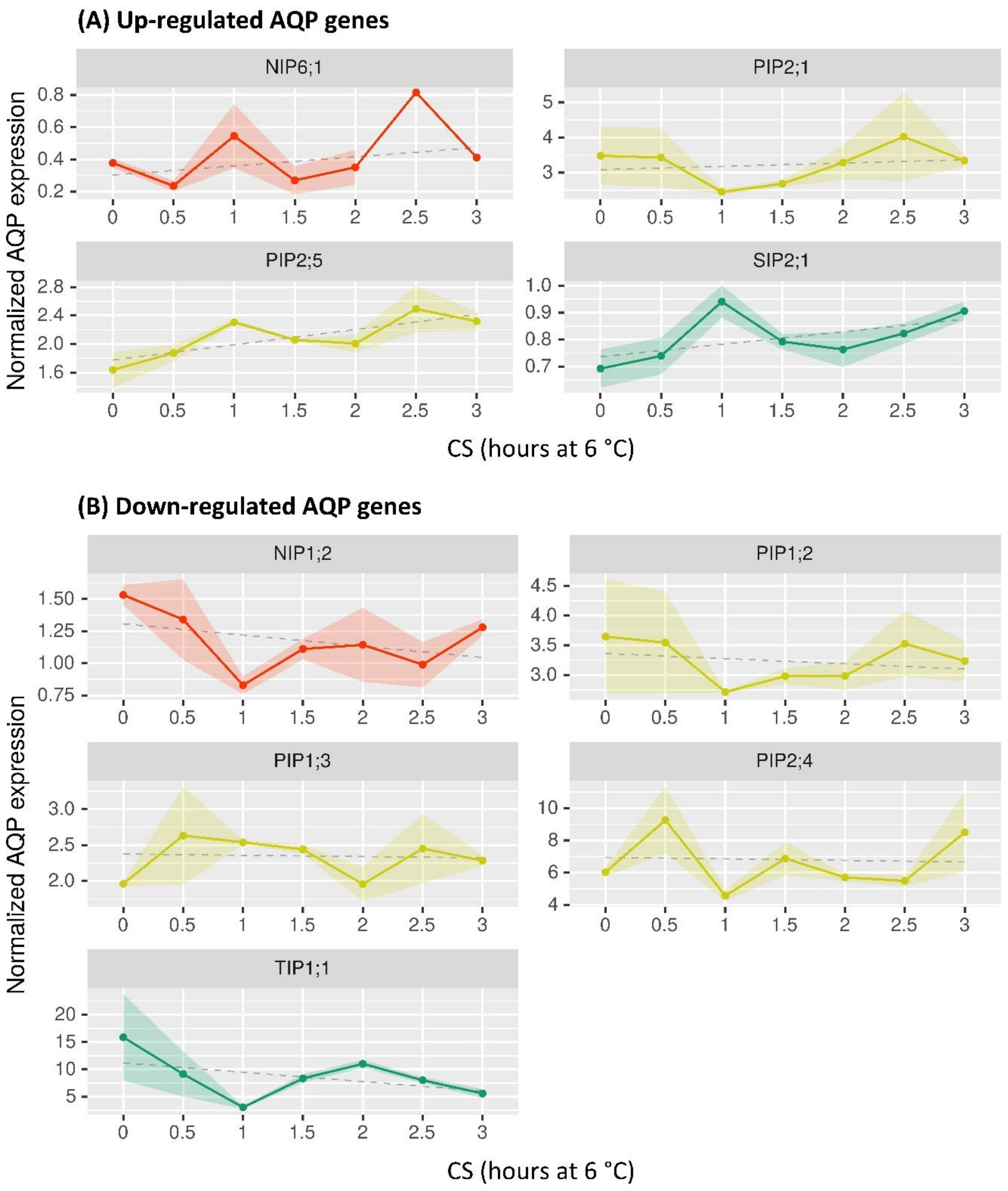
2.4. Profiling of BpeAQP Transcriptome
3. Material and Methods
3.1. Identification of BpeAQP Family Members and Subfamily Classification
3.2. Bioinformatics Analysis
3.3. Construction of Homology-Based Tertiary Protein Structure
3.4. Differential Expression Profile of BpeAQP Gene Family (RNA-Seq Analysis)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AQP | Aquaporin |
| ar/R | “Aromatic/arginine” selectivity filter |
| FPs | Froger’s positions |
| GIP | GlpF-like intrinsic protein |
| Gly | Glycine |
| GRAVY | Grand average of hydropathicity |
| HIP | Hybrid intrinsic protein |
| H2O2 | Hydrogen peroxide |
| LIP | Large intrinsic Protein |
| MW | Molecular weight |
| NH3 | Ammonia |
| NIP | Nodulin 26-like intrinsic protein |
| NPA | “Asparagine–proline–alanine” motif |
| pI | Theoretical isoelectric point |
| PIP | Plasma membrane intrinsic protein |
| Si | Silicic acid |
| SIP | Small basic intrinsic protein |
| TIP | Tonoplast intrinsic protein |
| TMH | Trans-alpha helical transmembrane region |
| TMM | Trimmed mean of M-values |
| XIP | X-intrinsic protein |
References
- Gomes, D.; Agasse, A.; Thiébaud, P.; Delrot, S.; Gerós, H.; Chaumont, F. Aquaporins are multifunctional water and solute transporters highly divergent in living organisms. Biochim. Biophys. Acta 2009, 1788, 1213–1228. [Google Scholar] [CrossRef]
- Johanson, U.; Karlsson, M.; Johansson, I.; Gustavsson, S.; Sjövall, S.; Fraysse, L.; Weig, A.R.; Kjellbom, P. The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol. 2001, 126, 1358–1369. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Yang, S.; Wang, Y.; Bendahmane, M.; Fu, X. Genome-wide identification and characterization of aquaporin gene family in Beta vulgaris. PeerJ 2017, 5, e3747. [Google Scholar] [CrossRef]
- Reddy, P.S.; Rao, T.S.R.B.; Sharma, K.K.; Vadez, V. Genome-wide identification and characterization of the aquaporin gene family in Sorghum bicolor (L.). Plant Gene 2015, 1, 18–28. [Google Scholar] [CrossRef]
- Faize, M.; Fumanal, B.; Luque, F.; Ramírez-Tejero, J.; Zou, Z.; Qiao, X.; Faize, L.; Gousset-Dupont, A.; Roeckel-Drevet, P.; Label, P.; et al. Genome wild analysis and molecular understanding of the aquaporin diversity in olive trees (Olea Europaea, L.). Int. J. Mol. Sci. 2020, 11, 4183. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.B.; Sankararamakrishnan, R. Genome-wide analysis of major intrinsic proteins in the tree plant Populus trichocarpa: Characterization of XIP subfamily of aquaporins from evolutionary perspective. BMC Plant Biol. 2009, 9, 134. [Google Scholar] [CrossRef] [PubMed]
- Park, W.; Scheffler, B.E.; Bauer, P.J.; Campbell, B.T. Identification of the family of aquaporin genes and their expression in upland cotton (Gossypium hirsutum L.). BMC Plant Biol. 2010, 10, 142. [Google Scholar] [CrossRef] [PubMed]
- Sonah, H.; Deshmukh, R.K.; Labbé, C.; Bélanger, R.R. Analysis of aquaporins in Brassicaceae species reveals high-level of conservation and dynamic role against biotic and abiotic stress in canola. Sci. Rep. 2017, 7, 2771. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, R.; Sonah, H.; Belanger, R. Plant Aquaporins: Genome-wide identification, transcriptomics, proteomics, and advanced analytical tools. Front. Plant Sci. 2016, 7, 1896. [Google Scholar] [CrossRef]
- Maurel, C.; Boursiac, Y.; Luu, D.T.; Santoni, V.; Shahzad, Z.; Verdoucq, L. Aquaporins in plants. Physiol. Rev. 2015, 95, 1321–1358. [Google Scholar] [CrossRef]
- Lopez, D.; Ben Amira, M.; Brown, D.; Muries, B.; Brunel-Michac, N.; Bourgerie, S.; Porcheron, B.; Lemoine, R.; Chrestin, H.; Mollison, E.; et al. The Hevea brasiliensis XIP aquaporin subfamily: Genomic, structural and functional characterizations with relevance to intensive latex harvesting. Plant Mol. Biol. 2016, 91, 375–396. [Google Scholar] [CrossRef]
- Danielson, J.Å.; Johanson, U. Unexpected complexity of the aquaporin gene family in the moss Physcomitrella patens. BMC Plant Biol. 2008, 8, 45. [Google Scholar] [CrossRef] [PubMed]
- Khabudaev, K.V.; Petrova, D.P.; Grachev, M.A.; Likhoshway, Y.V. A new subfamily LIP of the major intrinsic proteins. BMC Genom. 2014, 15, 173. [Google Scholar] [CrossRef]
- Lopez, D.; Bronner, G.; Brunel, N.; Auguin, D.; Bourgerie, S.; Brignolas, F.; Carpin, S.; Tournaire-Roux, C.; Maurel, C.; Fumanal, B.; et al. Insights into Populus XIP aquaporins: Evolutionary expansion, protein functionality, and environmental regulation. J. Exp. Bot. 2012, 63, 2217–2230. [Google Scholar] [CrossRef]
- Venkatesh, J.; Yu, J.W.; Gaston, D.; Park, S.W. Molecular evolution and functional divergence of X-intrinsic protein genes in plants. Mol. Genet. Genom. 2015, 290, 443–460. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, R.K.; Vivancos, J.; Ramakrishnan, G.; Guérin, V.; Carpentier, G.; Sonah, H.; Labbé, C.; Isenring, P.; Belzile, F.; Bélanger, R.R. A precise spacing between the NPA domains of aquaporins is essential for silicon permeability in plants. Plant J. 2015, 83, 489–500. [Google Scholar] [CrossRef]
- De Groot, B.L.; Grubmüller, H. Water permeation across biological membranes: Mechanism and dynamics of aquaporin-1 and GlpF. Science 2001, 294, 2353–2357. [Google Scholar] [CrossRef]
- Cheng, A.; van Hoek, A.N.; Yeager, M.; Verkman, A.S.; Mitra, A.K. Three-dimensional organization of a human water channel. Nature 1997, 387, 627–630. [Google Scholar] [CrossRef]
- Tornroth-Horsefield, S.; Wang, Y.; Hedfalk, K.; Johanson, U.; Karlsson, M.; Tajkhorshid, E.; Neutze, R.; Kjellbom, P. Structural mechanism of plant aquaporin gating. Nature 2006, 439, 688–694. [Google Scholar] [CrossRef] [PubMed]
- Murata, K.; Mitsuoka, K.; Hirai, T.; Walz, T.; Agre, P.; Heymann, J.B.; Engel, A.; Fujiyoshi, Y. Structural determinants of water permeation through aquaporin. Nature 2000, 407, 599–605. [Google Scholar] [CrossRef]
- Savage, D.F.; O’Connell, J.D.; Miercke, L.J.W.; Finer-Moore, J.; Stroud, R.M. Structural context shapes the aquaporin selectivity filter. Proc. Natl. Acad. Sci. USA 2010, 107, 17164–17169. [Google Scholar] [CrossRef]
- Sui, H.; Han, B.G.; Lee, J.K.; Walian, P.; Jap, B.K. Structural basis of transport-specific transport through the AQP1 transport channel. Nature 2000, 414, 872–878. [Google Scholar] [CrossRef]
- Froger, A.; Tallur, B.; Thomas, D.; Delamarche, C. Prediction of functional residues in water channels and related proteins. Protein Sci. 1998, 7, 1458–1468. [Google Scholar] [CrossRef]
- Hove, R.M.; Bhave, M. Plant aquaporins with non-aqua functions: Deciphering the signature sequences. Plant Mol. Biol. 2011, 75, 413–430. [Google Scholar] [CrossRef]
- Fox, A.R.; Maistriaux, L.C.; Chaumont, F. Toward understanding of the high number of plant aquaporin isoforms and multiple regulation mechanisms. Plant Sci. 2017, 264, 179–187. [Google Scholar] [CrossRef]
- Li, G.; Santoni, V.; Maurel, C. Plant aquaporins: Roles in plant physiology. Biochim. Biophys. Acta (BBA) Gen. Subj. 2014, 1840, 1574–1582. [Google Scholar] [CrossRef]
- Singh, R.K.; Deshmukh, R.; Muthamilarasan, M.; Rani, R.; Prasad, M. Versatile roles of aquaporin in physiological processes and stress tolerance in plants. Plant Physiol. Biochem. 2020, 149, 178–189. [Google Scholar] [CrossRef]
- Ishibashi, K.; Kondo, S.; Hara, S.; Morishita, Y. The evolutionary aspects of aquaporin family. Am. J. Physiol. Reg. I 2011, 300, R566–R576. [Google Scholar] [CrossRef]
- Bienert, G.P.; Chaumont, F. Aquaporin-facilitated transmembrane diffusion of hydrogen peroxide. Biochim. Biophys. Acta (BBA) Gen. Subj. 2014, 1840, 1596–1604. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Wang, X.; Li, P.; Wang, H.; Ji, H.; Xie, J.; Qiu, Q.; Shen, D.; Dong, H. Plant Aquaporin AtPIP1;4 Links Apoplastic H2O2 Induction to disease immunity pathways. Plant Physiol. 2016, 171, 1635–1650. [Google Scholar] [CrossRef]
- Li, G.; Chen, T.; Zhang, Z.; Li, B.; Tian, S. Roles of aquaporins in plant-pathogen interaction. Plants 2020, 9, 1134. [Google Scholar] [CrossRef]
- Salojärvi, J.; Smolander, O.P.; Nieminen, K.; Rajaraman, S.; Safronov, O.; Safdari, P.; Lamminmäki, A.; Immanen, J.; Lan, T.; Tanskanen, J. Genome sequencing and population genomic analyses provide insights into the adaptive landscape of silver birch. Nat. Genet. 2017, 49, 904–912. [Google Scholar] [CrossRef]
- Chen, S.; Lin, X.; Zhang, D.; Li, Q.; Chen, S. Genome-Wide Analysis of NAC Gene Family in Betula pendula. Forests 2019, 10, 741. [Google Scholar] [CrossRef]
- Hänninen, H.; Tanino, K. Tree seasonality in a warming climate. Trends Plant Sci. 2011, 16, 412–416. [Google Scholar] [CrossRef]
- Li, C.Y.; Junttila, O.; Heino, P.; Palva, E.T. Low temperature sensing in silver birch (Betula pendula Roth) ecotypes. Plant Sci. 2004, 167, 165–171. [Google Scholar] [CrossRef]
- Li, C.Y.; Welling, A.; Puhakainen, T.; Viherä-Aarnio, A.; Ernstsen, A.; Junttila, O.; Heino, P.; Palva, E.T. Differential responses of silver birch (Betula pendula) ecotypes to short-day photoperiod and low temperature. Tree Physiol. 2005, 25, 1563–1569. [Google Scholar] [CrossRef]
- Taulavuori, K.M.J.; Taulavuori, E.B.; Oddvar, S.; Nilsen, J.S.; Igeland, B.; Laine, K.M. Dehardening of mountain birch (Betula pubescens ssp. czerepanovii) ecotypes at elevated winter temperatures. New Phytol. 2004, 162, 427–436. [Google Scholar] [CrossRef]
- Gambetta, G.A.; Knipfer, T.; Fricke, W.; McElrone, A.J. Aquaporins and root water uptake. In Plant Aquaporins. Signalling and Communication in Plants; Chaumont, F., Tyerman, S., Eds.; Springer: Cham, Switzerland, 2017; pp. 133–153. [Google Scholar]
- Prado, K.; Maurel, C. Regulation of leaf hydraulics: From molecular to whole plant levels. Front. Plant Sci. 2013, 4, 255–269. [Google Scholar] [CrossRef]
- Charrier, G.; Ameglio, T. The timing of leaf fall affects cold acclimation by inter-actions with air temperature through water and carbohydrate contents. Environ. Exp. Bot. 2011, 72, 351–357. [Google Scholar] [CrossRef]
- Chen, S.; Wang, Y.; Yu, L.; Zheng, T.; Wang, S.; Yue, Z.; Jiang, J.; Kumari, S.; Zheng, C.; Tang, H.; et al. Genome sequence and evolution of Betula platyphylla. Hortic Res. 2021, 8, 37. [Google Scholar] [CrossRef]
- Yue, C.; Hongli, C.; Lu, W.; Yanhua, Z.; Xinyuan, H.; Jianming, Z.; Xinchao, W.; Yajun, Y. Molecular cloning and expression analysis of tea plant aquaporin (AQP) gene family. Plant Physiol. Biochem. 2014, 83, 65–76. [Google Scholar] [CrossRef]
- Wei, Q.; Ma, Q.; Ma, Z.; Zhou, G.; Feng, F.; Le, S.; Lei, C.; Gu, Q. Genome-wide identification and characterization of sweet orange (Citrus sinensis) aquaporin genes and their expression in two citrus cultivars differing in drought tolerance. Tree Genet. Genomes 2019, 15, 17. [Google Scholar] [CrossRef]
- Yaguinuma, D.H.; dos Santos, T.B.; de Souza, S.G.H.; Vieira, L.G.E.; Ribas, A.F. Genome-wide identification, evolution, and expression profile of aquaporin genes in Coffea canephora in response to water deficit. Plant Mol. Biol. Rep. 2020. [Google Scholar] [CrossRef]
- Rodrigues, M.I.; Takeda, A.A.; Bravo, J.P.; Maia, I.G. The eucalyptus tonoplast intrinsic protein (TIP) gene subfamily: Genomic organization, structural features, and expression profiles. Front. Plant Sci. 2016, 7, 1810. [Google Scholar] [CrossRef] [PubMed]
- Feltrim, D.; Pereira, L.; de Santana Costa, M.G.; Balbuena, T.S.; Mazzafera, P. Stem aquaporins and surfactant-related genes are differentially expressed in two Eucalyptus species in response to water stress. Plant Stress 2021, 1, 100003. [Google Scholar] [CrossRef]
- Li, W.; Zhang, D.; Zhu, G.; Mi, X.; Guo, W. Combining genome-wide and transcriptome-wide analyses reveal the evolutionary conservation and functional diversity of aquaporins in cotton. BMC Genom. 2019, 20, 538. [Google Scholar] [CrossRef]
- Zou, Z.; Gong, J.; An, F.; Xie, G.; Wang, J.; Mo, Y.; Yang, L. Genome-wide identification of rubber tree (Hevea brasiliensis Muell. Arg.) aquaporin genes and their response to ethephon stimulation in the laticifer, a rubber-producing tissue. BMC Genom. 2015, 16, 1001. [Google Scholar] [CrossRef]
- Zou, Z.; Yang, L.; Gong, J.; Mo, Y.; Wang, J.; Cao, J.; An, F.; Xie, G. Genome-wide identification of Jatropha curcas aquaporin genes and the comparative analysis provides insights into the gene family expansion and evolution in Hevea brasiliensis. Front. Plant Sci. 2016, 7, 395. [Google Scholar] [CrossRef]
- Liu, H.; Yang, L.; Xin, M.; Ma, F.; Liu, J. Gene-wide analysis of aquaporin gene family in Malus domestica and Heterologous expression of the gene MpPIP2;1 confers drought and salinity tolerance in Arabidposis thaliana. Int. J. Mol. Sci. 2019, 20, 3710. [Google Scholar] [CrossRef]
- Shelden, M.C.; Howitt, S.M.; Kaiser, B.N.; Tyerman, S.D. Identification and functional characterisation of aquaporins in the grapevine, Vitis vinifera. Funct. Plant Biol. 2009, 36, 1065–1078. [Google Scholar] [CrossRef]
- Zou, Z.; Yang, J. Genome-wide comparison reveals divergence of cassava and rubber aquaporin family genes after the recent whole-genome duplication. BMC Genom. 2019, 20, 380. [Google Scholar] [CrossRef]
- Cohen, D.; Bogeat-Bulot, M.B.; Vialet-Chabrand, S.; Merret, R.; Courty, P.E.; Moretti, S.; Bizet, F.; Guilliot, A.; Hummel, I. Developmental and environmental regulation of aquaporin gene expression across Populus species: Divergence or redundancy? PLoS ONE 2013, 8, e55506. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Flagel, L.E.; Paterson, A.H.; Rapp, R.A.; Soltis, D.E.; Soltis, P.S.; Wendel, J.F. Evolutionary genetics of genome merger and doubling in plants. Annu. Rev. Genet. 2008, 42, 443–461. [Google Scholar] [CrossRef] [PubMed]
- Kayum, M.A.; Park, J.I.; Nath, U.K.; Biswas, M.K.; Kim, H.T.; Nou, I.S. Genome-wide expression profiling of aquaporin genes confer responses to abiotic and biotic stresses in Brassica rapa. BMC Plant Biol. 2017, 17, 23. [Google Scholar] [CrossRef]
- Gu, Z.; Cavalcanti, A.; Chen, F.; Bouman, P.; Li, W. Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol. Biol. Evol. 2002, 19, 256–262. [Google Scholar] [CrossRef]
- Abascal, F.; Irisarri, I.; Zardoya, R. Diversity and evolution of membrane intrinsic proteins. Biochim. Biophys. Acta 2014, 1840, 1468–1481. [Google Scholar] [CrossRef]
- Deshmukh, R.K.; Sonah, H.; Singh, N.K. Intron gain, a dominant evolutionary process supporting high levels of gene expression in rice. J. Plant Biochem. Biotechnol. 2016, 25, 142–146. [Google Scholar] [CrossRef]
- Narayan, R. The role of genomic constraints upon evolutionary changes in genome size and chromosome organization. Ann. Bot. 1998, 82, 57–66. [Google Scholar] [CrossRef][Green Version]
- Severing, E.I.; van Dijk, A.D.J.; Stiekema, W.J.; van Ham, R.C.H.J. Comparative analysis indicates that alternative splicing in plants has a limited role in functional expansion of the proteome. BMC Genom. 2009, 10, 154. [Google Scholar] [CrossRef] [PubMed]
- Magadum, S.; Banerjee, U.; Murugan, P.; Gangapur, D.; Ravikesavan, R. Gene duplication as a major force in evolution. J. Genet. 2013, 92, 55–161. [Google Scholar] [CrossRef]
- Roy, S.W.; Penny, D. Patterns of intron loss and gain in plants: Intron loss-dominated evolution and genome-wide comparison of O. sativa and A. thaliana. Mol. Biol. Evol. 2006, 24, 171–181. [Google Scholar] [CrossRef]
- Samadder, P.; Sivamani, E.; Lu, J.; Li, X.; Qu, R. Transcriptional and post-transcriptional enhancement of gene expression by the 5′ UTR intron of rice rubi3 gene in transgenic rice cells. Mol. Genet. Genom. 2008, 279, 429–439. [Google Scholar] [CrossRef]
- Shaul, O. How introns enhance gene expression. Int. J. Biochem. Cell Biol. 2017, 91, 145–155. [Google Scholar] [CrossRef]
- Fetter, K.; van Wilder, V.; Moshelion, M.; Chaumont, F. Interactions between plasma membrane aquaporins modulate their water channel activity. Plant Cell 2004, 16, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Yaneff, A.; Sigaut, L.; Marquez, M.; Alleva, K.; Pietrasanta, L.I.; Amodeo, G. Heteromerization of PIP aquaporins affects their intrinsic permeability. Proc. Natl. Acad. Sci. USA 2014, 111, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Bienert, M.D.; Diehn, T.A.; Richet, N.; Chaumont, F.; Bienert, G.P. Heterotetramerization of plant PIP1 and PIP2 aquaporins Is an evolutionary ancient feature to guide PIP1 plasma membrane localization and function. Front. Plant Sci. 2018, 9, 382. [Google Scholar] [CrossRef]
- Mom, R.; Muries, B.; Benoit, P.; Robert-Paganin, J.; Réty, S.; Venisse, J.S.; Padua, A.; Label, P.; Auguin, D. Voltage-gating of aquaporins, a putative conserved safety mechanism during ionic stresses. FEBS Lett. 2021, 595, 41–57. [Google Scholar] [CrossRef]
- Kozlowski, L.P. IPC–isoelectric point calculator. Biol. Direct 2016, 11, 55. [Google Scholar] [CrossRef]
- Chaumont, F.; Moshelion, M.; Daniels, M.J. Regulation of plant aquaporin activity. Biol Cell 2005, 97, 749–764. [Google Scholar] [CrossRef] [PubMed]
- Shivaraj, S.M.; Sharma, Y.; Chaudhary, J.; Rajora, N.; Sharma, S.; Thakral, V.; Ram, H.; Sonah, H.; Singla-Pareek, S.L.; Sharma, T.R.; et al. Dynamic role of aquaporin transport system under drought stress in plants. Environ. Exp. Bot. 2021, 184, 104367. [Google Scholar] [CrossRef]
- Kapilan, R.; Vaziri, M.; Zwiazek, J.J. Regulation of aquaporins in plants under stress. Biol. Res. 2018, 51, 4. [Google Scholar] [CrossRef]
- Anderberg, H.I.; Kjellbom, P.; Johanson, U. Annotation of Selaginella moellendorffli major intrinsic proteins and the evolution of the protein family in terrestrial plants. Front. Plant Sci. 2012, 3, 33. [Google Scholar] [CrossRef]
- Yuan, D.; Li, W.; Hua, Y.P.; King, G.J.; Xu, F.S.; Shi, L. Genome-wide identification and characterization of the aquaporin gene family and transcriptional responses to boron deficiency in Brassica napus. Front. Plant Sci. 2017, 8, 1336. [Google Scholar] [CrossRef] [PubMed]
- Heckwolf, M.; Pater, D.; Hanson, D.T.; Kaldenhoff, R. The Arabidopsis thaliana aquaporin AtPIP1:2 is a physiologically relevant CO2 transport facilitator. Plant J. 2011, 67, 734–737. [Google Scholar] [CrossRef]
- Groszmann, M.; Osborn, H.L.; Evans, J.R. Carbon dioxide and water transport through plant aquaporins. Plant Cell Environ. 2017, 40, 938–961. [Google Scholar] [CrossRef]
- Zwiazek, J.J.; Xu, H.; Tan, X.; Navarro-Ródenas, A.; Morte, A. Significance of oxygen transport through aquaporins. Sci. Rep. 2017, 7, 40411. [Google Scholar] [CrossRef]
- Otto, B.; Uehlein, N.; Sdorra, S.; Fischer, M.; Ayaz, M.; Belastegui-Macadam, X.; Heckwolf, M.; Lachnit, M.; Pede, N.; Priem, N. Aquaporin tetramer composition modifies the function of tobacco aquaporins. J. Biol. Chem. 2010, 285, 31253–31260. [Google Scholar] [CrossRef] [PubMed]
- Mori, I.C.; Rhee, J.; Shibasaka, M.; Sasano, S.; Kaneko, T.; Horie, T.; Katsuhara, M. CO2 transport by PIP2 aquaporins in barley. Plant Cell Physiol. 2014, 55, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Smirnoff, N.; Arnaud, D. Hydrogen peroxide metabolism and functions in plants. New Phytol. 2019, 221, 1197–1214. [Google Scholar] [CrossRef]
- Wang, H.; Schoebel, S.; Schmitz, F.; Donga, H. Characterization of aquaporin-driven hydrogen peroxide transport. Biochim. Biophys. Acta 2020, 1862, 183065. [Google Scholar] [CrossRef]
- Wudick, M.M.; Li, X.; Valentini, V.; Geldner, N.; Chory, J.; Lin, J.; Maurel, C.; Luu, D.T. Subcellular redistribution of root aquaporins induced by hydrogen peroxide. Mol. Plant 2015, 8, 1103–1114. [Google Scholar] [CrossRef] [PubMed]
- Grondin, A.; Rodrigues, O.; Verdoucq, L.; Merlot, S.; Leonhardt, N.; Maurel, C. Aquaporins contribute to ABA-triggered stomatal closure through OST1-mediated phosphorylation. Plant Cell 2015, 27, 1945–1954. [Google Scholar] [CrossRef] [PubMed]
- Rhee, J.; Horie, T.; Sasano, S.; Nakahara, Y.; Katsuhara, M. Identification of an H2O2 permeable PIP aquaporin in barley and a serine residue promoting H2O2 transport. Physiol. Plant 2017, 159, 120–128. [Google Scholar] [CrossRef]
- Wallace, I.S.; Roberts, D.M. Homology modeling of representative subfamilies of Arabidopsis major intrinsic proteins. Classification based on the aromatic/arginine selectivity filter. Plant Physiol. 2004, 135, 1059–1068. [Google Scholar] [CrossRef]
- Regon, P.; Panda, P.; Kshetrimayum, E.; Panda, S.K. Genome-wide comparative analysis of tonoplast intrinsic protein (TIP) genes in plants. Funct. Integr. Genom. 2014, 14, 617–629. [Google Scholar] [CrossRef]
- Soto, G.; Alleva, K.; Mazzella, M.A.; Amodeo, G.; Muschietti, J.P. AtTIP1;3 and AtTIP5;1, the only highly expressed Arabidopsis pollen-specific aquaporins, transport water and urea. FEBS Lett. 2008, 582, 4077–4082. [Google Scholar] [CrossRef]
- Tyerman, S.D.; Wignes, J.A.; Kaiser, B.N. Root hydraulic and aquaporin responses to N availability. In Plant Aquaporins: From Transport to Signaling; Springer: Berlin, Germany, 2017; pp. 207–236. [Google Scholar] [CrossRef]
- Liu, L.H.; Ludewig, U.; Gassert, B.; Frommer, W.B.; von Wirén, N. Urea transport by nitrogen-regulated tonoplast intrinsic proteins in arabidopsis. Plant Physiol. 2003, 133, 1220–1228. [Google Scholar] [CrossRef]
- Jahn, T.P.; Jahn, T.P.; Møller, A.L.B.; Zeuthen, T.; Holm, L.M.; Klaerke, D.A.; Mohsin, B.; Kühlbrandt, W.; Schjoerring, J.K. Aquaporin homologues in plants and mammals transport ammonia. FEBS Lett. 2004, 574, 31–36. [Google Scholar] [CrossRef]
- Beitz, E.; Wu, B.; Holm, L.M.; Schultz, J.E.; Zeuthen, T. Point mutations in the aromatic/arginine region in aquaporin 1 allow passage of urea, glycerol, ammonia, and protons. Proc. Natl. Acad. Sci. USA 2006, 103, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Kirscht, A.; Kaptan, S.S.; Bienert, G.P.; Chaumont, F.; Nissen, P.; de Groot, B.L.; Kjellbom, P.; Gourdon, P.; Johanson, U. Crystal structure of an ammonia-permeable aquaporin. PLoS Biol. 2016, 14, 1–19. [Google Scholar] [CrossRef]
- Bienert, G.P.; Møller, A.L.B.; Kristiansen, K.A.; Schulz, A.; Møller, I.M.; Schjoerring, J.K.; Jahn, T.P. Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J. Biol. Chem. 2007, 282, 1183–1192. [Google Scholar] [CrossRef] [PubMed]
- Dynowski, M.; Mayer, M.; Moran, O.; Ludewig, U. Molecular determinants of ammonia and urea conductance in plant aquaporin homologs. FEBS Lett. 2008, 582, 2458–2462. [Google Scholar] [CrossRef] [PubMed]
- Azad, A.K.; Yoshikawa, N.; Ishikawa, T.; Sawa, Y.; Shibata, H. Substitution of a single amino acid residue in the aromatic/arginine selectivity filter alters the transport profiles of tonoplast aquaporin homologs. Biochim. Biophys. Acta Biomembr. 2012, 1818, 1–11. [Google Scholar] [CrossRef]
- Bienert, G.P.; Bienert, M.D.; Jahn, T.P.; Boutry, M.; Chaumont, F. Solanaceae XIPs are plasma membrane aquaporins that facilitate the transport of many uncharged substrates. Plant J. 2011, 66, 306–317. [Google Scholar] [CrossRef]
- Bienert, M.D.; Muries, B.; Crappe, D.; Chaumont, F.; Bienert, G.P. Overexpression of X Intrinsic Protein 1;1 in Nicotiana tabacum and Arabidopsis reduces boron allocation to shoot sink tissues. Plant Direct 2019, 3, e00143. [Google Scholar] [CrossRef]
- Anderberg, H.I.; Danielson, J.Å.H.; Johanson, U. Algal MIPs, high diversity and conserved motifs. BMC Evol. Biol. 2011, 11, 110. [Google Scholar] [CrossRef] [PubMed]
- Pommerrenig, B.; Diehn, T.A.; Bienert, G.P. Metalloido-porins: Essentiality of Nodulin 26-like intrinsic proteins in metalloid transport. Plant Sci. 2015, 238, 212–227. [Google Scholar] [CrossRef]
- Mitani-Ueno, N.; Yamaji, N.; Zhao, F.J.; Ma, J.F. The aromatic/arginine selectivity filter of NIP aquaporins plays a critical role in substrate selectivity for silicon, boron, and arsenic. J. Exp. Bot. 2011, 62, 4391–4398. [Google Scholar] [CrossRef]
- Fu, D.; Libson, A.; Miercke, L.J.; Weitzman, C.; Nollert, P.; Krucinski, J.; Stroud, R.M. Structure of a glycerol-conducting channel and the basis for its selectivity. Science 2000, 290, 481–486. [Google Scholar] [CrossRef]
- Ma, J.F.; Yamaji, N. Silicon uptake and accumulation in higher plants. Trends Plant Sci. 2006, 11, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Takano, J.; Wada, M.; Ludewig, U.; Schaaf, G.; von Wirén, N.; Fujiwara, T. The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell 2006, 18, 1498–1509. [Google Scholar] [CrossRef]
- Bienert, G.P.; Schüssler, M.D.; Jahn, T.P. Metalloids: Essential, beneficial or toxic? Major intrinsic proteins sort it out. Trends Biochem. Sci. 2008, 33, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Wallace, I.S.; Roberts, D.M. Distinct transport selectivity of two structural subclasses of the nodulin-like intrinsic protein family of plant aquaglyceroporin channel. Biochemistry 2005, 44, 16826–16834. [Google Scholar] [CrossRef]
- Tanaka, M.; Wallace, I.S.; Takano, J.; Roberts, D.M.; Fujiwara, T. NIP6;1 is a boric acid channel for preferential transport of boron to growing shoot tissues in Arabidopsis. Plant Cell 2008, 20, 2860–2875. [Google Scholar] [CrossRef]
- Li, T.; Choi, W.G.; Wallace, I.S.; Baudry, J.; Roberts, D.M. Arabidopsis thaliana NIP7;1: An anther-specific boric acid transporter of the aquaporin superfamily regulated by an unusual tyrosine in helix 2 of the transport pore. Biochemistry 2011, 50, 6633–6641. [Google Scholar] [CrossRef]
- Currie, H.A.; Perry, C.C. Silica in plants: Biological, biochemical and chemical studies. Ann. Bot. 2007, 100, 1383–1389. [Google Scholar] [CrossRef]
- Deshmukh, R.; Bélanger, R.R. Molecular evolution of aquaporins and silicon influx in plants. Funct. Ecol. 2016, 30, 1277–1285. [Google Scholar] [CrossRef]
- Ma, J.F.; Tamai, K.; Yamaji, N.; Mitani, N.; Konishi, S.; Katsuhara, M.; Ishiguro, M.; Murata, Y.; Yano, M. A silicon transporter in rice. Nature 2006, 440, 688–691. [Google Scholar] [CrossRef]
- Mitani, N.; Chiba, Y.; Yamaji, N.; Ma, J.F. Identification and characterization of maize and barley Lsi2-like silicon efflux transporters reveals a distinct silicon uptake system from that in rice. Plant Cell 2009, 21, 2133–2142. [Google Scholar] [CrossRef]
- Gu, R.; Chen, X.; Zhou, Y.; Yuan, L. Isolation and characterization of three maize aquaporin genes, ZmNIP2;1, ZmNIP2;4 and ZmTIP4;4 involved in urea transport. BMB Rep. 2012, 45, 96–101. [Google Scholar] [CrossRef]
- Epstein, E. The anomaly of silicon in plant biology. Proc. Natl. Acad. Sci. USA 1994, 91, 11–17. [Google Scholar] [CrossRef]
- Hodson, M.J.; White, P.J.; Mead, A.; Broadley, M.R. Phylogenetic variation in the silicon composition of plants. Ann. Bot. 2005, 96, 1027–1046. [Google Scholar] [CrossRef]
- Hub, J.S.; de Groot, B.L. Mechanism of selectivity in aquaporins and aquaglyceroporins. Proc. Natl. Acad. Sci. USA 2008, 105, 1198–1203. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, F.; Suga, S.; Uemura, T.; Sato, M.H.; Maeshima, M. Novel type aquaporin SIPs are mainly localized to the ER membrane and show cell-specific expression in Arabidopsis thaliana. FEBS Lett. 2005, 579, 5814–5820. [Google Scholar] [CrossRef] [PubMed]
- Azad, A.K.; Ahmed, J.; Alum, M.A.; Hasan, M.M.; Ishikawa, T.; Sawa, Y.; Katsuhara, M. Genome-wide characterization of major intrinsic proteins in four grass plants and their non-Aqua transport selectivity profiles with comparative perspective. PLoS ONE 2016, 11, e0157735. [Google Scholar]
- Yoo, Y.J.; Lee, H.K.; Han, W.; Kim, D.H.; Lee, M.H.; Jeon, J.; Lee, D.W.; Lee, J.; Lee, Y.; Lee, J. Interactions between transmembrane helices within monomers of the aquaporin AtPIP2;1 play a crucial role in tetramer formation. Mol. Plant 2016, 9, 1004–1017. [Google Scholar] [CrossRef]
- Uehlein, N.; Otto, B.; Hanson, D.T.; Fischer, M.; McDowell, N.; Kaldenhoff, R. Function of Nicotiana tabacum aquaporins as chloroplast gas pores challenges the concept of membrane CO2 permeability. Plant Cell 2008, 20, 648–657. [Google Scholar] [CrossRef] [PubMed]
- Soto, G.; Fox, R.; Ayub, N.; Alleva, K.; Guaimas, F.; Erijman, E.J.; Mazzella, A.; Amodeo, G.; Muschietti, J. TIP5;1 is an aquaporin specifically targeted to pollen mitochondria and is probably involved in nitrogen remobilization in Arabidopsis thaliana. Plant J. 2010, 64, 1038–1047. [Google Scholar] [CrossRef]
- Maurel, C.; Verdoucq, L.; Luu, D.T.; Santoni, V. Plant aquaporins: Membrane channels with multiple integrated functions. Annu. Rev. Plant Biol. 2008, 59, 595–624. [Google Scholar] [CrossRef] [PubMed]
- Wudick, M.M.; Luu, D.T.; Maurel, C. A look inside: Localization patterns and functions of intracellular plant aquaporins. New Phytol. 2009, 184, 289–302. [Google Scholar] [CrossRef]
- Sanghera, G.S.; Wani, S.H.; Hussain, W.; Singh, N.B. Engineering cold stress tolerance in crop plants. Curr. Genom. 2011, 12, 30–43. [Google Scholar] [CrossRef]
- Browse, J.; Xin, Z.G. Temperature sensing and cold acclimation. Curr. Opin. Plant Biol. 2001, 4, 241–246. [Google Scholar] [CrossRef]
- Li, C.; Puhakainen, T.; Welling, A.; Vihera-Aarnio, A.; Ernstsen, A.; Junttila, O.; Heino, P.; Palva, E.T. Cold acclimation in silver birch (Betula pendula). Development of freezing tolerance in different tissues and climatic ecotypes. Physiol. Plant 2002, 116, 478–488. [Google Scholar] [CrossRef]
- Afzal, Z.; Howton, T.C.; Sun, Y.; Mukhtar, M.S. The roles of aquaporins in plant stress responses. Dev. Biol. 2016, 4, 9. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Hou, X.; Huang, C.; Yan, Y.; Tie, W.; Ding, Z.; Wei, Y.; Liu, J.; Miao, H.; Lu, Z.; et al. Genome-wide identification and expression analyses of aquaporin gene family during development and abiotic stress in banana. Int. J. Mol. Sci. 2015, 16, 19728–19751. [Google Scholar] [CrossRef] [PubMed]
- Ahamed, A.; Murai-Hatano, M.; Ishikawa-Sakurai, J.; Hayashi, H.; Kawamura, Y.; Uemura, M. Cold stress-induced acclimation in rice is mediated by root-specific aquaporins. Plant Cell Physiol. 2012, 53, 1445–1456. [Google Scholar] [CrossRef]
- Rahman, A.; Kawamura, Y.; Maeshima, M.; Rahman, A.; Uemura, M. Plasma membrane aquaporin members pips act in concert to regulate cold acclimation and freezing tolerance responses in Arabidopsis thaliana. Plant Cell Physiol. 2020, 61, 787–802. [Google Scholar] [CrossRef]
- Shekhawat, U.K.; Ganapathi, T.R. Overexpression of a native plasma membrane aquaporin for development of abiotic stress tolerance in banana. Plant Biotechnol. J. 2013, 11, 942–952. [Google Scholar]
- Huang, C.; Zhou, S.; Hu, W.; Deng, X.; Wei, S.; Yang, G.; He, G. The wheat aquaporin gene TaAQP7 confers tolerance to cold stress in transgenic tobacco. Z. Naturforschung. C 2014, 69, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Muries, B.; Mom, R.; Benoit, P.; Brunel-Michac, N.; Cochard, H.; Drevet, P.; Petel, G.; Badel, E.; Fumanal, B.; Gousset-Dupont, A.; et al. Aquaporins and water control in drought-stressed poplar leaves: A glimpse into the extraxylem vascular territories. Environ. Exp. Bot. 2019, 162, 25–37. [Google Scholar] [CrossRef]
- Hynynen, J.P.; Niemistö, A.; Viherä-Aarnio, A.; Brunner, A.; Hein, S.; Velling, P. Silviculture of birch (Betula pendula Roth and Betula pubescens Ehrh.) in northern Europe. For. An. Int. J. For. Res. 2010, 83, 103–119. [Google Scholar] [CrossRef]
- Maurel, C.; Tacnet, F.; Güclü, J.; Guern, J.; Ripoche, P. Purified vesicles of tobacco cell vacuolar and plasma membranes exhibit dramatically different water permeability and water channel activity. Proc. Natl. Acad. Sci. USA 1997, 94, 7103–7108. [Google Scholar] [CrossRef] [PubMed]
- Ludevid, D.; Höfte, H.; Himelblau, E.; Chrispeels, M.J. The expression pattern of the tonoplast intrinsic protein γ-TIP in Arabidopsis thaliana is correlated with cell enlargement. Plant Physiol. 1992, 100, 1633–1639. [Google Scholar] [CrossRef]
- Chaumont, F.; Barrieu, F.; Herman, E.M.; Chrispeels, M.J. Characterization of a maize tonoplast aquaporin expressed in zones of cell division and elongation. Plant Physiol. 1998, 117, 1143–1152. [Google Scholar] [CrossRef] [PubMed]
- Loqué, D.; Ludewig, U.; Yuan, L.; von Wirén, N. Tonoplast intrinsic proteins AtTIP2;1 and AtTIP2;3 facilitate NH3 transport into the vacuole. Plant Physiol. 2005, 137, 671–680. [Google Scholar] [CrossRef] [PubMed]
- De Rosa, A.; Watson-Lazowski, A.; Evans, J.R.; Groszmann, M. Genome-wide identification and characterisation of Aquaporins in Nicotiana tabacum and their relationships with other Solanaceae species. BMC Plant Biol. 2020, 20, 266. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Bendahmane, M.; Fu, X. Genome-wide identification and characterization of aquaporins and their role in the flower opening processes in carnation (Dianthus caryophyllus). Molecules 2018, 23, 1895. [Google Scholar] [CrossRef]
- Azad, A.K.; Katsuhara, M.; Sawa, Y.; Ishikawa, T.; Shibata, H. Characterization of four plasma membrane aquaporins in tulip petals: A putative homolog is regulated by phosphorylation. Plant Cell Physiol. 2008, 49, 1196–1208. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ma, N.; Xue, J.; Li, Y.; Liu, X.; Dai, F.; Jia, W.; Luo, Y.; Gao, J. Rh-PIP2; 1, a rose aquaporin gene, is involved in ethylene-regulated petal expansion. Plant Physiol. 2008, 148, 894–907. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Steinmetz, L.M.; Gu, X.; Scharfe, C.; Davis, R.W.; Li, W.-H. Role of duplicate genes in genetic robustness against null mutations. Nature 2003, 421, 63–66. [Google Scholar] [CrossRef]
- DeLuna, A.; Vetsigian, K.; Shoresh, N.; Hegreness, M.; Colón-González, M.; Chao, S.; Kishony, R. Exposing the fitness contribution of duplicated genes. Nat. Genet. 2008, 40, 676–681. [Google Scholar] [CrossRef]
- Reuscher, S.; Akiyama, M.; Mori, C.; Aoki, K.; Shibata, D.; Shiratake, K. genome-wide identification and expression analysis of aquaporins in tomato. PLoS ONE 2013, 8, e79052. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Zou, Z.; Gong, J.; Huang, Q.; Mo, Y.; Yang, L.; Xie, G. Gene Structures, evolution, classification and expression profiles of the aquaporin gene family in castor bean (Ricinus communis L.). PLoS ONE 2015, 10, e0141022. [Google Scholar] [CrossRef]
- Sonnhammer, E.L.L.; von Heijne, G.; Krogh, A. A hidden Markov model for predicting transmembrane helices in protein sequences. In Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology, Montreal, QC, Canada, 28 June–1 July 1998; AAAI Press: Menlo Park, CA, USA, 1998; pp. 175–182. [Google Scholar]
- Hirokawa, T.; Boon Chieng, S.; Mitaku, S. SOSUI: Classification and secondary structure prediction system for membrane proteins. Bioinformatics 1998, 14, 378–379. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Berka, K.; Hanák, O.; Sehnal, D.; Banáš, P.; Navratilova, V.; Jaiswal, D.; Ionescu, C.M.; Svobodová Vareková, R.; Kocá, J.; Otyepka, M. MOLE online 2.0: Interactive web-based analysis of biomacromolecular channels. Nucleic Acids Res. 2012, 40, W222–W227. [Google Scholar] [CrossRef]
- Chang, S.; Puryear, J.; Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 1993, 11, 113–116. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; von Mering, C.; Bork, P. Fast genome-wide functional annotation through orthology assignment by eggNOG-Mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Forslund, K.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Mende, D.R.; Sunagawa, S.; Kuhn, M. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2015, 44, D286–D293. [Google Scholar] [CrossRef] [PubMed]
- Peterson, R.A.; Cavanaugh, J.E. Ordered quantile normalization: A semiparametric transformation built for the cross-validation era. J. Appl. Stat. 2019, 47, 1–16. [Google Scholar] [CrossRef]
- Robinson, D.; Hayes, A.; Couch, S. Broom: Convert Statistical Objects into Tidy Tibbles. Available online: CRAN.R-project.org/package=broom (accessed on 1 April 2021).
- Le, S.; Josse, J.; Husson, F. FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Kassambara, A.; Mundt, F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. Available online: CRAN.R-project.org/package=factoextra. (accessed on 1 April 2021).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-0-387-98141-3. [Google Scholar]
- Firke, S. Janitor: Simple Tools for Examining and Cleaning Dirty Data. Available online: CRAN.R-project.org/package=janitor (accessed on 1 April 2021).
- Hothorn, T.; Bretz, F.; Westfall, P. Simultaneous Inference in General Parametric Models. Biom. J. 2008, 50, 346–363. [Google Scholar] [CrossRef]
- Daróczi, G.; Tsegelskyi, R. Pander: An R ‘Pandoc’ Writer. Available online: CRAN.R-project.org/package=pander (accessed on 1 April 2021).
- Kassambara, A. Rstatix: Pipe-Friendly Framework for Basic Statistical Tests. 2020. Available online: CRAN.R-project.org/package=rstatix (accessed on 1 April 2021).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; D’Agostino McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practicaland powerful approach to multiple testing. J. R. Statist. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]

| a Loci | Size | b MW | bpI | b GRAVY | c TMH | d SubCL | e NPA | f ar/R SF | g Froger’s | h Predicted Transport | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Proposed Gene Name/Locus | (aa) | (kDa) | LB | LE | Residues | Substrate | |||||
| Plasma membrane Intrinsic Proteins (PIPs) | |||||||||||
| BpePIP1;1/Bpev01.c0190.g0072.m0001 | 287 | 30.86 | 9.26 | 0.314 | 6 (5) * | PM | NPA | NPA | F-H-T-R | Q-S-A-F-W | Boron H2O2 Urea |
| BpePIP1;2/Bpev01.c0170.g0038.m0001 | 286 | 30.51 | 8.76 | 0.417 | 6 | PM-Vac | NPA | NPA | F-H-T-R | G-S-A-F-W | Boron H2O2 Urea CO2 |
| BpePIP1;3/Bpev01.c1699.g0001.m0001 | 288 | 30.89 | 8.61 | 0.384 | 6 | PM-Vac | NPA | NPA | F-H-T-R | Q-S-A-F-W | Boron H2O2 Urea CO2 |
| BpePIP1;4/Bpev01.c0027.g0083.m0001 | 286 | 30.69 | 9 | 0.357 | 6 (5) * | PM-Vac | NPA | NPA | F-H-T-R | Q-S-A-F-W | Boron H2O2 Urea CO2 |
| BpePIP2;1/Bpev01.c0658.g0014.m0001 | 281 | 29.79 | 8.83 | 0.48 | 6 | PM-Vac | NPA | NPA | F-H-T-R | M-S-A-F-W | H2O2 Urea |
| BpePIP2;2/Bpev01.c0577.g0022.m0001 | 278 | 29.86 | 6.72 | 0.527 | 6 | PM-Vac | NPA | NPA | F-H-T-R | M-S-V-F-W | Urea |
| BpePIP2;3/Bpev01.c0577.g0023.m0001 | 278 | 29.91 | 6.2 | 0.44 | 6 | PM-Vac | NPA | NPA | F-H-T-R | M-S-A-F-W | Urea |
| BpePIP2;4/Bpev01.c0042.g0019.m0001 | 287 | 30.41 | 8.68 | 0.498 | 6 | PM-Vac-ER | NPA | NPA | F-H-T-R | N-S-A-F-W | H2O2 Urea |
| BpePIP2;5/Bpev01.c0552.g0003.m0001 | 286 | 30.79 | 8.25 | 0.424 | 6 | PM | NPA | NPA | F-H-T-R | Q-S-A-F-W | H2O2 Urea |
| BpePIP2;6/Bpev01.c0483.g0002.m0001 | 287 | 30.44 | 8.93 | 0.506 | 6 | PM-Vac-Gol | NPA | NPA | F-H-T-R | Q-S-A-F-W | H2O2 Urea |
| Tonoplast Intrinsic Proteins (TIPs) | |||||||||||
| BpeTIP1;1/Bpev01.c0278.g0002.m0001 | 252 | 26.39 | 5.8 | 0.624 | 6 | PM-Vac | NPA | NPA | H-I-A-V | T-S-A-Y-W | H2O2 Urea |
| BpeTIP1;2/Bpev01.c0396.g0015.m0001 | 252 | 25.81 | 4.95 | 0.843 | 6 | Vac-PM | NPA | NPA | H-I-A-V | T-S-A-Y-W | Urea |
| BpeTIP1;3/Bpev01.c2330.g0012.m0001 | 252 | 26.15 | 5 | 0.755 | 6 | PM-Vac | NPA | NPA | H-I-A-V | T-S-A-Y-W | Urea |
| BpeTIP2;1/Bpev01.c0665.g0011.m0001 | 247 | 25.19 | 6.15 | 0.866 | 6 | PM-Vac | NPA | NPA | H-I-G-R | T-S-A-F-W | H2O2 Urea |
| BpeTIP2;2/Bpev01.c0120.g0064.m0001 | 250 | 25.44 | 5.11 | 0.913 | 6 | Vac-PM | NPA | NPA | H-I-G-R | T-S-A-Y-W | H2O2 Ammonia Urea |
| BpeTIP3;1/Bpev01.c1026.g0003.m0001 | 253 | 27.11 | 6.7 | 0.874 | 6 | PM-Vac-ER | NPA | NPA | H-I-A-R | T-A-A-Y-W | H2O2 Urea |
| BpeTIP4;1/Bpev01.c0921.g0003.m0001 | 247 | 25.91 | 5.72 | 0.774 | 6 (7) * | Vac-PM | NPA | NPA | H-I-A-R | T-S-A-Y-W | Urea |
| BpeTIP5;1/Bpev01.c0477.g0022.m0001 | 248 | 25.24 | 6.39 | 0.75 | 6 | Chlo-Vac | NPA | NPA | N-V-G-C | V-A-A-Y-W | H2O2 Urea |
| Uncharacterized X Intrinsic Proteins (XIPs) | |||||||||||
| BpeXIP2;1 Redefined sequence | 321 | 30.08 | 8.35 | 0.623 | 6 | PM-Vac-ER | NPV | NPA | I-T-A-R | V-C-P-F-W | H2O2 Urea |
| BpeXIP1;1/Bpev01.c1577.g0026.m0001 | 305 | 32.84 | 5.67 | 0.724 | 6 | PM-ER-Chlo-Vac-Pero | NPI | NPA | V-I-V-R | V-C-P-F-W | Urea |
| BpeXIP1;2/Bpev01.c1577.g0028.m0001 | 305 | 32.77 | 6.29 | 0.764 | 6 | PM-Vac-ER-Gol | NPI | NPA | V-I-V-R | V-C-P-L-W | - |
| BpeXIP1;3/Bpev01.c2937.g0004.m0001 | 305 | 32.76 | 6.06 | 0.704 | 6 | PM-ER-Vac-Chlo | NPI | NPA | V-I-V-R | V-C-P-F-W | - |
| Nodulin-26 like Intrinsic Proteins (NIPs) | |||||||||||
| BpeNIP1;1/Bpev01.c0145.g0009.m0001 | 270 | 28.47 | 8.33 | 0.715 | 6 | PM-Vac | NPA | NPA | W-V-A-R | F-S-A-Y-I | Ammonia Urea |
| BpeNIP1;2/Bpev01.c0230.g0002.m0001 | 282 | 29.73 | 9.25 | 0.425 | 6 | PM-Golg-ER-Vac | NPA | NPA | W-V-A-R | F-S-A-Y-L | Ammonia Urea |
| BpeNIP2;1/Bpev01.c0281.g0063.m0001 | 282 | 29.5 | 8.43 | 0.405 | 6 | PM-Vac-ER | NPA | NPA | G-S-G-R | L-T-A-Y-L | Boron Urea |
| BpeNIP4;1/Bpev01.c1162.g0002.m0001 | 269 | 28.57 | 9.05 | 0.67 | 6 | PM-ER | NPA | NPA | W-V-A-R | F-S-A-Y-I | Ammonia Urea |
| BpeNIP5;1/Bpev01.c1045.g0004.m0001 | 247 | 25.94 | 8.91 | 0.737 | 6 (5) * | Vac-PM-Mito-ER | NPS | NPV | A-I-G-R | F-T-A-Y-L | Urea |
| BpeNIP5;2/Bpev01.c1084.g0010.m0001 | 255 | 26.39 | 5.39 | 0.938 | 6 (5) * | PM-Vac | NPS | NPV | A-I-A-R | F-T-A-Y-M | Boron Urea |
| BpeNIP6;1/Bpev01.c0044.g0051.m0001 | 305 | 31.46 | 8.51 | 0.41 | 6 | PM-Vac-ER-Gol | NPA | NPV | S-I-A-R | F-T-A-Y-L | Boron Urea |
| BpeNIP7;1/Bpev01.c0330.g0006.m0001 | 272 | 29.12 | 8.13 | 0.508 | 6 (5) * | PM-ER-Vac-Gol | NPA | NPA | A-V-G-R | Y-S-A-Y-M | - |
| Small basic Intrinsic Proteins (SIPs) | |||||||||||
| BpeSIP1;1/Bpev01.c0082.g0002.m0001 | 239 | 25.34 | 9.76 | 0.781 | 6 | Vac-PM-ER-Gol-Chlo | NPT | NPA | V-T-P-N | F-A-A-Y-W | - |
| BpeSIP1;2/Bpev01.c0387.g0011.m0001 | 231 | 24.49 | 10.05 | 0.775 | 6 | PM-Chlo-Gol-Vac | NPS | NPA | A-T-P-N | F-A-A-Y-W | - |
| BpeSIP2;1/Bpev01.c0212.g0010.m0001 | 240 | 26.27 | 9.4 | 0.626 | 6 (4) * | Vac-PM-ER | NPL | NPA | S-K-G-S | I-V-A-Y-W | - |
| Aquaporins | a size (X—Y—Z) | b Channel | b ar/R Bottleneck | b Bottleneck | c Hydropathy | d Charge | e Polarity | f Mutability | Lipophilicity | Solubility | j Ionizable | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (Å) | Length (Å) | Radius (Å) | Radius (Å) | gLogP | hLogD | iLogS | ||||||
| Plasma membrane Intrinsic Proteins (PIPs) | ||||||||||||
| BpePIP1;1 | 41.358—46.156—64.972 | 50.3 | 1.2 | 1.1 | 0.7 | 0 | 9.08 | 86 | 0.61 | 0.41 | −0.25 | 2 |
| BpePIP1;2 | 75.341—52.171—52.769 | 52.7 | 0.8 | 0.8 | 1.45 | 2 | 6.45 | 85 | 1.04 | 0.86 | −0.6 | 2 |
| BpePIP1;3 | 41.338—49.248—63.209 | 49 | 1.2 | 1.1 | 1.01 | −1 | 6.75 | 87 | 0.71 | 0.55 | −0.29 | 3 |
| BpePIP1;4 | 79.903—52.414—52.769 | 40.5 | 1.2 | 0.6 | 1.46 | 1 | 5.41 | 83 | 0.83 | 0.74 | −0.53 | 1 |
| BpePIP2;1 | 77.283—50.592—50.381 | 51.3 | 0.7 | 0.5 | 1.38 | 2 | 6.56 | 90 | 0.92 | 0.76 | −0.47 | 2 |
| BpePIP2;2 | 73.807—57.060—51.485 | 52.2 | 0.7 | 0.6 | 1.07 | 0 | 9.75 | 87 | 0.94 | 0.69 | −0.49 | 2 |
| BpePIP2;3 | 73.807—57.060—51.485 | 52.4 | 0.9 | 0.5 | 0.84 | 1 | 9.59 | 87 | 0.83 | 0.57 | −0.31 | 3 |
| BpePIP2;4 | 76.112—54.225—51.485 | 48.6 | 0.7 | 0.6 | 0.99 | 0 | 10.36 | 88 | 0.92 | 0.66 | −0.45 | 2 |
| BpePIP2;5 | 76.894—50.699—50.542 | 47.4 | 0.9 | 0.6 | 0.78 | −1 | 12.73 | 87 | 0.94 | 0.58 | −0.42 | 3 |
| BpePIP2;6 | 79.375—57.747—50.542 | 48.9 | 0.7 | 0.5 | 1.04 | 1 | 11.23 | 88 | 1.03 | 0.74 | −0.58 | 3 |
| Tonoplast Intrinsic Proteins (TIPs) | ||||||||||||
| BpeTIP1;1 | 72.798—51.488—50.744 | 43.2 | 1.3 | 0.5 | 0.93 | −1 | 8.19 | 91 | 0.83 | 0.66 | −0.44 | 1 |
| BpeTIP1;2 | 72.546—44.617—51.466 | 60.5 | 1.3 | 0.4 | 0.92 | 0 | 6.22 | 84 | 0.88 | 0.7 | −0.35 | 2 |
| BpeTIP1;3 | 40.145—37.773—54.582 | 58.4 | 0.9 | 0.6 | 1.14 | 0 | 4.49 | 91 | 0.67 | 0.67 | −0.37 | 1 |
| BpeTIP2;1 | 40.061—37.320—52.603 | 54.3 | 1.7 | 0.4 | 0.5 | 1 | 9.59 | 88 | 0.53 | 0.41 | −0.32 | 1 |
| BpeTIP2;2 | 38.810—37.320—50.132 | 40 | 1.6 | 0.6 | 0.98 | 1 | 7.58 | 84 | 0.61 | 0.5 | −0.4 | 1 |
| BpeTIP3;1 | 41.075—36.911—53.559 | 52.9 | 1.7 | 0.6 | 1.18 | 1 | 6.4 | 85 | 0.84 | 0.7 | −0.54 | 3 |
| BpeTIP4;1 | 41.339—38.620—53.616 | 60.6 | 1.5 | 0.6 | 0.89 | 1 | 9.55 | 84 | 0.8 | 0.63 | −0.5 | 1 |
| BpeTIP5;1 | 72.604—38.620—53.617 | 43.5 | 2.3 | 0.5 | 0.3 | 1 | 5.27 | 88 | 0.37 | 0.23 | −0.01 | 1 |
| Uncharacterized X Intrinsic Proteins (XIPs) | ||||||||||||
| BpeXIP2;1 | 46.806—47.507—65.542 | 59.3 | 1.7 | 1.1 | 0.99 | 0 | 6.89 | 85 | 0.74 | 0.56 | −0.34 | 2 |
| BpeXIP1;1 | 77.536—59.089—66.029 | 57.8 | 1.8 | 0.7 | 0.95 | 2 | 5.72 | 87 | 0.46 | 0.27 | −0.13 | 2 |
| BpeXIP1;2 | 40.311—46.757—67.465 | 48.8 | 2 | 1.3 | 1.1 | 2 | 6.23 | 88 | 0.74 | 0.52 | −0.29 | 2 |
| BpeXIP1;3 | 76.853—56.997—54.194 | 48 | 2.4 | 0.4 | 1.16 | 1 | 8.73 | 86 | 0.88 | 0.63 | −0.27 | 3 |
| Nodulin-26 like Intrinsic Proteins (NIPs) | ||||||||||||
| BpeNIP1;1 | 47.170—47.961—60.868 | 45.4 | 1.2 | 0.7 | 0.94 | 1 | 3.99 | 91 | 0.64 | 0.52 | −0.29 | 1 |
| BpeNIP1;2 | 48.080—48.913—64.173 | 34.8 | 1.4 | 1.2 | 0.41 | 1 | 6.97 | 93 | 0.28 | 0.09 | 0.15 | 1 |
| BpeNIP2;1 | 45.851—47.139—66.412 | 51.6 | 2.6 | 0.8 | 0.51 | 2 | 7.02 | 92 | 0.47 | 0.31 | 0.09 | 2 |
| BpeNIP4;1 | 50.644—47.660—62.545 | 52 | 1.2 | 0.8 | 0.89 | 2 | 6.13 | 85 | 0.71 | 0.56 | −0.35 | 2 |
| BpeNIP5;1 | 46.077—36.449—63.012 | 55.5 | 2.1 | 0.7 | 0.13 | 0 | 8.43 | 86 | 0.6 | 0.4 | −0.22 | 2 |
| BpeNIP5;2 | 45.644—34.894—57.891 | 52.5 | 2 | 0.9 | 0.5 | 0 | 9.26 | 90 | 0.63 | 0.32 | −0.08 | 2 |
| BpeNIP6;1 | 46.270—46.230—62.913 | 45.2 | 1.7 | 0.8 | 0.61 | 0 | 7.6 | 85 | 0.47 | 0.32 | −0.11 | 2 |
| BpeNIP7;1 | 48.706—48.254—67.609 | 60.8 | 2.5 | 1 | 0.16 | 0 | 12.76 | 82 | 0.52 | 0.26 | −0.01 | 2 |
| Small basic Intrinsic Proteins (SIPs) | ||||||||||||
| BpeSIP1;1 | 76.118—45.635—48.786 | 48.4 | 1,2 | 0.9 | 0.22 | 0 | 2.03 | 91 | 0.33 | 0.33 | −0.07 | N/A |
| BpeSIP1;2 | 41.601—38.523—52.141 | 46.8 | 0.7 | 0.6 | −0.11 | 3 | 8.57 | 87 | 0.53 | 0.2 | −0.03 | 3 |
| BpeSIP2;1 | 76.963—48.686—44.305 | 51.3 | 0.7 | 0.5 | 0.37 | 2 | 8.27 | 84 | 0.47 | 0.2 | 0.06 | 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Venisse, J.-S.; Õunapuu-Pikas, E.; Dupont, M.; Gousset-Dupont, A.; Saadaoui, M.; Faize, M.; Chen, S.; Chen, S.; Petel, G.; Fumanal, B.; et al. Genome-Wide Identification, Structure Characterization, and Expression Pattern Profiling of the Aquaporin Gene Family in Betula pendula. Int. J. Mol. Sci. 2021, 22, 7269. https://doi.org/10.3390/ijms22147269
Venisse J-S, Õunapuu-Pikas E, Dupont M, Gousset-Dupont A, Saadaoui M, Faize M, Chen S, Chen S, Petel G, Fumanal B, et al. Genome-Wide Identification, Structure Characterization, and Expression Pattern Profiling of the Aquaporin Gene Family in Betula pendula. International Journal of Molecular Sciences. 2021; 22(14):7269. https://doi.org/10.3390/ijms22147269
Chicago/Turabian StyleVenisse, Jean-Stéphane, Eele Õunapuu-Pikas, Maxime Dupont, Aurélie Gousset-Dupont, Mouadh Saadaoui, Mohamed Faize, Song Chen, Su Chen, Gilles Petel, Boris Fumanal, and et al. 2021. "Genome-Wide Identification, Structure Characterization, and Expression Pattern Profiling of the Aquaporin Gene Family in Betula pendula" International Journal of Molecular Sciences 22, no. 14: 7269. https://doi.org/10.3390/ijms22147269
APA StyleVenisse, J.-S., Õunapuu-Pikas, E., Dupont, M., Gousset-Dupont, A., Saadaoui, M., Faize, M., Chen, S., Chen, S., Petel, G., Fumanal, B., Roeckel-Drevet, P., Sellin, A., & Label, P. (2021). Genome-Wide Identification, Structure Characterization, and Expression Pattern Profiling of the Aquaporin Gene Family in Betula pendula. International Journal of Molecular Sciences, 22(14), 7269. https://doi.org/10.3390/ijms22147269






