Structure, Biosynthesis, and Biological Activity of Succinylated Forms of Bacteriocin BacSp222
Abstract
1. Introduction
2. Results and Discussion
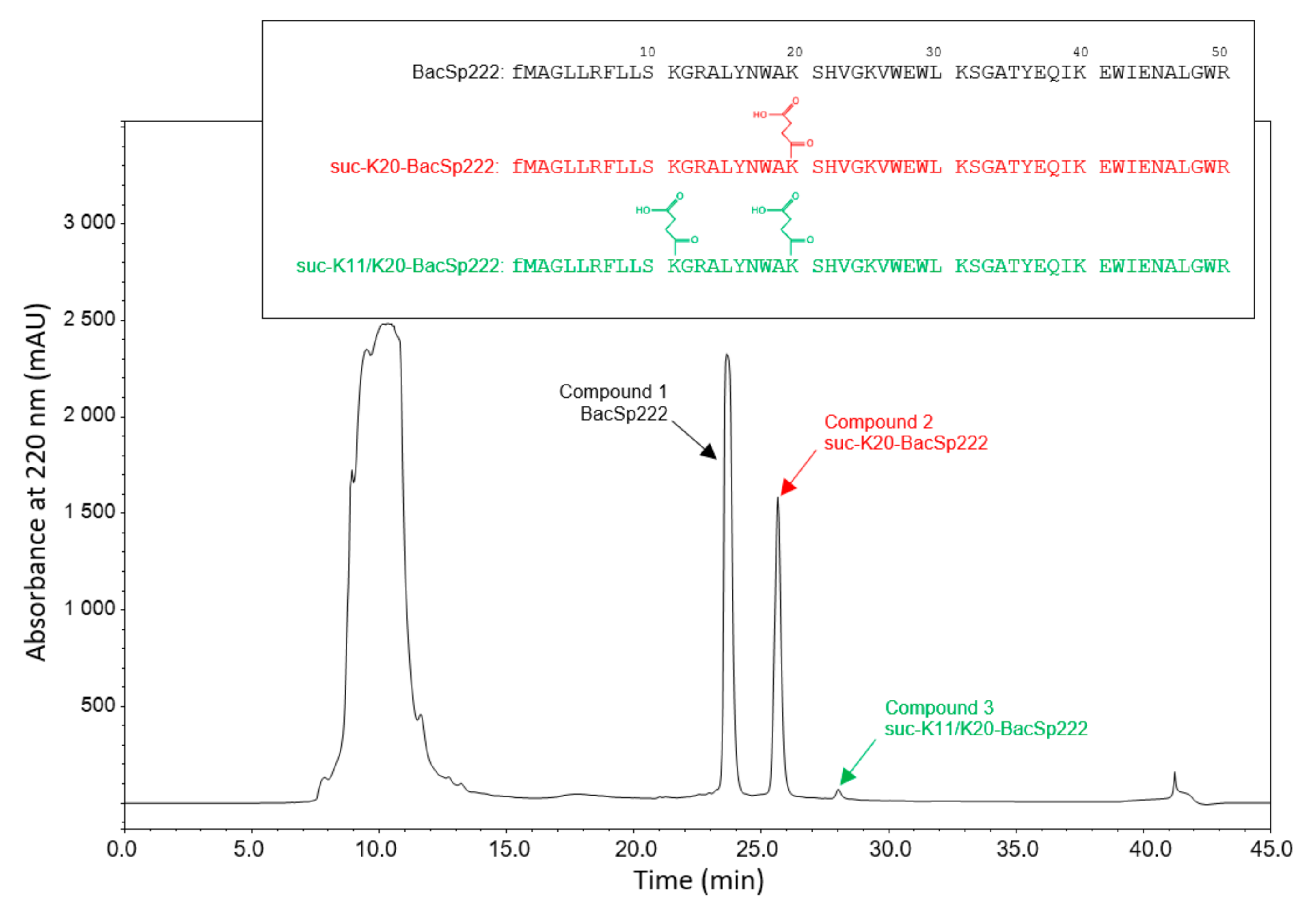
2.1. Staphylococcus pseudintermedius 222 Produces Native BacSp222 and Its Two Posttranslationally Modified Forms: Suc-K20-BacSp222 and suc-K11/K20-BacSp222
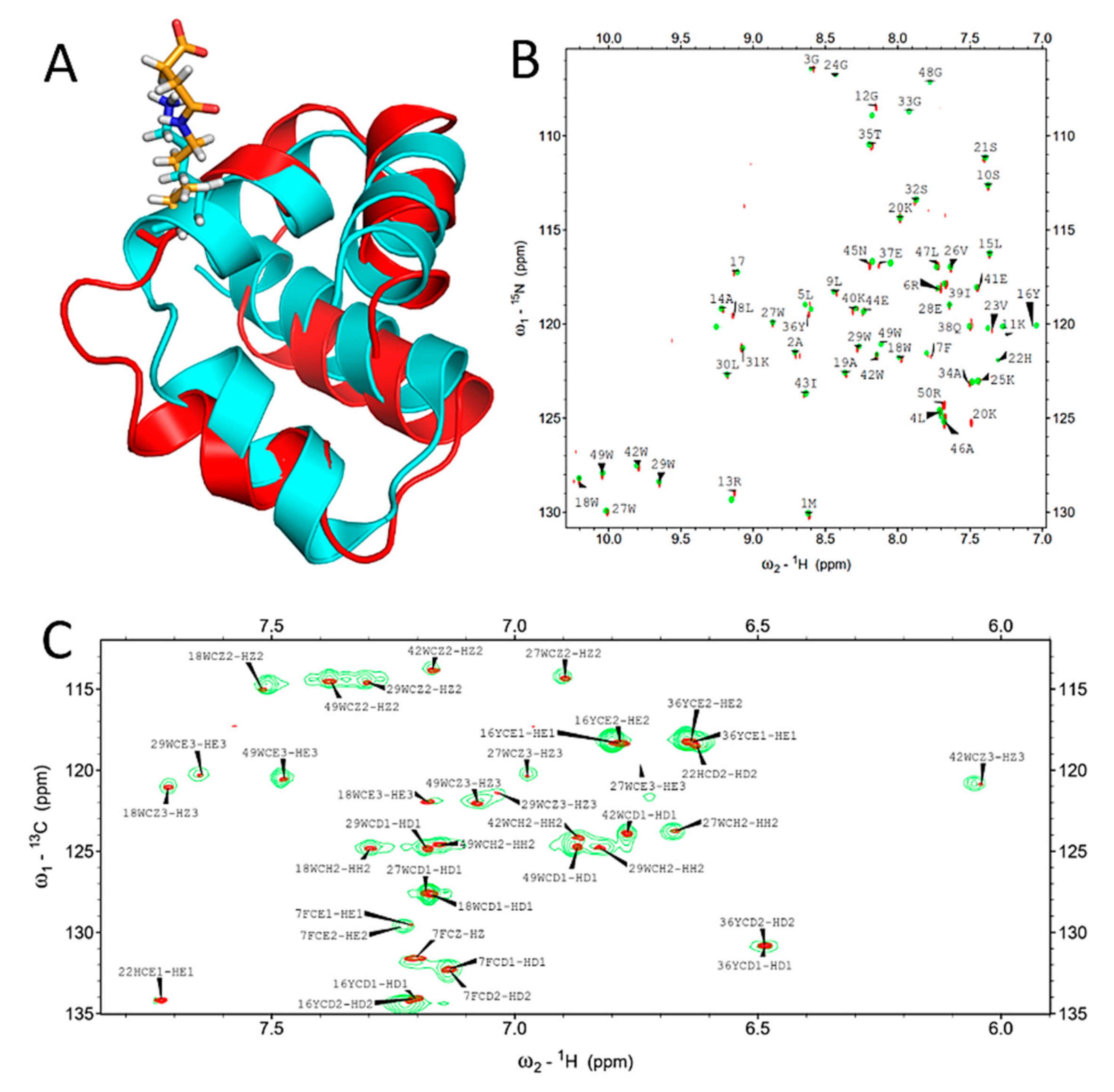
2.2. Succinylation Does Not Significantly Influence the Overall Structure of BacSp222
2.3. Succinylation Decreases the Antibacterial Activity of BacSp222 Bacteriocin
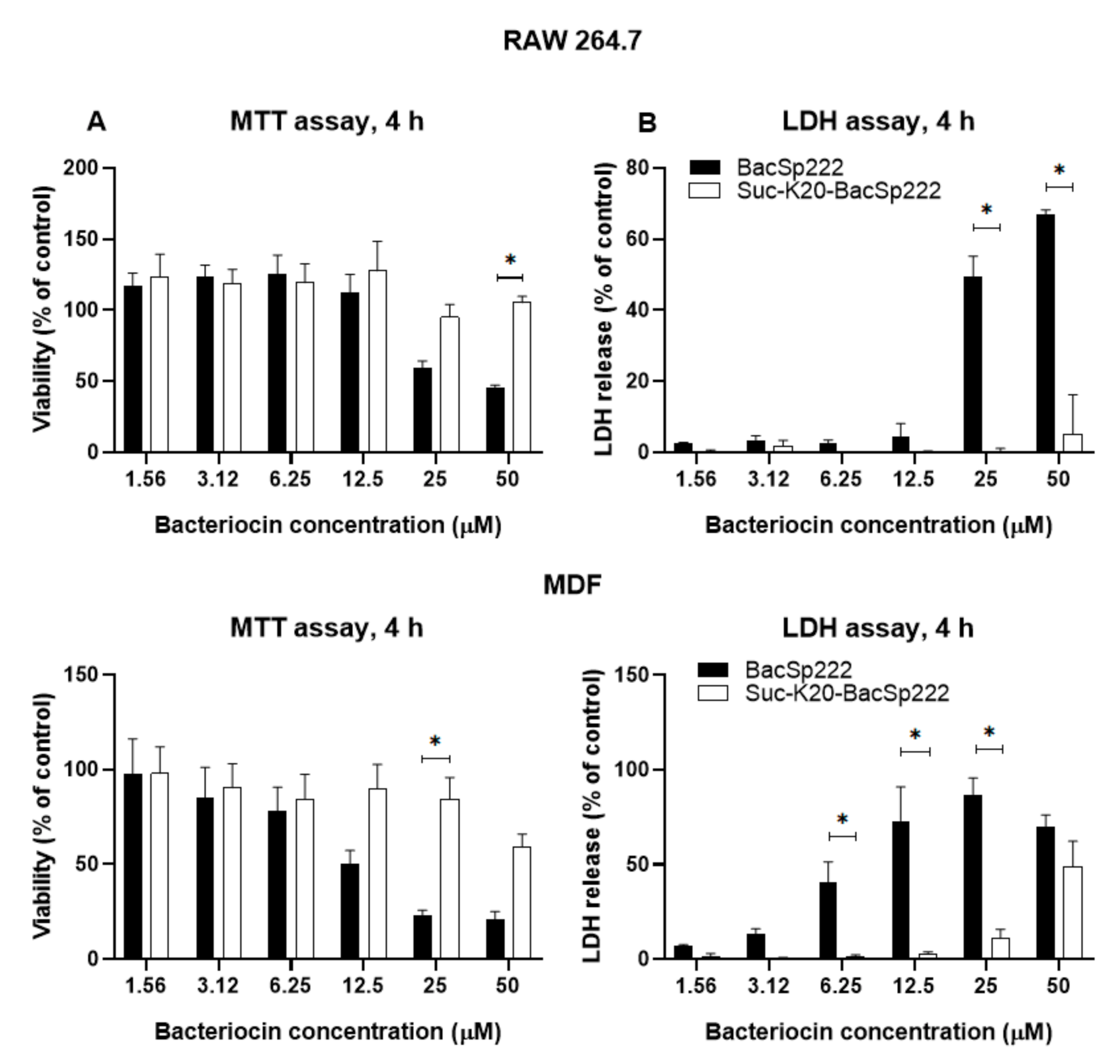
2.4. Succinylation limits BacSp222 Cytotoxic Activity Against Eukaryotic Cells
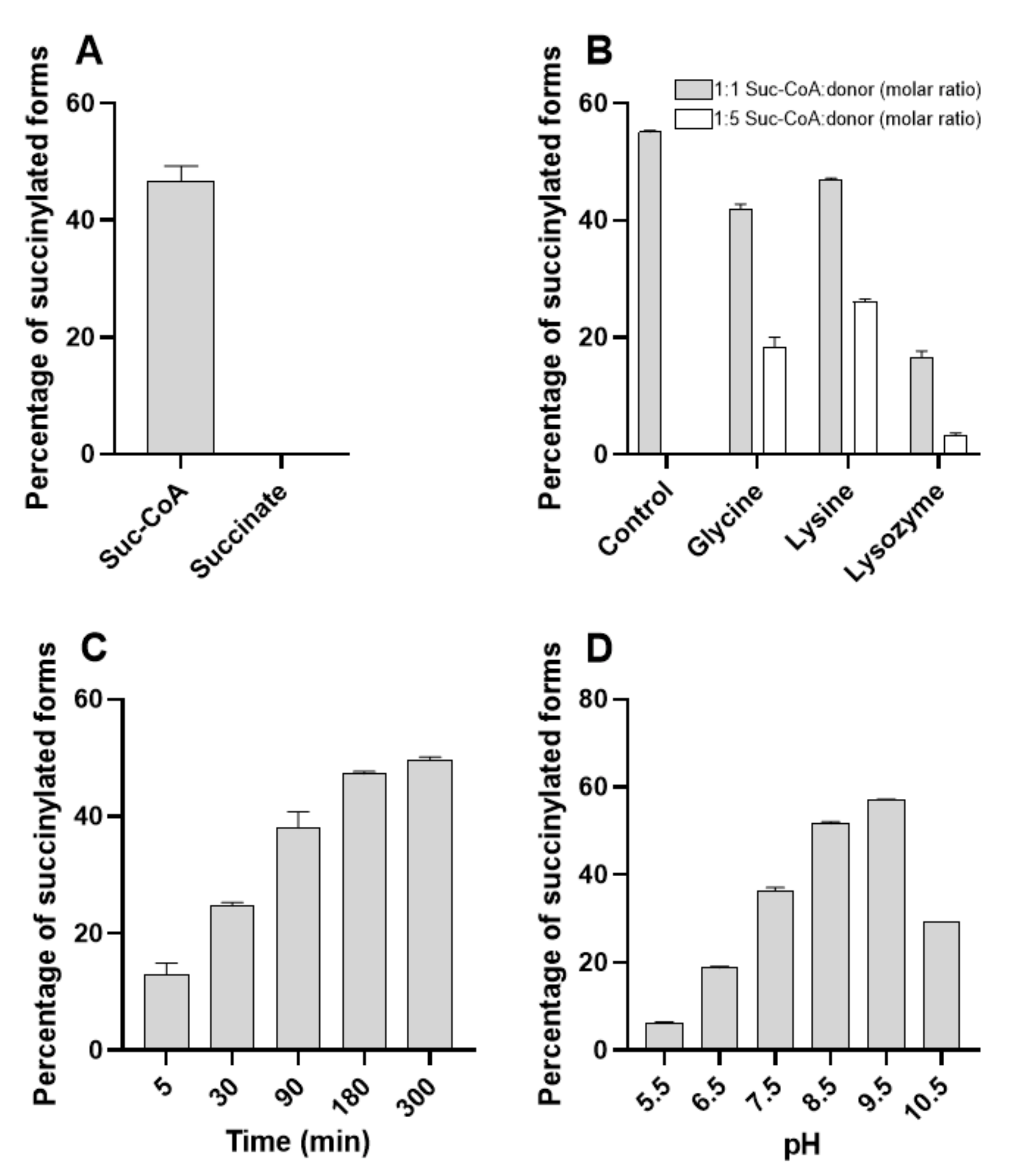
2.5. BacSp222 Succinylation Is a Nonenzymatic Process Dependent On Suc-CoA and pH, Whereas Desuccinylation Requires NAD+ and Cytoplasmic Enzymes
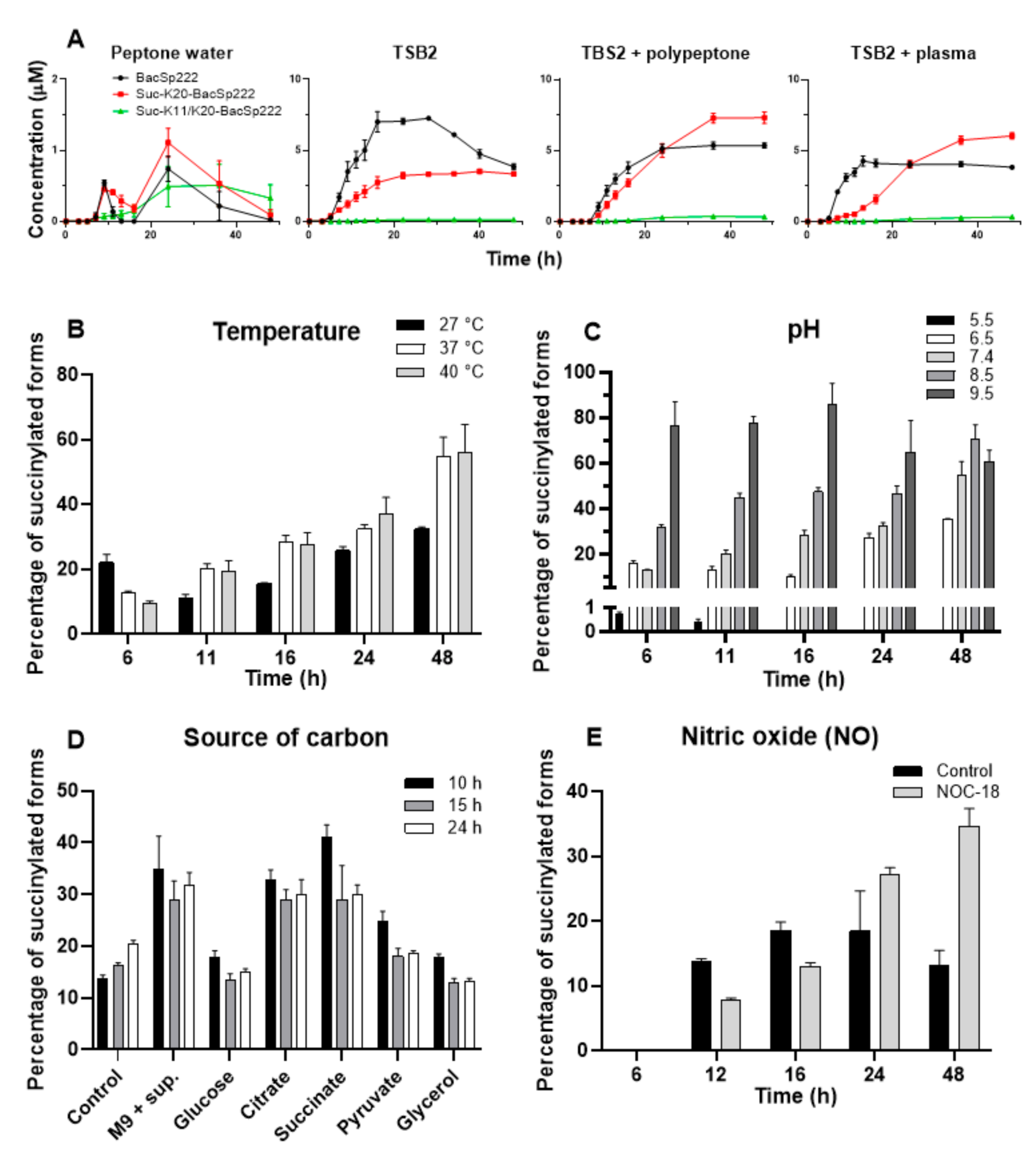
2.6. Environmental Factors Affect the Level of Excreted Succinylated BacSp222 Forms
3. Materials and Methods
3.1. Bacterial Strains and Culture Conditions
3.2. Eukaryotic Cells and Culture Conditions
3.3. Isolation of BacSp222 and Its Succinylated Forms
3.4. NMR Spectroscopy and Structural Calculations
3.5. CD Spectroscopy
3.6. Biochemical Techniques
3.7. Antibacterial Radial Diffusion Assay
3.8. Analysis of the Effect of Bacteriocin on Eukaryotic Cells
3.9. In Vitro Analysis of Succinylation and Desuccinylation of BacSp222
3.10. Analysis of the Influence of Environmental Factors on the Level of Succinylated Forms of BacSp222
3.11. Determination of Intracellular pH in Bacteria
3.12. Statistics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Z.; Tan, M.; Xie, Z.; Dai, L.; Chen, Y.; Zhao, Y. Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 2011, 7, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Alleyn, M.; Breitzig, M.; Lockey, R.; Kolliputi, N. The dawn of succinylation: A posttranslational modification. Am. J. Physiol. Physiol. 2018, 314, C228–C232. [Google Scholar] [CrossRef] [PubMed]
- Sreedhar, A.; Wiese, E.K.; Hitosugi, T. Enzymatic and metabolic regulation of lysine succinylation. Genes Dis. 2020, 7, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Weinert, B.T.; Schölz, C.; Wagner, S.A.; Iesmantavicius, V.; Su, D.; Daniel, J.A.; Choudhary, C. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013, 4, 842–851. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Wang, J.; Cheng, Z.; Gao, P.; Sun, J.; Chen, X.; Chen, C.; Wang, Y.; Wang, Z. Quantitative global proteome and lysine succinylome analyses provide insights into metabolic regulation and lymph node metastasis in gastric cancer. Sci. Rep. 2017, 7, 42053. [Google Scholar] [CrossRef]
- Li, X.; Hu, X.; Wan, Y.; Xie, G.; Li, X.; Chen, D.; Cheng, Z.; Yi, X.; Liang, S.; Tan, F. Systematic identification of the lysine succinylation in the protozoan parasite Toxoplasma Gondii. J. Proteome Res. 2014, 13, 6087–6095. [Google Scholar] [CrossRef]
- Yang, Y.; Gibson, G.E. Succinylation links metabolism to protein functions. Neurochem. Res. 2019, 44, 2346–2359. [Google Scholar] [CrossRef]
- Wagner, G.R.; Bhatt, D.P.; O’Connell, T.M.; Thompson, J.W.; Dubois, L.G.; Backos, D.S.; Yang, H.; Mitchell, G.A.; Ilkayeva, O.R.; Stevens, R.D.; et al. A class of reactive acyl-coa species reveals the non-enzymatic origins of protein acylation. Cell Metab. 2017, 25, 823–837. [Google Scholar] [CrossRef]
- Gläser, L.; Kuhl, M.; Jovanovic, S.; Fritz, M.; Vögeli, B.; Erb, T.J.; Becker, J.; Wittmann, C. A common approach for absolute quantification of short chain CoA thioesters in prokaryotic and eukaryotic microbes. Microb. Cell Factories 2020, 19. [Google Scholar] [CrossRef]
- Park, J.; Chen, Y.; Tishkoff, D.X.; Peng, C.; Tan, M.; Dai, L.; Xie, Z.; Zhang, Y.; Zwaans, B.M.M.; Skinner, M.E.; et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell 2013, 50, 919–930. [Google Scholar] [CrossRef]
- Zhang, K.; Xiong, Y.; Sun, W.; Wang, G.; Liu, W. Global proteomic analysis reveals widespread lysine succinylation in rice seedlings. Int. J. Mol. Sci. 2019, 20, 5911. [Google Scholar] [CrossRef]
- Xie, L.; Liu, W.; Li, Q.; Chen, S.; Xu, M.; Huang, Q.; Zeng, J.; Zhou, M.; Xie, J. First succinyl-proteome profiling of extensively drug-resistant mycobacterium tuberculosis revealed involvement of succinylation in cellular physiology. J. Proteome Res. 2015, 14, 107–119. [Google Scholar] [CrossRef]
- Zhou, H.; Finkemeier, I.; Guan, W.; Tossounian, M.A.; Wei, B.; Young, D.; Huang, J.; Messens, J.; Yang, X.; Zhu, J.; et al. Oxidative stress-triggered interactions between the succinyl- and acetyl-proteomes of rice leaves. Plant Cell Environ. 2018, 41, 1139–1153. [Google Scholar] [CrossRef]
- Yang, M.; Wang, Y.; Chen, Y.; Cheng, Z.; Gu, J.; Deng, J.; Bi, L.; Chen, C.; Mo, R.; Wang, X.; et al. Succinylome analysis reveals the involvement of lysine succinylation in metabolism in pathogenic mycobacterium tuberculosis. Mol. Cell. Proteom. 2015, 14, 796–811. [Google Scholar] [CrossRef]
- Zhao, Y.; Han, Y.; Sun, Y.; Wei, Z.; Chen, J.; Niu, X.; An, Q.; Zhang, L.; Qi, R.; Gao, X. Comprehensive succinylome profiling reveals the pivotal role of lysine succinylation in energy metabolism and quorum sensing of Staphylococcus epidermidis. Front. Microbiol. 2021, 11, 632367. [Google Scholar] [CrossRef]
- Xie, Z.; Dai, J.; Dai, L.; Tan, M.; Cheng, Z.; Wu, Y.; Boeke, J.D.; Zhao, Y. Lysine succinylation and lysine malonylation in histones. Mol. Cell. Proteom. 2012, 11, 100–107. [Google Scholar] [CrossRef]
- Colak, G.; Xie, Z.; Zhu, A.Y.; Dai, L.; Lu, Z.; Zhang, Y.; Wan, X.; Chen, Y.; Cha, Y.H.; Lin, H.; et al. Identification of lysine succinylation substrates and the succinylation regulatory enzyme CobB in Escherichia coli. Mol. Cell. Proteom. 2013, 12, 3509–3520. [Google Scholar] [CrossRef]
- Kidwai, S.A.; Ansari, A.A.; Salahuddin, A. Effect of succinylation (3 carboxypropionylation) on the conformation and immunological activity of ovalbumin. Biochem. J. 1976, 155, 171–180. [Google Scholar] [CrossRef]
- Habeeb, A.F.S.A. Quantitation of conformational changes on chemical modification of proteins: Use of succinylated proteins as a model. Arch. Biochem. Biophys. 1967, 121, 652–664. [Google Scholar] [CrossRef]
- Nandi, S.K.; Rakete, S.; Nahomi, R.B.; Michel, C.; Dunbar, A.; Fritz, K.S.; Nagaraj, R.H. Succinylation is a gain-of-function modification in human lens αb-crystallin. Biochemistry 2019, 58, 1260–1274. [Google Scholar] [CrossRef]
- Wladyka, B.; Piejko, M.; Bzowska, M.; Pieta, P.; Krzysik, M.; Mazurek, Ł.; Guevara-Lora, I.; Bukowski, M.; Sabat, A.J.; Friedrich, A.W.; et al. A peptide factor secreted by Staphylococcus pseudintermedius exhibits properties of both bacteriocins and virulence factors. Sci. Rep. 2015, 5, 14569. [Google Scholar] [CrossRef]
- Nowakowski, M.; Jaremko, Ł.; Wladyka, B.; Dubin, G.; Ejchart, A.; Mak, P. Spatial attributes of the four-helix bundle group of bacteriocins—The high-resolution structure of BacSp222 in solution. Int. J. Biol. Macromol. 2018, 107, 2715–2724. [Google Scholar] [CrossRef]
- Mak, P. Staphylococcal bacteriocins. In Pet-to-Man Traveling Staphylococci: A World in Progress; Savini, V., Ed.; Elsevier: Amsterdam, The Netherlands, 2018. [Google Scholar]
- Pieta, P.; Majewska, M.; Su, Z.; Grossutti, M.; Wladyka, B.; Piejko, M.; Lipkowski, J.; Mak, P. Physicochemical studies on orientation and conformation of a new bacteriocin BacSp222 in a planar phospholipid bilayer. Langmuir 2016, 32, 5653–5662. [Google Scholar] [CrossRef]
- Cross, A.S. What is a virulence factor? Crit. Care 2008, 12. [Google Scholar] [CrossRef]
- Briaud, P.; Carroll, R.K. Extracellular vesicle biogenesis and functions in gram-positive bacteria. Infect. Immun. 2020, 88. [Google Scholar] [CrossRef]
- Perez, R.H.; Zendo, T.; Sonomoto, K. Circular and leaderless bacteriocins: Biosynthesis, mode of action, applications, and prospects. Front. Microbiol. 2018, 9, 2085. [Google Scholar] [CrossRef]
- Xu, H.D.; Shi, S.P.; Wen, P.P.; Qiu, J.D. SuccFind: A novel succinylation sites online prediction tool via enhanced characteristic strategy. Bioinformatics 2015, 31, 3748–3750. [Google Scholar] [CrossRef]
- Jia, J.; Liu, Z.; Xiao, X.; Liu, B.; Chou, K.C. PSuc-Lys: Predict lysine succinylation sites in proteins with PseAAC and ensemble random forest approach. J. Theor. Biol. 2016, 394, 223–230. [Google Scholar] [CrossRef]
- Jia, J.; Liu, Z.; Xiao, X.; Liu, B.; Chou, K.C. ISuc-PseOpt: Identifying lysine succinylation sites in proteins by incorporating sequence-coupling effects into pseudo components and optimizing imbalanced training dataset. Anal. Biochem. 2016, 497, 48–56. [Google Scholar] [CrossRef]
- Hasan, M.; Kurata, H. GPSuc: Global prediction of generic and species-specific succinylation sites by aggregating multiple sequence features. PLoS ONE 2018, 13, e0200283. [Google Scholar] [CrossRef]
- Hasan, M.M.; Yang, S.; Zhou, Y.; Mollah, M.N.H. SuccinSite: A computational tool for the prediction of protein succinylation sites by exploiting the amino acid patterns and properties. Mol. BioSyst. 2016, 12, 786–795. [Google Scholar] [CrossRef] [PubMed]
- Ning, Q.; Zhao, X.; Bao, L.; Ma, Z.; Zhao, X. Detecting succinylation sites from protein sequences using ensemble support vector machine. BMC Bioinform. 2018, 19, 237. [Google Scholar] [CrossRef]
- Xu, Y.; Ding, Y.X.; Ding, J.; Lei, Y.H.; Wu, L.Y.; Deng, N.Y. ISuc-PseAAC: Predicting lysine succinylation in proteins by incorporating peptide position-specific propensity. Sci. Rep. 2015, 5, 10184. [Google Scholar] [CrossRef]
- Zeng, F.; Pang, H.; Chen, Y.; Zheng, H.; Li, W.; Ramanathan, S.; Hoare, R.; Monaghan, S.J.; Lin, X.; Jian, J. First succinylome profiling of Vibrio alginolyticus reveals key role of lysine succinylation in cellular metabolism and virulence. Front. Cell. Infect. Microbiol. 2021, 10, 626574. [Google Scholar] [CrossRef] [PubMed]
- Cochrane, S.A.; Surgenor, R.R.; Khey, K.M.W.; Vederas, J.C. Total synthesis and stereochemical assignment of the antimicrobial lipopeptide cerexin A1. Org. Lett. 2015, 17, 5428–5431. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.W.; Jaskolla, T.W.; Bochmann, S.; Kötter, P.; Wichelhaus, T.; Karas, M.; Stein, T.; Entian, K.D. Entianin, a novel subtilin-like lantibiotic from Bacillus subtilis subsp. spizizenii DSM 15029T with high antimicrobial activity. Appl. Environ. Microbiol. 2011, 77, 1698–1707. [Google Scholar] [CrossRef] [PubMed]
- Bowers, A.A.; Walsh, C.T.; Acker, M.G. Genetic Interception and structural characterization of thiopeptide cyclization precursors from Bacillus cereus. J. Am. Chem. Soc. 2010, 132, 12182–12184. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.E.; Hallett, M.B. Neutrophil cell shape change: Mechanism and signalling during cell spreading and phagocytosis. Int. J. Mol. Sci. 2019, 20, 1383. [Google Scholar] [CrossRef]
- Ohba, H.; Nishikawa, M.; Kimura, M.; Yamasaki, N.; Moriwaki, S.; Itoh, K. Cytotoxicity Induced by Erythrina variegata serine proteinase inhibitors in tumor hematopoietic stem cell lines. Biosci. Biotechnol. Biochem. 1998, 62, 1166–1170. [Google Scholar] [CrossRef][Green Version]
- Simon, E.J.; Shemin, D. The preparation of s-succinyl coenzyme A. J. Am. Chem. Soc. 1953, 75. [Google Scholar] [CrossRef]
- Sadhukhan, S.; Liu, X.; Ryu, D.; Nelson, O.D.; Stupinski, J.A.; Li, Z.; Chen, W.; Zhang, S.; Weiss, R.S.; Locasale, J.W.; et al. Metabolomics-assisted proteomics identifies succinylation and SIRT5 as important regulators of cardiac function. Proc. Natl. Acad. Sci. USA 2016, 113, 4320–4325. [Google Scholar] [CrossRef]
- Wagner, G.R.; Payne, R.M. Widespread and enzyme-independent Nε-acetylation and Nε-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J. Biol. Chem. 2013, 288, 29036–29045. [Google Scholar] [CrossRef]
- O’Neil, M.J. The Merck Index: An Encyclopedia of Chemicals, Drugs, and Biologicals, 15th ed.; Royal Society of Chemistry: Cambridge, UK, 2013. [Google Scholar]
- Bonar, E.; Bukowski, M.; Chlebicka, K.; Madry, A.; Bereznicka, A.; Kosecka-Strojek, M.; Dubin, G.; Miedzobrodzki, J.; Mak, P.; Wladyka, B. Human skin microbiota-friendly lysostaphin. Int. J. Biol. Macromol. 2021, 183, 852–860. [Google Scholar] [CrossRef]
- Aasen, I.M.; Møretrø, T.; Katla, T.; Axelsson, L.; Storrø, I. Influence of complex nutrients, temperature and ph on bacteriocin production by Lactobacillus sakei CCUG 42687. Appl. Microbiol. Biotechnol. 2000, 53, 159–166. [Google Scholar] [CrossRef]
- Padan, E.; Bibi, E.; Ito, M.; Krulwich, T.A. Alkaline PH homeostasis in bacteria: New insights. Biochim. Biophys. Acta Biomembr. 2005, 1717, 67–88. [Google Scholar] [CrossRef]
- Krulwich, T.A.; Sachs, G.; Padan, E. Molecular aspects of bacterial PH sensing and homeostasis. Nat. Rev. Microbiol. 2011, 9, 330–343. [Google Scholar] [CrossRef]
- Vallari, D.S.; Jackowski, S.; Rock, C.O. Regulation of pantothenate kinase by coenzyme A and its thioesters. J. Biol. Chem. 1987, 262, 2468–2471. [Google Scholar] [CrossRef]
- Kosono, S.; Tamura, M.; Suzuki, S.; Kawamura, Y.; Yoshida, A.; Nishiyama, M.; Yoshida, M. Changes in the acetylome and succinylome of bacillus subtilis in response to carbon source. PLoS ONE 2015, 10, e0131169. [Google Scholar] [CrossRef]
- Shibuta, S.; Mashimo, T.; Ohara, A.; Zhang, P.; Yoshiya, I. Intracerebroventricular administration of a nitric oxide-releasing compound, NOC-18, produces thermal hyperalgesia in rats. Neurosci. Lett. 1995, 187, 103–106. [Google Scholar] [CrossRef]
- Grosser, M.R.; Paluscio, E.; Thurlow, L.R.; Dillon, M.M.; Cooper, V.S.; Kawula, T.H.; Richardson, A.R. Genetic requirements for Staphylococcus aureus nitric oxide resistance and virulence. PLoS Pathog. 2018, 14, e1006907. [Google Scholar] [CrossRef]
- Wishart, D.S.; Bigam, C.G.; Yao, J.; Abildgaard, F.; Dyson, H.J.; Oldfield, E.; Markley, J.L.; Sykes, B.D. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR 1995, 6, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Braunschweiler, L.; Ernst, R.R. Coherence transfer by isotropic mixing: Application to proton correlation spectroscopy. J. Magn. Reason. 1983, 53, 521–528. [Google Scholar] [CrossRef]
- Jeener, J.; Meier, B.H.; Bachmann, P.; Ernst, R.R. Investigation of exchange processes by two-dimensional NMR spectroscopy. J. Chem. Phys. 1979, 71, 4546–4553. [Google Scholar] [CrossRef]
- Bodenhausen, G.; Ruben, D.J. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 1980, 69, 185–189. [Google Scholar] [CrossRef]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef]
- Goddard, T.D.; Kneller, D.G. Sparky—NMR Assignment and Integration Software. Available online: https://www.cgl.ucsf.edu/home/sparky/ (accessed on 9 June 2021).
- Güntert, P.; Mumenthaler, C.; Wüthrich, K. Torsion angle dynamics for NMR structure calculation with the new program dyana. J. Mol. Biol. 1997, 273, 283–298. [Google Scholar] [CrossRef]
- Shen, Y.; Bax, A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR 2013, 56, 227–241. [Google Scholar] [CrossRef]
- Schwieters, C.D.; Kuszewski, J.J.; Tjandra, N.; Marius Clore, G. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reason. 2003, 160, 65–73. [Google Scholar] [CrossRef]
- Rosato, A.; Tejero, R.; Montelione, G.T. Quality assessment of protein NMR structures. Curr. Opin. Struct. Biol. 2013, 23, 715–724. [Google Scholar] [CrossRef]
- Micsonai, A.; Wien, F.; Kernya, L.; Lee, Y.H.; Goto, Y.; Réfrégiers, M.; Kardos, J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. USA 2015, 112, E3095–E3103. [Google Scholar] [CrossRef]
- Micsonai, A.; Wien, F.; Bulyáki, É.; Kun, J.; Moussong, É.; Lee, Y.H.; Goto, Y.; Réfrégiers, M.; Kardos, J. BeStSel: A web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res. 2018, 46, W315–W322. [Google Scholar] [CrossRef]
- Wladyka, B.; Wielebska, K.; Wloka, M.; Bochenska, O.; Dubin, G.; Dubin, A.; Mak, P. Isolation, biochemical characterization, and cloning of a bacteriocin from the poultry-associated Staphylococcus aureus strain CH-91. Appl. Microbiol. Biotechnol. 2013, 97, 7229–7239. [Google Scholar] [CrossRef]
- Diribe, O.; Thomas, S.; AbuOun, M.; Fitzpatrick, N.; la Ragione, R. Genotypic relatedness and characterization of Staphylococcus pseudintermedius associated with post-operative surgical infections in dogs. J. Med. Microbiol. 2015, 64, 1074–1081. [Google Scholar] [CrossRef][Green Version]
- AAT Bioquest. Four Parameter Logistic (4PL) Curve Calculator. Available online: https://www.aatbio.com/tools/four-parameter-logistic-4pl-curve-regression-online-calculator (accessed on 20 April 2021).

| Bacteria | Residual Antibacterial Activity in Relation to the Activity of Unmodified BacSp222(Percent ± SD) | |
|---|---|---|
| Suc-K20-BacSp222 | Suc-K11/K20-BacSp222 | |
| Bacillus subtilis ATCC 6633 | 72.24 ± 4.42 | 32.66 ± 6.78 |
| Lactococcus lactis ŁOCK 0871 strain 239 | 80.32 ± 3.67 | 48.95 ± 6.21 |
| Micrococcus luteus ATCC 4698 | 92.97 ± 0.56 | 41.97 ± 4.77 |
| Staphylococcus aureus ATCC 25923 | 33.20 ± 5.42 | N.a. |
| Staphylococcus epidermidis ATCC 35547 | 64.19 ± 7.80 | 18.00 ± 7.55 |
| Staphylococcus intermedius ATCC 29663 | 3.24 ± 2.51 | N.a. |
| Staphylococcus pseudintermedius 222 | N.a. | N.a. |
| Staphylococcus pseudintermedius 22221 | 49.51 ± 11.44 | N.a. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Śmiałek, J.; Nowakowski, M.; Bzowska, M.; Bocheńska, O.; Wlizło, A.; Kozik, A.; Dubin, G.; Mak, P. Structure, Biosynthesis, and Biological Activity of Succinylated Forms of Bacteriocin BacSp222. Int. J. Mol. Sci. 2021, 22, 6256. https://doi.org/10.3390/ijms22126256
Śmiałek J, Nowakowski M, Bzowska M, Bocheńska O, Wlizło A, Kozik A, Dubin G, Mak P. Structure, Biosynthesis, and Biological Activity of Succinylated Forms of Bacteriocin BacSp222. International Journal of Molecular Sciences. 2021; 22(12):6256. https://doi.org/10.3390/ijms22126256
Chicago/Turabian StyleŚmiałek, Justyna, Michał Nowakowski, Monika Bzowska, Oliwia Bocheńska, Agnieszka Wlizło, Andrzej Kozik, Grzegorz Dubin, and Paweł Mak. 2021. "Structure, Biosynthesis, and Biological Activity of Succinylated Forms of Bacteriocin BacSp222" International Journal of Molecular Sciences 22, no. 12: 6256. https://doi.org/10.3390/ijms22126256
APA StyleŚmiałek, J., Nowakowski, M., Bzowska, M., Bocheńska, O., Wlizło, A., Kozik, A., Dubin, G., & Mak, P. (2021). Structure, Biosynthesis, and Biological Activity of Succinylated Forms of Bacteriocin BacSp222. International Journal of Molecular Sciences, 22(12), 6256. https://doi.org/10.3390/ijms22126256






