Pathogenic Roles of Autoantibodies and Aberrant Epigenetic Regulation of Immune and Connective Tissue Cells in the Tissue Fibrosis of Patients with Systemic Sclerosis
Abstract
1. Introduction
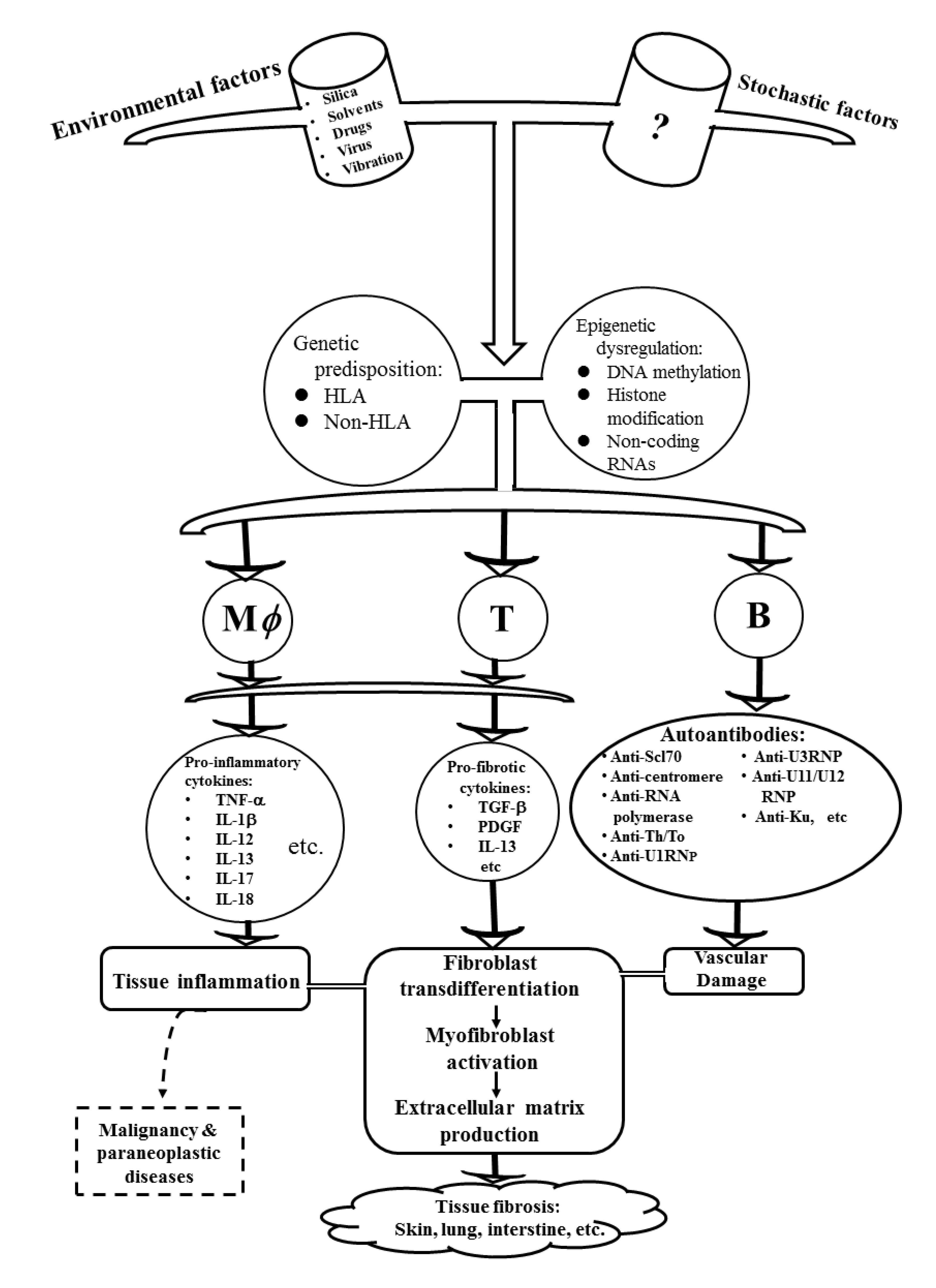
2. The Pathogenic Factors Contributing to the Development of SSc
2.1. Genetic Predisposition in Patients with SSc
2.2. Environmental Risk Factors and Their Modes of Action Associated with SSc
2.3. Aberrant Epigenetic Regulations in SSc
2.3.1. Abnormal DNA Methylation in the Immune-Related Cells of Patients with SSc
2.3.2. Abnormal Histone Modifications in the Immune-Related Cells of Patients with SSc
2.3.3. Enhanced DNA Hypomethylation in the Dermal Fibroblasts of SSc Patients
3. Cellular and Molecular Mechanisms for Tissue Fibrosis in Patients with SSc
3.1. Pathophysiology of Myofibroblasts and Other Connective Tissue Cell Lineages in Patients with SSc
3.1.1. Aberrant Ontogenesis of Mesenchymal Stem Cells (MSCs) and Abnormal Cellular Physiology of Their Descendant Vascular Smooth Muscle and Endothelial Cells in Patients with SSc
3.1.2. Histological Characteristics and the Biochemical Constituents in Tissue Fibrosis of Patients with SSc
3.2. Tissue Fibrosis-Related Cytokines and Their Signaling Pathways in SSc
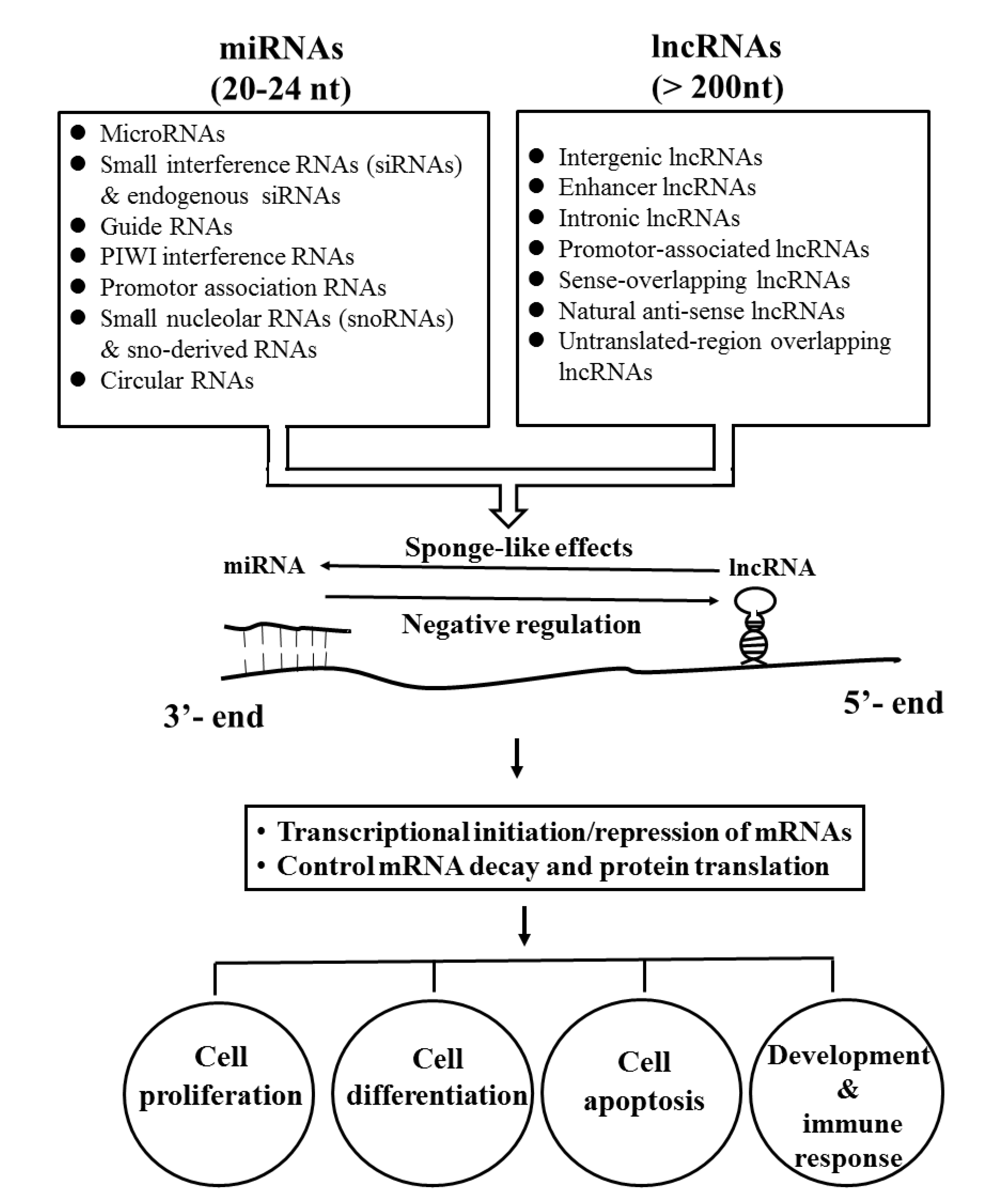
3.3. Tissue Fibrosis-Related ncRNAs and Their Signaling Pathways in Patients with SSc
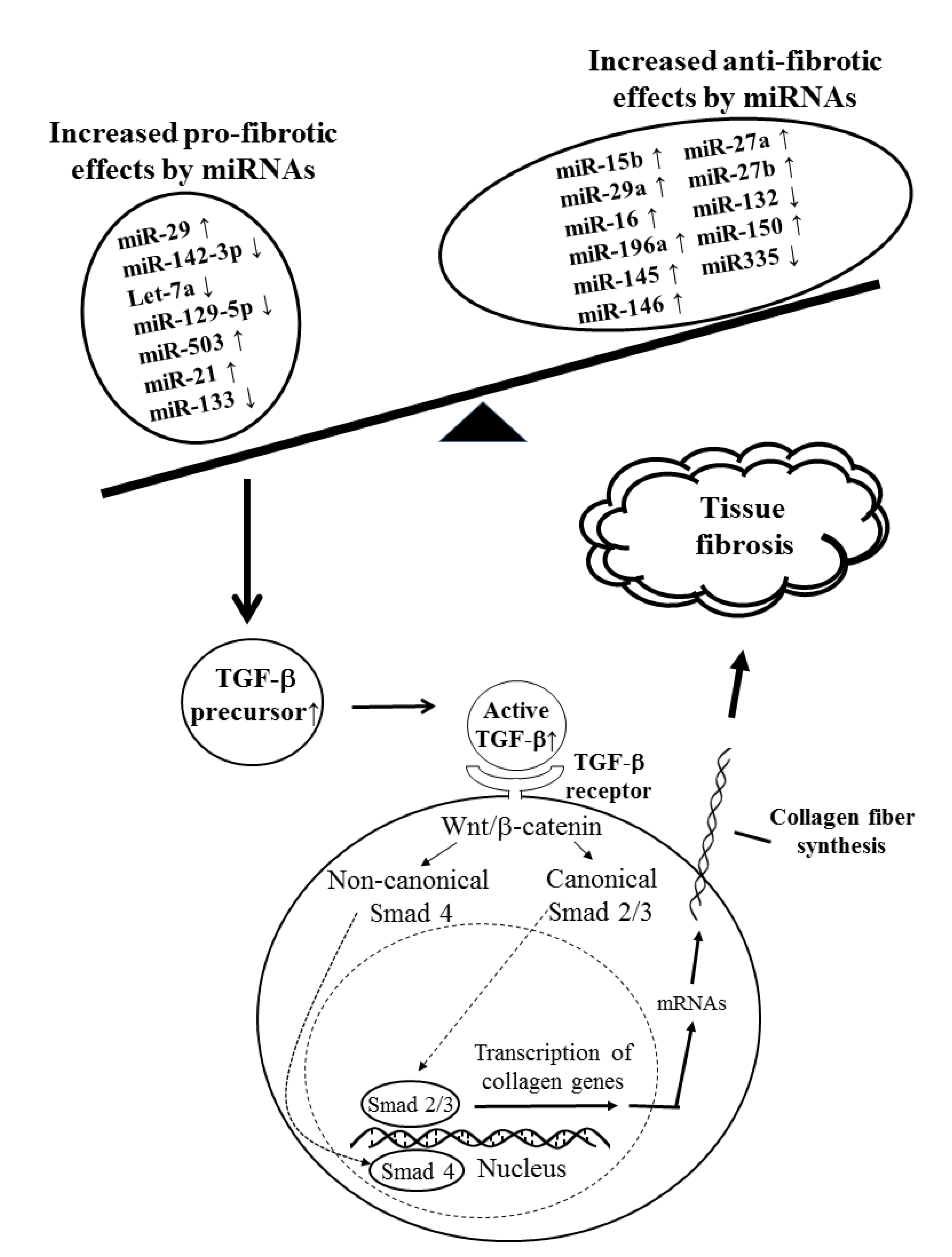
3.3.1. Tissue Fibrosis-Related miRs in SSc
3.3.2. Tissue Fibrosis Relevant Long Non-Coding RNA in SSc
4. Potential Biomarkers and New Therapeutic Strategy for Patients with SSc
5. Conclusions and Prospects
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| anti-CENP | anti-centromere protein antibody |
| anti-ICAM-1 | anti-intercellular adhesion molecule-1 antibody |
| anti-PDGFR | anti-platelet derived growth factor receptor antibody |
| anti-TOPO-1 | anti-topoisomerase-1 antibody (anti-Scl-70) |
| A-MSC | adipocyte derived mesenchymal stem cell |
| AS | anti-sense non-coding RNA |
| BM-MSC | bone marrow derived mesenchymal stem cell |
| CIR | cartilage injury related long non-coding RNA |
| COL | collagen |
| CTGF | connective tissue growth factor |
| DC | dendritic cell |
| EC | endothelial cell |
| ECM | extracellular matrix |
| EndoMT | trans-differentiation from endothelial cell to mesenchymal cell |
| EV | extracellular vesicle |
| FB | fibroblast |
| HIF | hypoxia-induced factors |
| HPASMC | human pulmonary arterial smooth muscle cell |
| IL | interleukin |
| ILD | interstitial lung disease |
| lncRNA | long non-coding RNA |
| MFB | myofibroblast |
| miR | microRNA |
| MMP | matrix metalloproteinase |
| mRNA | messenger RNA |
| MSC | mesenchymal stem cell |
| NLRP-3 | neuronal apoptosis inhibitor protein, leucine-rich repeat, pyrin domain containing protein 3 |
| NRIR | a negative regulator of interferon response |
| PDGF | platelet derived growth factor |
| S100A4 | S100 calcium-binding protein A4 |
| SNAI2 | Snail superfamily of C2H2-type zinc finger transcription factor 2 |
| SPARC | secreted protein and rich in cysteine |
| SSc | systemic sclerosis |
| SSc-dMVEC | dermal microvascular endothelial cell obtained from systemic sclerosis |
| Tang | angiogenic T cell |
| TIMP | tissue inhibitor of metalloproteinase |
| TGF-β | transforming growth factor-β |
| Th | helper T cell |
| TWIST1 | Twist related protein 1 or class A basic helix–loop–helix protein 38 (bHLHa38) |
| VE | vascular endothelium |
| VSMC | vascular smooth muscle cell |
References
- Harris, M.L.; Rosen, A. Autoimmunity in scleroderma: The origin, pathogenic role, and clinical significance of autoantibodies. Curr. Opin. Rheumatol. 2003, 15, 778–784. [Google Scholar] [CrossRef] [PubMed]
- Nihtyanova, S.I.; Denton, C.P. Autoantibodies as predictive tools in systemic sclerosis. Nat. Rev. Rheumatol. 2010, 6, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Eckes, B.; Moinzadeh, P.; Sengle, G.; Hunzelmann, N.; Krieg, T. Molecular and cellular basis of scleroderma. J. Mol. Med. 2014, 92, 913–924. [Google Scholar] [CrossRef] [PubMed]
- Ciechomska, M.; van Laar, J.; O’Reilly, S. Current frontiers in systemic sclerosis pathogenesis. Exp. Dermatol. 2015, 24, 401–406. [Google Scholar] [CrossRef]
- Denton, C.P.; Khanna, D. Systmic sclerosis. Lancet 2017, 390, 1685–1699. [Google Scholar] [CrossRef]
- Furue, M.; Mitoma, C.; Mitoma, H.; Tsuji, G.; Chiba, T.; Nakahara, T.; Uchi, H.; Kadono, T. Pathogenesis of systemic sclerosis-current concept and emerging treatments. Immunol. Res. 2017, 65, 790–797. [Google Scholar] [CrossRef]
- Brown, M.; O’Reilly, S. The immunopathogenesis of fibrosis in systemic sclerosis. Clin. Exp. Immunol. 2019, 195, 310–321. [Google Scholar] [CrossRef]
- Yoshizaki, A. Pathogenic roles of B lymphocytes in systemic sclerosis. Immunol. Lett. 2018, 195, 76–82. [Google Scholar] [CrossRef]
- Varga, J.; Abraham, D. Systemic sclerosis: A prototypic multisystem fibrotic disorder. J. Clin. Invest. 2007, 117, 557–567. [Google Scholar] [CrossRef]
- Yamamoto, T. Scleroderma-pathophysiology. Eur. J. Dermatol. 2009, 19, 14–24. [Google Scholar] [CrossRef]
- Gabrielli, A.; Avvedimento, E.V.; Krieg, T. Scleroderma. N. Engl. J. Med. 2009, 360, 1989–2003. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, S.; Wei, J.; Varga, J. Understanding fibrosis in systemic sclerosis: Shifting paradigms, emerging opportunities. Nat. Rev. Rheumatol. 2011, 8, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Manetti, M.; Pratesi, S.; Romano, E.; Bellando-Randone, S.; Rosa, I.; Guiducci, S.; Fioretto, B.S.; Ibba-Manneschi, L.; Maggi, E.; Matucci-Cerinic, M. Angiogenic T cell expansion correlates with severity of peripheral vascular damage in systemic sclerosis. PLoS ONE 2017, 12, e0183102. [Google Scholar] [CrossRef] [PubMed]
- Truchetet, M.E.; Demoures, B.; Eduardo Guimaraes, J.; Bertrand, A.; Laurent, P.; Jolivel, V.; Douchet, I.; Jacquemin, C.; Khoryati, L.; Daffau, P.; et al. Platelets induce thymic stromal lymphopoietin production by endothelial cells: Contribution to fibrosis in human systemic sclerosis. Arthritis Rheumatol. 2016, 68, 2784–2794. [Google Scholar] [CrossRef] [PubMed]
- Benyamine, A.; Magalon, J.; Sabatier, F.; Lyonnet, L.; Robert, S.; Dumoulin, C.; Morange, S.; Mazodier, K.; Kaplanski, G.; Reynaud-Gaubert, M.; et al. Natural killer cells exhibit a peculiar phenotypic profile in systemic sclerosis and are potent inducer of endothelial microparticles release. Front. Immunol. 2018, 9, 1665. [Google Scholar] [CrossRef] [PubMed]
- Steen, V.D.; Powell, D.L.; Medsger, T.A., Jr. Clinical correlations and prognosis based on serum autoantibodies in patients with systemic sclerosis. Arthritis Rheum. 1988, 31, 196–203. [Google Scholar] [CrossRef]
- Hénault, J.; Tremblay, M.; Clément, I.; Raymond, Y.; Senécal, J.L. Direct binding of anti-DNA topoisomerase I autoantibodies to the cell surface of fibroblasts in patients with systemic sclerosis. Arthritis Rheum. 2004, 50, 3265–3274. [Google Scholar] [CrossRef]
- Tan, E.M.; Rodnan, G.P.; Garcia, I.; Moroi, Y.; Fritzler, M.J.; Peebles, C. Diversity of anti-nuclear antibodies in progressive systemic sclerosis: Anti-centromere antibodies and its relationship to CREST syndrome. Arthritis Rheum. 1980, 23, 617–625. [Google Scholar] [CrossRef]
- Weiner, E.S.; Earnshaw, W.C.; Senécal, J.L.; Bordwell, B.; Johnson, P.; Rothfield, N.F. Clinical associations of anti-centromere antibodies and antibodies to topoisomerase I: A study of 355 patients. Arthritis Rheum. 1988, 31, 378–385. [Google Scholar] [CrossRef]
- Ferri, C.; Valentini, G.; Cozzi, F.; Sebastiani, M.; Michelassi, C.; La Montagna, G.; Bullo, A.; Cazzato, M.; Tirri, E.; Storino, F.; et al. Systemic sclerosis: Demographic, clinical, and serological features and survival in 1,012 Italian patients. Medicine 2002, 81, 139–153. [Google Scholar] [CrossRef]
- Rothfield, N.F. Autoantibodies in scleroderma. Rheum. Dis. Clin. North Am. 1992, 18, 483–498. [Google Scholar] [PubMed]
- Harvey, G.R.; McHugh, N.J. Serological abnormalities in systemic sclerosis. Curr. Opin. Rheumatol. 1999, 11, 495–502. [Google Scholar] [CrossRef]
- Arnett, F.C.; Reveille, J.D.; Goldstein, R.; Pollard, K.M.; Leaird, K.; Smith, E.A.; Leroy, C.; Fritzler, M.J. Autoantibodies to fibrillarin in systemic sclerosis (scleroderma). An immunogenetic, serologic and clinical analysis. Arthritis Rheum 1996, 39, 1151–1560. [Google Scholar] [CrossRef]
- Okano, Y.; Steen, V.D.; Mesger, T.A., Jr. Autoantibody to U3 nucleolar ribonucleoprotein (fibrillarin) in patients with systemic sclerosis. Arthritis Rheum. 1992, 35, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Sacks, D.G.; Okano, Y.; Steen, V.D.; Curtiss, E.; Shapiro, L.S.; Mesger, T.A., Jr. Isolated pulmonary hypertension in systemic sclerosis with diffuse cutaneous involvement: Association with serum anti-U3RNP antibody. J. Rheumatol. 1996, 23, 639–642. [Google Scholar] [PubMed]
- Yang, J.M.; Hildebrandt, B.; Luderschmidt, C.; Pollard, K.M. Human scleroderma sera contain autoantibodies to protein components specific to the U3 small nucleolar RNP complex. Arthritis Rheum. 2003, 48, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Gilliam, A.C.; Steitz, J.A. Rare scleroderma autoantibodies to the U11 small nuclear ribonucleoprotein and to the trimethylguanosine cap of U small nuclear RNAs. Proc. Natl. Acad. Sci. USA 1993, 90, 6781–6785. [Google Scholar] [CrossRef]
- Fertig, N.; Domsic, R.T.; Rodriguez-Reyna, T.; Kuwana, M.; Lucas, M.; Medsger, T.A., Jr.; Feghali-Bostwick, C.A. Anti-U11/U12 RNP antibodies in systemic sclerosis: A new serologic marker associated with pulmonary fibrosis. Arthritis Rheum. 2009, 61, 958–965. [Google Scholar] [CrossRef]
- Ulanet, D.B.; Wigley, F.M.; Gelber, A.C.; Rosen, A. Autoantibodies against B23, a nucleolar phosphoprotein, occur in scleroderma and are associated with pulmonary hypertension. Arthritis Rheum. 2003, 49, 85–92. [Google Scholar] [CrossRef]
- Chung, L.; Utz, P.J. Antibodies in scleroderma: Direct pathogenicity and phenotypic associations. Curr. Rheumatol. Rep. 2004, 6, 156–163. [Google Scholar] [CrossRef]
- Hoa, S.; Hudson, M.; Troyanov, Y.; Proudman, S.; Walker, J.; Stevens, W.; Nikpour, M.; Assassi, S.; Mayes, M.D.; Wang, M.; et al. Single-specificity and anti-Ku antibodies in an international cohort of 2140 systemic sclerosis subjects: Clinical associations. Medicine 2016, 95, e4713. [Google Scholar] [CrossRef]
- Liaskos, C.; Marou, E.; Simopoulou, T.; Barmakoudi, M.; Efthymiou, G.; Scheper, T.; Meyer, W.; Bogdanos, D.P.; Sakkas, L.I. Disease-related autoantibody profile in patients with systemic sclerosis. Autoimmunity 2017, 50, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Mitri, G.M.; Lucas, M.; Fertig, N.; Steen, V.D.; Medsger, T.A., Jr. A comparison between anti-Th/To- and anticentromere antibody-positive systemic sclerosis patients with limited cutaneous involvement. Arthritis Rheum. 2003, 48, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Mahler, M.; Fritzler, M.J.; Satoh, M. Autoantibodies to the mitochondrial RNA processing (MRP) complex also known as Th/To autoantigen. Autoimmun Rev. 2015, 14, 254–257. [Google Scholar] [CrossRef] [PubMed]
- Renaudineau, Y.; Revelen, R.; Levy, Y.; Salojin, K.; Gilburg, B.; Shoenfeld, Y.; Youinou, P. Anti-endothelial cell antibodies in systemic sclerosis. Clin. Diagn. Lab. Immunol. 1999, 6, 156–160. [Google Scholar] [CrossRef] [PubMed]
- Chizzolini, C.; Raschi, E.; Rezzonico, R.; Testoni, G.; Mallone, R.; Gabrielli, A.; Facchini, A.; Del Papa, N.; Borghi, M.O.; Dayer, J.M.; et al. Autoantibodies to fibroblasts induce a proadhesive and proinflammatory fibroblast phenotype in patients with systemic sclerosis. Arthritis Rheum. 2002, 46, 1602–1613. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Hayakawa, I.; Hasegawa, M.; Fujimoto, M.; Takehara, K. Function blocking autoantibodies against matrix metalloproteinase-1 in patients with systemic sclerosis. J. Invest. Dermatol. 2003, 120, 542–547. [Google Scholar] [CrossRef]
- Goldblatt, F.; Gordon, T.P.; Waterman, S.A. Antibody-mediated gastrointestinal dysmotility in scleroderma. Gastroenterology 2002, 123, 1144–1150. [Google Scholar] [CrossRef]
- Svegliati, S.; Amico, D.; Spadoni, T.; Fischetti, C.; Finke, D.; Moroncini, G.; Paolini, C.; Tonnini, C.; Grieco, A.; Rovinelli, M.; et al. Agonistic anti-PDGF receptor autoantibodies from patients with systemic sclerosis impact human pulmonary artery smooth muscle cells function in vitro. Front. Immunol. 2017, 8, 75. [Google Scholar]
- Malia, R.G.; Greaves, M.; Rowlands, L.M.; Lawrence, A.C.; Hume, A.; Rowell, N.R.; Moult, J.; Holt, C.M.; Lindsey, N.; Hughes, P. Anticardiolipin antibodies in systemic sclerosis: Immunological and clinical associations. Clin. Exp. Immunol. 1988, 73, 456–460. [Google Scholar]
- Altorok, N.; Wang, Y.; Bashar, K. Endothelial dysfunction in systemic sclerosis. Curr. Opin. Rheumatol. 2014, 26, 615–620. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Tan, F.K.; Milewicz, D.M.; Guo, X.; Bona, C.A.; Arnett, F.C. Autoantibodies to fibrillin-1 activate normal human fibroblasts in culture through the TGF-β pathway to recapitulate the “scleroderma phenotype”. J. Immunol. 2005, 175, 4555–4560. [Google Scholar] [CrossRef] [PubMed]
- Tan, F.K.; Arnett, F.C.; Antohi, S.; Saito, S.; Mirarchi, A.; Spiera, H.; Sasaki, T.; Shoichi, O.; Takeuchi, K.; Pandey, J.P.; et al. Autoantibodies to the extracellular matrix microfibrillar protein, fibrillin-1, in patients with scleroderma and other connective tissue diseases. J. Immunol. 1999, 163, 1066–1072. [Google Scholar]
- Shen, C.-Y.; Li, K.-J.; Lai, P.-H.; Yu, C.-L.; Hsieh, S.-C. Anti-CENP-B and anti-TOPO-1-containing sera from systemic sclerosis-related diseases with Raynaud’s phenomenon induce vascular endothelial cell senescence not via classical p53-p21 pathway. Clin. Rheumatol. 2018, 37, 749–756. [Google Scholar] [CrossRef]
- Bossini-Castillo, L.; López-Isac, E.; Mayes, M.D.; Martin, J. Genetics of systemic sclerosis. Semin. Immunopathol. 2015, 37, 443–451. [Google Scholar] [CrossRef]
- Korman, B.D.; Criswell, L.A. Recent advances in the genetics of systemic sclerosis: Toward biological and clinical significance. Curr. Rheumatol. Rep. 2015, 17, 21. [Google Scholar] [CrossRef] [PubMed]
- Chairta, P.; Nicolaou, P.; Christodoulou, K. Genomic and genetic studies of systemic sclerosis: A systematic review. Human Immunol. 2017, 78, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Tsou, P.-S.; Sawalha, A.H. Unfolding the pathogenesis of scleroderma through genomics and epigenomics. J. Autoimmun. 2017, 83, 73–94. [Google Scholar] [CrossRef] [PubMed]
- Angiolilli, C.; Marut, W.; van der Kroef, M.; Chouri, E.; Reedquist, K.A.; Radstake, T.R.D.J. New insights into the genetics and epigenetics of systemic sclerosis. Nat. Rev. Rheumatol. 2018, 14, 657–673. [Google Scholar] [CrossRef]
- Dolcino, M.; Pelosi, A.; Fiore, P.F.; Patuzzo, G.; Tinazzi, E.; Lunardi, C.; Puccetti, A. Gene profiling in patients with systemic sclerosis reveals the presence of oncogenic gene signatures. Front. Immunol. 2018, 9, 449. [Google Scholar] [CrossRef]
- Maria, A.T.J.; Partouche, L.; Goulabchand, R.; Rivière, S.; Rozier, P.; Bourgier, C.; Le Quellec, A.; Morel, J.; Noël, D.; Guilpain, P. Intriguing relationships between cancer and systemic sclerosis: Role of the immune system and other contributors. Front. Immunol. 2019, 9, 3112. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.A.; Rosen, A.; Hummers, L.; Wigley, F.; Casciola-Rosen, L. Close temporal relationship between onset of cancer and scleroderma in patients with RNA polymerase I/III antibodies. Arthritis Rheum. 2010, 62, 2787–2795. [Google Scholar] [CrossRef] [PubMed]
- Joseph, C.G.; Darrah, E.; Shah, A.A.; Skora, A.D.; Casciola-Rosen, L.A.; Wigley, F.M.; Boin, F.; Fava, A.; Thoburn, C.; Kinde, I.; et al. Association of the autoimmune disease scleroderma with an immunologic response to cancer. Science 2014, 343, 152–157. [Google Scholar] [CrossRef]
- Moinzadeh, P.; Fonseca, C.; Hellmich, M.; Shah, A.A.; Chighizola, C.; Denton, C.P.; Ong, V.H. Association of anti-RNA polymerase III autoantibodies and cancer in scleroderma. Arthritis Res. Ther. 2014, 16, R53. [Google Scholar] [CrossRef] [PubMed]
- Mora, G.F. Systemic sclerosis: Environmental factors. J. Rheumatol. 2009, 36, 2383–2396. [Google Scholar] [CrossRef] [PubMed]
- Dospinescu, P.; Jones, G.T.; Basu, N. Environmental risk factors in systemic sclerosis. Curr. Opin. Rheumatol. 2013, 25, 179–183. [Google Scholar] [CrossRef]
- Marie, I.; Gehanno, J.-F.; Bubenheim, M.; Duval-Modeste, A.B.; Joly, P.; Dominique, S.; Bravard, P.; Noël, D.; Cailleux, A.F.; Weber, J.; et al. Prospective study to evaluate the association between systemic sclerosis and occupational exposure and review of the literature. Autoimmun. Rev. 2014, 13, 151–156. [Google Scholar] [CrossRef]
- Marie, I.; Gehanno, J.-F. Environmental risk factors of systemic sclerosis. Semin. Immunopathol. 2015, 37, 463–473. [Google Scholar] [CrossRef]
- De Martinis, M.; Ciccarelli, F.; Sirufo, M.M.; Ginaldi, L. An overview of environmental risk factors in systemic sclerosis. Expert Rev. Clin. Immunol. 2016, 12, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Walecka, I.; Roszkiewicz, M.; Malewska, A. Potential occupational and environmental factors in SSc onset. Ann. Agric. Environ. Med. 2018, 25, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Dias de Rojas, F.; Castro Garcia, M.; Abaitua Borda, I.; Alonso Gordo, J.M.; Posada de la Paz, M.; Kilbourne, E.M.; Tabuenca Oliver, J.M. The association of oil ingestion with toxic oil syndrome in two convents. Am. J. Epidemiol. 1987, 125, 907–911. [Google Scholar] [CrossRef] [PubMed]
- Marie, I. Systmic sclerosis and exposure to heavy metals. Autoimmun. Rev. 2019, 18, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Kanherkar, R.R.; Bhatia-Dey, N.; Csoka, A.B. Epigenetics across the human lifespan. Front. Cell. Dev. Biol. 2014, 2, 49. [Google Scholar] [CrossRef] [PubMed]
- Portela, A.; Esteller, M. Epigenetic modifications and human disease. Nat. Biotechnol. 2010, 28, 1057–1068. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Tajima, S.; Suetake, I.; Takeshita, K.; Nakagawa, A.; Kimura, H. Domain structure of the Dnmt 1, Dnmt 3a and Dnmt 3b DNA methyltransferases. Adv. Exp. Med. Biol. 2016, 945, 63–86. [Google Scholar]
- Wu, X.; Zhang, Y. TET-mediated active DNA demethylation: Mechanism, function, and beyond. Nat. Rev. Genet. 2017, 18, 517–534. [Google Scholar] [CrossRef] [PubMed]
- Peterson, C.L.; Laniel, M.A. Histone and histone modifications. Curr. Biol. 2004, 14, R546–R551. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Sabari, B.R.; Garcia, B.A.; Allis, C.D.; Zhao, Y. SnapShot: Histone modifications. Cell 2014, 159, 458. [Google Scholar] [CrossRef]
- Tsou, P.-S. Epigenetic control of scleroderma: Current knowledge and future perspectives. Curr. Rheumatol. Rep. 2019, 21, 69. [Google Scholar] [CrossRef]
- Bartel, D.P. microRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Han, P.; Chang, C.-P. Long non-coding RNA and chromatin remodeling. RNA Biol. 2015, 12, 1094–1098. [Google Scholar] [CrossRef] [PubMed]
- Rashid, F.; Shah, A.; Shan, G. Long non-coding RNAs in the cytoplasm. Genom. Proteom. Bioinform. 2016, 14, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, C. Coding or non-coding, the converging concepts of RNAs. Front. Genet. 2019, 10, 496. [Google Scholar] [CrossRef]
- Quinn, J.J.; Ilik, I.A.; Qu, K.; Georqiev, P.; Chu, C.; Akhtar, A.; Chang, H.Y. Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification. Nat. Biotechnol. 2014, 32, 933–940. [Google Scholar] [CrossRef]
- Bayoumi, A.S.; Sayed, A.; Broskova, Z.; Teoh, J.-P.; Wilson, J.; Su, H.; Tang, Y.-L.; Kim, I. Cross-talk between long non-coding RNAs and microRNAs in health and disease. Int. J. Mol. Sci. 2016, 17, 356. [Google Scholar] [CrossRef]
- Yamamura, S.; Imai-Sumida, M.; Tanaka, Y.; Dahiya, R. Interaction and cross-talk between non-coding RNAs. Cell. Mol. Life Sci. 2018, 75, 467–484. [Google Scholar] [CrossRef]
- Lei, W.; Luo, Y.; Lei, W.; Luo, Y.; Yan, K.; Zhao, S.; Li, Y.; Qui, X.; Zhou, Y.; Long, H.; et al. Abnormal DNA methylation in CD4+T cells from patients with systemic lupus erythematosus, systemic sclerosis and dermatomyositis. Scand. J. Rheumatol. 2009, 38, 369–374. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Wang, Q.; Sun, X.H.; Liu, R.Z.; Shu, Y.; Kanekura, T.; Huang, J.H.; Li, Y.P.; Wang, J.C.; Zhao, M.; et al. DNA hypermethylation of the forkhead box protein 3 (FOXP3) promoter in CD4+T cells of patients with systemic sclerosis. Br. J. Dermatol 2014, 171, 39–47. [Google Scholar] [CrossRef]
- Almanzar, G.; Klein, M.; Schmalzing, M.; Hilliqardt, D.; El Hajj, N.; Kneitz, H.; Wild, V.; Rosenwald, A.; Benoit, S.; Hamm, H.; et al. Disease manifestation and inflammatory activity as modulators of Th17/Treg balance and RORC/FoxP3 methylation in systemic sclerosis. Int. Arch. Allergy Immunol. 2016, 171, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Zhu, C.; Mi, W.; Chen, T.; Zhao, H.; Zuo, X.; Luo, H.; Li, Q.-Z. Integration of genome-wide DNA methylation and transcription uncovered aberrant methylation-regulated genes and pathways in the peripheral blood mononuclear cells of systemic sclerosis. Int. J. Rheumatol. 2018, 2018, 7342472. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Yang, Y.; Luo, Y.Y.; Yin, Y.X.; Wang, Q.; Li, Y.P.; Kanekura, T.; Wang, J.C.; Liang, G.P.; Zhao, M.; et al. Aberrant histone modification in peripheral blood B cells from patients with systemic sclerosis. Clin. Immunol. 2013, 149, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Ciechomska, M.; O’Reilly, S.; Przyborski, S.; Oakley, F.; Bogunia-Kubik, K.; van Laar, J.M. Histone demthylation and Toll-like receptor 8-dependent cross-talk in monocytes promotes transdifferentiation of fibroblasts in systemic sclerosis via Fra-2. Arthritis Rheumatol. 2016, 68, 1493–1504. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, C.; Distler, J.H. Epigenetic factors as drivers of fibrosis in systemic sclerosis. Epigenomics 2017, 9, 463–477. [Google Scholar] [CrossRef]
- Hattori, M.; Yokoyama, Y.; Hattori, T.; Moteqi, S.; Amano, H.; Hatada, I.; Ishikawa, O. Global DNA hypomethylation and hypoxia-induced expression of the ten eleven translocation (TET) family, TET1, in scleroderma fibroblasts. Exp. Dermatol. 2015, 24, 841–846. [Google Scholar] [CrossRef] [PubMed]
- Altorok, N.; Tsou, P.-S.; Coit, P.; Khanna, D.; Sawalha, A.H. Genome-wide DNA methylation analysis in dermal fibroblasts from patients with diffuse and limited systemic sclerosis reveals common and subset-specific DNA methylation aberrancies. Ann. Rheum. Dis. 2015, 74, 1612–1620. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.; Laqares, D. Matrix stiffness: The conductor of organ-fibrosis. Curr. Rheumatol. Rep. 2018, 20, 2. [Google Scholar] [CrossRef]
- Gyftaki-Venieri, D.; Abraham, D.; Ponticos, M. Insights into myofibroblasts and their activation in scleroderma: Opportunities for therapy? Curr. Opin. Rheumatol. 2018, 30, 581–587. [Google Scholar] [CrossRef]
- Ebmeier, S.; Horsley, V. Origin of fibrosing cells in systemic sclerosis. Curr. Opin. Rheumatol. 2015, 27, 555–562. [Google Scholar] [CrossRef]
- Korman, B. Evolving insights into the cellular and molecular pathogenesis of fibrosis in systemic sclerosis. Transl. Res. 2019, 209, 77–89. [Google Scholar] [CrossRef]
- Shook, B.A.; Wasko, R.R.; Rivera Gonzalez, G.C.; Salazar-Gatzimas, E.; Lόpez-Giráldez, F.; Dash, B.C.; Muñoz-Rojas, A.R.; Aultman, K.D.; Zwick, R.K.; Lei, V.; et al. Myofibroblast proliferation and heterogeneity are supported by macrophages during skin repair. Science 2018, 362, eaar2971. [Google Scholar] [CrossRef] [PubMed]
- Chia, J.J.; Lu, T.T. Update on macrophages and innate immunity in scleroderma. Curr. Opin. Rheumatol. 2015, 27, 530–536. [Google Scholar] [CrossRef] [PubMed]
- Laurent, P.; Sisirak, V.; Lazaro, E.; Richez, C.; Duffau, P.; Blanco, P.; Truchetet, M.E.; Contin-Bordes, C. Innate immunity in systemic sclerosis: Recent advances. Front. Immunol. 2018, 9, 1702. [Google Scholar] [CrossRef] [PubMed]
- Pincha, N.; Hajam, E.Y.; Badarinath, K.; Batta, S.P.R.; Masudi, T.; Dey, R.; Andreasen, P.; Kawakami, T.; Samuel, R.; George, R.; et al. PAI1 mediates fibroblast-mast cell interactions in skin fibrosis. J. Clin. Invest. 2018, 128, 1807–1819. [Google Scholar] [CrossRef] [PubMed]
- Mauqeri, N.; Capobianco, A.; Rovere-Querini, P.; Ramirez, G.A.; Tombetti, E.; Valle, P.D.; Monno, A.; D’Apiilberti, V.; Gasparri, A.M.; Franchini, S.; et al. Platelet microparticles sustain autophagy-associated activation of neutrophils in systemic sclerosis. Sci. Transl. Med. 2018, 10, eaao3089. [Google Scholar] [CrossRef]
- Dumoitier, N.; Chaigne, B.; Régent, A.; Lofek, S.; Mhibik, M.; Dorfmüller, P.; Terrier, B.; London, J.; Bérezné, A.; Tama, N.; et al. Scleroderma peripheral B lymphocytes secrete interleukin-6 and transforming growth factor beta and activate fibroblasts. Arthritis Rheumatol. 2017, 69, 1078–1089. [Google Scholar] [CrossRef]
- Choi, M.Y.; Fritzler, M.J. Progress in understanding the diagnostic and pathogenic role of autoantibodies associated with systemic sclerosis. Curr. Opin. Rheumatol. 2016, 28, 586–594. [Google Scholar] [CrossRef]
- Li, G.; Larregina, A.T.; Domsic, R.T.; Stolz, D.B.; Medsger, T.A., Jr.; Lafyatis, R.; Fuschiotti, P. Skin-resident effector memory CD8 (+) CD28 (-) T cells exhibit a profibrotic phenotype in patients with systemic sclerosis. J. Invest. Dermatol. 2017, 137, 1042–1050. [Google Scholar] [CrossRef]
- Fuschiotti, P.; Larregina, A.T.; Ho, J.; Feghali-Bostwick, C.; Medsger, T.A., Jr. Interleukin-13-producing CD8+T cells mediate dermal fibrosis in patients with systemic sclerosis. Arthritis Rheum. 2013, 65, 236–246. [Google Scholar] [CrossRef]
- Roan, F.; Stoklasek, T.A.; Whalen, E.; Molitor, J.A.; Blustone, J.A.; Buckner, J.H.; Ziegler, S.F. CD4+ group 1 innate lymphoid cells (ILC) form a functionally distinct ILC subset that is increased in systemic sclerosis. J. Immunol. 2016, 196, 2051–2062. [Google Scholar] [CrossRef]
- Mendoza, F.A.; Piera-Velazquez, S.; Farber, J.L.; Feghali-Bostwick, C.; Jiménez, S.A. Endothelial cells expressing endothelial and mesenchymal cell gene products in lung tissue from patients with systemic sclerosis-associated interstitial lung disease. Arthritis Rheumatol. 2016, 68, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Ntelis, K.; Bogdanos, D.; Dimitroulas, T.; Sakkas, L.; Daoussis, D. Platelets in systemic sclerosis: The missing link connecting vasculopathy, autoimmunity, and fibrosis? Curr. Rheumatol. Rep. 2019, 21, 15. [Google Scholar] [CrossRef] [PubMed]
- Korman, B.; Marangoni, R.G.; Lord, G.; Olefsky, J.; Troutellotte, W.; Varga, J. Adipocyte-specific repression of PPAR-gamma by NCoR contributes to scleroderma skin fibrosis. Arthritis Res. Ther. 2018, 20, 145. [Google Scholar] [CrossRef] [PubMed]
- McCoy, S.S.; Reed, T.J.; Berthier, C.C.; Tsou, P.-S.; Liu, J.; Gudjonsson, J.E.; Khanna, D.; Kahlenberg, J.M. Scleroderma keratinocytes promote fibroblast activation independent of transforming growth factor beta. Rheumatology 2017, 56, 1970–1981. [Google Scholar] [CrossRef]
- Di Benedetto, P.; Panzera, N.; Cipriani, P.; Mastroiaco, V.; Tessitore, A.; Liakouli, V.; Ruscitti, P.; Berardicurti, O.; Carubbi, F.; Guggino, G.; et al. Mesenchymal stem cells of systemic sclerosis patients, derived from different sources, show a porfibrotic microRNA profiling. Sci. Rep. 2019, 9, 7144. [Google Scholar] [CrossRef]
- Hegner, B.; Schaub, T.; Catar, R.; Kusch, A.; Wagner, P.; Essin, K.; Lange, C.; Riemerkasten, G.; Draqun, D. Intrinsic deregulation of vascular smooth muscle and myofibroblast differentiation in mesenchymal stromal cells from patients with systemic sclerosis. PLoS ONE 2016, e0153101. [Google Scholar] [CrossRef]
- Manetti, M.; Romano, E.; Rosa, I.; Guiducci, S.; Bellando-Randone, S.; De Paulis, A.; Ibba-Manneschi, L.; Matucci-Cerinic, M. Endothelial-to-mesenchymal transition contributes to endothelial dysfunction and dermal fibrosis in systemic sclerosis. Ann. Rheum. Dis. 2017, 76, 924–934. [Google Scholar] [CrossRef]
- Zhao, Y.; Feng, G.; Wang, Y.; Yue, Y.; Zhao, W. Regulation of apoptosis by long non-coding RNA HIF1A-AS1 in VSMCs: Implications for TAA pathogenesis. Int. J. Clin. Exp. Pathol. 2014, 7, 7643–7652. [Google Scholar] [PubMed]
- Gelse, K.; Pöschl, E.; Aigner, T. Collagens: Structure, function and biosynthesis. Adv. Drug Deliv. Rev. 2003, 55, 1531–1546. [Google Scholar] [CrossRef]
- Birk, D.E.; Fitch, J.M.; Babiarz, J.P.; Doane, K.J.; Linsenmayer, T.F. Collagen fibrillogenesis in vitro: Interaction of types I and V collagen regulates fibril diameter. J. Cell Sci. 1990, 95 Pt 4, 649–657. [Google Scholar]
- Chanut-Delalande, H.; Bonod-Bidaud, C.; Cogne, S.; Malbouyres, M.; Ramirez, F.; Fichard, A.; Ruggiero, F. Development of a functional skin matrix requires deposition of collagne V heterotrimers. Mol. Cell. Biol. 2004, 6049–6057. [Google Scholar] [CrossRef] [PubMed]
- Roulet, M.; Ruggiero, F.; Karsenty, G.; LeGuellec, D. A comprehensive study of the spatial and temporal expression of the col5a1 gene in mouse embryos: A clue for understanding collagen V function in developing connective tissues. Cell Tissue Res. 2007, 327, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Parra, E.R.; Aguiar, A.C., Jr.; Teodoro, W.R.; de Souza, R.; Yoshinari, N.H.; Capelozzi, V.L. Collagen V and vascular injury promote lung architectural changes in systemic sclerosis. Clin. Respir. J. 2009, 3, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Parra, E.R.; Teodoro, W.R.; de Morais, J.; Katayama, M.L.H.; de Souza, R.; Yoshinari, N.H.; Capelozzi, V.L. Increased mRNA expression of collagen V gene in pulmonary fibrosis of systemic sclerosis. Eur. J. Clin. Invest. 2010, 40, 110–120. [Google Scholar] [CrossRef]
- Martin, P.; Teodoro, W.R.; Velosa, A.P.P.; de Morais, J.; Carrasco, S.; Christmann, R.B.; Goldenstein-Schainberg, C.; Parra, E.R.; Katayama, M.L.; Sotto, M.N.; et al. Abnormal collagen V deposition in dermis corrleates with skin thickening and disease activity in systemic sclerosis. Autoimmun. Rev. 2012, 11, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Dooley, A.; Shi-Wen, X.; Aden, N.; Tranah, T.; Desai, N.; Denton, C.P.; Abraham, D.J.; Bruckdorfer, R. Modulation of collegen type I, fibronectin and dermal fibroblast function and activity in systemic sclerosis by the anti-oxidant epigallocatechin-3-gallate. Rheumatology 2010, 49, 2024–2036. [Google Scholar] [CrossRef]
- Verrechia, F.; Mauviel, A.; Farge, D. Transforming growth factor-β signaling through the Smad proteins: Role in systemic sclerosis. Autoimmun. Rev. 2006, 5, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Nikolic-Paterson, D.J.; Lan, H.Y. TGF-β: The master regulator of fibrosis. Nat. Rev. Nephrol. 2016, 12, 325–338. [Google Scholar] [CrossRef]
- Tang, P.M.-K.; Tang, P.C.-T.; Chung, J.Y.-F.; Lan, H.-Y. TGF-β1 signaling in kidney disease: From Smads to long non-coding RNAs. Noncoding RNA Res. 2017, 2, 68–73. [Google Scholar] [CrossRef]
- Hu, H.-H.; Chen, D.-Q.; Wang, Y.-N.; Feng, Y.-L.; Cao, G.; Vaziri, N.D.; Zhao, Y.-Y. New insights into TGF-β/Smad signaling in tissue fibrosis. Chem Biol. Interact. 2018, 292, 76–83. [Google Scholar] [CrossRef]
- Trojanowska, M. Role of PDGF in fibrotic diseases and systemic sclerosis. Rheumatology 2008, 47 (Suppl. 5), v2–v4. [Google Scholar] [CrossRef]
- Artlett, C.M. The IL-1 family of cytokines. Do they have a role in scleroderma fibrosis? Immunol. Lett. 2018, 195, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Kotsiou, O.S.; Gourgoulianis, K.I.; Zarogiannis, S.G. IL-33/ST2 Axis in Organ Fibrosis. Front. Immunol. 2018, 02432. [Google Scholar] [CrossRef]
- Xu, D.; Barbour, M.; Jiang, H.R.; Mu, R. Role of IL-33/ST2 signaling pathway in systemic sclerosis and other fibrotic diseases. Clin. Exp. Rheumatol. 2019, 37, S141–S146. [Google Scholar]
- O’Reilly, S.; Ciechomska, M.; Fullard, N.; Prizyborski, S.; van Laar, J.M. IL-13 mediates collagen deposition via STAT6 and microRNA-135b: A role for epigenetics. Sci. Rep. 2016, 6, 25066. [Google Scholar] [CrossRef]
- Nquyen, J.K.; Austin, E.; Huang, A.; Mamalis, A.; Jagdeo, J. The IL-4/IL-13 axis in skin fibrosis and scarring: Mechanistic concepts and therapeutic tragets. Arch. Dermatol. Res. 2020, 312, 81–92. [Google Scholar]
- Wang, J.-H.; Zhao, L.; Pan, X.; Chen, N.-N.; Chen, J.; Gong, Q.L.; Su, F.; Yan, J.; Zhang, Y.; Zhang, S.-H. Hypoxia-stimulated cardiac fibroblast production of IL-6 promotes myocardial fibrosis via the TGF-β1 signaling pathway. Lab. Invest. 2016, 96, 1035. [Google Scholar] [CrossRef] [PubMed]
- Robak, E.; Gerlicz-Kowalczuk, Z.; Dziankowska-Bartkowiak, B.; Wozniacka, A.; Bogaczewicz, J. Serum concentrations of IL-17A, IL-17B, IL-17E and IL-17F in patients with systemic sclerosis. Arch. Med. Sci. 2019, 15, 706–712. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, T.; Jinnin, M.; Yamane, K.; Honda, N.; Kajihara, I.; Makino, T.; Masuguchi, S.; Fukushima, S.; Okamoto, Y.; Hasegawa, M.; et al. Impaired IL-17 signaling pathway contributes to the increased collagen expression in scleroderma fibroblasts. J. Immunol. 2012, 188, 3573–3583. [Google Scholar] [CrossRef]
- Ahmed, S.; Misra, D.P.; Agarwal, V. Interleukin-17 pathways in systemic sclerosis-associated fibrosis. Rheumatol. Int. 2019, 39, 1135–1143. [Google Scholar] [CrossRef]
- Sawamura, S.; Jinnin, M.; Inoue, K.; Yamane, K.; Honda, N.; Kajihara, I.; Makino, T.; Masuguchi, S.; Fukushima, S.; Ihn, H. Regulatory mechanisms of collagen expression by interleukin-22 signaling in scleroderma fibroblasts. J. Dermatol. Sci. 2018, 90, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Peng, H.; Chen, P.; Zhou, Y. The immunomodulatory role of interleukin-35 in fibrotic diseases. Exp. Rev. Clin. Immunol. 2019, 15, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Maring, J.A.; Trojanowska, M.; ten Dijke, P. Endoglin in fibrosis and scleroderma. Int. Rev. Cell. Mol. Biol. 2012, 297, 295–308. [Google Scholar] [PubMed]
- Carvalheiro, T.; Malvar Fernández, B.; Ottria, A.; Giovannone, B.; Marut, W.; Reedquist, K.A.; Garcia, S.; Radstake, T.R. Extracellular SPARC cooperates with TGF-β signaling to induce pro-fibrotic activation of systemic sclerosis patient dermal fibroblasts. Rheumatology 2020. [Google Scholar] [CrossRef]
- Huang, X.-L.; Zhang, L.; Duan, Y.; Wang, Y.-J.; Zhao, J.-H.; Wang, J. E3 ubiquitin ligase: A potential regulator in fibrosis and systemic sclerosis. Cell. Immunol 2016, 306–307, 1–8. [Google Scholar] [CrossRef]
- Wyman, A.E.; Atamas, S.P. Sirtuins and accelerated aging in scleroderma. Curr. Rheumatol. Rep. 2018, 20, 16. [Google Scholar] [CrossRef]
- Aslani, S.; Sobhani, S.; Gharibdoost, F.; Famshide, A.; Mahmoudi, M. Epigenetics and pathogenesis of systemic sclerosis, the ins and outs. Human Immunol. 2018, 79, 178–187. [Google Scholar] [CrossRef]
- Henry, T.W.; Mendoza, F.A.; Jimenez, S.A. Role of microRNA in the pathogenesis of systemic sclerosis tissue fibrosis and vasculopathy. Autoimmun. Rev. 2019, 18, 102396. [Google Scholar] [CrossRef]
- Wermuth, P.J.; Piera-Velazquez, S.; Jimenez, S.A. Exosomes isolated from serum of systemic sclerosis patients display alterations in their content of profibrotic and antifibrotic microRNA and induce a profibrotic phenotype in cultured normal dermal fibroblasts. Clin. Exp. Rheumatol. 2017, 35 (Suppl. 106), 21–30. [Google Scholar]
- Colletti, M.; Galardi, A.; De Santis, M.; Guidelli, G.M.; Di Giannatale, A.; Di Luigi, L.; Antinozzi, C. Exosomes in systemic sclerosis: Messengers between immune, vascular and fibrotic components? Int. J. Mol. Sci. 2019, 20, 4337. [Google Scholar] [CrossRef]
- Li, Y.; Huang, J.; Guo, M.; Zuo, X. MicroRNAs regulating signaling pathways: Potential biomarkers in systemic sclerosis. Genom. Proteom. Bioinform. 2015, 13, 234–241. [Google Scholar] [CrossRef]
- Luo, Y.; Wang, Y.; Shu, Y.; Lu, Q.; Xiao, R. Epigenetic mechanisms: An emerging role in pathogenesis and its therapeutic potential in systemic sclerosis. Int. J. Biochem. Cell Biol. 2015, 67, 92–100. [Google Scholar] [CrossRef]
- Wermuth, P.J.; Piera-Valazquez, S.; Rosenbloom, J.; Jimenez, S.A. Existing and novel biomarkers for precision medicine in systemic sclerosis. Nat. Rev. Rheumatol. 2018, 14, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.-H.; Xie, M.; Wu, S.-D.; Zhang, J.; Huang, C.Z. Identification and interaction analysis of key genes and microRNAs in systemic sclerosis by bioinformatics approaches. Curr. Med. Sci. 2019, 39, 645–652. [Google Scholar] [CrossRef]
- He, Y.; Liu, H.; Wang, S.; Chen, Y. In silico detection and characterization of microRNAs and their target genes in microRNA microarray datasets from patients with systemic sclerosis-interstitial lung disease. DNA Cell Biol. 2019, 38, 933–944. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.-C.; Pan, H.-F.; Leng, R.-X.; Wang, D.-G.; Li, X.-P.; Li, X.-M.; Ye, D.-Q. Emerging role of long noncoding RNAs in autoimmune diseases. Autoimmun. Rev. 2015, 14, 798–805. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhou, T.; Yu, X.; Xue, Z.; Shen, N. The role of long non-coding RNAs in rheumatic diseases. Nat. Rev. Rheumatol. 2017, 13, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Melissari, M.-T.; Grote, P. Roles of long non-coding RNAs in physiology and disease. Eur. J. Physiol. 2016, 468, 945–958. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, L.; Ding, Y.; Lu, X.; Zhang, G.; Yang, J.; Zheng, H.; Wang, H.; Jiang, Y.; Xu, L. lncRNA structural characteristics in epigenetic regulation. Int. J. Mol. Sci. 2017, 18, 2659. [Google Scholar] [CrossRef]
- Wang, J.-Y.; Yang, Y.; Ma, Y.; Wang, F.; Xue, A.; Zhu, J.; Yang, H.; Chen, Q.; Chen, M.; Ye, L.; et al. Potential regulatory role of lncRNA-miRNA-mRNA axis in osteosarcoma. Biomed. Pharmacother. 2020, 121, 109627. [Google Scholar] [CrossRef]
- Xu, F.; Jin, L.; Jin, Y.; Nie, Z.; Zheng, H. Long noncoding RNAs in autoimmune diseases. J. Biomed. Mater. Res. Part A 2019, 107, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Teimuri, S.; Hosseini, A.; Rezaenasab, A.; Ghaedi, K.; Ghoveud, E.; Etemadifar, M.; Nasr-Esfahani, M.H.; Megraw, T.L. Integrative analysis of lncRNAs in Th17 cell lineage to discover new potential biomarkers and therapeutic targets in autoimmune diseases. Mol. Ther. Nucleic Acids 2018, 12, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Jinnin, M.; Nakamura, K.; Harada, M.; Kudo, H.; Nakayama, W.; Inoue, K.; Nakashima, T.; Honda, N.; Fukushima, S.; et al. Long non-coding RNA TSIX is upregulated in scleroderma dermal fibroblasts and controls collagen mRNA stabilization. Exp. Dermatol. 2016, 25, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Mariotti, B.; Servaas, N.H.; Rossato, M.; Taimassia, N.; Cassatella, M.A.; Cossu, M.; Beretta, L.; van der Kroef, M.; Radstake, T.R.D.J.; Bazzoni, F. The long non-coding RNA NRIR drives IFN-response in monocytes: Implication for systemic sclerosis. Front. Immunol. 2019, 10, 100. [Google Scholar] [CrossRef]
- Dolcino, M.; Trinazzi, E.; Puccetti, A.; Lunardi, C. In systemic sclerosis, a unique long non-coding RNA regulates genes and pathways involved in the three main features of the disease (vasculopapthy, fibrosis and autoimmunity) and in carcinogenesis. J. Clin. Med. 2019, 8, 320. [Google Scholar] [CrossRef]
- Messemaker, T.C.; Chadli, L.; Cai, G.; Goelela, V.S.; Boonstra, M.; Dorjée, A.L.; Andersen, S.N.; Mikkers, H.M.M.; van’t Hof, P.; Mei, H.; et al. Antisense long non-coding RNAs are dysregulated in skin tissue of patients with systemic sclerosis. J. Invest. Dermatol. 2018, 138, 826–835. [Google Scholar] [CrossRef]
- Takata, M.; Pachera, E.; Frank-Bertoncelj, M.; Kozlova, A.; Jüngel, A.; Whitefield, M.L.; Assassi, S.; Calcagni, M.; de Vries-Bouwastra, J.; Huizinga, T.W.; et al. OTUD6B-AS1 might be a novel regulator of apoptosis in systemic sclerosis. Front. Immunol. 2019, 10, 1100. [Google Scholar] [CrossRef] [PubMed]
- Robbins, P.D.; Dorronsoro, A.; Booker, C.N. Regulation of chronic inflammatory and immune processes by extracellular vesicles. J. Clin. Invest. 2016, 126, 1173–1180. [Google Scholar] [CrossRef]
- Stypinska, B.; Wajda, A.; Walczuk, E.; Olesinska, M.; Lewandowska, A.; Walczyk, M.; Paradowska-Gorycka, A. The serum cell-free microRNA expression profile in MCTD, SLE, SSc and RA patients. J. Clin. Med. 2020, 9, 161. [Google Scholar] [CrossRef]
- Chouri, E.C.; Servaas, N.H.; Bekker, C.P.J.; Affandi, A.J.; Cossu, M.; Hillen, M.R.; Angiolilli, C.; Mertens, J.S.; van den Hoogen, L.L.; Silva-Cardoso, C.; et al. Serum microRNA screening and functional studies reveal miR-483-5p as a potential driver of fibrosis in systemic sclerosis. J. Autoimmun. 2018, 89, 162–170. [Google Scholar] [CrossRef]
- Rusek, M.; Michalska-Jakubus, M.; Kowal, M.; Beltowski, J.; Krasowska, D. A novel miRNA-4484 is up-regulated on microarray and associated with increased MMP-21 expression in serum of systemic sclerosis patients. Sci. Rep. 2019, 9, 14264. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Fan, P.S.; Kahaleh, B. Association between enhanced type I collagen expression and epigenetic repression of FLI1 gene in scleroderma fibroblasts. Arthritis Rheum. 2006, 54, 2271–2279. [Google Scholar] [CrossRef] [PubMed]
- Hemmatazad, H.; Rodrigues, H.M.; Maurer, B.; Brentano, F.; Pileckyte, M.; Distler, J.H.W.; Gay, R.E.; Michel, B.A.; Gay, S.; Huber, L.C. Histone deacetylase 7, a potential target for the antifibrotic treatment of systemic sclerosis. Arthritis Rheum. 2009, 60, 1519–1529. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.W.Y.; Chang, C.-B.; Tung, C.-H.; Sun, J.; Suen, J.-L.; Wu, S.-F. Low-dose 5-aza-2’-deoxycytidine pretreatment inhibits experimental autoimmune encephalomyelitis by induction of regulatory T cells. Mol. Med. 2014, 20, 248–256. [Google Scholar] [CrossRef]
- Dees, C.; Schlottmann, I.; Funke, R.; Distler, A.; Palumbo-Zerr, K.; Zerr, P.; Lin, N.-Y.; Beyer, C.; Distler, O.; Schett, G.; et al. The Wnt antagonists DKK1 and SFRP1 are downregulated by promoter hypermethylation in systemic sclerosis. Ann. Rheum. Dis. 2014, 73, 1232–1239. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, R. Epigenetics in autoimmune diseases: Pathogenesis and prospects for therapy. Autoimmun. Rev. 2015, 14, 854–863. [Google Scholar] [CrossRef]
- Wang, Z. The principles of MiRNA-masking anti-sense oligonucleotides technology. Methods Mol. Biol. 2011, 676, 43–49. [Google Scholar]
- Jeffries, M.A. Epigenetic editing: How cutting-edge targeted epigenetic modification might provide novel avenues for autoimmune disease therapy. Clin. Immunol. 2018, 196, 49–58. [Google Scholar] [CrossRef]
- Brown, J.M.; Wasson, M.-C.D.; Marcato, P. The missing lnc: The potential of targeting triple-negative breast cancer and cancer stem cells by inhibiting long non-coding RNAs. Cells 2020, 9, 763. [Google Scholar] [CrossRef]
- O’Reilly, S. Epigenetic modulation as a therapy in systemic sclerosis. Rheumatology 2019, 58, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.F.; Li, S.H.; Liu, Y.; Luo, Y.T. Long noncoding RNA CIR promotes chondrocyte extracellular matrix degradation in osteoarthritis by acting as a sponge for miR-27b. Cell Physiol. Biochem. 2017, 43, 602–610. [Google Scholar] [CrossRef] [PubMed]


| Autoantibody | Clinical Manifestation | References |
|---|---|---|
| Anti-topoisomerase 1 (anti-Scl-70) | Pulmonary fibrosis Cardiac involvement Malignancy Raynaud’s phenomenon | [16,17,18] |
| Anti-centromere proteins B and C | Raynaud’s phenomenon Ischemic digital loss Sicca syndrome | [18,19,20] |
| Anti-RNA polymerase III | Skin fibrosis Renal crisis | [21,22] |
| Anti-U3-RNP (fibrillarin) | Pulmonary arterial hypertension Cardiac involvement Skeletal muscle involvement | [23,24,25,26] |
| Anti-U11/U12-RNP | Pulmonary fibrosis | [27,28] |
| Anti-B23 | Pulmonary hypertension Lung diseases | [29,30] |
| Anti-Ku | Muscle and joint involvement | [31,32] |
| Anti-Th/To-RNP | Lung diseases Renal crisis Small-bowel involvement | [33,34] |
| Anti-endothelial cells | Skin and lung fibrosis | [35] |
| Anti-fibroblast | Skin and lung fibrosis | [36] |
| Anti-metalloproteinase 1 | Extracellular matrix deposition | [37] |
| Anti-M3-muscarinic receptor | Gastrointestinal dysmotility Sicca | [38] |
| Anti-PDGFR | Tissue fibrosis | [39] |
| Anti-cardiolipin/phospholipid | Vasculopathy | [40] |
| Anti-ICAM-1 | Endothelial dysfunction | [41] |
| Anti-fibrillin-1 | Tissue fibrosis | [42,43] |
| Fibrosis-Related Cytokines/Molecules | Signaling/Modes of Action |
|---|---|
| [I] Pro-fibrogenic cytokines: | |
| TGF-β [117,118,119,120] | Smad 2/3, Wnt/β-catenin |
| PDGF [121] | |
| IL-1 family (IL-1, IL-33, IL-36) [122,123,124] | |
| IL-4/IL-13 [125,126] | STAT6, miR-135b |
| IL-6 [127] | |
| IL-17B, IL-17E, IL-17F [128,129,130] | |
| IL-18 [122] | |
| IL-22 [131] | let-7a ↓→ collagen I ↑ |
| IL-33 [122] | ST2 (suppressor of tumorigenicity 2 receptor) |
| [II] Fibrosis-related molecules: | |
| Endoglin (co-receptor for TGF-β signaling) [133] | Smad 2/3, Wnt/β-catenin |
| SPARC (secreted protein acidic and rich in cysteine) [134] | Smad 2/3, Wnt/β-catenin |
| E3 ubiquitin ligase [135] | Ubiquitin-mediated degradation of TGF-β/Smad signaling pathway |
| [III] Anti-fibrinogenic cytokines: | |
| IL-17A [129] | miR-129-5p↑→CTGF *↓ |
| IL-35 [132] | |
| [IV] Anti-fibrogenic molecules: | |
| Sirtuins (histone deacetylase) [126] | TGF-β inducing signaling↓ |
| mTOR signaling↓ | |
| Oxidative stress↓ | |
| Cell senescence marker p-21↓ | |
| lncRNA | Expression Level | Tissue or Cell Type | Target mRNA | Pathology |
|---|---|---|---|---|
| TSIX [149] | ↑ | Dermal fibroblast, skin tissue and serum | Type I collagen mRNA stabilization | Fibrosis |
| NRIR [150] | ↓ | Peripheral blood monocytes | Type 1 IFN and its stimulated mRNA | Autoimmuity, Inflammation |
| ncRNA00201 [151] | ↓ | Peripheral blood mononuclear cells | EGFR Enb B1 S1P1 ALK1 Endothelins RhoA MAPK Class I-PI3K mTOR TGF-βR MyD88 TLRs RAC | Autoimmunity, Vasculopathy, Fibrosis, & Carcinogenesis |
| OTUD6B-AS1 [49,152,153] | ↓ | Skin tissue, Fibroblast *, HPASMC | Cyclin D1 | Fibrosis, Vasculopathy |
| CTBP1-AS2 [49,152] | ↑ | Skin tissue | ND | ND |
| AGAP2-AS1 [49] | ↑ | Skin tissue | ND | ND |
| HIFA-AS1 [108] | ↑ | Vascular smooth muscle cells | Bcl-2 Caspase 3 and 8 | Vasculopathy |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsai, C.-Y.; Hsieh, S.-C.; Wu, T.-H.; Li, K.-J.; Shen, C.-Y.; Liao, H.-T.; Wu, C.-H.; Kuo, Y.-M.; Lu, C.-S.; Yu, C.-L. Pathogenic Roles of Autoantibodies and Aberrant Epigenetic Regulation of Immune and Connective Tissue Cells in the Tissue Fibrosis of Patients with Systemic Sclerosis. Int. J. Mol. Sci. 2020, 21, 3069. https://doi.org/10.3390/ijms21093069
Tsai C-Y, Hsieh S-C, Wu T-H, Li K-J, Shen C-Y, Liao H-T, Wu C-H, Kuo Y-M, Lu C-S, Yu C-L. Pathogenic Roles of Autoantibodies and Aberrant Epigenetic Regulation of Immune and Connective Tissue Cells in the Tissue Fibrosis of Patients with Systemic Sclerosis. International Journal of Molecular Sciences. 2020; 21(9):3069. https://doi.org/10.3390/ijms21093069
Chicago/Turabian StyleTsai, Chang-Youh, Song-Chou Hsieh, Tsai-Hung Wu, Ko-Jen Li, Chieh-Yu Shen, Hsien-Tzung Liao, Cheng-Han Wu, Yu-Min Kuo, Cheng-Shiun Lu, and Chia-Li Yu. 2020. "Pathogenic Roles of Autoantibodies and Aberrant Epigenetic Regulation of Immune and Connective Tissue Cells in the Tissue Fibrosis of Patients with Systemic Sclerosis" International Journal of Molecular Sciences 21, no. 9: 3069. https://doi.org/10.3390/ijms21093069
APA StyleTsai, C.-Y., Hsieh, S.-C., Wu, T.-H., Li, K.-J., Shen, C.-Y., Liao, H.-T., Wu, C.-H., Kuo, Y.-M., Lu, C.-S., & Yu, C.-L. (2020). Pathogenic Roles of Autoantibodies and Aberrant Epigenetic Regulation of Immune and Connective Tissue Cells in the Tissue Fibrosis of Patients with Systemic Sclerosis. International Journal of Molecular Sciences, 21(9), 3069. https://doi.org/10.3390/ijms21093069





