Anti-Fibrotic Effect of Synthetic Noncoding Decoy ODNs for TFEB in an Animal Model of Chronic Kidney Disease
Abstract
:1. Introduction
2. Results
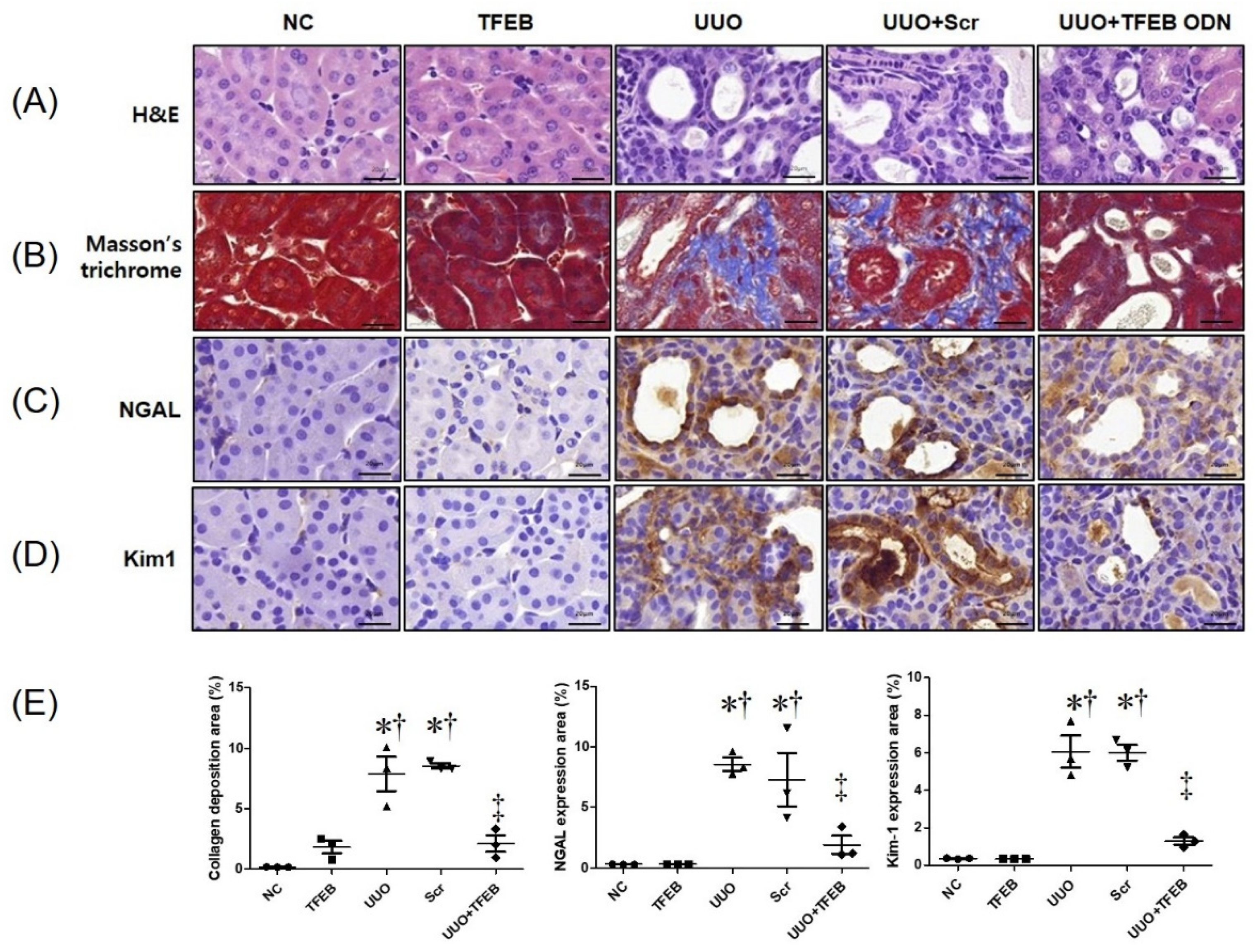
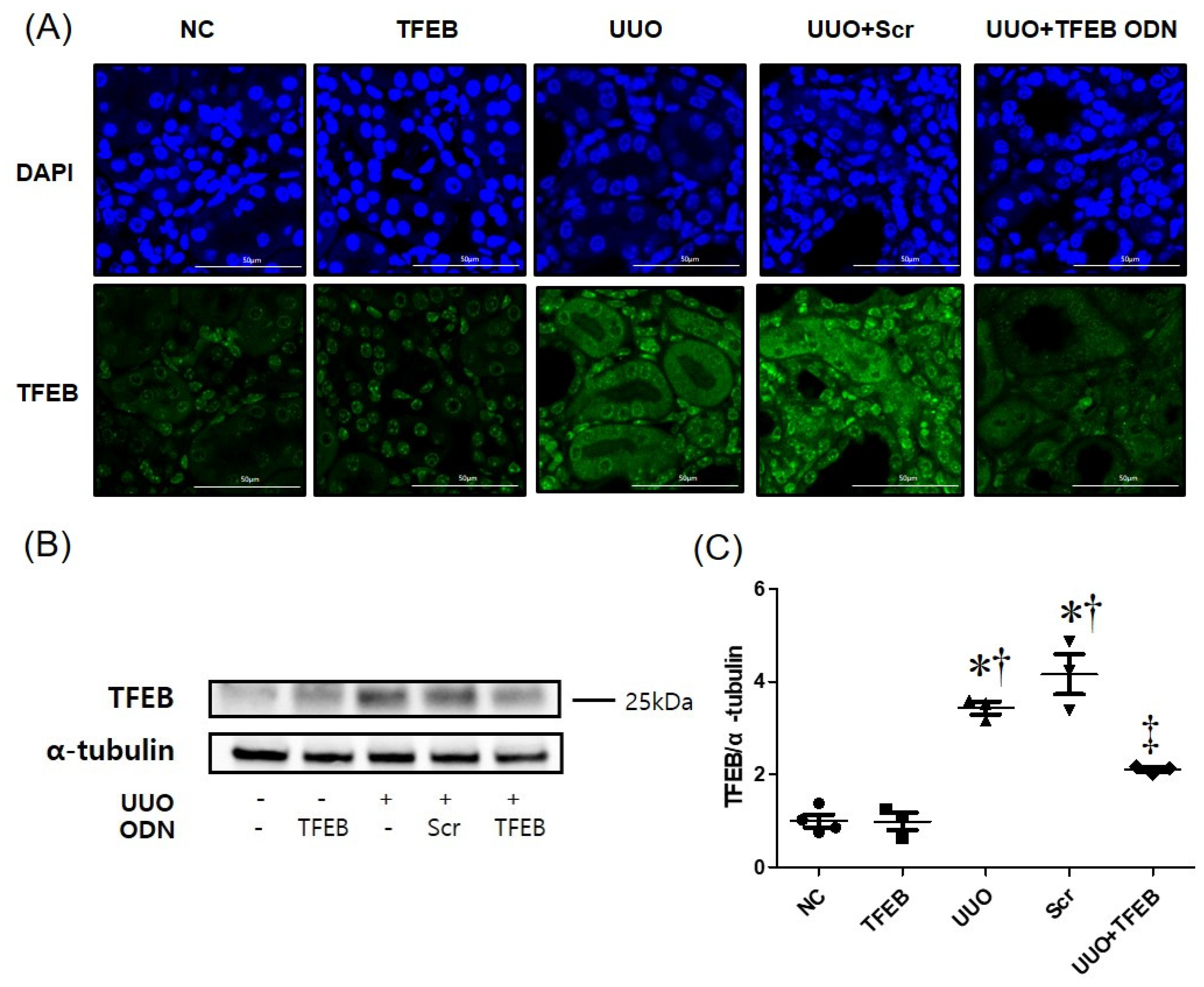
2.1. TFEB Decoy ODN Attenuats Morphological Changes in UUO-induced Renal Fibrosis
2.2. TFEB Synthetic ODN Attenuates UUO-Induced Tubular Injury in Kidney
2.3. TFEB ODN Attenuates Inflammation in a UUO-induced Mouse Model
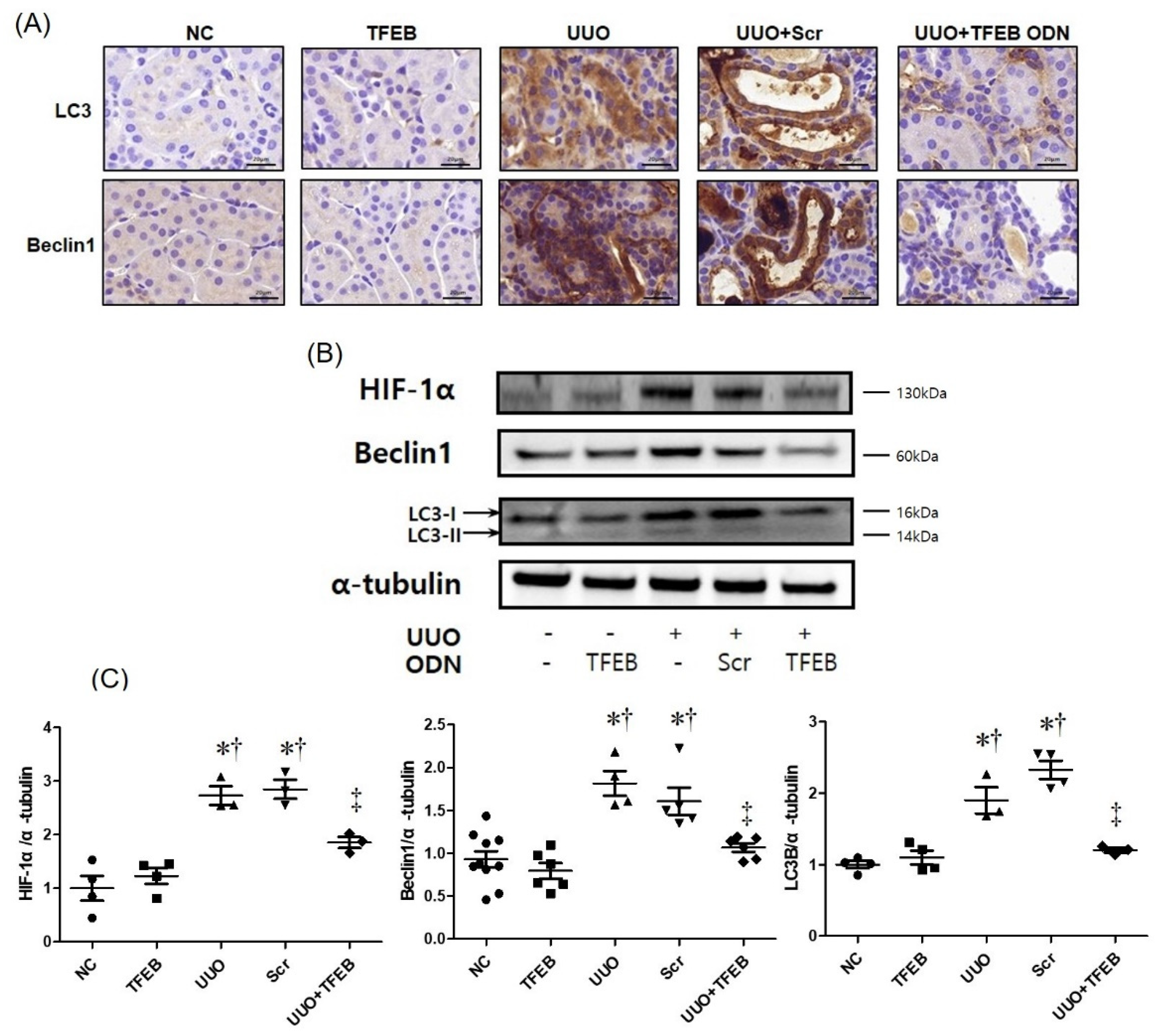
2.4. TFEB Decoy ODN Decreases Autophagic Activity in UUO-Induced Renal Injury
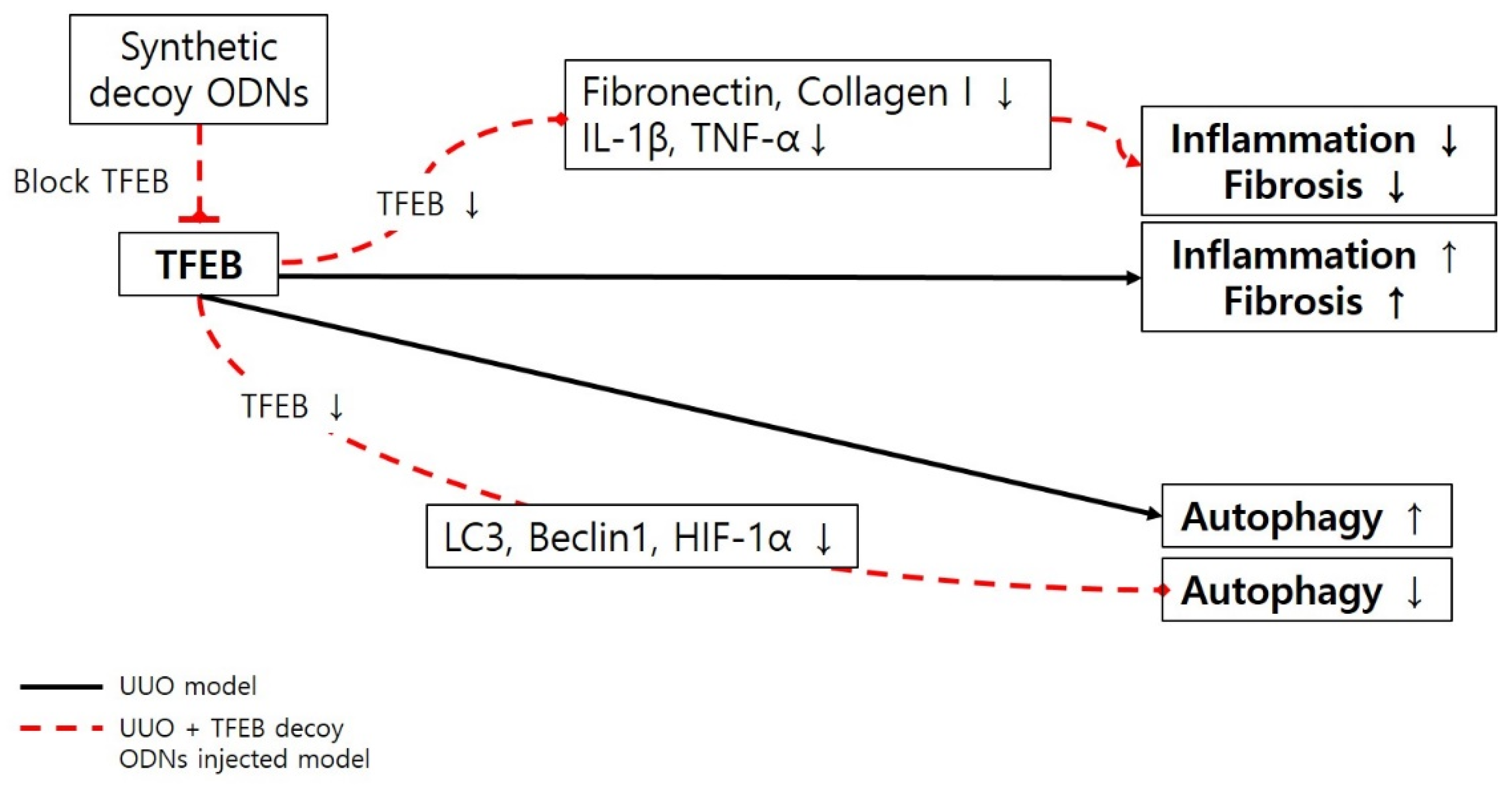
3. Discussion
4. Materials and Methods
4.1. Synthesis of Ring-type TFEB Decoy ODNs and Scrambled ODNs
4.2. Animal Model
4.3. Histological Analysis
4.4. Immunofluorescence Staining
4.5. Immunohistochemical (IHC) Staining
4.6. Western Blot Analysis
4.7. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Levey, A.S.; Atkins, R.; Coresh, J.; Cohen, E.P.; Collins, A.J.; Eckardt, K.U.; Nahas, M.E.; Jaber, B.L.; Jadoul, M.; Levin, A.; et al. Chronic kidney disease as a global public health problem: Approaches and initiatives—A position statement from Kidney Disease Improving Global Outcomes. Kidney Int. 2007, 72, 247–259. [Google Scholar] [CrossRef] [Green Version]
- Bohle, A.; Strutz, F.; Müller, G.A. On the pathogenesis of chronic renal failure in primary glomerulopathies: A view from the interstitium. Exp. Nephrol. 1994, 2, 205–210. [Google Scholar] [PubMed]
- Nath, K.A. Tubulointerstitial changes as a major determinant in the progression of renal damage. Am. J. Kidney Dis. 1992, 20, 1–17. [Google Scholar] [CrossRef]
- Boor, P.; Ostendorf, T.; Floege, J. Renal fibrosis: Novel insights into mechanisms and therapeutic targets. Nat. Rev. Nephrol. 2010, 6, 643–656. [Google Scholar] [CrossRef] [PubMed]
- Kendall, R.T.; Feghali-Bostwick, C.A. Fibroblasts in fibrosis: Novel roles and mediators. Front. Pharmacol. 2014, 5, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Ricardo, S.D.; Samuel, C.S. Enhancing the Therapeutic Potential of Mesenchymal Stromal Cell-Based Therapies with an Anti-Fibrotic Agent for the Treatment of Chronic Kidney Disease. Int. J. Mol. Sci. 2022, 23, 6035. [Google Scholar] [CrossRef]
- Zeisberg, E.M.; Potenta, S.; Xie, L.; Zeisberg, M.; Kalluri, R. Discovery of endothelial to mesenchymal transition as a source for carcinoma-associated fibroblasts. Cancer Res. 2007, 67, 10123–10128. [Google Scholar] [CrossRef] [Green Version]
- Zeisberg, E.M.; Tarnavski, O.; Zeisberg, M.; Dorfman, A.L.; McMullen, J.R.; Gustafsson, E.; Chandraker, A.; Yuan, X.; Pu, W.T.; Roberts, A.B.; et al. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat. Med. 2007, 13, 952–961. [Google Scholar] [CrossRef]
- Piera-Velazquez, S.; Mendoza, F.A.; Jimenez, S.A. Endothelial to Mesenchymal Transition (EndoMT) in the Pathogenesis of Human Fibrotic Diseases. J. Clin. Med. 2016, 5, 45. [Google Scholar] [CrossRef]
- Isono, M.; Chen, S.; Hong, S.W.; Iglesias-de la Cruz, M.C.; Ziyadeh, F.N. Smad pathway is activated in the diabetic mouse kidney and Smad3 mediates TGF-beta-induced fibronectin in mesangial cells. Biochem. Biophys. Res. Commun. 2002, 296, 1356–1365. [Google Scholar] [CrossRef]
- Zhou, T.; He, X.; Cheng, R.; Zhang, B.; Zhang, R.R.; Chen, Y.; Takahashi, Y.; Murray, A.R.; Lee, K.; Gao, G.; et al. Implication of dysregulation of the canonical wingless-type MMTV integration site (WNT) pathway in diabetic nephropathy. Diabetologia 2012, 55, 255–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farías-Basulto, A.; Martínez-Ramírez, H.R.; Gómez-García, E.F.; Cueto-Manzano, A.M.; Cortés-Sanabria, L.; Hernández-Ramos, L.E.; Ramírez-López, G.; Mendoza-Carrera, F. Circulating Levels of Soluble Klotho and Fibroblast Growth Factor 23 in Diabetic Patients and Its Association with Early Nephropathy. Arch. Med. Res. 2018, 49, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wu, Y.; Yu, F.; Hu, W.; Fang, X.; Hao, W. The implication of Numb-induced Notch signaling in endothelial-mesenchymal transition of diabetic nephropathy. J. Diabetes Complicat. 2018, 32, 889–899. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.P.; Chang, S.Y.; Liao, M.C.; Lo, C.S.; Chenier, I.; Luo, H.; Chiasson, J.L.; Ingelfinger, J.R.; Chan, J.S.D.; Zhang, S.L. Hedgehog Interacting Protein Promotes Fibrosis and Apoptosis in Glomerular Endothelial Cells in Murine Diabetes. Sci. Rep. 2018, 8, 5958. [Google Scholar] [CrossRef]
- Srivastava, S.P.; Zhou, H.; Setia, O.; Liu, B.; Kanasaki, K.; Koya, D.; Dardik, A.; Fernandez-Hernando, C.; Goodwin, J. Loss of endothelial glucocorticoid receptor accelerates diabetic nephropathy. Nat. Commun. 2021, 12, 2368. [Google Scholar] [CrossRef]
- Srivastava, S.P.; Zhou, H.; Setia, O.; Dardik, A.; Fernandez-Hernando, C.; Goodwin, J. Podocyte Glucocorticoid Receptors Are Essential for Glomerular Endothelial Cell Homeostasis in Diabetes Mellitus. J. Am. Heart Assoc. 2021, 10, e019437. [Google Scholar] [CrossRef]
- Li, J.; Liu, H.; Takagi, S.; Nitta, K.; Kitada, M.; Srivastava, S.P.; Takagaki, Y.; Kanasaki, K.; Koya, D. Renal protective effects of empagliflozin via inhibition of EMT and aberrant glycolysis in proximal tubules. JCI Insight 2020, 5, e129034. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, S.P.; Goodwin, J.E.; Kanasaki, K.; Koya, D. Inhibition of Angiotensin-Converting Enzyme Ameliorates Renal Fibrosis by Mitigating DPP-4 Level and Restoring Antifibrotic MicroRNAs. Genes 2020, 11, 211. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.Y.; Zhang, X.B.; Zhao, Y.F.; Qu, K.; Yu, X.Y. Research Progress of Chinese Herbal Medicine Intervention in Renal Interstitial Fibrosis. Front. Pharmacol. 2022, 13, 900491. [Google Scholar] [CrossRef]
- García-Alvarez, J.; Ramirez, R.; Checa, M.; Nuttall, R.K.; Sampieri, C.L.; Edwards, D.R.; Selman, M.; Pardo, A. Tissue inhibitor of metalloproteinase-3 is up-regulated by transforming growth factor-beta1 in vitro and expressed in fibroblastic foci in vivo in idiopathic pulmonary fibrosis. Exp. Lung Res. 2006, 32, 201–214. [Google Scholar] [CrossRef]
- Klinkhammer, B.M.; Goldschmeding, R.; Floege, J.; Boor, P. Treatment of Renal Fibrosis-Turning Challenges into Opportunities. Adv. Chronic Kidney Dis. 2017, 24, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.J.; An, H.J.; Gwon, M.G.; Gu, H.; Bae, S.; Lee, S.J.; Kim, Y.A.; Leem, J.; Park, K.K. Anti-Fibrotic Effect of Synthetic Noncoding Oligodeoxynucleotide for Inhibiting mTOR and STAT3 via the Regulation of Autophagy in an Animal Model of Renal Injury. Molecules 2022, 27, 766. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Zhu, Y.; Sun, Y.; Li, X. IL-6 accelerates renal fibrosis after acute kidney injury via DNMT1-dependent FOXO3a methylation and activation of Wnt/β-catenin pathway. Int. ImmunoPharmacol. 2022, 109, 108746. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Zheng, C.; Zhu, L.; Song, Z.; Dai, L.; Hu, Q.; Wang, L.; Chen, Y.; Xiong, J. Contribution of TFEB-mediated autophagy to tubulointerstitial fibrosis in mice with adenine-induced chronic kidney disease. BioMed. Pharmacother. 2021, 133, 110949. [Google Scholar] [CrossRef]
- Rossing, P.F.P.; Feldt-Rasmussen, B. Brenner and Rector’s the Kidney, 10th ed.; Elsevier: Amsterdam, The Netherlands, 2016. [Google Scholar]
- Rossing, P. Clinical perspective-evolving evidence of mineralocorticoid receptor antagonists in patients with chronic kidney disease and type 2 diabetes. Kidney Int. Suppl. 2022, 12, 27–35. [Google Scholar] [CrossRef]
- Srivastava, S.P.; Goodwin, J.E.; Kanasaki, K.; Koya, D. Metabolic reprogramming by N-acetyl-seryl-aspartyl-lysyl-proline protects against diabetic kidney disease. Br. J. Pharmacol. 2020, 177, 3691–3711. [Google Scholar] [CrossRef]
- Feng, J.; Lu, C.; Dai, Q.; Sheng, J.; Xu, M. SIRT3 Facilitates Amniotic Fluid Stem Cells to Repair Diabetic Nephropathy through Protecting Mitochondrial Homeostasis by Modulation of Mitophagy. Cell. Physiol. Biochem. 2018, 46, 1508–1524. [Google Scholar] [CrossRef] [Green Version]
- Morita, M.; Kanasaki, K. Sodium-glucose cotransporter-2 inhibitors for diabetic kidney disease: Targeting Warburg effects in proximal tubular cells. Diabetes Metab. 2020, 46, 353–361. [Google Scholar] [CrossRef]
- Wei, Q.; Su, J.; Dong, G.; Zhang, M.; Huo, Y.; Dong, Z. Glycolysis inhibitors suppress renal interstitial fibrosis via divergent effects on fibroblasts and tubular cells. Am. J. Physiol. Renal Physiol. 2019, 316, F1162–F1172. [Google Scholar] [CrossRef]
- Awal, H.B.; Nandula, S.R.; Domingues, C.C.; Dore, F.J.; Kundu, N.; Brichacek, B.; Fakhri, M.; Elzarki, A.; Ahmadi, N.; Safai, S.; et al. Linagliptin, when compared to placebo, improves CD34+ve endothelial progenitor cells in type 2 diabetes subjects with chronic kidney disease taking metformin and/or insulin: A randomized controlled trial. Cardiovasc. Diabetol. 2020, 19, 72. [Google Scholar] [CrossRef]
- Kanlaya, R.; Thongboonkerd, V. Persistent Escherichia coli infection in renal tubular cells enhances calcium oxalate crystal-cell adhesion by inducing ezrin translocation to apical membranes via Rho/ROCK pathway. Cell Mol. Life Sci. 2022, 79, 381. [Google Scholar] [CrossRef] [PubMed]
- Ida-Naitoh, M.; Tokuyama, H.; Futatsugi, K.; Yasuda, M.; Adachi, K.; Kanda, T.; Tanabe, Y.; Wakino, S.; Itoh, H. Proximal-tubule molecular relay from early Protein diaphanous homolog 1 to late Rho-associated protein kinase 1 regulates kidney function in obesity-induced kidney damage. Kidney Int. 2022. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.A.; Kim, H.J.; Gwon, M.G.; Gu, H.; An, H.J.; Bae, S.; Leem, J.; Jung, H.J.; Park, K.K. Inhibitory Effects of STAT3 Transcription Factor by Synthetic Decoy ODNs on Autophagy in Renal Fibrosis. Biomedicines 2021, 9, 331. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J. Autophagy: From phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 2007, 8, 931–937. [Google Scholar] [CrossRef]
- Levine, B.; Kroemer, G. Autophagy in the pathogenesis of disease. Cell 2008, 132, 27–42. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, R.F., Jr.; Thakur, S.A.; Holian, A. Silica binding and toxicity in alveolar macrophages. Free Radic. Biol. Med. 2008, 44, 1246–1258. [Google Scholar] [CrossRef] [Green Version]
- Biswas, R.; Trout, K.L.; Jessop, F.; Harkema, J.R.; Holian, A. Imipramine blocks acute silicosis in a mouse model. Part Fibre Toxicol. 2017, 14, 36. [Google Scholar] [CrossRef] [Green Version]
- Napolitano, G.; Ballabio, A. TFEB at a glance. J. Cell Sci. 2016, 129, 2475–2481. [Google Scholar] [CrossRef] [Green Version]
- Sardiello, M.; Palmieri, M.; di Ronza, A.; Medina, D.L.; Valenza, M.; Gennarino, V.A.; Di Malta, C.; Donaudy, F.; Embrione, V.; Polishchuk, R.S.; et al. A gene network regulating lysosomal biogenesis and function. Science 2009, 325, 473–477. [Google Scholar] [CrossRef] [Green Version]
- Settembre, C.; Di Malta, C.; Polito, V.A.; Garcia Arencibia, M.; Vetrini, F.; Erdin, S.; Erdin, S.U.; Huynh, T.; Medina, D.; Colella, P.; et al. TFEB links autophagy to lysosomal biogenesis. Science 2011, 332, 1429–1433. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.H.; Kim, G.; Han, D.H.; Lee, M.; Kim, I.; Kim, B.; Kim, K.H.; Song, Y.M.; Yoo, J.E.; Wang, H.J.; et al. Ezetimibe ameliorates steatohepatitis via AMP activated protein kinase-TFEB-mediated activation of autophagy and NLRP3 inflammasome inhibition. Autophagy 2017, 13, 1767–1781. [Google Scholar] [CrossRef] [PubMed]
- He, G.; Karin, M. NF-κB and STAT3—Key players in liver inflammation and cancer. Cell Res. 2011, 21, 159–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, C.; Isaka, Y.; Takabatake, Y.; Tanaka, H.; Koike, M.; Shibata, M.; Uchiyama, Y.; Takahara, S.; Imai, E. Participation of autophagy in renal ischemia/reperfusion injury. Biochem. Biophys. Res. Commun. 2008, 368, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Sansanwal, P.; Yen, B.; Gahl, W.A.; Ma, Y.; Ying, L.; Wong, L.J.; Sarwal, M.M. Mitochondrial autophagy promotes cellular injury in nephropathic cystinosis. J. Am. Soc. Nephrol. 2010, 21, 272–283. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.Y.; An, H.J.; Kim, W.H.; Gwon, M.G.; Gu, H.; Park, Y.Y.; Park, K.K. Anti-fibrotic Effects of Synthetic Oligodeoxynucleotide for TGF-β1 and Smad in an Animal Model of Liver Cirrhosis. Mol. Ther. Nucleic Acids 2017, 8, 250–263. [Google Scholar] [CrossRef] [Green Version]
- Cao, C.C.; Ding, X.Q.; Ou, Z.L.; Liu, C.F.; Li, P.; Wang, L.; Zhu, C.F. In vivo transfection of NF-kappaB decoy oligodeoxynucleotides attenuate renal ischemia/reperfusion injury in rats. Kidney Int. 2004, 65, 834–845. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.H.; Lee, E.S.; Cha, S.H.; Park, J.H.; Park, J.S.; Chang, Y.C.; Park, K.K. Transcriptional regulation of NF-kappaB by ring type decoy oligodeoxynucleotide in an animal model of nephropathy. Exp. Mol. Pathol. 2009, 86, 114–120. [Google Scholar] [CrossRef]
- Lee, W.R.; Kim, K.H.; An, H.J.; Park, Y.Y.; Kim, K.S.; Lee, C.K.; Min, B.K.; Park, K.K. Effects of chimeric decoy oligodeoxynucleotide in the regulation of transcription factors NF-κB and Sp1 in an animal model of atherosclerosis. Basic Clin. Pharmacol. Toxicol. 2013, 112, 236–243. [Google Scholar] [CrossRef]
- Yang, H.C.; Zuo, Y.; Fogo, A.B. Models of chronic kidney disease. Drug Discov. Today Dis. Models 2010, 7, 13–19. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Ruan, S.; Wu, X.; Chen, H.; Zheng, K.; Fu, B. Autophagy and apoptosis in tubular cells following unilateral ureteral obstruction are associated with mitochondrial oxidative stress. Int. J. Mol. Med. 2013, 31, 628–636. [Google Scholar] [CrossRef]
- Kawaoka, K.; Doi, S.; Nakashima, A.; Yamada, K.; Ueno, T.; Doi, T.; Masaki, T. Valproic acid attenuates renal fibrosis through the induction of autophagy. Clin. Exp. Nephrol. 2017, 21, 771–780. [Google Scholar] [CrossRef] [Green Version]
- Boelens, M.C.; Wu, T.J.; Nabet, B.Y.; Xu, B.; Qiu, Y.; Yoon, T.; Azzam, D.J.; Twyman-Saint Victor, C.; Wiemann, B.Z.; Ishwaran, H.; et al. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell 2014, 159, 499–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, X.; Yuan, Q.; Chen, Y.; Huang, Z.; Fang, X.; Zhang, H.; Peng, L.; Xiao, P. LncRNA ENST00000453774.1 contributes to oxidative stress defense dependent on autophagy mediation to reduce extracellular matrix and alleviate renal fibrosis. J. Cell. Physiol. 2019, 234, 9130–9143. [Google Scholar] [CrossRef]
- Jiang, P.; Mizushima, N. Autophagy and human diseases. Cell Res. 2014, 24, 69–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, A.M.; Ryter, S.W.; Levine, B. Autophagy in human health and disease. N. Engl. J. Med. 2013, 368, 1845–1846. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Bravo-San Pedro, J.M.; Levine, B.; Green, D.R.; Kroemer, G. Pharmacological modulation of autophagy: Therapeutic potential and persisting obstacles. Nat. Rev. Drug Discov. 2017, 16, 487–511. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Nartiss, Y.; Steipe, B.; McQuibban, G.A.; Kim, P.K. ROS-induced mitochondrial depolarization initiates PARK2/PARKIN-dependent mitochondrial degradation by autophagy. Autophagy 2012, 8, 1462–1476. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.A.; Wu, V.C.; Wang, C.Y. Autophagy in Chronic Kidney Diseases. Cells 2019, 8, 61. [Google Scholar] [CrossRef] [Green Version]
- Xu, G.; Yue, F.; Huang, H.; He, Y.; Li, X.; Zhao, H.; Su, Z.; Jiang, X.; Li, W.; Zou, J.; et al. Defects in MAP1S-mediated autophagy turnover of fibronectin cause renal fibrosis. Aging 2016, 8, 977–985. [Google Scholar] [CrossRef] [Green Version]
- Livingston, M.J.; Ding, H.F.; Huang, S.; Hill, J.A.; Yin, X.M.; Dong, Z. Persistent activation of autophagy in kidney tubular cells promotes renal interstitial fibrosis during unilateral ureteral obstruction. Autophagy 2016, 12, 976–998. [Google Scholar] [CrossRef] [Green Version]
- Frangou, E.; Chrysanthopoulou, A.; Mitsios, A.; Kambas, K.; Arelaki, S.; Angelidou, I.; Arampatzioglou, A.; Gakiopoulou, H.; Bertsias, G.K.; Verginis, P.; et al. REDD1/autophagy pathway promotes thromboinflammation and fibrosis in human systemic lupus erythematosus (SLE) through NETs decorated with tissue factor (TF) and interleukin-17A (IL-17A). Ann. Rheum. Dis. 2019, 78, 238–248. [Google Scholar] [CrossRef] [PubMed]
- San-Miguel, B.; Crespo, I.; Sánchez, D.I.; González-Fernández, B.; Ortiz de Urbina, J.J.; Tuñón, M.J.; González-Gallego, J. Melatonin inhibits autophagy and endoplasmic reticulum stress in mice with carbon tetrachloride-induced fibrosis. J. Pineal Res. 2015, 59, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Chien, C.T.; Shyue, S.K.; Lai, M.K. Bcl-xL augmentation potentially reduces ischemia/reperfusion induced proximal and distal tubular apoptosis and autophagy. Transplantation 2007, 84, 1183–1190. [Google Scholar] [CrossRef]
- Kaushal, G.P. Autophagy protects proximal tubular cells from injury and apoptosis. Kidney Int. 2012, 82, 1250–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.L.; Zhang, J.; Cui, L.Y.; Yang, S. Autophagy activation attenuates renal ischemia-reperfusion injury in rats. Exp. Biol. Med. 2015, 240, 1590–1598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, M.; Wei, Q.; Dong, G.; Komatsu, M.; Su, Y.; Dong, Z. Autophagy in proximal tubules protects against acute kidney injury. Kidney Int. 2012, 82, 1271–1283. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Huang, H.; Jin, Y.; Shen, K.; Chen, X.; Xu, Z.; Jin, B.; Pan, H. Role of TFEB in autophagic modulation of ischemia reperfusion injury in mice kidney and protection by urolithin A. Food Chem. Toxicol. 2019, 131, 110591. [Google Scholar] [CrossRef]
- Velentzas, P.D.; Velentzas, A.D.; Mpakou, V.E.; Antonelou, M.H.; Margaritis, L.H.; Papassideri, I.S.; Stravopodis, D.J. Detrimental effects of proteasome inhibition activity in Drosophila melanogaster: Implication of ER stress, autophagy, and apoptosis. Cell Biol. Toxicol. 2013, 29, 13–37. [Google Scholar] [CrossRef]
- Kim, W.Y.; Nam, S.A.; Song, H.C.; Ko, J.S.; Park, S.H.; Kim, H.L.; Choi, E.J.; Kim, Y.S.; Kim, J.; Kim, Y.K. The role of autophagy in unilateral ureteral obstruction rat model. Nephrology 2012, 17, 148–159. [Google Scholar] [CrossRef]
- Han, K.; Zhou, H.; Pfeifer, U. Inhibition and restimulation by insulin of cellular autophagy in distal tubular cells of the kidney in early diabetic rats. Kidney Blood Press. Res. 1997, 20, 258–263. [Google Scholar] [CrossRef]
- Chevalier, R.L.; Forbes, M.S.; Thornhill, B.A. Ureteral obstruction as a model of renal interstitial fibrosis and obstructive nephropathy. Kidney Int. 2009, 75, 1145–1152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linkermann, A.; Chen, G.; Dong, G.; Kunzendorf, U.; Krautwald, S.; Dong, Z. Regulated cell death in AKI. J. Am. Soc. Nephrol. 2014, 25, 2689–2701. [Google Scholar] [CrossRef] [PubMed]
- Kers, J.; Leemans, J.C.; Linkermann, A. An Overview of Pathways of Regulated Necrosis in Acute Kidney Injury. Semin. Nephrol. 2016, 36, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Wang, Y.; Li, H.; Fan, J.; Shen, J.; Yu, X.; Zhou, Y.; Mao, H. ATG5-mediated autophagy suppresses NF-κB signaling to limit epithelial inflammatory response to kidney injury. Cell Death Dis. 2019, 10, 253. [Google Scholar] [CrossRef] [Green Version]
- Lu, H.; Fan, Y.; Qiao, C.; Liang, W.; Hu, W.; Zhu, T.; Zhang, J.; Chen, Y.E. TFEB inhibits endothelial cell inflammation and reduces atherosclerosis. Sci. Signal. 2017, 10, eaah4214. [Google Scholar] [CrossRef]
- Jung, H.J.; Kim, H.J.; Park, K.K. Potential Roles of Long Noncoding RNAs as Therapeutic Targets in Renal Fibrosis. Int. J. Mol. Sci. 2020, 21, 2698. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Xiao, T.; Li, F.; Li, Y.; Zeng, O.; Liu, M.; Liang, B.; Li, Z.; Chu, C.; Yang, J. Hydrogen sulfide reduced renal tissue fibrosis by regulating autophagy in diabetic rats. Mol. Med. Rep. 2017, 16, 1715–1722. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Hu, X.; Wang, J.; Xu, W.; Yi, C.; Ma, R.; Jiang, H. Inhibition of autophagy via activation of PI3K/Akt/mTOR pathway contributes to the protection of hesperidin against myocardial ischemia/reperfusion injury. Int. J. Mol. Med. 2018, 42, 1917–1924. [Google Scholar] [CrossRef] [Green Version]
- Ding, Y.; Kim, S.; Lee, S.Y.; Koo, J.K.; Wang, Z.; Choi, M.E. Autophagy regulates TGF-β expression and suppresses kidney fibrosis induced by unilateral ureteral obstruction. J. Am. Soc. Nephrol. 2014, 25, 2835–2846. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.H.; Jackson, S.; Seaman, M.; Brown, K.; Kempkes, B.; Hibshoosh, H.; Levine, B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999, 402, 672–676. [Google Scholar] [CrossRef]
- Füllgrabe, J.; Klionsky, D.J.; Joseph, B. The return of the nucleus: Transcriptional and epigenetic control of autophagy. Nat. Rev. Mol. Cell Biol. 2014, 15, 65–74. [Google Scholar] [CrossRef] [PubMed]


| Decoy | Sequence |
|---|---|
| Scr | 5′-GAATTCAATTCAGGGTACGGCAAAAAATTGCCGTACCCTGAATT-3′ |
| TFEB | 5′-GAATTCCCGCCACGTGGTCAAAAGACCACGTGGCGGGAA TTCCCCGACCACGTGGC AAAAGCCACGTGGTCGGG-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.-J.; Kim, Y.-A.; Park, K.-K. Anti-Fibrotic Effect of Synthetic Noncoding Decoy ODNs for TFEB in an Animal Model of Chronic Kidney Disease. Int. J. Mol. Sci. 2022, 23, 8138. https://doi.org/10.3390/ijms23158138
Lee S-J, Kim Y-A, Park K-K. Anti-Fibrotic Effect of Synthetic Noncoding Decoy ODNs for TFEB in an Animal Model of Chronic Kidney Disease. International Journal of Molecular Sciences. 2022; 23(15):8138. https://doi.org/10.3390/ijms23158138
Chicago/Turabian StyleLee, Sun-Jae, Young-Ah Kim, and Kwan-Kyu Park. 2022. "Anti-Fibrotic Effect of Synthetic Noncoding Decoy ODNs for TFEB in an Animal Model of Chronic Kidney Disease" International Journal of Molecular Sciences 23, no. 15: 8138. https://doi.org/10.3390/ijms23158138
APA StyleLee, S.-J., Kim, Y.-A., & Park, K.-K. (2022). Anti-Fibrotic Effect of Synthetic Noncoding Decoy ODNs for TFEB in an Animal Model of Chronic Kidney Disease. International Journal of Molecular Sciences, 23(15), 8138. https://doi.org/10.3390/ijms23158138






