microRNAs in the Antitumor Immune Response and in Bone Metastasis of Breast Cancer: From Biological Mechanisms to Therapeutics
Abstract
1. Introduction
miRNA Biogenesis and Functions
2. Breast Cancer: Metastasis and Role of the Immune System
2.1. Breast Cancer
2.2. Bone Metastasis
2.3. Breast Cancer and Immunity
3. microRNAs in Immune Escaping and Metastasis
3.1. miRNAs and Cancer-Related Immunity
3.1.1. miRNA and Immune Checkpoint Regulation
3.1.2. NK-Mediated Immune Surveillance and miRNAs Involved in Immune Escaping
3.2. miRNA in Metastasis
miRNA in Bone Metastasis from Breast Cancer
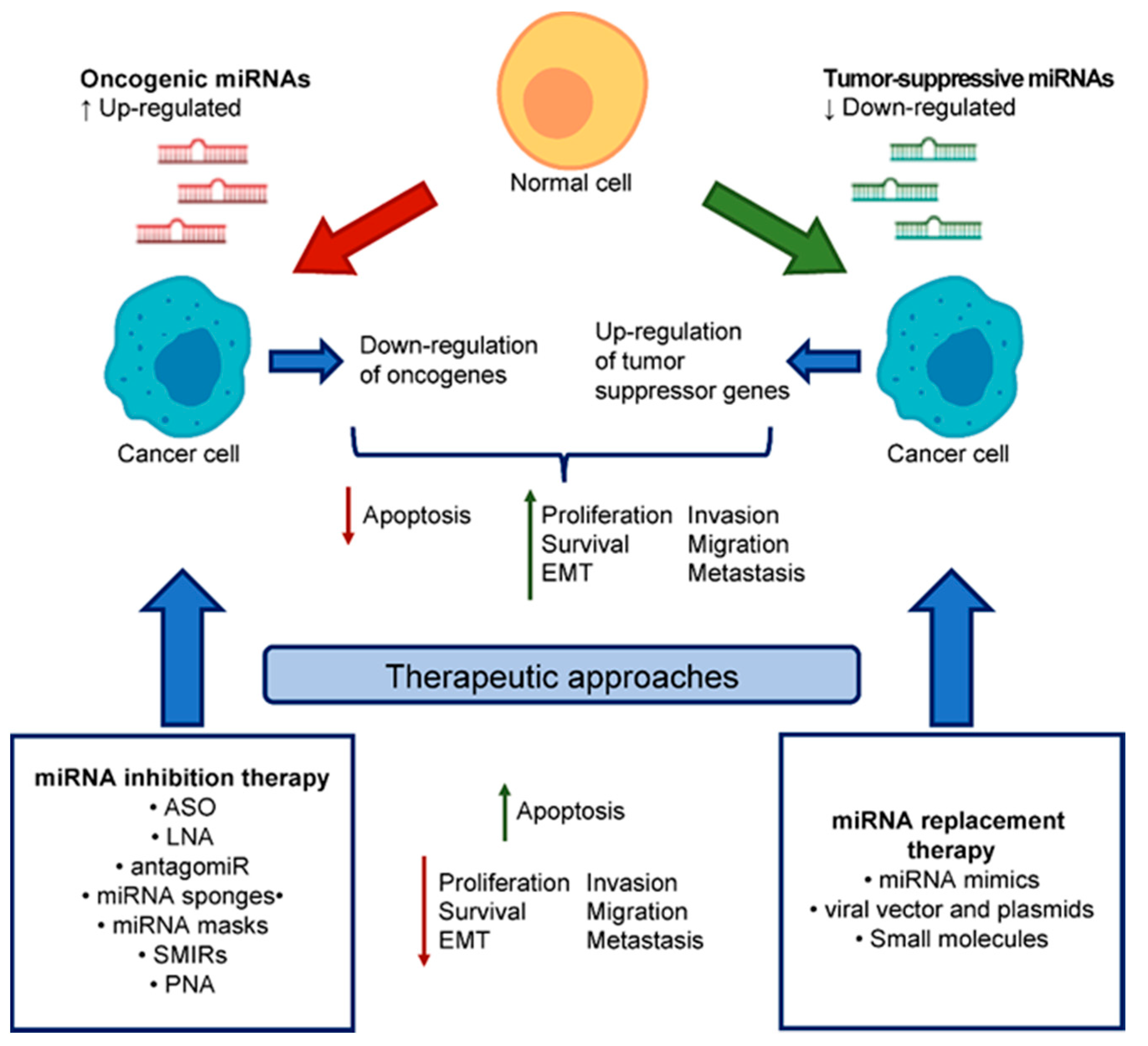
4. miRNA-Based Therapeutics
4.1. miRNA Replacement Therapy
4.2. miRNA Inhibition Therapy
4.3. PNA: Novel Class of MiRNA Inhibitors
5. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| miRNA | Micro RNA |
| ER | Oestrogen Receptor |
| PR | Progesteron Receptor |
| HER2 | Human Epidermal Growth Factor Receptor 2 |
| TNBC | Triple Negative Breast Cancer |
| MDSC | Myeloid-Derived Suppressor Cell |
| NK | Natural Killer |
| SRE | Skeletal-related Event |
| DTC | Disseminated tumor cell |
| TIL | Tumor infiltrating lymphocyte |
| DC | Dendritic Cell |
| APC | Antigen Presenting Cell |
| MHC | Major Histocompatibility Complex |
| TIIC | Tumor-Infiltrating Immune Cell |
| OS | Overall Survival |
| TAM | Tumor-Associate Macrophage |
| PD-L1 | Programmed cell Death-Ligand 1 |
| im-miRNAs | immune-modulatory miRNAs |
| TNF | Tumor Necrosis Factor |
| MICA | Major Histocompatibility Complex Class I–Related molecule A |
| MICB | Major Histocompatibility Complex Class I–Related molecule B |
| ULB1-6 | Unique Long 16 Binding Protein 1-6 |
| TRAIL | TNF-related apoptosis-inducing ligand |
| HDACi | Histone Deacetylase Inhibitors |
| EMT | Epithelial-Mesenchymal Transition |
| HOXD10 | Homeobox D10 |
| LIFR | Leukemia Inhibitory Factor Receptor |
| AMO | Anti-miRNA Oligonucleotides |
| LNA | Locked Nucleic Acid |
| SMIR | Small Molecule Inhibitors of miRNAs |
| PNA | Peptide Nucleic Acids |
| CPP | Cell-penetrating Peptide |
| PLGA | poly(lactic-co-glycolic acid) |
References
- Gnoni, A.; Brunetti, O.; Longo, V.; Calabrese, A.; Argentiero, A.; Roberto, C.; Giovanni, S.A.; Antonella, L. Immune system and bone microenvironment: Rationale for targeted cancer therapies. Oncotarget 2020, 11, 480–487. [Google Scholar] [CrossRef][Green Version]
- Clements, M.E.; Johnson, R.W. Breast Cancer Dormancy in Bone. Curr. Osteoporos. Rep. 2019, 17, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Bottani, M.; Banfi, G.; Lombardi, G. Circulating miRNAs as Diagnostic and Prognostic Biomarkers in Common Solid Tumors: Focus on Lung, Breast, Prostate Cancers, and Osteosarcoma. J. Clin. Med. 2019, 8, 1661. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F.; Monod, J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961, 3, 318–356. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Faraldi, M.; Gomarasca, M.; Banfi, G.; Lombardi, G. Free Circulating miRNAs Measurement in Clinical Settings: The Still Unsolved Issue of the Normalization. Adv. Clin. Chem. 2018, 87, 113–139. [Google Scholar] [CrossRef]
- Lujambio, A.; Calin, G.A.; Villanueva, A.; Ropero, S.; Sanchez-Cespedes, M.; Blanco, D.; Montuenga, L.M.; Rossi, S.; Nicoloso, M.S.; Faller, W.J.; et al. A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl. Acad. Sci. USA 2008, 105, 13556–13561. [Google Scholar] [CrossRef]
- Hammond, S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015, 87, 3–14. [Google Scholar] [CrossRef]
- Bernstein, E.; Kim, S.Y.; Carmell, M.A.; Murchison, E.P.; Alcorn, H.; Li, M.Z.; Mills, A.A.; Elledge, S.J.; Anderson, K.V.; Hannon, G.J. Dicer is essential for mouse development. Nat. Genet. 2003, 35, 215–217. [Google Scholar] [CrossRef]
- Murchison, E.P.; Partridge, J.F.; Tam, O.H.; Cheloufi, S.; Hannon, G.J. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA 2005, 102, 12135–12140. [Google Scholar] [CrossRef]
- Suh, M.R.; Lee, Y.; Kim, J.Y.; Kim, S.K.; Moon, S.H.; Lee, J.Y.; Cha, K.Y.; Chung, H.M.; Yoon, H.S.; Moon, S.Y.; et al. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 2004, 270, 488–498. [Google Scholar] [CrossRef] [PubMed]
- Bottani, M.; Banfi, G.; Lombardi, G. Perspectives on miRNAs as Epigenetic Markers in Osteoporosis and Bone Fracture Risk: A Step Forward in Personalized Diagnosis. Front. Genet. 2019, 10, 1044. [Google Scholar] [CrossRef] [PubMed]
- Ardekani, A.M.; Naeini, M.M. The Role of MicroRNAs in Human Diseases. Avicenna J. Med. Biotechnol. 2010, 2, 161–179. [Google Scholar] [PubMed]
- Wang, J.; Chen, J.; Sen, S. MicroRNA as Biomarkers and Diagnostics. J. Cell. Physiol. 2016, 231, 25–30. [Google Scholar] [CrossRef]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Wang, H.; Peng, R.; Wang, J.; Qin, Z.; Xue, L. Circulating microRNAs as potential cancer biomarkers: The advantage and disadvantage. Clin. Epigenetics 2018, 10, 59. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar] [CrossRef]
- Yang, Q.; Diamond, M.P.; Al-Hendy, A. The emerging role of extracellular vesicle-derived miRNAs: Implication in cancer progression and stem cell related diseases. J. Clin. Epigenet 2016, 2, 13. [Google Scholar]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- Fromm, B.; Billipp, T.; Peck, L.E.; Johansen, M.; Tarver, J.E.; King, B.L.; Newcomb, J.M.; Sempere, L.F.; Flatmark, K.; Hovig, E.; et al. A Uniform System for the Annotation of Vertebrate microRNA Genes and the Evolution of the Human microRNAome. Annu. Rev. Genet. 2015, 49, 213–242. [Google Scholar] [CrossRef]
- Li, Z.; Qiu, Y.; Lu, W.; Jiang, Y.; Wang, J. Immunotherapeutic interventions of Triple Negative Breast Cancer. J. Transl. Med. 2018, 16, 147. [Google Scholar] [CrossRef] [PubMed]
- Hammerl, D.; Smid, M.; Timmermans, A.M.; Sleijfer, S.; Martens, J.W.M.; Debets, R. Breast cancer genomics and immuno-oncological markers to guide immune therapies. Semin. Cancer Biol. 2018, 52, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.C.; Hu, Z.Q.; Long, J.H.; Zhu, G.M.; Wang, Y.; Jia, Y.; Zhou, J.; Ouyang, Y.; Zeng, Z. Clinical Implications of Tumor-Infiltrating Immune Cells in Breast Cancer. J. Cancer 2019, 10, 6175–6184. [Google Scholar] [CrossRef] [PubMed]
- Verma, C.; Kaewkangsadan, V.; Eremin, J.M.; Cowley, G.P.; Ilyas, M.; El-Sheemy, M.A.; Eremin, O. Natural killer (NK) cell profiles in blood and tumour in women with large and locally advanced breast cancer (LLABC) and their contribution to a pathological complete response (PCR) in the tumour following neoadjuvant chemotherapy (NAC): Differential restoration of blood profiles by NAC and surgery. J. Transl. Med. 2015, 13, 180. [Google Scholar] [CrossRef]
- Gentles, A.J.; Newman, A.M.; Liu, C.L.; Bratman, S.V.; Feng, W.; Kim, D.; Nair, V.S.; Xu, Y.; Khuong, A.; Hoang, C.D.; et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015, 21, 938–945. [Google Scholar] [CrossRef]
- Jiang, X.; Shapiro, D.J. The immune system and inflammation in breast cancer. Mol. Cell. Endocrinol. 2014, 382, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.J.; Guise, T.A.; Yin, J.J.; Elliott, J.; Horwood, N.J.; Martin, T.J.; Gillespie, M.T. Breast cancer cells interact with osteoblasts to support osteoclast formation. Endocrinology 1999, 140, 4451–4458. [Google Scholar] [CrossRef] [PubMed]
- Gomis, R.R.; Gawrzak, S. Tumor cell dormancy. Mol. Oncol. 2017, 11, 62–78. [Google Scholar] [CrossRef]
- Ghajar, C.M. Metastasis prevention by targeting the dormant niche. Nat. Rev. Cancer 2015, 15, 238–247. [Google Scholar] [CrossRef]
- Goss, P.E.; Chambers, A.F. Does tumour dormancy offer a therapeutic target? Nat. Rev. Cancer 2010, 10, 871–877. [Google Scholar] [CrossRef]
- Malladi, S.; Macalinao, D.G.; Jin, X.; He, L.; Basnet, H.; Zou, Y.; de Stanchina, E.; Massague, J. Metastatic Latency and Immune Evasion through Autocrine Inhibition of WNT. Cell 2016, 165, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Dunn, G.P.; Bruce, A.T.; Ikeda, H.; Old, L.J.; Schreiber, R.D. Cancer immunoediting: From immunosurveillance to tumor escape. Nat. Immunol. 2002, 3, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, R.D.; Old, L.J.; Smyth, M.J. Cancer immunoediting: Integrating immunity’s roles in cancer suppression and promotion. Science 2011, 331, 1565–1570. [Google Scholar] [CrossRef] [PubMed]
- Pages, F.; Galon, J.; Dieu-Nosjean, M.C.; Tartour, E.; Sautes-Fridman, C.; Fridman, W.H. Immune infiltration in human tumors: A prognostic factor that should not be ignored. Oncogene 2010, 29, 1093–1102. [Google Scholar] [CrossRef]
- Bense, R.D.; Sotiriou, C.; Piccart-Gebhart, M.J.; Haanen, J.; van Vugt, M.; de Vries, E.G.E.; Schroder, C.P.; Fehrmann, R.S.N. Relevance of Tumor-Infiltrating Immune Cell Composition and Functionality for Disease Outcome in Breast Cancer. J. Natl. Cancer Inst. 2017, 109. [Google Scholar] [CrossRef]
- Loi, S. Tumor-infiltrating lymphocytes, breast cancer subtypes and therapeutic efficacy. Oncoimmunology 2013, 2, e24720. [Google Scholar] [CrossRef]
- Mahmoud, S.M.; Paish, E.C.; Powe, D.G.; Macmillan, R.D.; Grainge, M.J.; Lee, A.H.; Ellis, I.O.; Green, A.R. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J. Clin. Oncol. 2011, 29, 1949–1955. [Google Scholar] [CrossRef]
- Ali, H.R.; Provenzano, E.; Dawson, S.J.; Blows, F.M.; Liu, B.; Shah, M.; Earl, H.M.; Poole, C.J.; Hiller, L.; Dunn, J.A.; et al. Association between CD8+ T-cell infiltration and breast cancer survival in 12,439 patients. Ann. Oncol. 2014, 25, 1536–1543. [Google Scholar] [CrossRef]
- Beyer, M.; Schultze, J.L. Regulatory T cells in cancer. Blood 2006, 108, 804–811. [Google Scholar] [CrossRef]
- Bates, G.J.; Fox, S.B.; Han, C.; Leek, R.D.; Garcia, J.F.; Harris, A.L.; Banham, A.H. Quantification of regulatory T cells enables the identification of high-risk breast cancer patients and those at risk of late relapse. J. Clin. Oncol. 2006, 24, 5373–5380. [Google Scholar] [CrossRef]
- Savas, P.; Salgado, R.; Denkert, C.; Sotiriou, C.; Darcy, P.K.; Smyth, M.J.; Loi, S. Clinical relevance of host immunity in breast cancer: From TILs to the clinic. Nat. Rev. Clin. Oncol. 2016, 13, 228–241. [Google Scholar] [CrossRef] [PubMed]
- Sznol, M.; Chen, L. Antagonist antibodies to PD-1 and B7-H1 (PD-L1) in the treatment of advanced human cancer. Clin. Cancer Res. 2013, 19, 1021–1034. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, D.; Kaur, H. Cell-free miRNAs as non-invasive biomarkers in breast cancer: Significance in early diagnosis and metastasis prediction. Life Sci. 2020, 246, 117417. [Google Scholar] [CrossRef] [PubMed]
- Paladini, L.; Fabris, L.; Bottai, G.; Raschioni, C.; Calin, G.A.; Santarpia, L. Targeting microRNAs as key modulators of tumor immune response. J. Exp. Clin. Cancer Res. 2016, 35, 103. [Google Scholar] [CrossRef] [PubMed]
- Smyth, M.J.; Dunn, G.P.; Schreiber, R.D. Cancer immunosurveillance and immunoediting: The roles of immunity in suppressing tumor development and shaping tumor immunogenicity. Adv. Immunol. 2006, 90, 1–50. [Google Scholar] [CrossRef] [PubMed]
- Vesely, M.D.; Kershaw, M.H.; Schreiber, R.D.; Smyth, M.J. Natural innate and adaptive immunity to cancer. Annu. Rev. Immunol. 2011, 29, 235–271. [Google Scholar] [CrossRef]
- Tili, E.; Michaille, J.J.; Croce, C.M. MicroRNAs play a central role in molecular dysfunctions linking inflammation with cancer. Immunol. Rev. 2013, 253, 167–184. [Google Scholar] [CrossRef]
- Lujambio, A.; Lowe, S.W. The microcosmos of cancer. Nature 2012, 482, 347–355. [Google Scholar] [CrossRef]
- Eichmuller, S.B.; Osen, W.; Mandelboim, O.; Seliger, B. Immune Modulatory microRNAs Involved in Tumor Attack and Tumor Immune Escape. J. Natl. Cancer Inst. 2017, 109, 10. [Google Scholar] [CrossRef]
- Mattiske, S.; Suetani, R.J.; Neilsen, P.M.; Callen, D.F. The oncogenic role of miR-155 in breast cancer. Cancer Epidemiol. Biomark. Prev. 2012, 21, 1236–1243. [Google Scholar] [CrossRef]
- Zonari, E.; Pucci, F.; Saini, M.; Mazzieri, R.; Politi, L.S.; Gentner, B.; Naldini, L. A role for miR-155 in enabling tumor-infiltrating innate immune cells to mount effective antitumor responses in mice. Blood 2013, 122, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Liu, M.; Xu, Z.; Li, Y.; Guo, H.; Ge, Y.; Liu, Y.; Zheng, D.; Shi, J. A double feedback loop mediated by microRNA-23a/27a/24-2 regulates M1 versus M2 macrophage polarization and thus regulates cancer progression. Oncotarget 2016, 7, 13502–13519. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Cai, Y.; Zhang, D.; Sun, J.; Xu, C.; Zhao, W.; Jiang, W.; Pan, C. miR-195/miR-497 Regulate CD274 Expression of Immune Regulatory Ligands in Triple-Negative Breast Cancer. J. Breast Cancer 2018, 21, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Z.; Chen, C.; Liu, Y.; Si, Q.; Chuang, T.H.; Li, N.; Gomez-Cabrero, A.; Reisfeld, R.A.; Xiang, R.; et al. MicroRNA-19a-3p inhibits breast cancer progression and metastasis by inducing macrophage polarization through downregulated expression of Fra-1 proto-oncogene. Oncogene 2014, 33, 3014–3023. [Google Scholar] [CrossRef] [PubMed]
- Hong, B.S.; Ryu, H.S.; Kim, N.; Kim, J.; Lee, E.; Moon, H.; Kim, K.H.; Jin, M.S.; Kwon, N.H.; Kim, S.; et al. Tumor Suppressor miRNA-204-5p Regulates Growth, Metastasis, and Immune Microenvironment Remodeling in Breast Cancer. Cancer Res. 2019, 79, 1520–1534. [Google Scholar] [CrossRef]
- Stern-Ginossar, N.; Gur, C.; Biton, M.; Horwitz, E.; Elboim, M.; Stanietsky, N.; Mandelboim, M.; Mandelboim, O. Human microRNAs regulate stress-induced immune responses mediated by the receptor NKG2D. Nat. Immunol. 2008, 9, 1065–1073. [Google Scholar] [CrossRef]
- Codo, P.; Weller, M.; Meister, G.; Szabo, E.; Steinle, A.; Wolter, M.; Reifenberger, G.; Roth, P. MicroRNA-mediated down-regulation of NKG2D ligands contributes to glioma immune escape. Oncotarget 2014, 5, 7651–7662. [Google Scholar] [CrossRef]
- Tsukerman, P.; Stern-Ginossar, N.; Gur, C.; Glasner, A.; Nachmani, D.; Bauman, Y.; Yamin, R.; Vitenshtein, A.; Stanietsky, N.; Bar-Mag, T.; et al. MiR-10b downregulates the stress-induced cell surface molecule MICB, a critical ligand for cancer cell recognition by natural killer cells. Cancer Res. 2012, 72, 5463–5472. [Google Scholar] [CrossRef]
- Kim, J.; Yao, F.; Xiao, Z.; Sun, Y.; Ma, L. MicroRNAs and metastasis: Small RNAs play big roles. Cancer Metastasis Rev. 2018, 37, 5–15. [Google Scholar] [CrossRef]
- Ma, L. Role of miR-10b in breast cancer metastasis. Breast Cancer Res. 2010, 12, 210. [Google Scholar] [CrossRef]
- Ma, L.; Reinhardt, F.; Pan, E.; Soutschek, J.; Bhat, B.; Marcusson, E.G.; Teruya-Feldstein, J.; Bell, G.W.; Weinberg, R.A. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat. Biotechnol. 2010, 28, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Teruya-Feldstein, J.; Weinberg, R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007, 449, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Min, D.; Lv, X.B.; Wang, X.; Zhang, B.; Meng, W.; Yu, F.; Hu, H. Downregulation of miR-302c and miR-520c by 1,25(OH)2D3 treatment enhances the susceptibility of tumour cells to natural killer cell-mediated cytotoxicity. Br. J. Cancer 2013, 109, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Pan, J.; Du, C.; Si, W.; Yao, M.; Xu, L.; Zheng, H.; Xu, M.; Chen, D.; Wang, S.; et al. Silencing NKG2D ligand-targeting miRNAs enhances natural killer cell-mediated cytotoxicity in breast cancer. Cell Death Dis. 2017, 8, e2740. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wang, Q.; Wang, Z.; Jiang, J.; Yu, S.C.; Ping, Y.F.; Yang, J.; Xu, S.L.; Ye, X.Z.; Xu, C.; et al. Metastatic consequences of immune escape from NK cell cytotoxicity by human breast cancer stem cells. Cancer Res. 2014, 74, 5746–5757. [Google Scholar] [CrossRef] [PubMed]
- Breunig, C.; Pahl, J.; Kublbeck, M.; Miller, M.; Antonelli, D.; Erdem, N.; Wirth, C.; Will, R.; Bott, A.; Cerwenka, A.; et al. MicroRNA-519a-3p mediates apoptosis resistance in breast cancer cells and their escape from recognition by natural killer cells. Cell Death Dis. 2017, 8, e2973. [Google Scholar] [CrossRef]
- Ma, L.; Young, J.; Prabhala, H.; Pan, E.; Mestdagh, P.; Muth, D.; Teruya-Feldstein, J.; Reinhardt, F.; Onder, T.T.; Valastyan, S.; et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 2010, 12, 247–256. [Google Scholar] [CrossRef]
- Chen, D.; Sun, Y.; Wei, Y.; Zhang, P.; Rezaeian, A.H.; Teruya-Feldstein, J.; Gupta, S.; Liang, H.; Lin, H.K.; Hung, M.C.; et al. LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nat. Med. 2012, 18, 1511–1517. [Google Scholar] [CrossRef]
- Johnson, R.W.; Finger, E.C.; Olcina, M.M.; Vilalta, M.; Aguilera, T.; Miao, Y.; Merkel, A.R.; Johnson, J.R.; Sterling, J.A.; Wu, J.Y.; et al. Induction of LIFR confers a dormancy phenotype in breast cancer cells disseminated to the bone marrow. Nat. Cell Biol. 2016, 18, 1078–1089. [Google Scholar] [CrossRef]
- Jin, T.; Suk Kim, H.; Ki Choi, S.; Hye Hwang, E.; Woo, J.; Suk Ryu, H.; Kim, K.; Moon, A.; Kyung Moon, W. microRNA-200c/141 upregulates SerpinB2 to promote breast cancer cell metastasis and reduce patient survival. Oncotarget 2017, 8, 32769–32782. [Google Scholar] [CrossRef]
- Cai, J.; Guan, H.; Fang, L.; Yang, Y.; Zhu, X.; Yuan, J.; Wu, J.; Li, M. MicroRNA-374a activates Wnt/beta-catenin signaling to promote breast cancer metastasis. J. Clin. Invest. 2013, 123, 566–579. [Google Scholar] [CrossRef] [PubMed]
- Martello, G.; Rosato, A.; Ferrari, F.; Manfrin, A.; Cordenonsi, M.; Dupont, S.; Enzo, E.; Guzzardo, V.; Rondina, M.; Spruce, T.; et al. A MicroRNA targeting dicer for metastasis control. Cell 2010, 141, 1195–1207. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Ivan, C.; Yang, D.; Gharpure, K.M.; Wu, S.Y.; Pecot, C.V.; Previs, R.A.; Nagaraja, A.S.; Armaiz-Pena, G.N.; McGuire, M.; et al. Hypoxia-upregulated microRNA-630 targets Dicer, leading to increased tumor progression. Oncogene 2016, 35, 4312–4320. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, X.; Yang, M.; Kan, Q.; Duan, Z. miR3609 sensitizes breast cancer cells to adriamycin by blocking the programmed death-ligand 1 immune checkpoint. Exp. Cell Res. 2019, 380, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Tavazoie, S.F.; Alarcon, C.; Oskarsson, T.; Padua, D.; Wang, Q.; Bos, P.D.; Gerald, W.L.; Massague, J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 2008, 451, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, P.; Sun, T.; Li, D.; Xu, X.; Rui, Y.; Li, C.; Chong, M.; Ibrahim, T.; Mercatali, L.; et al. miR-126 and miR-126* repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nat. Cell Biol. 2013, 15, 284–294. [Google Scholar] [CrossRef]
- Park, S.M.; Gaur, A.B.; Lengyel, E.; Peter, M.E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008, 22, 894–907. [Google Scholar] [CrossRef]
- Dykxhoorn, D.M.; Wu, Y.; Xie, H.; Yu, F.; Lal, A.; Petrocca, F.; Martinvalet, D.; Song, E.; Lim, B.; Lieberman, J. miR-200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS ONE 2009, 4, e7181. [Google Scholar] [CrossRef]
- Yu, Y.; Luo, W.; Yang, Z.J.; Chi, J.R.; Li, Y.R.; Ding, Y.; Ge, J.; Wang, X.; Cao, X.C. miR-190 suppresses breast cancer metastasis by regulation of TGF-beta-induced epithelial-mesenchymal transition. Mol. Cancer 2018, 17, 70. [Google Scholar] [CrossRef]
- Jiang, Q.; He, M.; Ma, M.T.; Wu, H.Z.; Yu, Z.J.; Guan, S.; Jiang, L.Y.; Wang, Y.; Zheng, D.D.; Jin, F.; et al. MicroRNA-148a inhibits breast cancer migration and invasion by directly targeting WNT-1. Oncol. Rep. 2016, 35, 1425–1432. [Google Scholar] [CrossRef]
- Mohammadi-Yeganeh, S.; Paryan, M.; Arefian, E.; Vasei, M.; Ghanbarian, H.; Mahdian, R.; Karimipoor, M.; Soleimani, M. MicroRNA-340 inhibits the migration, invasion, and metastasis of breast cancer cells by targeting Wnt pathway. Tumour Biol. 2016, 37, 8993–9000. [Google Scholar] [CrossRef] [PubMed]
- Krzeszinski, J.Y.; Wei, W.; Huynh, H.; Jin, Z.; Wang, X.; Chang, T.C.; Xie, X.J.; He, L.; Mangala, L.S.; Lopez-Berestein, G.; et al. miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature 2014, 512, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.L.; Huang, W.D.; Li, B.; Chen, T.R.; Li, Z.X.; Zhao, C.L.; Li, H.Y.; Wu, Y.M.; Yan, W.J.; Xiao, J.R. microRNA-124 inhibits bone metastasis of breast cancer by repressing Interleukin-11. Mol. Cancer 2018, 17, 9. [Google Scholar] [CrossRef] [PubMed]
- Croset, M.; Pantano, F.; Kan, C.W.S.; Bonnelye, E.; Descotes, F.; Alix-Panabieres, C.; Lecellier, C.H.; Bachelier, R.; Allioli, N.; Hong, S.S.; et al. miRNA-30 Family Members Inhibit Breast Cancer Invasion, Osteomimicry, and Bone Destruction by Directly Targeting Multiple Bone Metastasis-Associated Genes. Cancer Res. 2018, 78, 5259–5273. [Google Scholar] [CrossRef] [PubMed]
- Darvin, P.; Toor, S.M.; Sasidharan Nair, V.; Elkord, E. Immune checkpoint inhibitors: Recent progress and potential biomarkers. Exp. Mol. Med. 2018, 50, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Muenst, S.; Schaerli, A.R.; Gao, F.; Daster, S.; Trella, E.; Droeser, R.A.; Muraro, M.G.; Zajac, P.; Zanetti, R.; Gillanders, W.E.; et al. Expression of programmed death ligand 1 (PD-L1) is associated with poor prognosis in human breast cancer. Breast Cancer Res. Treat. 2014, 146, 15–24. [Google Scholar] [CrossRef]
- Kao, S.C.; Cheng, Y.Y.; Williams, M.; Kirschner, M.B.; Madore, J.; Lum, T.; Sarun, K.H.; Linton, A.; McCaughan, B.; Klebe, S.; et al. Tumor Suppressor microRNAs Contribute to the Regulation of PD-L1 Expression in Malignant Pleural Mesothelioma. J. Thorac. Oncol. 2017, 12, 1421–1433. [Google Scholar] [CrossRef]
- Wang, X.; Li, J.; Dong, K.; Lin, F.; Long, M.; Ouyang, Y.; Wei, J.; Chen, X.; Weng, Y.; He, T.; et al. Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell. Signal. 2015, 27, 443–452. [Google Scholar] [CrossRef]
- Chen, L.; Gibbons, D.L.; Goswami, S.; Cortez, M.A.; Ahn, Y.H.; Byers, L.A.; Zhang, X.; Yi, X.; Dwyer, D.; Lin, W.; et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat. Commun. 2014, 5, 5241. [Google Scholar] [CrossRef]
- Xu, S.; Tao, Z.; Hai, B.; Liang, H.; Shi, Y.; Wang, T.; Song, W.; Chen, Y.; OuYang, J.; Chen, J.; et al. miR-424(322) reverses chemoresistance via T-cell immune response activation by blocking the PD-L1 immune checkpoint. Nat. Commun. 2016, 7, 11406. [Google Scholar] [CrossRef]
- Gong, A.Y.; Zhou, R.; Hu, G.; Liu, J.; Sosnowska, D.; Drescher, K.M.; Dong, H.; Chen, X.M. Cryptosporidium parvum induces B7-H1 expression in cholangiocytes by down-regulating microRNA-513. J. Infect. Dis. 2010, 201, 160–169. [Google Scholar] [CrossRef]
- Zhao, L.; Yu, H.; Yi, S.; Peng, X.; Su, P.; Xiao, Z.; Liu, R.; Tang, A.; Li, X.; Liu, F.; et al. The tumor suppressor miR-138-5p targets PD-L1 in colorectal cancer. Oncotarget 2016, 7, 45370–45384. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Xi, Q.; Wang, H.; Zhang, Z.; Liu, H.; Cheng, Y.; Guo, X.; Zhang, J.; Zhang, Q.; Zhang, L.; et al. miR-142-5p regulates tumor cell PD-L1 expression and enhances anti-tumor immunity. Biochem. Biophys. Res. Commun. 2017, 488, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Cao, W.; Wang, Z.; Zhang, B.; Liu, J. Regulation of cancer immune escape: The roles of miRNAs in immune checkpoint proteins. Cancer Lett. 2018, 431, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Lin, W.; Tang, X.; Li, S.; Guo, L.; Lin, Y.; Kwok, H.F. The Roles of microRNAs in Regulating the Expression of PD-1/PD-L1 Immune Checkpoint. Int. J. Mol. Sci. 2017, 18, 2540. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Nduom, E.K.; Kong, L.Y.; Hashimoto, Y.; Xu, S.; Gabrusiewicz, K.; Ling, X.; Huang, N.; Qiao, W.; Zhou, S.; et al. MiR-138 exerts anti-glioma efficacy by targeting immune checkpoints. Neuro-Oncology 2016, 18, 639–648. [Google Scholar] [CrossRef]
- Topalian, S.L.; Drake, C.G.; Pardoll, D.M. Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer Cell 2015, 27, 450–461. [Google Scholar] [CrossRef]
- Leach, D.R.; Krummel, M.F.; Allison, J.P. Enhancement of antitumor immunity by CTLA-4 blockade. Science 1996, 271, 1734–1736. [Google Scholar] [CrossRef]
- Met, O.; Wang, M.; Pedersen, A.E.; Nissen, M.H.; Buus, S.; Claesson, M.H. The effect of a therapeutic dendritic cell-based cancer vaccination depends on the blockage of CTLA-4 signaling. Cancer Lett. 2006, 231, 247–256. [Google Scholar] [CrossRef]
- Chen, X.; Shao, Q.; Hao, S.; Zhao, Z.; Wang, Y.; Guo, X.; He, Y.; Gao, W.; Mao, H. CTLA-4 positive breast cancer cells suppress dendritic cells maturation and function. Oncotarget 2017, 8, 13703–13715. [Google Scholar] [CrossRef]
- Richardsen, E.; Andersen, S.; Al-Saad, S.; Rakaee, M.; Nordby, Y.; Pedersen, M.I.; Ness, N.; Ingebriktsen, L.M.; Fassina, A.; Tasken, K.A.; et al. Low Expression of miR-424-3p is Highly Correlated with Clinical Failure in Prostate Cancer. Sci. Rep. 2019, 9, 10662. [Google Scholar] [CrossRef] [PubMed]
- Tao, Z.; Xu, S.; Ruan, H.; Wang, T.; Song, W.; Qian, L.; Chen, K. MiR-195/-16 Family Enhances Radiotherapy via T Cell Activation in the Tumor Microenvironment by Blocking the PD-L1 Immune Checkpoint. Cell. Physiol. Biochem. 2018, 48, 801–814. [Google Scholar] [CrossRef] [PubMed]
- Huffaker, T.B.; Lee, S.H.; Tang, W.W.; Wallace, J.A.; Alexander, M.; Runtsch, M.C.; Larsen, D.K.; Thompson, J.; Ramstead, A.G.; Voth, W.P.; et al. Antitumor immunity is defective in T cell-specific microRNA-155-deficient mice and is rescued by immune checkpoint blockade. J. Biol. Chem. 2017, 292, 18530–18541. [Google Scholar] [CrossRef]
- Lopez-Soto, A.; Huergo-Zapico, L.; Acebes-Huerta, A.; Villa-Alvarez, M.; Gonzalez, S. NKG2D signaling in cancer immunosurveillance. Int. J. Cancer 2015, 136, 1741–1750. [Google Scholar] [CrossRef] [PubMed]
- Schmiedel, D.; Mandelboim, O. NKG2D Ligands-Critical Targets for Cancer Immune Escape and Therapy. Front. Immunol. 2018, 9, 2040. [Google Scholar] [CrossRef] [PubMed]
- Ogasawara, K.; Lanier, L.L. NKG2D in NK and T cell-mediated immunity. J. Clin. Immunol. 2005, 25, 534–540. [Google Scholar] [CrossRef]
- Watson, N.F.; Spendlove, I.; Madjd, Z.; McGilvray, R.; Green, A.R.; Ellis, I.O.; Scholefield, J.H.; Durrant, L.G. Expression of the stress-related MHC class I chain-related protein MICA is an indicator of good prognosis in colorectal cancer patients. Int. J. Cancer 2006, 118, 1445–1452. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, N.; Yu, Y.; Zhou, L.; Niu, C.; Liu, Y.; Tian, H.; Lv, Z.; Han, F.; Cui, J. Prognostic value of MICA/B in cancers: A systematic review and meta-analysis. Oncotarget 2017, 8, 96384–96395. [Google Scholar] [CrossRef]
- Mogilyansky, E.; Rigoutsos, I. The miR-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013, 20, 1603–1614. [Google Scholar] [CrossRef]
- Kishikawa, T.; Otsuka, M.; Yoshikawa, T.; Ohno, M.; Takata, A.; Shibata, C.; Kondo, Y.; Akanuma, M.; Yoshida, H.; Koike, K. Regulation of the expression of the liver cancer susceptibility gene MICA by microRNAs. Sci. Rep. 2013, 3, 2739. [Google Scholar] [CrossRef]
- Heinemann, A.; Zhao, F.; Pechlivanis, S.; Eberle, J.; Steinle, A.; Diederichs, S.; Schadendorf, D.; Paschen, A. Tumor suppressive microRNAs miR-34a/c control cancer cell expression of ULBP2, a stress-induced ligand of the natural killer cell receptor NKG2D. Cancer Res. 2012, 72, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Himmelreich, H.; Mathys, A.; Wodnar-Filipowicz, A.; Kalberer, C.P. Post-transcriptional regulation of ULBP1 ligand for the activating immunoreceptor NKG2D involves 3’ untranslated region. Hum. Immunol. 2011, 72, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Lan, P.; Hou, Z.; Guan, Y.; Zhang, J.; Xu, W.; Tian, Z.; Zhang, C. Histone deacetylase inhibitor SAHA epigenetically regulates miR-17-92 cluster and MCM7 to upregulate MICA expression in hepatoma. Br. J. Cancer 2015, 112, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Somarelli, J.A.; Shetler, S.; Jolly, M.K.; Wang, X.; Bartholf Dewitt, S.; Hish, A.J.; Gilja, S.; Eward, W.C.; Ware, K.E.; Levine, H.; et al. Mesenchymal-Epithelial Transition in Sarcomas Is Controlled by the Combinatorial Expression of MicroRNA 200s and GRHL2. Mol. Cell. Biol. 2016, 36, 2503–2513. [Google Scholar] [CrossRef] [PubMed]
- Loh, H.Y.; Norman, B.P.; Lai, K.S.; Rahman, N.; Alitheen, N.B.M.; Osman, M.A. The Regulatory Role of MicroRNAs in Breast Cancer. Int. J. Mol. Sci. 2019, 20, 4940. [Google Scholar] [CrossRef] [PubMed]
- van den Beucken, T.; Koch, E.; Chu, K.; Rupaimoole, R.; Prickaerts, P.; Adriaens, M.; Voncken, J.W.; Harris, A.L.; Buffa, F.M.; Haider, S.; et al. Hypoxia promotes stem cell phenotypes and poor prognosis through epigenetic regulation of DICER. Nat. Commun. 2014, 5, 5203. [Google Scholar] [CrossRef]
- Faraldi, M.; Sansoni, V.; Perego, S.; Gomarasca, M.; Kortas, J.; Ziemann, E.; Banfi, G.; Lombardi, G. Study of the preanalytical variables affecting the measurement of clinically relevant free-circulating microRNAs: Focus on sample matrix, platelet depletion, and storage conditions. Biochem. Med. 2020, 30, 010703. [Google Scholar] [CrossRef]
- Roth, C.; Rack, B.; Muller, V.; Janni, W.; Pantel, K.; Schwarzenbach, H. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010, 12, R90. [Google Scholar] [CrossRef]
- Chen, W.; Cai, F.; Zhang, B.; Barekati, Z.; Zhong, X.Y. The level of circulating miRNA-10b and miRNA-373 in detecting lymph node metastasis of breast cancer: Potential biomarkers. Tumour Biol. 2013, 34, 455–462. [Google Scholar] [CrossRef]
- McGuire, A.; Brown, J.A.; Kerin, M.J. Metastatic breast cancer: The potential of miRNA for diagnosis and treatment monitoring. Cancer Metastasis Rev. 2015, 34, 145–155. [Google Scholar] [CrossRef]
- Papadaki, C.; Stratigos, M.; Markakis, G.; Spiliotaki, M.; Mastrostamatis, G.; Nikolaou, C.; Mavroudis, D.; Agelaki, S. Circulating microRNAs in the early prediction of disease recurrence in primary breast cancer. Breast Cancer Res. 2018, 20, 72. [Google Scholar] [CrossRef] [PubMed]
- Papadaki, C.; Stoupis, G.; Tsalikis, L.; Monastirioti, A.; Papadaki, M.; Maliotis, N.; Stratigos, M.; Mastrostamatis, G.; Mavroudis, D.; Agelaki, S. Circulating miRNAs as a marker of metastatic disease and prognostic factor in metastatic breast cancer. Oncotarget 2019, 10, 966–981. [Google Scholar] [CrossRef] [PubMed]
- Madhavan, D.; Zucknick, M.; Wallwiener, M.; Cuk, K.; Modugno, C.; Scharpff, M.; Schott, S.; Heil, J.; Turchinovich, A.; Yang, R.; et al. Circulating miRNAs as surrogate markers for circulating tumor cells and prognostic markers in metastatic breast cancer. Clin. Cancer Res. 2012, 18, 5972–5982. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.; Chin, A.R.; et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014, 25, 501–515. [Google Scholar] [CrossRef] [PubMed]
- Asaga, S.; Kuo, C.; Nguyen, T.; Terpenning, M.; Giuliano, A.E.; Hoon, D.S. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin. Chem. 2011, 57, 84–91. [Google Scholar] [CrossRef]
- Shah, M.Y.; Ferrajoli, A.; Sood, A.K.; Lopez-Berestein, G.; Calin, G.A. microRNA Therapeutics in Cancer–An Emerging Concept. EBioMedicine 2016, 12, 34–42. [Google Scholar] [CrossRef]
- Balacescu, O.; Visan, S.; Baldasici, O.; Balacescu, L.; Vlad, C.; Achimas-Cadariu, P. MiRNA-Based Therapeutics in Oncology, Realities, and Challenges. In Antisense Therapy; Sharad, S., Kapur, S., Eds.; Interchopen: London, Greater London, UK, 2018. [Google Scholar]
- Melo, S.; Villanueva, A.; Moutinho, C.; Davalos, V.; Spizzo, R.; Ivan, C.; Rossi, S.; Setien, F.; Casanovas, O.; Simo-Riudalbas, L.; et al. Small molecule enoxacin is a cancer-specific growth inhibitor that acts by enhancing TAR RNA-binding protein 2-mediated microRNA processing. Proc. Natl. Acad. Sci. USA 2011, 108, 4394–4399. [Google Scholar] [CrossRef]
- Bader, A.G.; Brown, D.; Winkler, M. The promise of microRNA replacement therapy. Cancer Res. 2010, 70, 7027–7030. [Google Scholar] [CrossRef]
- Aagaard, L.; Rossi, J.J. RNAi therapeutics: Principles, prospects and challenges. Adv. Drug Deliv. Rev. 2007, 59, 75–86. [Google Scholar] [CrossRef]
- Mae, M.; Langel, U. Cell-penetrating peptides as vectors for peptide, protein and oligonucleotide delivery. Curr. Opin. Pharm. 2006, 6, 509–514. [Google Scholar] [CrossRef]
- Hoyer, J.; Schatzschneider, U.; Schulz-Siegmund, M.; Neundorf, I. Dimerization of a cell-penetrating peptide leads to enhanced cellular uptake and drug delivery. Beilstein J. Org. Chem. 2012, 8, 1788–1797. [Google Scholar] [CrossRef] [PubMed]
- Wiggins, J.F.; Ruffino, L.; Kelnar, K.; Omotola, M.; Patrawala, L.; Brown, D.; Bader, A.G. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010, 70, 5923–5930. [Google Scholar] [CrossRef] [PubMed]
- Kaban, K.; Salva, E.; Akbuga, J. The effects of chitosan/miR-200c nanoplexes on different stages of cancers in breast cancer cell lines. Eur. J. Pharm. Sci. 2016, 95, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Gambari, R.; Fabbri, E.; Borgatti, M.; Lampronti, I.; Finotti, A.; Brognara, E.; Bianchi, N.; Manicardi, A.; Marchelli, R.; Corradini, R. Targeting microRNAs involved in human diseases: A novel approach for modification of gene expression and drug development. Biochem. Pharm. 2011, 82, 1416–1429. [Google Scholar] [CrossRef]
- Garzon, R.; Marcucci, G.; Croce, C.M. Targeting microRNAs in cancer: Rationale, strategies and challenges. Nat. Rev. Drug Discov. 2010, 9, 775–789. [Google Scholar] [CrossRef]
- Krutzfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005, 438, 685–689. [Google Scholar] [CrossRef]
- Gallo Cantafio, M.E.; Nielsen, B.S.; Mignogna, C.; Arbitrio, M.; Botta, C.; Frandsen, N.M.; Rolfo, C.; Tagliaferri, P.; Tassone, P.; Di Martino, M.T. Pharmacokinetics and Pharmacodynamics of a 13-mer LNA-inhibitor-miR-221 in Mice and Non-human Primates. Mol. Nucleic Acids 2016, 5, e326. [Google Scholar] [CrossRef]
- Ebert, M.S.; Neilson, J.R.; Sharp, P.A. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 2007, 4, 721–726. [Google Scholar] [CrossRef]
- Xiao, J.; Yang, B.; Lin, H.; Lu, Y.; Luo, X.; Wang, Z. Novel approaches for gene-specific interference via manipulating actions of microRNAs: Examination on the pacemaker channel genes HCN2 and HCN4. J. Cell Physiol. 2007, 212, 285–292. [Google Scholar] [CrossRef]
- Monroig Pdel, C.; Chen, L.; Zhang, S.; Calin, G.A. Small molecule compounds targeting miRNAs for cancer therapy. Adv. Drug Deliv. Rev. 2015, 81, 104–116. [Google Scholar] [CrossRef]
- Nielsen, P.E.; Egholm, M.; Berg, R.H.; Buchardt, O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 1991, 254, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Piva, R.; Spandidos, D.A.; Gambari, R. From microRNA functions to microRNA therapeutics: Novel targets and novel drugs in breast cancer research and treatment (Review). Int. J. Oncol. 2013, 43, 985–994. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, P.E. Peptide nucleic acids (PNA) in chemical biology and drug discovery. Chem. Biodivers. 2010, 7, 786–804. [Google Scholar] [CrossRef] [PubMed]
- Demidov, V.V.; Frank-Kamenetskii, M.D. Sequence-specific targeting of duplex DNA by peptide nucleic acids via triplex strand invasion. Methods 2001, 23, 108–122. [Google Scholar] [CrossRef]
- Oh, S.Y.; Ju, Y.; Park, H. A highly effective and long-lasting inhibition of miRNAs with PNA-based antisense oligonucleotides. Mol. Cells 2009, 28, 341–345. [Google Scholar] [CrossRef]
- Nastruzzi, C.; Cortesi, R.; Esposito, E.; Gambari, R.; Borgatti, M.; Bianchi, N.; Feriotto, G.; Mischiati, C. Liposomes as carriers for DNA-PNA hybrids. J. Control Release 2000, 68, 237–249. [Google Scholar] [CrossRef]
- Macadangdang, B.; Zhang, N.; Lund, P.E.; Marple, A.H.; Okabe, M.; Gottesman, M.M.; Appella, D.H.; Kimchi-Sarfaty, C. Inhibition of multidrug resistance by SV40 pseudovirion delivery of an antigene peptide nucleic acid (PNA) in cultured cells. PLoS ONE 2011, 6, e17981. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Reissmann, S. Cell penetration: Scope and limitations by the application of cell-penetrating peptides. J. Pept. Sci. 2014, 20, 760–784. [Google Scholar] [CrossRef]
- Fabani, M.M.; Abreu-Goodger, C.; Williams, D.; Lyons, P.A.; Torres, A.G.; Smith, K.G.; Enright, A.J.; Gait, M.J.; Vigorito, E. Efficient inhibition of miR-155 function in vivo by peptide nucleic acids. Nucleic Acids Res. 2010, 38, 4466–4475. [Google Scholar] [CrossRef]
- Brognara, E.; Fabbri, E.; Aimi, F.; Manicardi, A.; Bianchi, N.; Finotti, A.; Breveglieri, G.; Borgatti, M.; Corradini, R.; Marchelli, R.; et al. Peptide nucleic acids targeting miR-221 modulate p27Kip1 expression in breast cancer MDA-MB-231 cells. Int. J. Oncol. 2012, 41, 2119–2127. [Google Scholar] [CrossRef]
- Cutrona, G.; Carpaneto, E.M.; Ulivi, M.; Roncella, S.; Landt, O.; Ferrarini, M.; Boffa, L.C. Effects in live cells of a c-myc anti-gene PNA linked to a nuclear localization signal. Nat. Biotechnol. 2000, 18, 300–303. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, E.; Manicardi, A.; Tedeschi, T.; Sforza, S.; Bianchi, N.; Brognara, E.; Finotti, A.; Breveglieri, G.; Borgatti, M.; Corradini, R.; et al. Modulation of the biological activity of microRNA-210 with peptide nucleic acids (PNAs). ChemMedChem 2011, 6, 2192–2202. [Google Scholar] [CrossRef] [PubMed]
- Brognara, E.; Fabbri, E.; Bazzoli, E.; Montagner, G.; Ghimenton, C.; Eccher, A.; Cantu, C.; Manicardi, A.; Bianchi, N.; Finotti, A.; et al. Uptake by human glioma cell lines and biological effects of a peptide-nucleic acids targeting miR-221. J. Neurooncol. 2014, 118, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Milani, R.; Brognara, E.; Fabbri, E.; Manicardi, A.; Corradini, R.; Finotti, A.; Gasparello, J.; Borgatti, M.; Cosenza, L.C.; Lampronti, I.; et al. Targeting miR1555p and miR2213p by peptide nucleic acids induces caspase3 activation and apoptosis in temozolomideresistant T98G glioma cells. Int. J. Oncol. 2019, 55, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Quijano, E.; Liu, Y.; Bahal, R.; Scanlon, S.E.; Song, E.; Hsieh, W.C.; Braddock, D.E.; Ly, D.H.; Saltzman, W.M.; et al. Anti-tumor Activity of miniPEG-gamma-Modified PNAs to Inhibit MicroRNA-210 for Cancer Therapy. Mol. Nucleic Acids 2017, 9, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.X.; Wu, Q.N.; Zhang, Y.; Li, Y.Y.; Liao, D.Z.; Hou, J.H.; Fu, J.; Zeng, M.S.; Yun, J.P.; Wu, Q.L.; et al. Knockdown of miR-21 in human breast cancer cell lines inhibits proliferation, in vitro migration and in vivo tumor growth. Breast Cancer Res. 2011, 13, R2. [Google Scholar] [CrossRef]
- Wang, X.; Xiao, X.; Zhang, B.; Li, J.; Zhang, Y. A self-assembled peptide nucleic acid-microRNA nanocomplex for dual modulation of cancer-related microRNAs. Chem. Commun. 2019, 55, 2106–2109. [Google Scholar] [CrossRef]
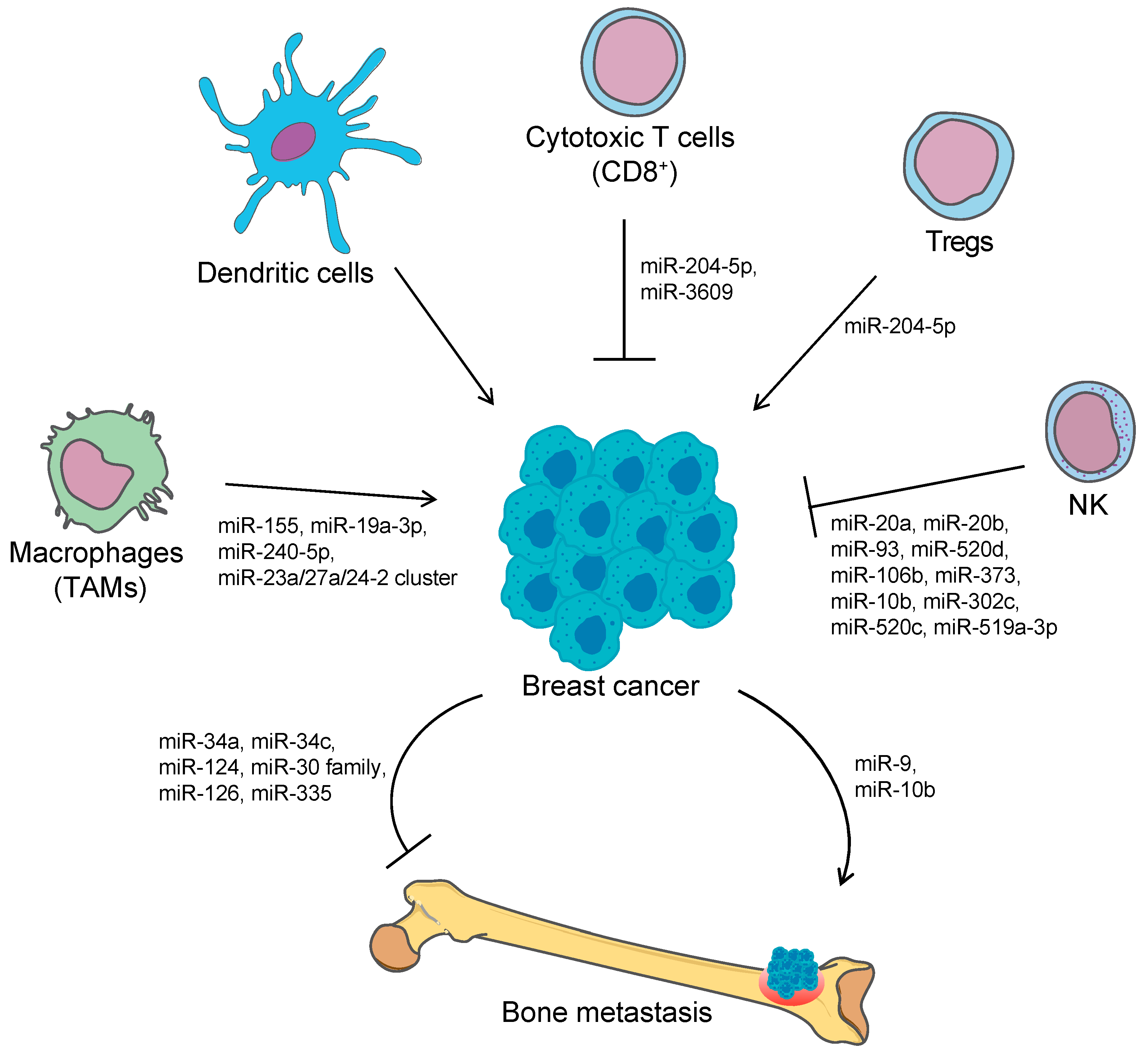
| miRNA | Cells/Tissue | Role | Target | Function | Reference |
|---|---|---|---|---|---|
| Oncogenic miRNAs | |||||
| miR-155 | BC and other tumors | im-miRNA | SPI1, Ship1 | Increases production of pro-tumor cytokines, reduces expression of activation markers, shifts from M1-like TAM to M2-like TAM | [51] |
| miR-23a/27a/24-2 cluster | BC | im-miRNA | JAK1, STAT-6, IRF4, PPAR-γ | Modulate macrophage polarization | [52] |
| miR-195, miR-497 | TNBC cells | im-miRNA | PD-L1 | Promote tumor progression, inhibit the immune response and promote tumor immune escape | [53] |
| miR-20a, miR-93, miR-520d, miR-106b, miR-373 | prostate, kidney, BC, glioblastoma cells | NK-mediated immune response evasion | MICA/B | Inhibit the NKG2DL-NKG2D pathway, escape from NK-mediated killing | [56,57] |
| miR-10b | Several cancer cell lines (prostate, colorectal, cervical cancer cells, BC cells) | NK-mediated immune response evasion | MICB | Inhibits NK-mediated cytotoxicity in vitro and contrasts tumor clearance in vivo | [58] |
| BC, pancreatic cancer, glioblastoma, other highly metastatic cancers | metastasis formation | TBX5, neurofibromin, KLF4 | Promotes proliferation, migration and invasion | [59,60] | |
| HOXD10 | Leads to the upregulation of the pro-metastatic gene RhoC with consequent promotion of tumor cells invasion and metastasis in BC | [61,62] | |||
| miR-302c, miR-520c | leukemia and BC cell lines | NK-mediated immune response evasion | MICA/B, ULBP2 | Mediate NK-dependent cell killing upon Vitamin D3 treatment | [63] |
| miR-20a, miR-20b, miR-93, miR-106b | BC cells | NK-mediated immune response evasion | MICA/B, ULBP2 | Affect the capacity of NK to recognize and kill tumor cells | [64] |
| miR-20a | BC stem cells | NK-mediated immune response evasion, metastasis formation | MICA/B | Reduces susceptibility to NK-mediated cell lysis, enhances the metastatic potential | [65] |
| miR-519a-3p | BC cells | NK-mediated immune response evasion | MICA, ULBP2, TRAIL-R2, caspase-7, caspase-8 | Impairs NK-mediated cytotoxicity, confers resistance to apoptosis, regulates tumor progression and evasion from immunosurveillance | [66] |
| miR-9 | BC | metastasis formation | E-cadherin, LIFR | Promotes EMT and metastasis formation through activation of the Hippo signaling pathway | [67,68,69] |
| miR-200c/141 cluster | TNBC | metastasis formation | SerpinB2 upregulation | Promotes tumorigenesis and metastasis in lungs and lymph nodes in vivo, through overexpression of SerpinB2 | [70] |
| miR-374a | BC | metastasis formation | PTEN, WNT5A | Promotes EMT, induces cell proliferation and metastasis by activating the Wnt/β-catenin pathway | [71] |
| miR-103/107, miR-630 | BC | metastasis formation | Dicer | Promote EMT, migration and metastasis | [72,73] |
| Tumor-suppressive miRNAs | |||||
| miR-19a-3p | BC | im-miRNA | Fra-1 | Promotes shift from M2- to M1-phenotype of TAMs both in vitro and in vivo, inhibition of metastasis | [54] |
| miR-240-5p | BC | im-miRNA | PIK3CB | Alters expression of cytokines, remodeling and reprogramming of the tumor microenvironment, promote tumor growth and metastasis in vivo | [55] |
| miR-3609 | BC cells | im-miRNA | PD-L1 | Blocks PD-L1 immune checkpoint and sensitize BC cells to adriamycin | [74] |
| miR-126, miR-335 | BC | metastasis suppressor | SOX4, tenascin C (miR-335) | Their restoration in BC cell lines reduces tumor growth and proliferation, inhibits cell invasion, migration and metastasis, inhibit lung and bone metastasis formation in vivo | [75] |
| miR-126, miR-126* | BC | metastasis suppressor | Prevent recruitment of MSC and monocytes at tumor microenvironment, contrasting tumor cell migration, invasion and metastasis | [76] | |
| miR-200 family, miR-205 | BC | metastasis formation | ZEB1, ZEB2 | Inhibit EMT, promote metastasis formation in lung and liver in vivo when overexpressed | [77,78] |
| miR-190 | BC | metastasis suppressor | SMAD2 | Inhibits EMT and metastasis formation by regulating TGFβ/SMAD2 signaling pathway | [79] |
| miR-148a | BC | metastasis suppressor | WNT1 | Inhibits cell migration, invasion and metastasis by inhibiting the Wnt/β-catenin pathway | [80] |
| miR-340 | BC cells | metastasis suppressor | c-MYC, CTNNB1, ROCK1 | Inhibits cell migration, invasion and metastasis by inhibiting the Wnt/β-catenin pathway | [81] |
| miR-34a, miR-34c | BC | metastasis suppressor | Tgif2 | Suppress osteoclastogenesis, bone resorption and bone metastasis | [82] |
| miR-124 | BC cells | metastasis suppressor | IL-11 | Suppresses bone metastasis by inhibiting osteoclast progenitor cells differentiation and survival | [83] |
| miR-30 family | ER−/PR− BC cells | metastasis suppressor | IL-8, IL-11, DKK1, RUNX2, CDH11, CTGF, ITGA5, ITGB3 | Inhibits tumor cell invasiveness, restore bone homeostasis in vitro, reduce bone metastasis in vivo | [84] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomarasca, M.; Maroni, P.; Banfi, G.; Lombardi, G. microRNAs in the Antitumor Immune Response and in Bone Metastasis of Breast Cancer: From Biological Mechanisms to Therapeutics. Int. J. Mol. Sci. 2020, 21, 2805. https://doi.org/10.3390/ijms21082805
Gomarasca M, Maroni P, Banfi G, Lombardi G. microRNAs in the Antitumor Immune Response and in Bone Metastasis of Breast Cancer: From Biological Mechanisms to Therapeutics. International Journal of Molecular Sciences. 2020; 21(8):2805. https://doi.org/10.3390/ijms21082805
Chicago/Turabian StyleGomarasca, Marta, Paola Maroni, Giuseppe Banfi, and Giovanni Lombardi. 2020. "microRNAs in the Antitumor Immune Response and in Bone Metastasis of Breast Cancer: From Biological Mechanisms to Therapeutics" International Journal of Molecular Sciences 21, no. 8: 2805. https://doi.org/10.3390/ijms21082805
APA StyleGomarasca, M., Maroni, P., Banfi, G., & Lombardi, G. (2020). microRNAs in the Antitumor Immune Response and in Bone Metastasis of Breast Cancer: From Biological Mechanisms to Therapeutics. International Journal of Molecular Sciences, 21(8), 2805. https://doi.org/10.3390/ijms21082805







