Arf6 Can Trigger Wave Regulatory Complex-Dependent Actin Assembly Independent of Arno
Abstract
1. Introduction
2. Results
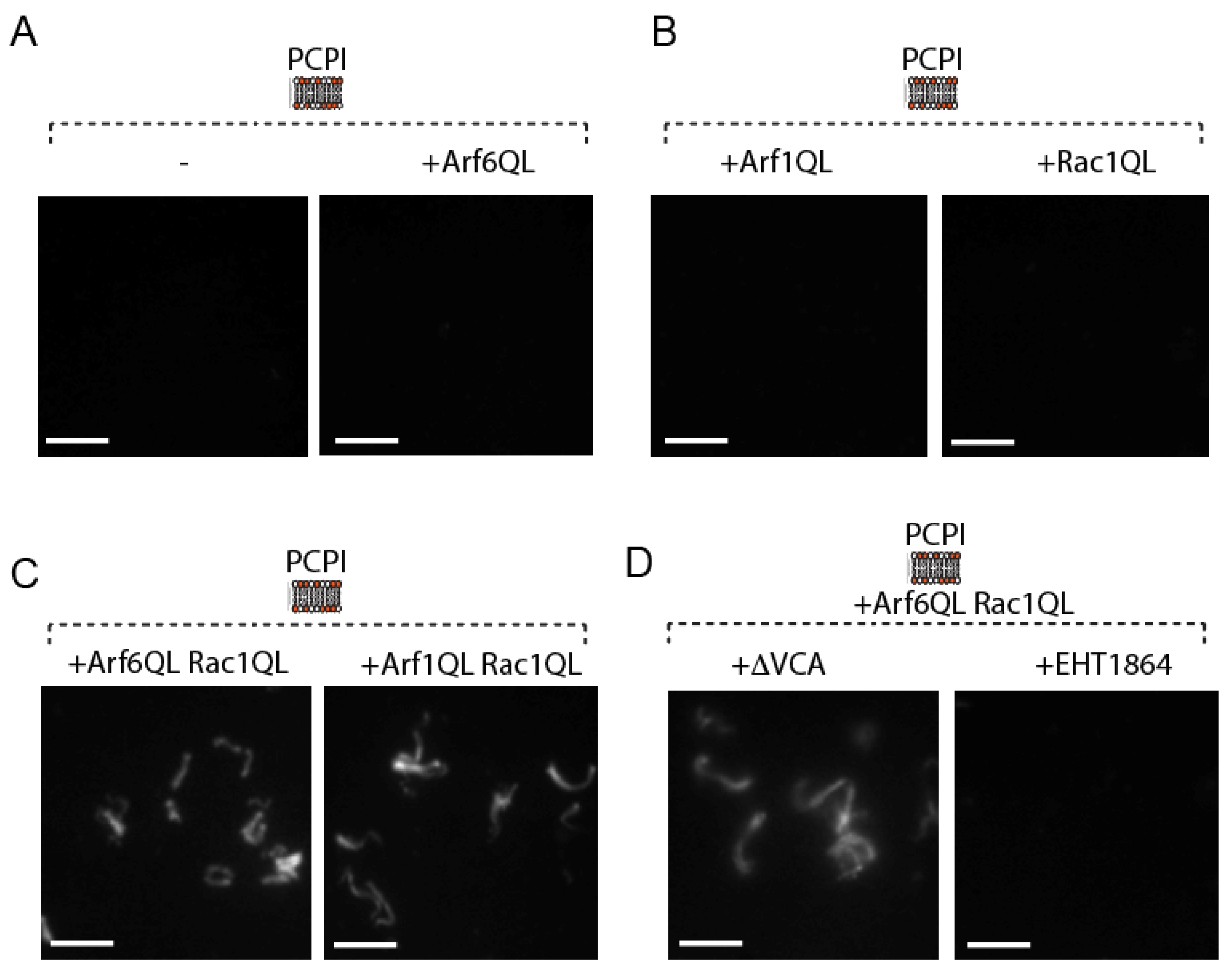
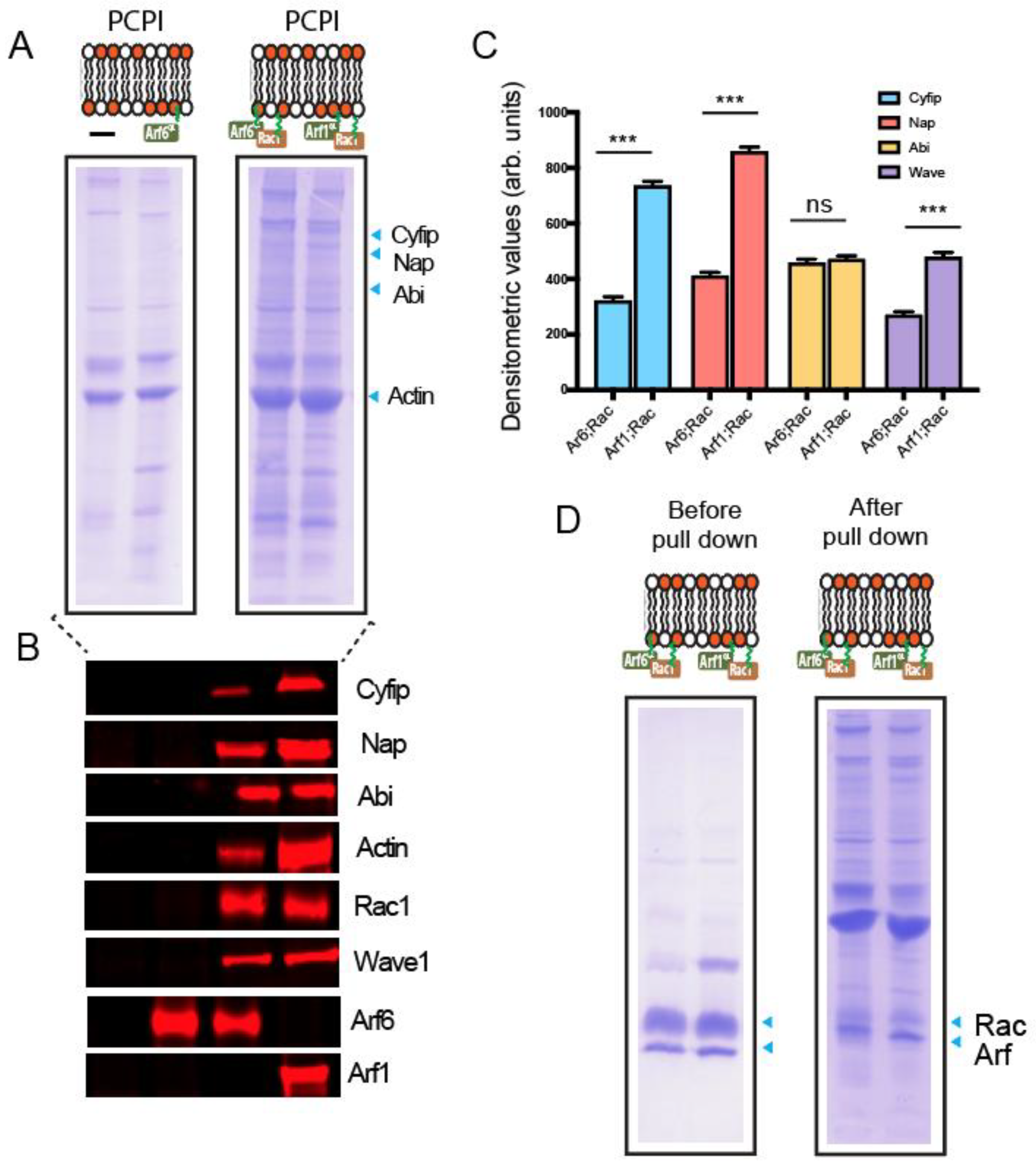
2.1. Constitutively Active Arf6 and Rac1 Cooperate in Triggering Actin Assembly In Vitro
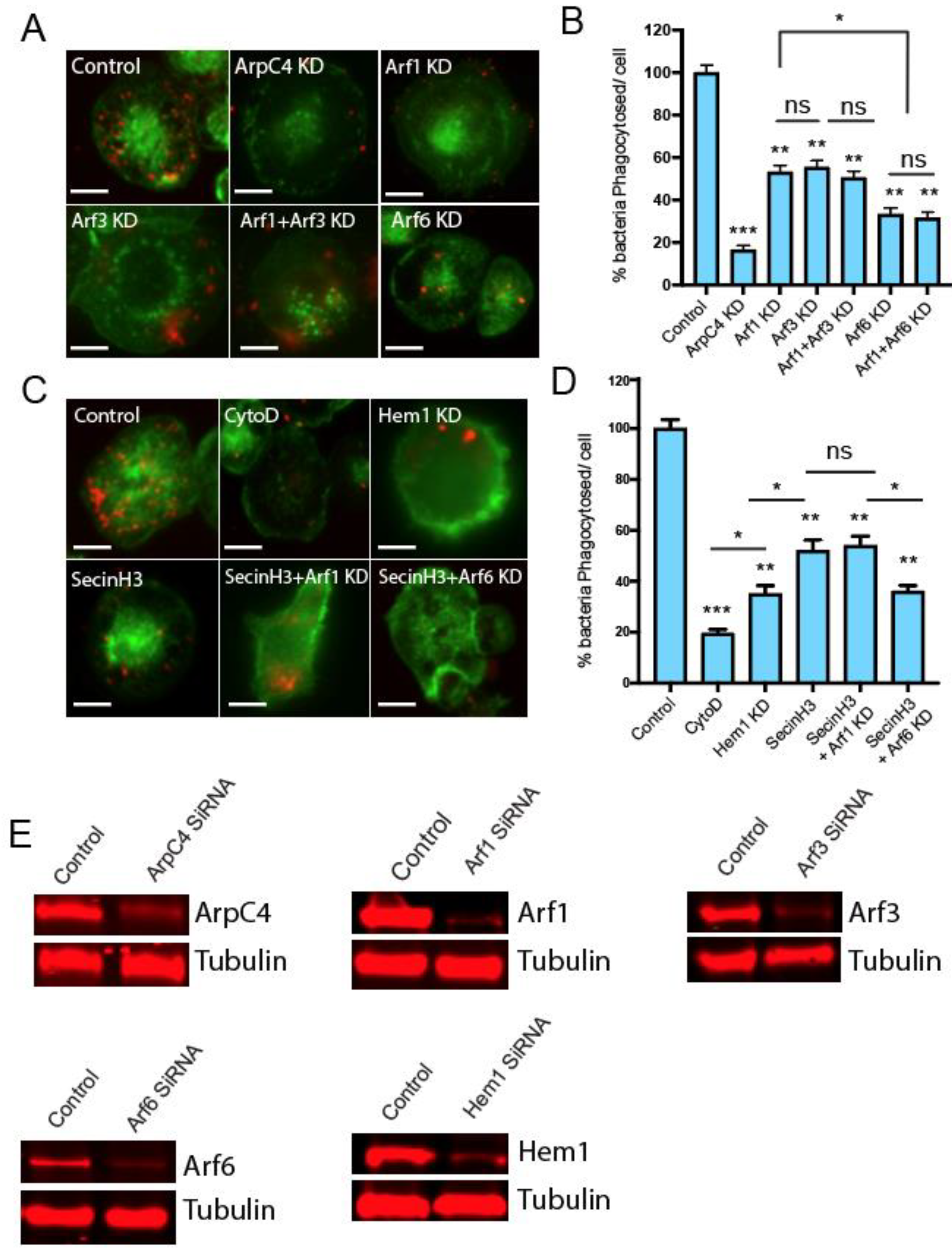
2.2. Arf6 Regulates Phagocytosis in Differentiated THP-1 Human Macrophages
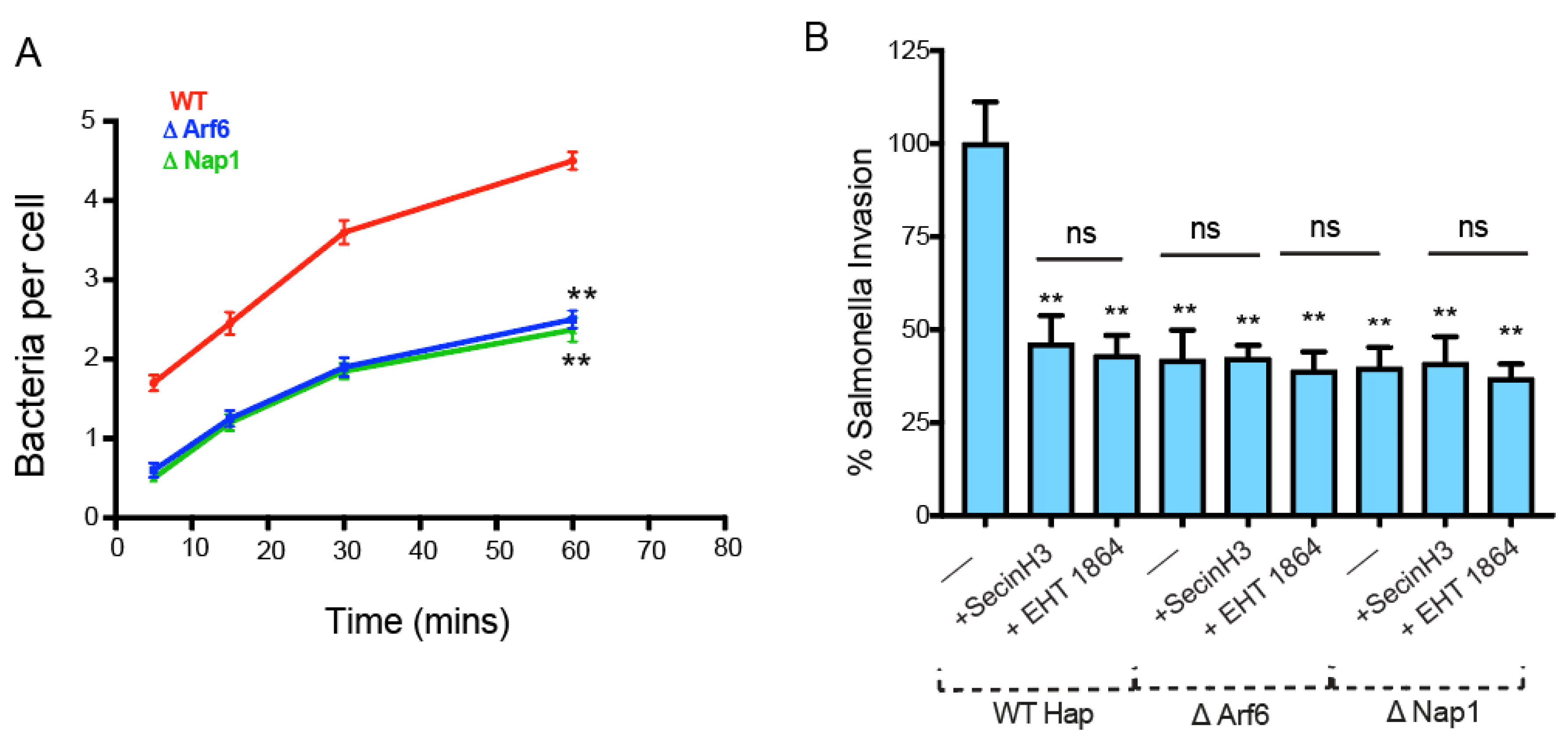
2.3. Arf6-Mediated Internalization of Salmonella is ARNO Dependent
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Antibodies
4.3. Plasmids
4.4. Cell Culture and Transfection
4.5. Salmonella Invasion of Non-Phagocytic Host Cells
4.6. Phagocytosis Assay
4.7. Actin-Based Motility and In Vitro Pull Downs
4.8. Immunoblotting and Densitometric Quantification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van Acker, T.; Tavernier, J.; Peelman, F. The Small GTPase Arf6: An Overview of Its Mechanisms of Action and of Its Role in Host⁻Pathogen Interactions and Innate Immunity. Int. J. Mol. Sci. 2019, 20, 2209. [Google Scholar] [CrossRef]
- D’Souza-Schorey, C.; Chavrier, P. ARF proteins: Roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 2006, 7, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, J.G. Multiple Roles for Arf6: Sorting, Structuring, and Signaling at the Plasma Membrane. J. Biol. Chem. 2003, 278, 41573–41576. [Google Scholar] [CrossRef] [PubMed]
- Gaschet, J.; Hsu, V.W. Distribution of ARF6 between membrane and cytosol is regulated by its GTPase cycle. J. Biol. Chem. 1999, 274, 20040–20045. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, J. Structural basis for activation of ARF GTPase: Mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell 1998, 95, 237–248. [Google Scholar] [CrossRef]
- Singh, V.; Davidson, A.C.; Hume, P.J.; Humphreys, D.; Koronakis, V. Arf GTPase interplay with Rho GTPases in regulation of the actin cytoskeleton. Small GTPases 2019, 10, 411–418. [Google Scholar] [CrossRef]
- Shi, A.; Grant, B.D. Interactions between Rab and Arf GTPases regulate endosomal phosphatidylinositol-4,5-bisphosphate during endocytic recycling. Small GTPases 2013, 4, 106–109. [Google Scholar] [CrossRef]
- Koronakis, V.; Hume, P.J.; Humphreys, D.; Liu, T.; Horning, O.; Jensen, O.N.; McGhie, E.J. WAVE regulatory complex activation by cooperating GTPases Arf and Rac1. Proc. Natl. Acad. Sci. USA 2011, 108, 14449–14454. [Google Scholar] [CrossRef]
- Myers, K.R.; Casanova, J.E. Regulation of actin cytoskeleton dynamics by Arf-family GTPases. Trends Cell Biol. 2008, 18, 184–192. [Google Scholar] [CrossRef]
- Humphreys, D.; Liu, T.; Davidson, A.C.; Hume, P.J.; Koronakis, V. The Drosophila Arf1 homologue Arf79F is essential for lamellipodium formation. J. Cell Sci. 2012, 125 (Pt 23), 5630–5635. [Google Scholar] [CrossRef]
- Boshans, R.L.; Szanto, S.; van Aelst, L.; D’Souza-Schorey, C. ADP-ribosylation factor 6 regulates actin cytoskeleton remodeling in coordination with Rac1 and RhoA. Mol. Cell. Biol. 2000, 20, 3685–3694. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, D.; Davidson, A.C.; Hume, P.J.; Makin, L.E.; Koronakis, V. Arf6 coordinates actin assembly through the WAVE complex, a mechanism usurped by Salmonella to invade host cells. Proc. Natl. Acad. Sci. USA 2013, 110, 16880–16885. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, D.; Davidson, A.; Hume, P.J.; Koronakis, V. Salmonella virulence effector SopE and Host GEF ARNO cooperate to recruit and activate WAVE to trigger bacterial invasion. Cell Host Microbe 2012, 11, 129–139. [Google Scholar] [CrossRef]
- Hume, P.J.; Humphreys, D.; Koronakis, V. WAVE regulatory complex activation. Methods Enzymol. 2014, 540, 363–379. [Google Scholar] [CrossRef] [PubMed]
- Uchida, H.; Kondo, A.; Yoshimura, Y.; Mazaki, Y.; Sabe, H. PAG3/Papalpha/KIAA0400, a GTPase-activating protein for ADP-ribosylation factor (ARF), regulates ARF6 in Fcγ receptor-mediated phagocytosis of macrophages. J. Exp. Med. 2001, 193, 955–966. [Google Scholar] [CrossRef] [PubMed]
- Someya, A.; Moss, J.; Nagaoka, I. The guanine nucleotide exchange protein for ADP-ribosylation factor 6, ARF-GEP100/BRAG2, regulates phagocytosis of monocytic phagocytes in an ARF6-dependent process. J. Biol. Chem. 2010, 285, 30698–30707. [Google Scholar] [CrossRef] [PubMed]
- Rougerie, P.; Miskolci, V.; Cox, D. Generation of membrane structures during phagocytosis and chemotaxis of macrophages: Role and regulation of the actin cytoskeleton. Immunol. Rev. 2013, 256, 222–239. [Google Scholar] [CrossRef]
- Zhang, Q.; Cox, D.; Tseng, C.C.; Donaldson, J.G.; Greenberg, S. A requirement for ARF6 in Fcγ receptor-mediated phagocytosis in macrophages. J. Biol. Chem. 1998, 273, 19977–19981. [Google Scholar] [CrossRef]
- Stuart, L.M.; Boulais, J.; Charriere, G.M.; Hennessy, E.J.; Brunet, S.; Jutras, I.; Goyette, G.; Rondeau, C.; Letarte, S.; Huang, H.; et al. A systems biology analysis of the Drosophila phagosome. Nature 2007, 445, 95–101. [Google Scholar] [CrossRef]
- Humphreys, D.; Singh, V.; Koronakis, V. Inhibition of WAVE Regulatory Complex Activation by a Bacterial Virulence Effector Counteracts Pathogen Phagocytosis. Cell Rep. 2016, 17, 697–707. [Google Scholar] [CrossRef]
- Volpicelli-Daley, L.A.; Li, Y.; Zhang, C.J.; Kahn, R.A. Isoform-selective effects of the depletion of ADP-ribosylation factors 1-5 on membrane traffic. Mol. Biol. Cell 2005, 16, 4495–4508. [Google Scholar] [CrossRef] [PubMed]
- Kondo, Y.; Hanai, A.; Nakai, W.; Katoh, Y.; Nakayama, K.; Shin, H.W. Arf1 and Arf3 are required for the integrity of recycling endosomes and the recycling pathway. Cell Struct. Funct. 2012, 37, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Santy, L.C.; Ravichandran, K.S.; Casanova, J.E. The DOCK180/Elmo complex couples ARNO-mediated Arf6 activation to the downstream activation of Rac1. Curr. Biol. 2005, 15, 1749–1754. [Google Scholar] [CrossRef]
- Garza-Mayers, A.C.; Miller, K.A.; Russo, B.C.; Nagda, D.V.; Goldberg, M.B. Shigella flexneri regulation of ARF6 activation during bacterial entry via an IpgD-mediated positive feedback loop. MBio 2015, 6, e02584. [Google Scholar] [CrossRef] [PubMed]
- Hume, P.J.; Singh, V.; Davidson, A.C.; Koronakis, V. Swiss Army Pathogen: The Salmonella Entry Toolkit. Front. Cell Infect. Microbiol. 2017, 7, 348. [Google Scholar] [CrossRef] [PubMed]
- Schafer, D.A.; D’Souza-Schorey, C.; Cooper, J.A. Actin assembly at membranes controlled by ARF6. Traffic 2000, 1, 892–903. [Google Scholar] [CrossRef]
- Beemiller, P.; Hoppe, A.D.; Swanson, J.A. A phosphatidylinositol-3-kinase-dependent signal transition regulates ARF1 and ARF6 during Fcγ receptor-mediated phagocytosis. PLoS Biol. 2006, 4, e162. [Google Scholar] [CrossRef]
- Egami, Y.; Fujii, M.; Kawai, K.; Ishikawa, Y.; Fukuda, M.; Araki, N. Activation-inactivation cycling of Rab35 and ARF6 is required for phagocytosis of zymosan in RAW264 macrophages. J. Immunol. Res. 2015, 2015, 429439. [Google Scholar] [CrossRef]
- Radhakrishna, H.; Alawar, O.S.; Khachikian, Z.; Donaldson, J.G. ARF6 requirement for Rac ruffling suggests a role for membrane trafficking in cortical actin rearrangements. J. Cell Sci. 1999, 112, 855–866. [Google Scholar]
- Shumilina, E.V.; khaitlina, S.Y.; Morachevskaya, E.A.; Negulyaev, Y.A. Non-hydrolyzable analog of GTP induces activity of Na+ channels via disassembly of cortical actin cytoskeleton. FEBS Lett. 2003, 547, 27–31. [Google Scholar] [CrossRef]
- Wiegandt, D.; Vieweg, S.; Hofmann, F.; Koch, D.; Li, F.; Wu, Y.; Itzen, A.; Muller, M.P.; Goody, R.S. Locking GTPases covalently in their functional states. Nat. Commun. 2015, 6, 7773. [Google Scholar] [CrossRef] [PubMed]
- Schlumberger, M.C.; Kappeli, R.; Wetter, M.; Muller, A.; Misselwitz, B.; Dilling, S.; Kremer, M.; Hardt, W. Two newly identified SipA domains (F1, F2) steer effector protein localization and contribute to Salmonella host cell manipulation. Mol. Microbiol. 2007, 65, 741–760. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, V.; Davidson, A.C.; Hume, P.J.; Koronakis, V. Arf6 Can Trigger Wave Regulatory Complex-Dependent Actin Assembly Independent of Arno. Int. J. Mol. Sci. 2020, 21, 2457. https://doi.org/10.3390/ijms21072457
Singh V, Davidson AC, Hume PJ, Koronakis V. Arf6 Can Trigger Wave Regulatory Complex-Dependent Actin Assembly Independent of Arno. International Journal of Molecular Sciences. 2020; 21(7):2457. https://doi.org/10.3390/ijms21072457
Chicago/Turabian StyleSingh, Vikash, Anthony C. Davidson, Peter J. Hume, and Vassilis Koronakis. 2020. "Arf6 Can Trigger Wave Regulatory Complex-Dependent Actin Assembly Independent of Arno" International Journal of Molecular Sciences 21, no. 7: 2457. https://doi.org/10.3390/ijms21072457
APA StyleSingh, V., Davidson, A. C., Hume, P. J., & Koronakis, V. (2020). Arf6 Can Trigger Wave Regulatory Complex-Dependent Actin Assembly Independent of Arno. International Journal of Molecular Sciences, 21(7), 2457. https://doi.org/10.3390/ijms21072457



