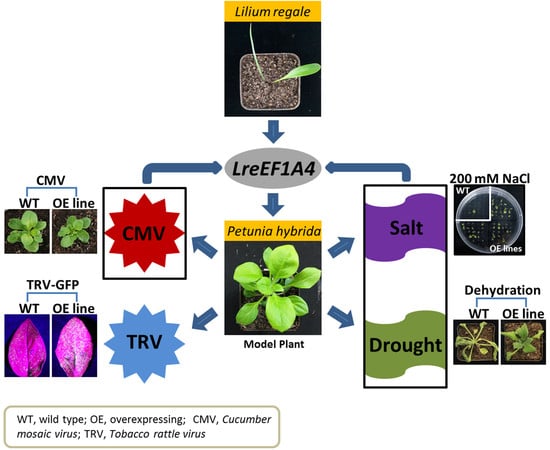
LreEF1A4, a Translation Elongation Factor from Lilium regale, Is Pivotal for Cucumber Mosaic Virus and Tobacco Rattle Virus Infections and Tolerance to Salt and Drought
Abstract
1. Introduction
2. Results
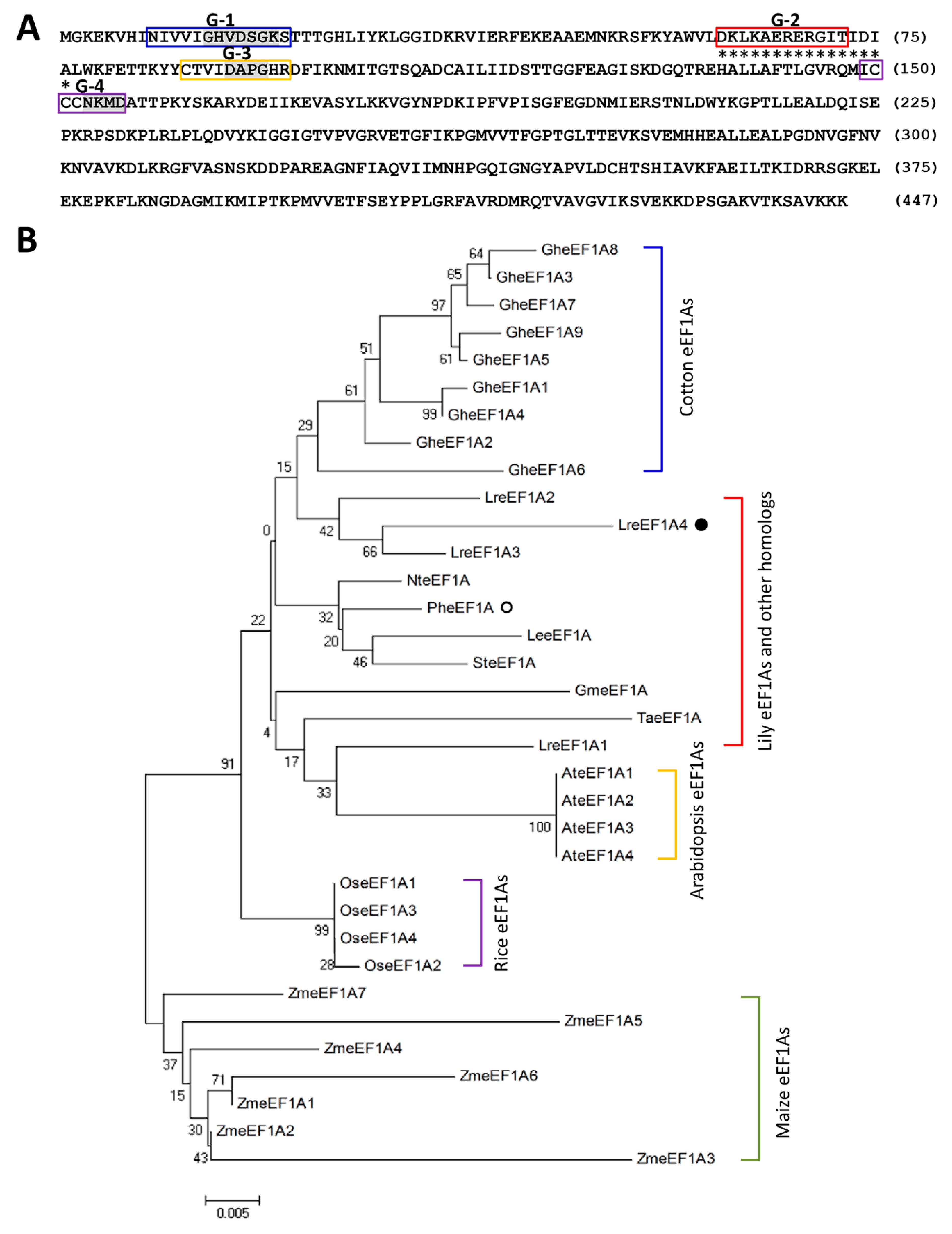
2.1. Isolation of Full-Length LreEF1A4 cDNA
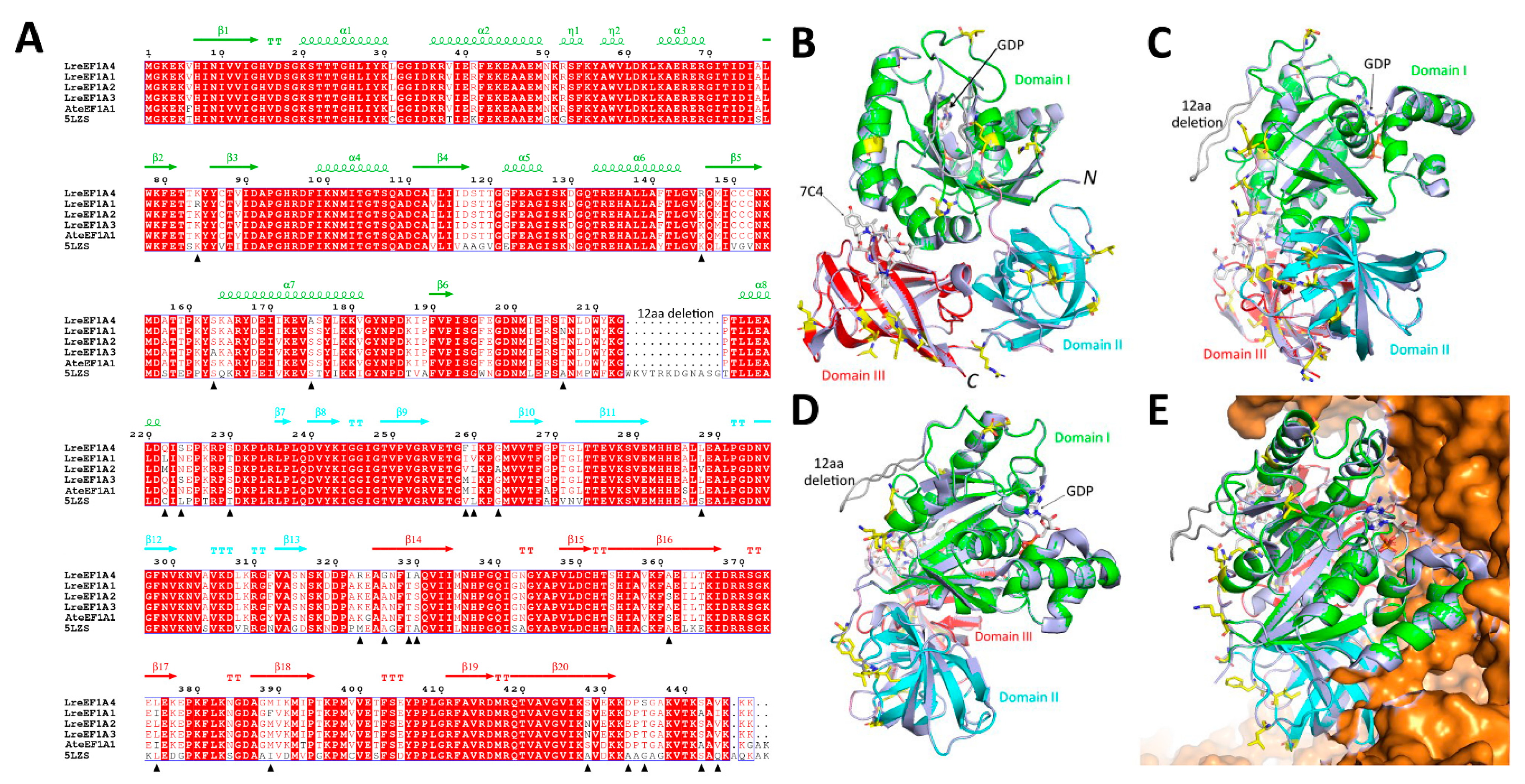
2.2. Protein Structure Analysis of LreEF1A4
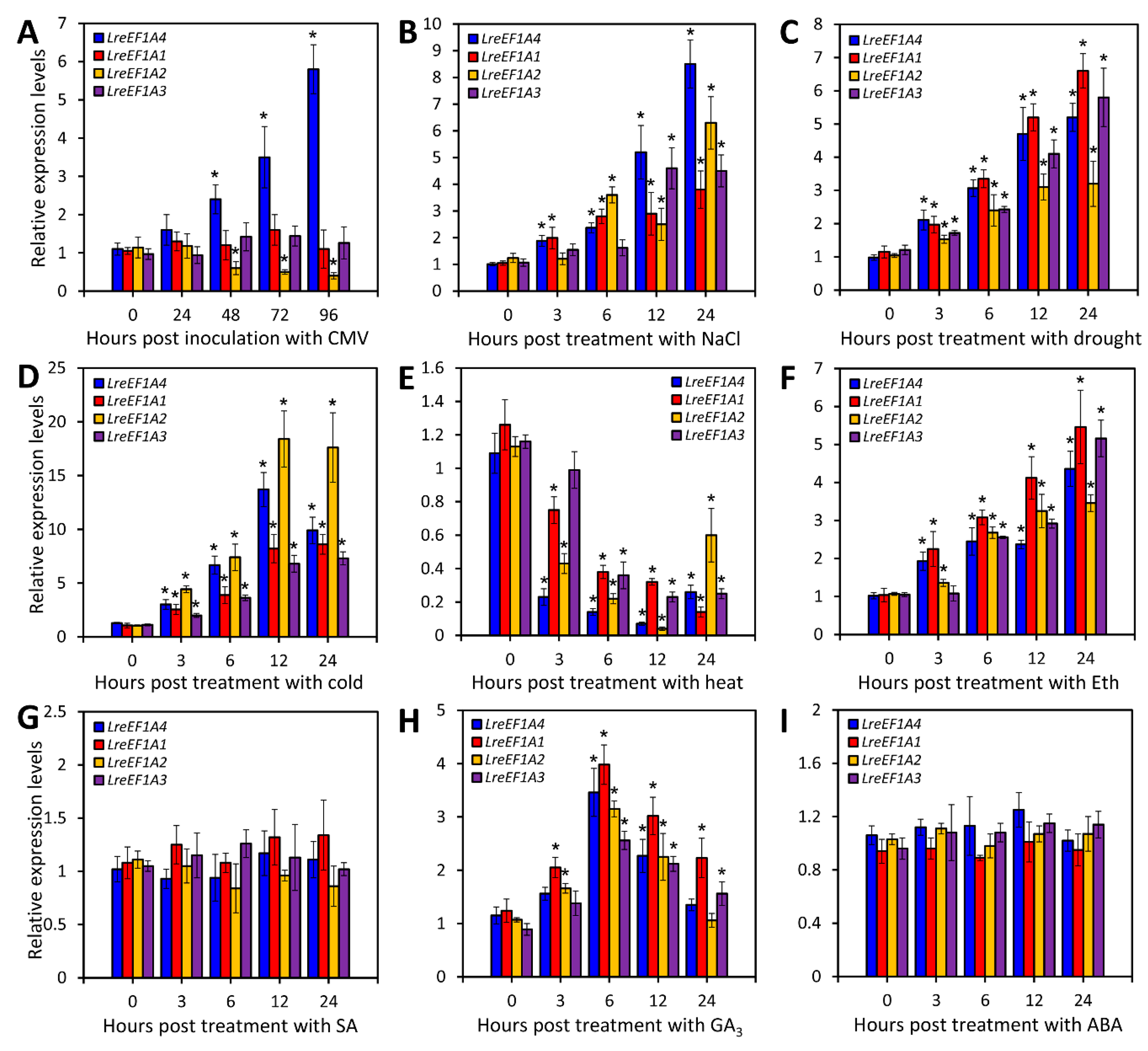
2.3. CMV Infection, Abiotic Stresses, and Hormone Treatments Change LreEF1As Expression
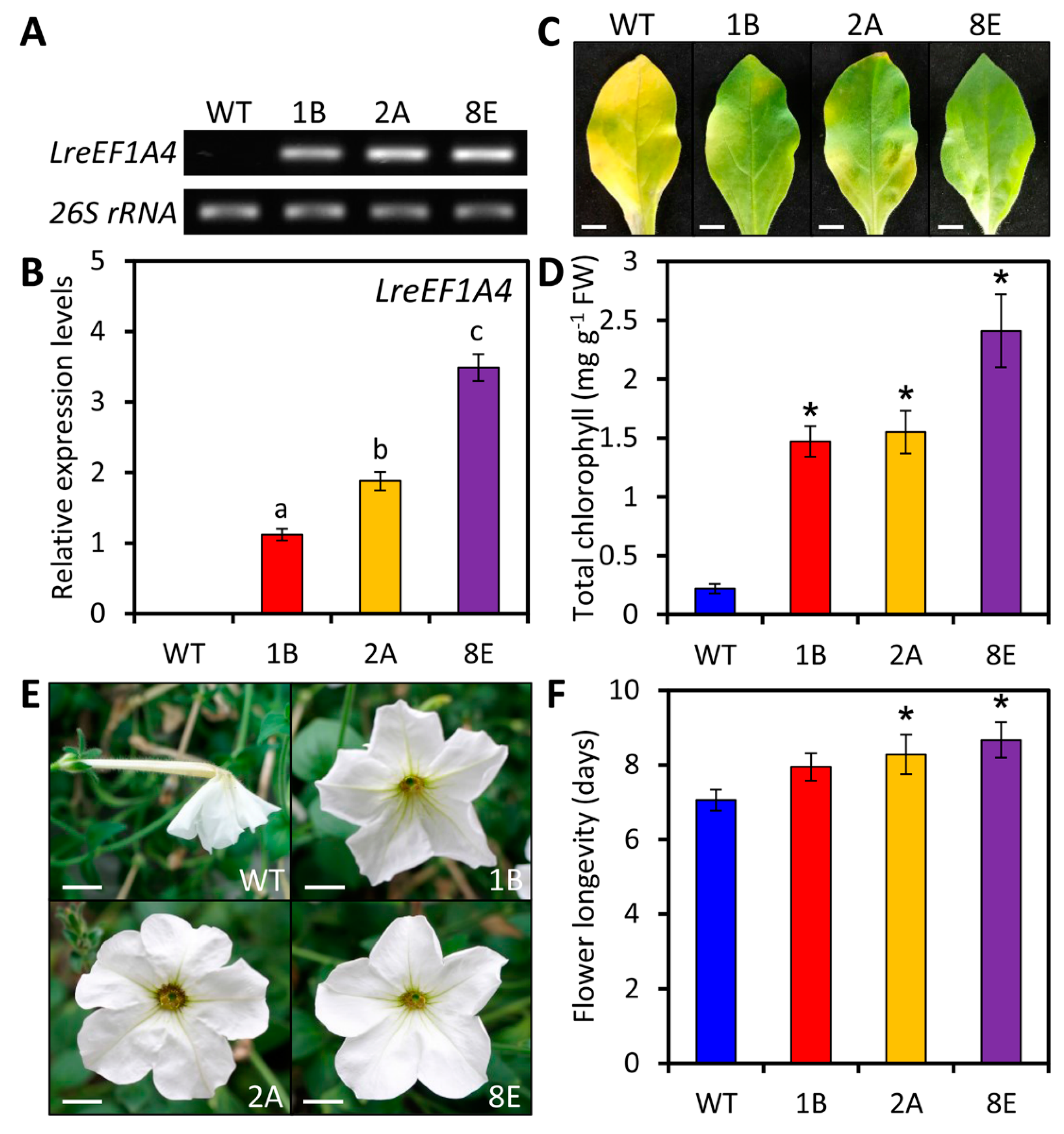
2.4. Ectopic Overexpression of LreEF1A4 Delays Leaf and Flower Senescence
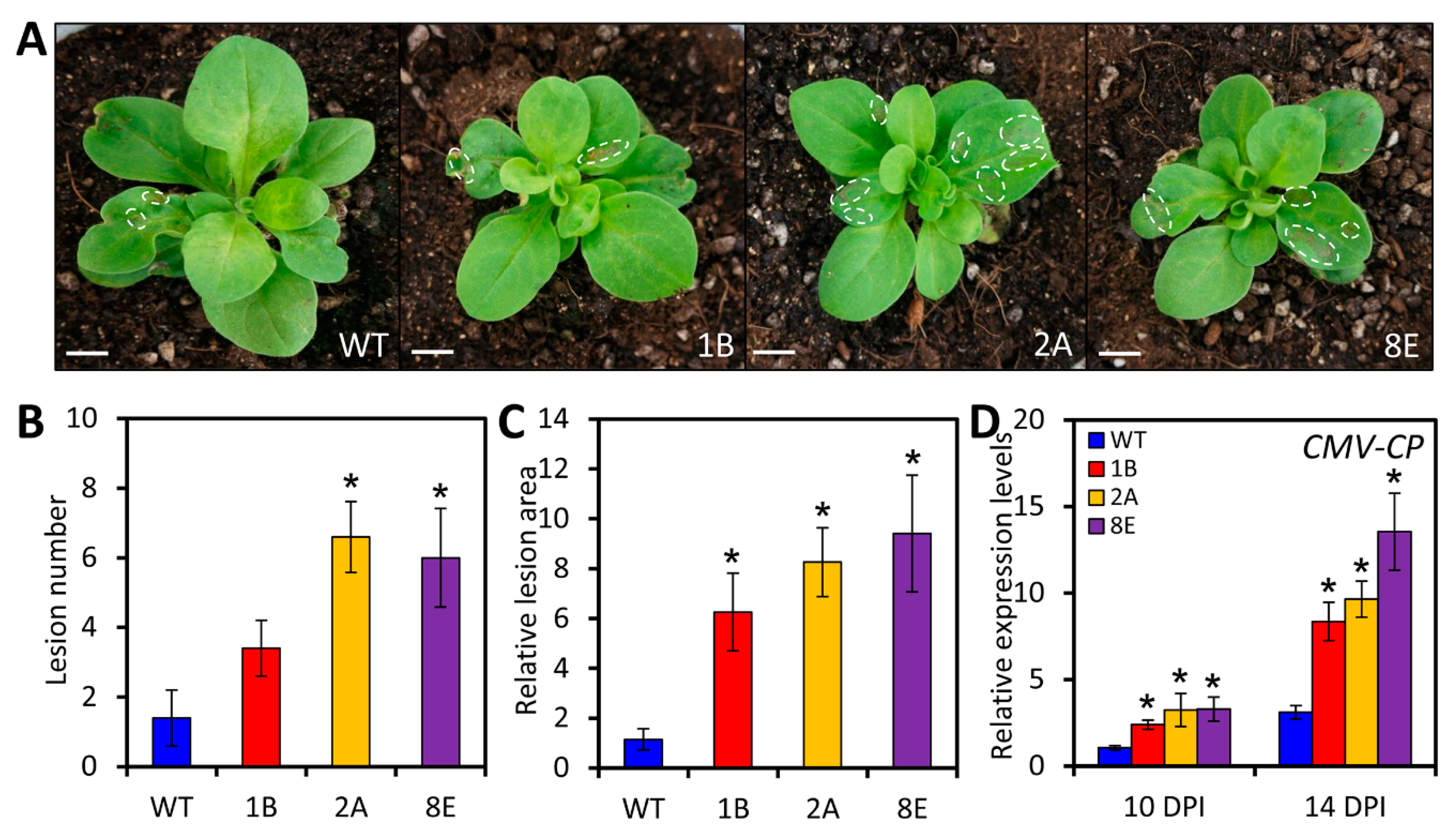
2.5. LreEF1A4 Affects Resistance to CMV
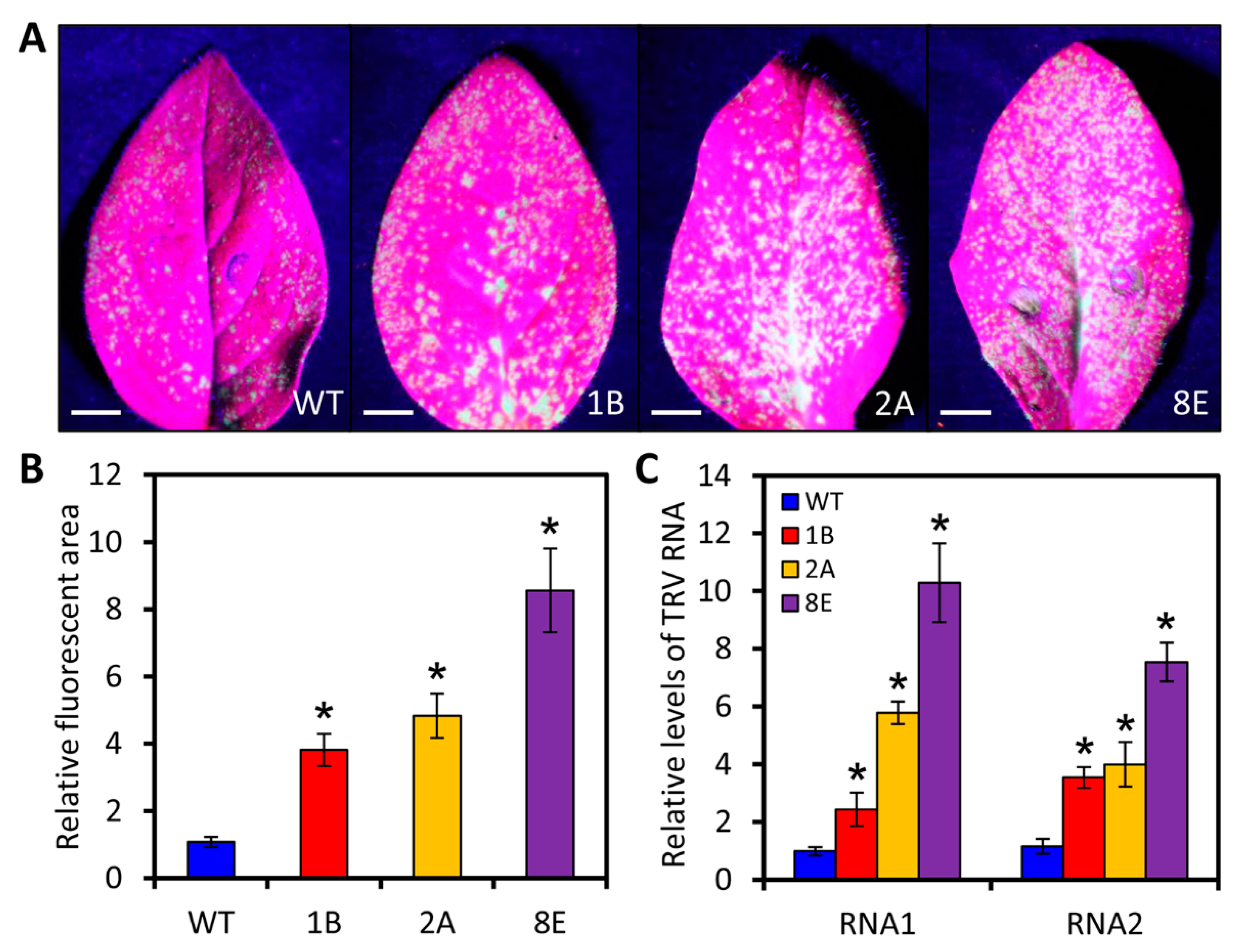
2.6. LreEF1A4 Affects Resistance to TRV
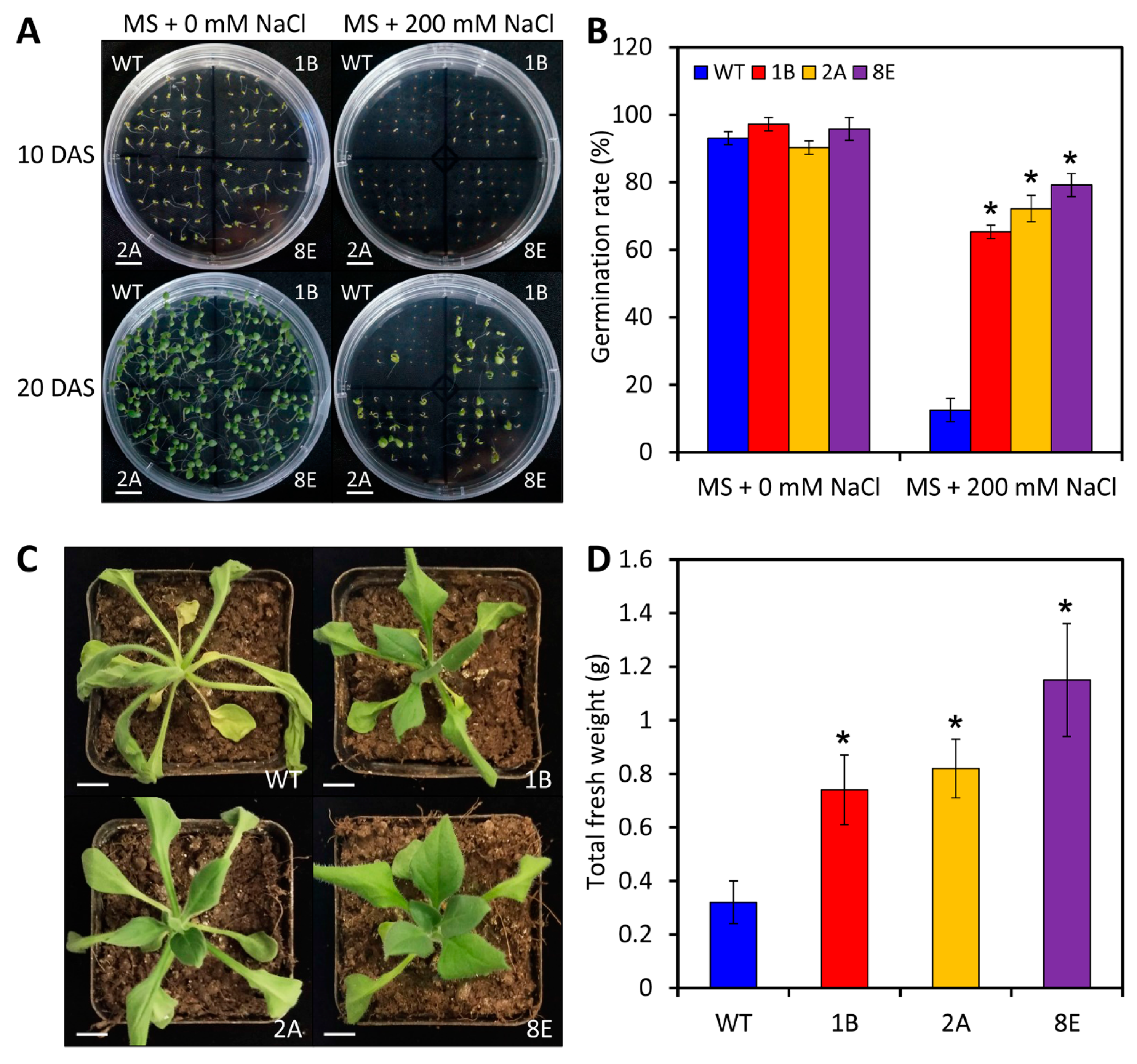
2.7. LreEF1A4 Participates in the Tolerance to Salt and Drought
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Viral Inoculation
4.3. Abiotic Stress and Hormone Treatments
4.4. Full-Length Cloning of Four LreEF1As
4.5. Sequence Analysis
4.6. Protein Structural Modelling
4.7. Semi-Quantitative RT-PCR and Quantitative Real-Time PCR
4.8. Plasmid Construction
4.9. Plant Transformation
4.10. Measurement of Chlorophyll Levels
4.11. VIGS Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| eEF1A | Eukaryotic translation elongation factor 1 alpha |
| SSH | Suppression subtractive hybridization |
| CMV | Cucumber mosaic virus |
| TRV | Tobacco rattle virus |
| Eth | Ethylene |
| SA | Salicylic acid |
| GA3 | Gibberellic acid |
| ABA | Abscisic acid |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| 26S rRNA | 26S ribosomal RNA |
| HPT | Hours post treatment |
| DPI | Days post inoculation |
| DAS | Days after sowing |
| WT | Wild type |
| rbcL | Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit |
| psaA | Photosystem I chlorophyll a apoprotein |
| SAG | Senescence-associated gene |
| CP | Coat protein |
| EV | Empty vector |
| SE | Standard error |
References
- Browne, G.J.; Proud, C.G. Regulation of peptide-chain elongation in mammalian cells. Eur. J. Biochem. 2002, 269, 5360–5368. [Google Scholar] [CrossRef]
- Infante, C.; Asensio, E.; Cañavate, J.P.; Manchado, M. Molecular characterization and expression analysis of five different elongation factor 1 alpha genes in the flatfish Senegalese sole (Solea senegalensis Kaup): Differential gene expression and thyroid hormones dependence during metamorphosis. BMC Mol. Biol. 2008, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Condeelis, J. Elongation factor la, translation and the cytoskeleton. Thends Biochem. Sci. 1995, 20, 169–170. [Google Scholar] [CrossRef]
- Fu, J.; Momčilović, I.; Prasad, P. Roles of protein synthesis elongation factor EF-Tu in heat tolerance in plants. J. Bot. 2012, 2012, 835836. [Google Scholar] [CrossRef]
- Le Sourd, F.; Boulben, S.; Le Bouffant, R.; Cormier, P.; Morales, J.; Belle, R.; Mulner-Lorillon, O. eEF1B: At the dawn of the 21st century. BBA-Gene Struct. Expr. 2006, 1759, 13–31. [Google Scholar] [CrossRef] [PubMed]
- Riis, B.; Rattan, S.I.; Clark, B.F.; Merrick, W.C. Eukaryotic protein elongation factors. Trends Biochem. Sci. 1990, 15, 420–424. [Google Scholar] [CrossRef]
- Durso, N.; Cyr, R. Beyond translation: Elongation factor-1α and the cytoskeleton. Protoplasma 1994, 180, 99–105. [Google Scholar] [CrossRef]
- Kaur, K.J.; Ruben, L. Protein translation elongation factor-1 alpha from Trypanosoma brucei binds calmodulin. J. Biol. Chem. 1994, 269, 23045–23050. [Google Scholar]
- Collings, D.A.; Wasteneys, G.O.; Miyazaki, M.; Williamson, R.E. Elongation factor 1α is a component of the subcortical actin bundles of characean algae. Cell Biol. Int. 1994, 18, 1019–1024. [Google Scholar] [CrossRef]
- Morita, K.; Bunai, F.; Numata, O. Roles of three domains of Tetrahymena eEF1A in bundling F-actin. Zool. Sci. 2008, 25, 22–29. [Google Scholar] [CrossRef]
- Yang, F.; Demma, M.; Warren, V.; Dharmawardhane, S.; Condeelis, J. Identification of an actin-binding protein from Dictyostelium as elongation factor 1a. Nature 1990, 347, 494–496. [Google Scholar] [CrossRef] [PubMed]
- Bunai, F.; Ando, K.; Ueno, H.; Numata, O. Tetrahymena eukaryotic translation elongation factor 1A (eEF1A) bundles filamentous actin through dimer formation. J. Biochem. 2006, 140, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Suda, M.; Fukui, M.; Sogabe, Y.; Sato, K.; Morimatsu, A.; Arai, R.; Motegi, F.; Miyakawa, T.; Mabuchi, I.; Hirata, D. Overproduction of elongation factor 1α, an essential translational component, causes aberrant cell morphology by affecting the control of growth polarity in fission yeast. Genes Cells 1999, 4, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Munshi, R.; Kandl, K.A.; Carr-Schmid, A.; Whitacre, J.L.; Adams, A.E.; Kinzy, T.G. Overexpression of translation elongation factor 1A affects the organization and function of the actin cytoskeleton in yeast. Genetics 2001, 157, 1425–1436. [Google Scholar] [PubMed]
- Yang, W.; Burkhart, W.; Cavallius, J.; Merrick, W.; Boss, W. Purification and characterization of a phosphatidylinositol 4-kinase activator in carrot cells. J. Biol. Chem. 1993, 268, 392–398. [Google Scholar]
- Ransom-Hodgkins, W.D.; Brglez, I.; Wang, X.; Boss, W.F. Calcium-regulated proteolysis of eEF1A. Plant Physiol. 2000, 122, 957–966. [Google Scholar] [CrossRef][Green Version]
- Toueille, M.; Saint-Jean, B.; Castroviejo, M.; Benedetto, J.P. The elongation factor 1A: A novel regulator in the DNA replication/repair protein network in wheat cells? Plant Physiol. Biochem. 2007, 45, 113–118. [Google Scholar] [CrossRef]
- Tarrant, D. Investigating the Effects of Increased Levels of the Translation Elongation Factor eEF1A within Eukaryotic Cells. Ph.D. Thesis, University of Kent, Canterbury, UK, 2015. [Google Scholar]
- Mazier, M.; Flamain, F.; Nicolaï, M.; Sarnette, V.; Caranta, C. Knock-down of both eIF4E1 and eIF4E2 genes confers broad-spectrum resistance against potyviruses in tomato. PLoS ONE 2011, 6, e29595. [Google Scholar] [CrossRef]
- Bastin, M.; Hall, T.C. Interaction of elongation factor 1 with aminoacylated brome mosaic virus and tRNA’s. J. Virol. 1976, 20, 117–122. [Google Scholar] [CrossRef]
- Joshi, R.L.; Ravel, J.M.; Haenni, A.L. Interaction of turnip yellow mosaic virus Val-RNA with eukaryotic elongation factor EF-1 [alpha]. Search for a function. EMBO J. 1986, 5, 1143–1148. [Google Scholar] [CrossRef]
- Yamaji, Y.; Kobayashi, T.; Hamada, K.; Sakurai, K.; Yoshii, A.; Suzuki, M.; Namba, S.; Hibi, T. In vivo interaction between tobacco mosaic virus RNA-dependent RNA polymerase and host translation elongation factor 1A. Virology 2006, 347, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Zeenko, V.V.; Ryabova, L.A.; Spirin, A.S.; Rothnie, H.M.; Hess, D.; Browning, K.S.; Hohn, T. Eukaryotic elongation factor 1A interacts with the upstream pseudoknot domain in the 3′ untranslated region of tobacco mosaic virus RNA. J. Virol. 2002, 76, 5678–5691. [Google Scholar] [CrossRef] [PubMed]
- Thivierge, K.; Cotton, S.; Dufresne, P.J.; Mathieu, I.; Beauchemin, C.; Ide, C.; Fortin, M.G.; Laliberté, J.F. Eukaryotic elongation factor 1A interacts with turnip mosaic virus RNA-dependent RNA polymerase and VPg-Pro in virus-induced vesicles. Virology 2008, 377, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Komoda, K.; Ishibashi, K.; Kawamura-Nagaya, K.; Ishikawa, M. Possible involvement of eEF1A in tomato spotted wilt virus RNA synthesis. Virology 2014, 468, 81–87. [Google Scholar] [CrossRef]
- Luan, H.; Shine, M.; Cui, X.; Chen, X.; Ma, N.; Kachroo, P.; Zhi, H.; Kachroo, A. The potyviral P3 protein targets eukaryotic elongation factor 1A to promote the unfolded protein response and viral pathogenesis. Plant Physiol. 2016, 172, 221–234. [Google Scholar] [CrossRef]
- Hwang, J.; Oh, C.S.; Kang, B.C. Translation elongation factor 1B (eEF1B) is an essential host factor for tobacco mosaic virus infection in plants. Virology 2013, 439, 105–114. [Google Scholar] [CrossRef]
- Yamaji, Y.; Sakurai, K.; Hamada, K.; Komatsu, K.; Ozeki, J.; Yoshida, A.; Yoshii, A.; Shimizu, T.; Namba, S.; Hibi, T. Significance of eukaryotic translation elongation factor 1A in tobacco mosaic virus infection. Arch. Virol. 2010, 155, 263–268. [Google Scholar] [CrossRef]
- Dunn, M.A.; Morris, A.; Jack, P.L.; Hughes, M.A. A low-temperature-responsive translation elongation factor 1α from barley (Hordeum vulgare L.). Plant Mol. Biol. 1993, 23, 221–225. [Google Scholar] [CrossRef]
- Bukovnik, U.; Fu, J.; Bennett, M.; Prasad, P.V.; Ristic, Z. Heat tolerance and expression of protein synthesis elongation factors, EF-Tu and EF-1α, in spring wheat. Funct. Plant Biol. 2009, 36, 234–241. [Google Scholar] [CrossRef]
- Shin, D.; Moon, S.J.; Park, S.R.; Kim, B.G.; Byun, M.O. Elongation factor 1α from A. thaliana functions as molecular chaperone and confers resistance to salt stress in yeast and plants. Plant Sci. 2009, 177, 156–160. [Google Scholar] [CrossRef]
- Wang, J.; Ma, S.; Li, W.; Wang, Q.; Cao, H.; Gu, J.; Lu, Y. Genetic variability and diversity of the main resources of lily assessed via phenotypic characters, pollen morphology, and ISSR markers. Genet. Mol. Res. 2016, 15, gmr.15027638. [Google Scholar] [CrossRef] [PubMed]
- Barba-Gonzalez, R.; Lim, K.B.; Zhou, S.; Ramanna, M.; Van Tuyl, J.M. Interspecific hybridization in lily: The use of 2n gametes in interspecific lily hybrids. Floric. Ornam. Plant Biotech. 2008, 5, 138–145. [Google Scholar]
- Yuan, L.; Liu, Q.; Liu, Q. Conservation, evaluation and enhancement of wild lily germplasm in China. Acta Hortic. 2010, 900, 53–57. [Google Scholar] [CrossRef]
- Li, H.; Liu, D.; He, H.; Zhang, N.; Ge, F.; Chen, C. Molecular cloning of a 14-3-3 protein gene from Lilium regale Wilson and overexpression of this gene in tobacco increased resistance to pathogenic fungi. Sci. Hortic. 2014, 168, 9–16. [Google Scholar] [CrossRef]
- Wang, X.Z.; Zhang, Y.L.; Niu, L.X. Identification of three kinds of viruses, and the preliminary evaluation on the resistance to the virus in six wild Lilium species from Qin-ba mountains under the nursery field. Sci. Agri. Sin. 2008, 11, 025. [Google Scholar]
- Sun, D.Y.; Zhang, X.G.; Li, S.H.; Jiang, C.Z.; Zhang, Y.L.; Niu, L.X. LrABCF1, a GCN-type ATP-binding cassette transporter from Lilium regale, is involved in defense responses against viral and fungal pathogens. Planta 2016, 244, 1185–1199. [Google Scholar] [CrossRef]
- Lim, J.; Rhee, H.; Kim, Y.; Lim, K.; Van Tuyl, J. Resistance to Fusarium oxysporum f. sp. lilii in Lilium. Acta Hortic. 2003, 620, 311–318. [Google Scholar] [CrossRef]
- Zhao, X.; Chen, X.; Li, D.; Liu, K. Resources and research situation of the genus Lilium in China. Acta Hortic. 1994, 414, 59–68. [Google Scholar] [CrossRef]
- Rao, J.; Liu, D.; Zhang, N.; He, H.; Ge, F.; Chen, C. Identification of genes differentially expressed in a resistant reaction to Fusarium oxysporum in Lilium regale by SSH. Ieri Procedia 2013, 5, 95–101. [Google Scholar] [CrossRef][Green Version]
- Cui, Q.; Liu, Q.; Gao, X.; Yan, X.; Jia, G. Transcriptome-based identification of genes related to resistance against Botrytis elliptica in Lilium regale. Can. J. Plant Sci. 2018, 98, 1058–1071. [Google Scholar] [CrossRef]
- Ransom-Hodgkins, W.D. The application of expression analysis in elucidating the eukaryotic elongation factor one alpha gene family in Arabidopsis thaliana. Mol. Genet. Genomics 2009, 281, 391–405. [Google Scholar] [CrossRef] [PubMed]
- Kidou, S.; Ejiri, S. Isolation, characterization and mRNA expression of four cDNAs encoding translation elongation factor 1A from rice (Oryza sativa L.). Plant Mol. Biol. 1998, 36, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, F.; Montandon, P.E.; Stutz, E. Two genes encoding the soybean translation elongation factor eEF-1α are transcribed in seedling leaves. Plant Mol. Biol. 1991, 17, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, R.; Sunabori, S.; Fukuda, H.; Komamine, A. A gene expressed preferentially in the globular stage of somatic embryogenesis encodes elongation-factor 1α in carrot. Eur. J. Biochem. 1992, 209, 157–162. [Google Scholar] [CrossRef]
- Pokalsky, A.; Hiatt, W.; Ridge, N.; Rasmussen, R.; Houck, C.; Shewmaker, C. Structure and expression of elongation factor 1α in tomato. Nucleic Acids Res. 1989, 17, 4661–4673. [Google Scholar] [CrossRef]
- Xu, W.L.; Wang, X.L.; Wang, H.; Li, X.B. Molecular characterization and expression analysis of nine cotton GhEF1A genes encoding translation elongation factor 1A. Gene 2007, 389, 27–35. [Google Scholar] [CrossRef]
- Carneiro, N.P.; Hughes, P.A.; Larkins, B.A. The eEFlA gene family is differentially expressed in maize endosperm. Plant Mol. Biol. 1999, 41, 801–814. [Google Scholar] [CrossRef]
- Kyrpides, N.C.; Woese, C.R. Universally conserved translation initiation factors. Proc. Natl. Acad. Sci. USA 1998, 95, 224–228. [Google Scholar] [CrossRef]
- Doxey, A.C.; Yaish, M.W.; Moffatt, B.A.; Griffith, M.; McConkey, B.J. Functional divergence in the Arabidopsis β-1, 3-glucanase gene family inferred by phylogenetic reconstruction of expression states. Mol. Biol. Evol. 2007, 24, 1045–1055. [Google Scholar] [CrossRef]
- Posas, F.; Chambers, J.R.; Heyman, J.A.; Hoeffler, J.P.; de Nadal, E.; Ariño, J.N. The transcriptional response of yeast to saline stress. J. Biol. Chem. 2000, 275, 17249–17255. [Google Scholar] [CrossRef]
- Sun, Y.L.; Hong, S.K. Molecular cloning and identification of genes encoding eukaryotic initiation factor family 1 (LceIF1, LceIF1A, and LceIF1B) in Leymus chinensis. In Proceedings of the 2012 International Conference on Biomedical Engineering and Biotechnology, Macao, China, 26–29 May 2012; pp. 381–384. [Google Scholar]
- Berberich, T.; Sugawara, K.; Harada, M.; Kusano, T. Molecular cloning, characterization and expression of an elongation factor 1α gene in maize. Plant Mol. Biol. 1995, 29, 611–615. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, S. Inducible expression of translation elongation factor 1A gene in rice seedlings in response to environmental stresses. Acta Bot. Sin. 1999, 41, 800–806. [Google Scholar]
- Gao, Y.; Ma, J.; Zheng, J.C.; Chen, J.; Chen, M.; Zhou, Y.B.; Fu, J.D.; Xu, Z.S.; Ma, Y.Z. The elongation factor GmEF4 is involved in the response to drought and salt tolerance in soybean. Int. J. Mol. Sci. 2019, 20, 3001. [Google Scholar] [CrossRef] [PubMed]
- Rausell, A.; Kanhonou, R.; Yenush, L.; Serrano, R.; Ros, R. The translation initiation factor eIF1A is an important determinant in the tolerance to NaCl stress in yeast and plants. Plant J. 2003, 34, 257–267. [Google Scholar] [CrossRef]
- Warren, K.; Wei, T.; Li, D.; Qin, F.; Warrilow, D.; Lin, M.H.; Sivakumaran, H.; Apolloni, A.; Abbott, C.M.; Jones, A. Eukaryotic elongation factor 1 complex subunits are critical HIV-1 reverse transcription cofactors. Proc. Natl. Acad. Sci. USA 2012, 109, 9587–9592. [Google Scholar] [CrossRef]
- Chen, Z.; Ye, J.; Ashraf, U.; Li, Y.; Wei, S.; Wan, S.; Zohaib, A.; Song, Y.; Chen, H.; Cao, S. MicroRNA-33a-5p modulates Japanese encephalitis virus replication by targeting eukaryotic translation elongation factor 1A1. J. Virol. 2016, 90, 3722–3734. [Google Scholar] [CrossRef]
- Wazwaz, A.T.H. Collapse of resistance to tomato yellow leaf curl virus in tomato upon silencing the elongation factor 1-alpha gene. Master of Science Thesis, Bethlehem University, Bethlehem, Palestine, 2013. [Google Scholar]
- Li, S.; Feng, S.; Wang, J.H.; He, W.R.; Qin, H.Y.; Dong, H.; Li, L.F.; Yu, S.X.; Li, Y.; Qiu, H.J. eEF1A interacts with the NS5A protein and inhibits the growth of classical swine fever virus. Viruses 2015, 7, 4563–4581. [Google Scholar] [CrossRef]
- Lino-Neto, T.; Tavares, R.; Palme, K.; Pais, M. Expression of glutathione peroxidase during Zantedeschia aethiopica spathe senescence and regreening. In The Chloroplast: From Molecular Biology to Biotechnology; Argyroudi-Akoyunoglou, J.H., Senger, H., Eds.; Springer: Dordrecht, The Netherlands, 1999; pp. 117–120. [Google Scholar]
- Mateyak, M.K.; Kinzy, T.G. eEF1A: Thinking outside the ribosome. J. Biol. Chem. 2010, 285, 21209–21213. [Google Scholar] [CrossRef]
- Rogers, H.; Munné-Bosch, S. Production and scavenging of reactive oxygen species and redox signaling during leaf and flower senescence: Similar but different. Plant Physiol. 2016, 171, 1560–1568. [Google Scholar] [CrossRef]
- Yin, J.; Chang, X.; Kasuga, T.; Bui, M.; Reid, M.S.; Jiang, C.Z. A basic helix-loop-helix transcription factor, PhFBH4, regulates flower senescence by modulating ethylene biosynthesis pathway in petunia. Hortic. Res. 2015, 2, 15059. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Zhang, X.; Zhang, Q.; Ji, X.; Jia, Y.; Wang, H.; Niu, L.; Zhang, Y. Comparative transcriptome profiling uncovers a Lilium regale NAC transcription factor, LrNAC35, contributing to defence response against cucumber mosaic virus and tobacco mosaic virus. Mol. Plant Pathol. 2019, 20, 1662–1681. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.P.; Valkonen, J.P. Genetic determinants of potato virus Y required to overcome or trigger hypersensitive resistance to PVY strain group O controlled by the gene Ny in potato. Mol. Plant Microbe In. 2013, 26, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhang, Z.; Zhang, H.; Wang, X.C.; Huang, R. Transcriptional modulation of ethylene response factor protein JERF3 in the oxidative stress response enhances tolerance of tobacco seedlings to salt, drought, and freezing. Plant Physiol. 2008, 148, 1953–1963. [Google Scholar] [CrossRef] [PubMed]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- van der Meer, I.M. Agrobacterium-mediated transformation of petunia leaf discs. In Plant Cell Culture Protocols; Loyola-Vargas, V.M., Vázquez-Flota, F., Eds.; Humana Press: Totowa, NJ, USA, 2006; pp. 265–272. [Google Scholar]
- Estrada-Melo, A.C.; Reid, M.S.; Jiang, C.Z. Overexpression of an ABA biosynthesis gene using a stress-inducible promoter enhances drought resistance in petunia. Hortic. Res. 2015, 2, 15013. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.; Chen, S.; Chen, F.; Guan, Z.; Fang, W. Morpho-anatomical and physiological responses of two Dendranthema species to waterlogging. Environ. Exp. Bot. 2010, 68, 122–130. [Google Scholar] [CrossRef]
- Chen, J.C.; Jiang, C.Z.; Gookin, T.; Hunter, D.; Clark, D.; Reid, M. Chalcone synthase as a reporter in virus-induced gene silencing studies of flower senescence. Plant Mol. Biol. 2004, 55, 521–530. [Google Scholar] [CrossRef]
- Jiang, C.Z.; Chen, J.C.; Reid, M.S. Virus-induced gene silencing in ornamental plants. In RNAi and Plant Gene Function Analysis; Kodama, H., Komamin, A., Eds.; Humana Press: Totowa, NJ, USA, 2011; pp. 81–96. [Google Scholar]
- Sun, D.Y.; Li, S.H.; Niu, L.X.; Reid, M.S.; Zhang, Y.L.; Jiang, C.Z. PhOBF1, a petunia ocs element binding factor, plays an important role in antiviral RNA silencing. J. Exp. Bot. 2017, 68, 915–930. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, D.; Ji, X.; Jia, Y.; Huo, D.; Si, S.; Zeng, L.; Zhang, Y.; Niu, L. LreEF1A4, a Translation Elongation Factor from Lilium regale, Is Pivotal for Cucumber Mosaic Virus and Tobacco Rattle Virus Infections and Tolerance to Salt and Drought. Int. J. Mol. Sci. 2020, 21, 2083. https://doi.org/10.3390/ijms21062083
Sun D, Ji X, Jia Y, Huo D, Si S, Zeng L, Zhang Y, Niu L. LreEF1A4, a Translation Elongation Factor from Lilium regale, Is Pivotal for Cucumber Mosaic Virus and Tobacco Rattle Virus Infections and Tolerance to Salt and Drought. International Journal of Molecular Sciences. 2020; 21(6):2083. https://doi.org/10.3390/ijms21062083
Chicago/Turabian StyleSun, Daoyang, Xiaotong Ji, Yong Jia, Dan Huo, Shiying Si, Lingling Zeng, Yanlong Zhang, and Lixin Niu. 2020. "LreEF1A4, a Translation Elongation Factor from Lilium regale, Is Pivotal for Cucumber Mosaic Virus and Tobacco Rattle Virus Infections and Tolerance to Salt and Drought" International Journal of Molecular Sciences 21, no. 6: 2083. https://doi.org/10.3390/ijms21062083
APA StyleSun, D., Ji, X., Jia, Y., Huo, D., Si, S., Zeng, L., Zhang, Y., & Niu, L. (2020). LreEF1A4, a Translation Elongation Factor from Lilium regale, Is Pivotal for Cucumber Mosaic Virus and Tobacco Rattle Virus Infections and Tolerance to Salt and Drought. International Journal of Molecular Sciences, 21(6), 2083. https://doi.org/10.3390/ijms21062083