Abstract
Pepper (Capsicum spp.) is one of the major vegetable crops grown worldwide largely appreciated for its economic importance and nutritional value. This crop belongs to the large Solanaceae family, which, among more than 90 genera and 2500 species of flowering plants, includes commercially important vegetables such as tomato and eggplant. The genus includes over 30 species, five of which (C. annuum, C. frutescens, C. chinense, C. baccatum, and C. pubescens) are domesticated and mainly grown for consumption as food and for non-food purposes (e.g., cosmetics). The main challenges for vegetable crop improvement are linked to the sustainable development of agriculture, food security, the growing consumers’ demand for food. Furthermore, demographic trends and changes to climate require more efficient use of plant genetic resources in breeding programs. Increases in pepper consumption have been observed in the past 20 years, and for maintaining this trend, the development of new resistant and high yielding varieties is demanded. The range of pathogens afflicting peppers is very broad and includes fungi, viruses, bacteria, and insects. In this context, the large number of accessions of domesticated and wild species stored in the world seed banks represents a valuable resource for breeding in order to transfer traits related to resistance mechanisms to various biotic stresses. In the present review, we report comprehensive information on sources of resistance to a broad range of pathogens in pepper, revisiting the classical genetic studies and showing the contribution of genomics for the understanding of the molecular basis of resistance.
Keywords:
Capsicum; resistance genes; fungal diseases; bacterial spot; viruses; insect; nematodes; QTL 1. Introduction
Pepper (Capsicum spp.) is a fruit vegetable originated in the American tropics and today widely consumed as fresh, dried, or processed products. Around the genus Capsicum, there is an increasing interest due to the amazing diversity in plant and fruit characteristics, which make this crop extremely versatile and suitable for innumerable uses. The consumption of pepper has been increased in the last 20 years with a production ranging from 19 to about 40 million tons and a surface area from 2.5 to about 3.8 million of hectares [1]. Further increases are expected due to the greater demand for high-value nutritional products by consumers. Indeed, pepper is a rich source of health-promoting compounds with important nutraceutical and anticancer properties. Despite this favorable trend, several pests and diseases threaten cultivation around the world representing a limiting factor for productivity [2].
The range of pathogens afflicting pepper is very broad and includes fungi (Phytophthora capsici, Rhizoctonia solani, Verticillium dahliae, Colletotrichum scovillei and truncatum, Leveillula taurica, Fusarium spp.), bacteria (e.g., Xanthomonas spp.), viruses such as Tospoviruses (e.g., Tomato spotted wilt orthotospovirus and Impatiens necrotic spot orthotospovirus), Potyviruses (e.g., Potato virus Y, Tobacco etch virus, Pepper mottle virus), Tobamoviruses (e.g., Tobacco mosaic virus, Tomato mosaic virus), Cucumoviruses (e.g., Cucumber mosaic virus), nematodes (Meloidogyne spp.) and insects (e.g., mites, aphids, Lepidoptera and thrips). Cultural methods and pesticides are applied to ensure a healthy and profitable pepper crop. Considering the increasing need for sustainable agriculture, the use of resistant plants represents the main strategy to protect pepper cultivation against biotic stresses [2,3,4]. As an example, the limitations imposed in recent years on the use of soil fumigants have led to the growth of interest in the introduction of resistance against soilborne pathogens such as Phytophthora spp. and Meloidogyne spp. in rootstocks and cultivars [5].
In the last decades, most of the pepper breeding programs have been addressed to the development of cultivars or hybrids against a wide range of pathogens and pests. Despite the efforts made, the exploitation of Capsicum germplasm (pre-breeding materials, landraces, wild relatives and closed related species) and its use in breeding programs for biotic stress resistance still represent challenging tasks [2]. Indeed, climate changes and the risk of a resistance breakdown, affect the durability of disease resistance, therefore, there is an urgent need to develop new resistant cultivars that can be adapted to varied pedoclimatic conditions. In this frame, gene pyramiding strategies can allow the accumulation of resistance genes in a single genotype and creates more durable and broad-spectrum mechanisms [6]. The strategy can be accomplished combining one or more alleles of major genes [7]. Pyramiding strategies have been successfully used for resistance to pathogens in several crops [8].
Some databases are available and refer to a global collection of several materials (wild and domesticated accessions, cultivars, breeding lines, and hybrids) as a source of resistance or tolerance to several pests and diseases. The most important public databases are Chile Variety Database [9], NPGS Germplasm Collection Genebanks from the USDA-ARS [10], World Vegetable Center database [11], The Centre for Genetic Resources, the Netherlands (CGN) of Wageningen University [12], National Bureau of Plant Genetic Resource (India) [13]. The present review aims to provide comprehensive information on the sources of resistance to a broad range of pathogens of pepper, revisiting the classical genetic studies and showing the contribution of genomics for the understanding of the molecular basis of resistance.
2. Fungal Diseases
2.1. Powdery Mildew
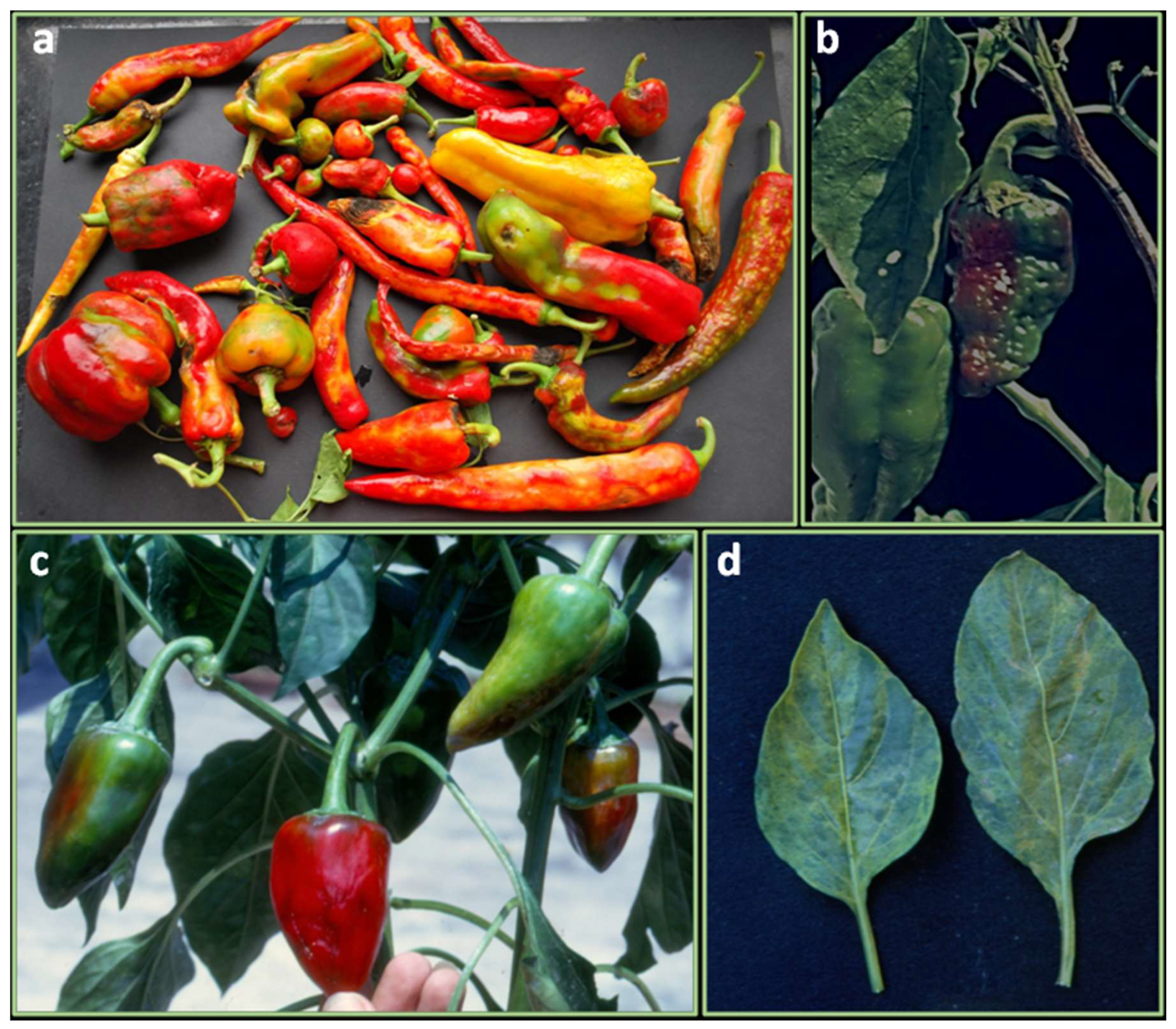
The powdery mildew of pepper occurs worldwide and is particularly severe in warm climates, dry or humid, where it causes severe yield losses. The disease, caused by Leveillula taurica (asexual stage: Oidiopsis taurica), appears as grayish white patches on the undersides of leaves and light green-yellow lesions on the upper leaf surface (Figure 1a). Genotypes from different Capsicum species have been reported to be immune or highly resistant to the fungus [14,15,16] (Table 1).

Figure 1.
Symptoms and damages caused by fungal disease in leaves, plants and fruits: (a) powdery mildew on leaf; (b) shriveled plants attacked by Phytophthora root rot; (c) anthracnose of fruit; (d) Verticillium wilt with discolored vascular tissue of infected stem; (e) Root and stem rot caused by Rhizoctonia solani.

Table 1.
Sources of resistance/tolerance to fungal and bacterial diseases in Capsicum spp.
At least three genes could be involved in the resistance to L. taurica in H3 cultivar [17]. The first attempt to map powdery mildew resistance was carried out by Lefebvre and colleagues, which described the quantitative nature of the resistance identifying a robust Quantitative Trait Locus (QTL) on chromosome 6 (Lt 6.1), and reported epistatic interactions explaining more than 50% of the genotypic variance [18] (Table 2). A single dominant locus, PMR1, located in a syntenic region of 4 Mb of pepper Chr 4 and responsible for the powdery mildew resistance has been reported [19]. Within this locus, two genes across the 622 predicted ones were found to share sequence similarity to the nucleotide-binding site leucine-rich repeat domain containing R proteins (NBS-LRR), which represent clusters of resistance genes in plants. Moreover, the authors identified six molecular markers [one Sequence Characterized Amplified Region (SCAR); five Single Nucleotide Polymorphysms (SNPs)] tightly linked to PMR1 and useful for marker-assisted selection (MAS) and pyramiding. Phylogenetic analysis based on Genotyping by Sequencing (GBS) data and InDel markers demonstrated a close relatedness of the PMR1 region from C. baccatum and C. annuum supporting the introgression of resistance from C. baccatum, possibly using C. chinense as a bridge species.

Table 2.
List of mapping populations, genetic map and Quantitative Trait Loci (QTLs) for biotic stress resistance in pepper.
A patent application using the resistant line PBC167 reports a QTL on linkage groups (LG) 1 and 8 explaining 57% of the variance [49]. Using the same line, the QTL was precisely mapped in an interval of ~40 cM on Chr 4 [20].
Functional studies allowed to determine two genes CaMlo1 and CaMlo2 as the responsible for pepper resistance to powdery mildew; the loss of function of these genes determine the reduction of disease susceptibility [50].
2.2. Phytophthora Root Rot and Foliar Blight
The disease is caused by Phytophthora capsici, one of the most destructive pathogens of pepper particularly where the soil is wet, and temperatures remain low (15–23°C) [51]. The oomycete can also cause stem and fruit rot, wilting, stunting, dumping-off, plant death as well as stem and leaf blight (Figure 1b). Separate and independent resistant systems have evolved for each P. capsici disease syndrome and independent resistance genes have been detected [52,53]. More than 45 physiological races have been identified within the Phytophthora root-rot and foliar blight [54]; for each physiological race, different R genes for given disease stages are involved [55]. Therefore, pyramiding multiple genes is essential as it occurs for the P. infestans pathosystem in the closely related potato (S. tuberosum) [56]. The characterization of pathogen races is traditionally performed utilizing a set of differential hosts which are not always affordable for breeding purposes due to reproduction barriers [57]. Recently, novel races have been identified based on the differential reactions of a set of New Mexico Recombinant Inbred lines (NMRILs) [54]. Identification of pathogen races using NMRILs suggests supplementing the term “race” by the term “virulence phenotype”, to designate the virulence of P. capsici isolates to the various host resistance genes [58]. Several C. annum accessions resistant to P. capsici and carrying a single dominant gene [59], or multiple genes with additive or epistatic effects [60] have been identified (Table 1). Among them, Serrano Criollo de Morelos (CM334) has the highest resistance level to all the disease stages [51]. Host-parasite coevolutionary relationships have been demonstrated [58]. Indeed, both P. capsici and resistant germplasm sources have the same geographical origin. Two main chromosomal regions deputed for the resistance to P. capsici were identified through a comparative mapping strategy involving three intraspecific C. annuum populations [61] (Table 2). From the alignment of the relative genetic maps, a common major QTL was positioned on Chr 5. Afterward, 16 chromosomal intervals containing single or clusters of resistance QTLs for root rot and (or) leaf blight, were identified by using a RIL mapping population [62]. Moreover, the authors reported five QTLs with an effect on the resistance to root rot using an intraspecific F2, highlighting the complex polygenic nature of the resistance to P. capsici. In the same year, a Random Amplification of Polymorphic DNA (RAPD) marker (OpD04.717) able to amplify a single band in genotypes with high levels of resistance and linked to the major QTL Phyto.5.2 has been identified [51]. In 2006, Sugita and collaborators detected a major QTL (Phyt-1) on Chr 5 and two minor QTLs on Chr 1 and 11 explaining over 80% and less than 10% of the phenotypic variance, respectively35. Phyt-1 was in the same chromosomal region of other major QTLs (Phyto-P; Phyt.5.1; Phyt.5.2) identified on Chr 5 previously [51,61,62]. Chromosome 5 was confirmed as the main region involved in the pathogen responses [63]. Kim and colleagues reported seven QTLs, four of which (66.3% of the phenotypic variation) were related to the root rot resistance, while three (45% of variation), were related to dumping off resistance [64]. The authors confirmed the existence on Chr 5 of a major QTL stable across several P. capsici populations and isolates [51,61,63,65]. Markers for rapid analysis of resistant genotypes were developed by sequencing the Bacterial Artificial Chromosome (BAC) clones of the Restriction Fragment Length Polymorphism (RFLP) markers closely linked to the major QTLs [64]. An intraspecific RIL (Table 2), was used to map QTLs for resistance against different P. capsici isolates in two different studies. The first identified 15 QTLs, seven of which located on Chr 5 explained a phenotypic variation from ~5% to ~50% [66]. The second, allowed to detect 4 QTLs evidencing three main-effect loci related to P. capsici resistance. A QTL located on Chr 5 explained over 60% of the heritability of additive effect, being the major-effect gene involved in the pathogen defense response [67]. Using the same RIL population, bulked segregant analysis (BSA) combined with RAPD and Amplified Fragment Length Polymorphism (AFLP) markers allowed to develop a co-dominant SCAR marker (SA133_4) linked to root rot resistance in the region of QTLs contributing to obtain a stable resistance on Chr 5 [66]. Combining BSA and microarray analysis (Affymetrix GeneChips), SNP markers tightly linked to the major QTL on Chr 5 were identified [66,68]. Among them, the marker Phyto5SAR showed the highest logarithm of the odds (LOD) value at the QTL on chromosome 5. Phyto5SAR was in a region containing clusters of resistance genes (NBS-LRR) and a systemic acquired resistance-related gene (SAR 8.2A) both associated with plant defense responses. Moreover, within this region, the reliable marker Phyto5NBS1 able to discriminate among susceptible and resistant lines with over 90% accuracy, was designed. A key QTL cluster on chromosome 5 (Pc5.1), exhibiting broad-spectrum resistance to P. capsici and conferring resistance against at least 12 P. capsici isolates worldwide collected was identified [69]. This broad-spectrum QTL showed robust effects in different genetic backgrounds and represented the major target for breeders. Additive and epistatic QTLs have been identified using three isolates of P. capsici in 63 F6-RILs confirming the Chr 5 as the main region of interest for resistance to root rot [70]. In the same population was identified the gene CaDMR1 encoding for a homoserine kinase, as the candidate responsible for the major QTL on Chr 5 for resistance to P. capsici [71]. Recently, Bulk Segregant Analysis (BSA) combined with Specific locus amplified fragment sequencing (SLAF-seq) allowed to identify PhR10: a single dominant gene positioned on the long arm of Chr 10 and responsible for the resistance to race 3 (Byl4) [72]. Functional studies reported that cell death was mediated by the increased reactive oxidative species (ROS) production due to the silencing of the methionine sulfoxide reductase B2 gene (CaMsrB2), suggesting that the regulation of pathogen defense responses and oxidative stresses is controlled by ROS accumulation or reduction, respectively [73]. Furthermore, the silencing of CaRGA2, a resistance gene analog developed in C. annuum CM334 lead to the induction of susceptible disease symptoms after the infection, accompanied by a proliferation of P. capsici in pepper tissues [74]. The results of the two studies suggest that gene suppression renders the plants unable to activate the resistance response increasing susceptibility. In 2016, genomic studies allowed to identify over a thousand genes differentially expressed in the resistant line PI201234 among them, 211 were involved in defense responses based on the gene annotations [75]. Validation tests in the resistant Qiemen line, allowed to identify seven genes responsible for many functions related to the prevention of infection (cell wall modification, symptom development, and phytohormone signaling pathways and phytoalexin biosynthesis). The accession PI201234 was used to develop a population of 794 F2 individuals by crossing to the susceptible Shanghaiyuan variety [75]. A single dominant gene, CaPhyto, on Chr 5 and two candidate genes, Capana05g000764 and Capana05g000769, were found to underly the resistance to race 2. A microsatellite marker (ZL6726) positioned at a distance of 1.5 cM from CaPhyto, was validated to be reliable for selecting phenotypes of resistance to the same P. capsici race. Several other molecular markers associated with resistance to P. capsici have been reported in chile pepper for more rapid selection [51,64,66,68,72,75].
2.3. Anthracnose or Ripe Rot of Pepper
Anthracnose causes serious losses of fruits in pre- and post-harvest stages [32,76]. Occasionally, it also damages stem and foliage. The typical fruit symptoms appear as circular water-soaked spots with concentric rings of black acervuli developing beneath the skin (Figure 1c). The spots are often numerous and coalesce, causing softening and rotting of fruits [27]. Anthracnose can be caused by a wide range of Colletotrichum species. To date, 24 species infecting pepper have been identified, of which the most common pathogenic are: C. scovillei (previously identified as C. acutatum), C. truncatum (syn. C. capsici) and C. siamense (previously identified as C. gloeosporioides). The latter is less virulent [76,77]. Within these three Colletotrichum species, different pathotypes have been identified based on the qualitative and quantitative reaction of fruits at different maturity stages on a set of chilli species and related accessions (Table 1) [27,28]. A major QTL conferring resistance to C. siamense and C. truncatum and three minor resistant QTLs against C. siamense were identified (Table 2) [29]. A single recessive gene conferring resistance to C. truncatum was mapped in an interspecific population derived from C. annuum cv. Bangchang (susceptible) × C. chinense acc. PBC932 (resistant) [30]. The inheritance model was then confirmed in introgression populations [78]. For C. scovillei, two major QTLs on Chr 8 and 9, and sixteen with minor effects were detected [79]. Furthermore, five major QTLs located on Chr 5 and conferring resistance to both matured green and matured red fruits, as well as four with minor-effect specific only for the green mature stage have been identified [80]. The first identified sources of resistance in C. baccatum (PBC80, PBC81) and C. chinense (PBC932) [81], have been extensively used to introgress the resistance in different susceptible C. annuum background through conventional breeding and embryo rescue technique [82,83]. PBC80 possesses recessive (co4) or dominant (Co5) genes located on Chr 12 and Chr 9, respectively. The first was identified in mature green fruit while Co5 was detected in ripe fruit [84]. PBC932 possesses recessive genes (co1, co2 and co3) located on Chr 5 [78]. Two accessions of C. baccatum var. pendulum (UENF 1718 e UENF 1797) were found very promising to be introduced in breeding programs [31]. Moreover, two SNP maps were constructed from two chilli populations including C. annuum Bangchang x C. chinense PBC932, and C. baccatum PBC80 x CA1316. The validated SNPs are using in anthracnose breeding programs [85]. Recently, sources of resistance to C. truncatum and C. siamense, under both field and in vitro conditions, have been identified in C. annuum accessions (Table 1) [32]. Breeding for resistance to races would broaden the resistance base of chilli cultivars through gene pyramiding of multiple resistance genes [77].
2.4. Vascular Diseases
Verticillium wilt represents a serious threat to the pepper production worldwide [98]. The disease is mainly caused by the soilborne fungus Verticillium dahliae and at a minor extent by V. alboatrum. Both pathogens penetrate plants directly or through wounds and spread acropetally through the xylem, causing browning of the vascular tissue, stunting, foliar epinasty, chlorosis and necrosis, wilting and death of the entire plant (Figure 1d). Resistance in peppers is not common in commercial cultivars and is difficult to identify in germplasm sources [99]. Recently, Gurung and colleagues [25], identified eight resistant accessions (Table 1) out of 397 analyzed, of which two (Grif 9073 and PI 439297) conferred resistance also to Phytophthora root rot. Although no genetic mapping studies are reported in Capsicum for Verticillium, molecular markers for assisted breeding have been developed based on the homology with the tomato resistance genes, Ve (Ve1 and Ve2) [100]. Like Ve gene in tomato, the homolog chilli CaVe gene is located on Chr 9 [100] and, through recognition of the Ave1 effector [101], confers resistance to race 1. Based on the polymorphism between susceptible and resistant accessions in the coding region of CaVe, a CAPS marker able to identify Verticillum resistant genotypes with the accuracy of 48% was developed [100]. The other vascular disease is caused by Fusarium, which determines crop yield losses ranging from 10% to 80% [102]. Several isolates within the Fusarium species complex have been linked to pepper wilt. Among them, F. oxysporum [103], F. solani [33], F. oxysporum f. sp. vasinfectum [104], F. redolens (previously classified as F. oxysporum var. redolens) [105], and F. oxysporum f. sp. capsici [106], are the prevailing ones worldwide. F. verticillioides (syn. F. moniliforme) and F. pallidoroseum cause pepper wilting in some parts of India [107]. Maruti and collaborators [33], screening 56 restorer lines and 38 F1 hybrids in controlled laboratory conditions, found one genotype (P3) moderately resistant. Moreover, two hybrids, viz., JNA2 × ACB1 × 9608D and Rajaput × P3, showed resistance under sick pot culture conditions. Resistant C. annuum genotypes to F. solani were also obtained using chemical mutagens such as Ethyl Methane Sulphonate (EMS) [108]. Manu and colleagues [109], studying three crosses viz., SNK x P3, KA2 x P3, and RAJPUT x P3, concluded that the inheritance of F. solani resistance was monogenic and dominant. Therefore, heterosis breeding is recommended, to boost the yield in sites where this soil-borne pathogen is widespread. Good sources of resistance to F. oxysporum, F. verticillioides and F. pallidoroseum were found in various C. annuum accessions [34,110,111] (Table 1).
2.5. Rhizoctonia Solani
Rhizoctonia solani (teleomorph Thanatephorus cucumeris) is a destructive soil-borne pathogen that causes several syndromes such as seedling damping-off, root rot, stem rot or canker (Figure 1e) [112]. A wide genetic range of resistance to the most aggressive New Mexican isolate of R. solani (PWB-25) was found in accessions belonging to four Capsicum species (C. annuum, C. baccatum, C. chinense and C. frutescens) [26,37]. In particular, two C. baccatum genotypes (PI439410 and PI5556119) were the most resistant to post-emergence inoculation. Nevertheless, the C. annuum accessions, Long Chilli (a Korean hybrid) and PI167061, had 67 and 71% resistant individuals, respectively, and could be useful for introducing R. solani resistance in C. annuum breeding schemes. The investigation of the sources of resistance to Fusarium spp., P. capsici and R. solani was performed in 44 genotypes retrieved from the INIFAP-CEBAJ germplasm bank as well in 141 accessions of C. annuum collected in different regions of Mèxico [26]. In total, 26 accessions resistant to Fusarium spp., six to R. solani and two (BG107 and BG102) to P. capsici, were identified. The latters showed mechanisms of resistance to the mixture of all the three pathogens, turning up to be a source of potentially useful genes to be used in breeding programs addressed to the control of wilt diseases.
3. Bacterial diseases
3.1. Bacterial Spot of Pepper
Bacterial spot is one of the major problems for the cultivation of pepper in tropical and subtropical regions and is principally due to four Xanthomonas (hereafter Xs.) species: Xs. euvesicatoria, Xs. perforans, Xs. gardneri, and Xs. Vesicatoria [38].
All parts of the plants are damaged by Xs. On the leaves, it causes small, water-soaked, black spots. The spots can coalesce and form large yellow areas that later become necrotic (Figure 2a). On the stem, elongated, raised cankers appear. On green fruits, small, circular, water-soaked, slightly raised lesions are produced. As the disease progresses, spots become brown, roughened, raised with cracked. Yield is reduced because of the scabbed lesions on fruits, which makes fruits unmarketable. The dropping of leaves reduces productivity and exposes fruits to the formation of sunscald.

Figure 2.
Bacterial diseases in pepper plants: (a) bacterial spots on plantlet leaves before transplant; (b) extensive wilting in pepper cultivation caused by Ralstonia spp.
Nine pepper races (P0-P8) have been identified among Xs. strains worldwide [113], and five non-allelic dominant genes (Bs1, Bs2, Bs3, Bs4, and Bs7) were reported to control hypersensitive reaction to Xs. according to the gene-for-gene hypothesis. These genes were found in PI163192 (Bs1, C. annuum), PI260535 (Bs2, C. chacoense), PI271322 (Bs3, C. annuum), PI235047 (Bs4, C. pubescens) and UNEF1556 (Bs7, C. baccatum var. pendulum [39]. Moreover, two recessive genes (bs5 and bs6), that govern a non-hypersensitive resistance and act additively with each other, were discovered in PI271322, Pep13 and PI163192 (C. annuum) [38,40]. One or more of the HR genes have been transferred in near-isogenic lines developed in the Early Calwonder background. Tai and colleagues [114], performed a high-resolution genetic mapping of Bs2 identifying tightly linked molecular markers in C. annuum near-isogenic lines holding introgressions from C. chacoense PI260435. A year later, AFLP markers tightly linked to the Bs3 were identified at a genetic resolution of 0.13 cM [115]. Another marker able to detect a functional nucleotide polymorphism in the Bs3 promoter (PR-Bs3) was also found by Romer and collaborators [116]. Although Bs1, Bs2 and Bs3 have been introgressed in several commercial pepper cultivars, mutations in the respective avirulence genes (avrBs1, avrBs2, avrBs3), occurring in the race P6, rendered useless the resistance making this strain highly virulent [117]. It has been demonstrated that the combination of Bs5 and Bs6 conferred an additive effect, leading to complete resistance against P6 [117]. Kompetitive Allele-Specific PCR (KASP) genotyping system has been used to develop markers linked to the Bs3 locus [35]. The developed markers were able to detect susceptible or resistant alleles due to preferential amplification of the transcriptional start site in the promoter region. This approach increased the robustness and throughput of screening resistance loci.
Functional studies evidenced the role of the C. annuum peroxidase gene, CaPO2 in the resistance against Xs. [118]. Knock-down of the CaPO2 gene mediated by virus-induced gene silencing evidenced plants highly susceptible to the Xs. infection as well as reduction of hydrogen peroxide (H2O2) and hypersensitive cell death. On the contrary, overexpression of CaPO2 exhibited disease resistance, accumulation of H2O2 accompanied by cell death [118]. These results evidence the role of CaPO2 in the hypersensitivy mechanism of defence against Xs. in pepper.
Moreover, CaMLO2 has been found to play a role in the Xs. resistance. Kim and collaborators [36], demonstrated that the silencing of CaMLO2 enhanced the resistance against virulent Xs., evidencing the reduced bacterial growth through the boost of reactive oxygen species burst.
3.2. Bacterial Wilt
Bacterial wilt (BW) of pepper, is the most devastating soil-borne disease in tropics and in the warmer climates throughout the world [119]. Young plants are rapidly infected and destroyed after the infection (Figure 2b). The older plants first show wilting of the youngest leaves during warm or hot weather day conditions, and after a temporary recovery under cooler temperatures can permanently wither. In the cross-section, plant vascular bundles show a brown discoloration and ooze a white bacterial exudate. Pepper may also show latent infections [120].
BW is caused by Ralstonia solanacearum, phylotype I, R. pseudosolanacearum, phylotype I and III, and R. syzyngii subsp. indonesiensis phylotype IV [121]. The three species were previously grouped in R. solanacearum species complex (RSSC) and classified into “races” and “biovars” [119,122,123,124]. Virulent isolates were reported in North America and in Japan on pepper cultivars, previously known as resistant [125].
Sources of resistance were found in several cultivated and domesticated pepper accessions (Table 1).
The inheritance of BW resistance has been established to be controlled by two to five genes with additive effects [45]. The quantitative nature of resistance has been confirmed in studies reporting up to six QTL analysis with additive effects and digenic interactions [87].
A major QTL responsible for resistance to Ralstonia was found on Chr 1 (named Bw1) [86]. The SSR marker CAMS451 was reported to be tightly associated being mapped in the center of this QTL. Although BW-resistance is thought to be polygenically controlled, the use of this linkage marker may improve the efficiency of breeding BW-resistant cultivars [86].
Recently, the resequencing of the two C. annuum cultivars, YCM334 and Tean, allowed to identify novel SNPs and insertions/deletions (Indels) associated with the BW-resistance [46]. The authors detected 10 genes involved in the resistance mechanism including disease resistance proteins, polyprotein, LRR like receptor kinase, N-like protein, CC (coiled-coil)-NBS-LRR, and putative phosphatidylinositol 4-kinase. In 2017, Mou and collaborators identified a further gene, CaHDZ27, encoding for a Homeodomain-Leucine Zipper I transcription factors [126], in BW-resistant plants. Gene silencing significantly reduced the resistance down-regulating as well as other defense-related genes (CaHIR1, CaACO1, CaPR1, CaPR4, CaPO2, and CaBPR1). On the contrary, the transient overexpression boosted cell death mediated by the hypersensitive response.
4. Viral Diseases
4.1. Thrips-Transmitted Viruses
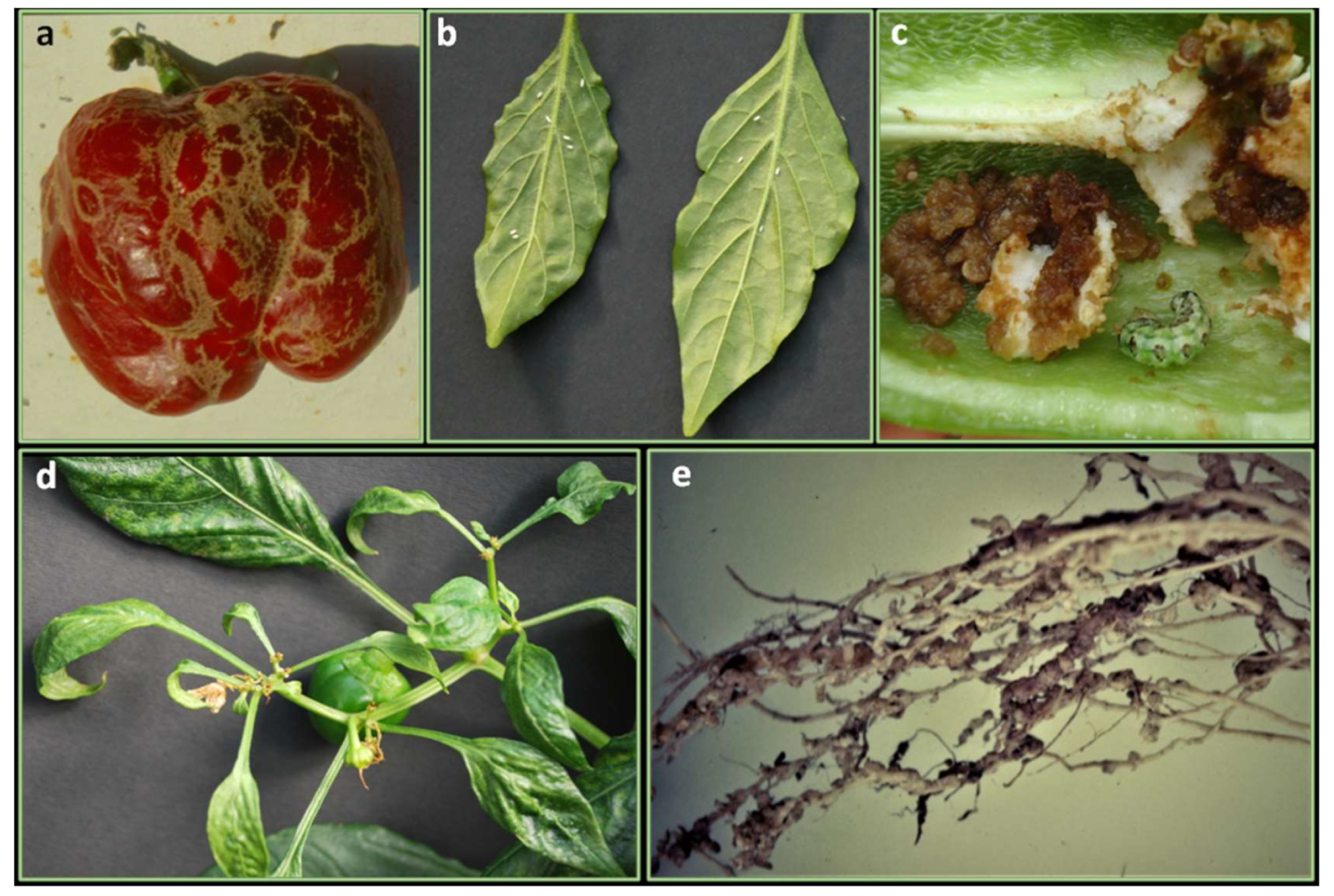
Orthotospoviruses are a group of virus causing serious damages to a wide range of hosts, being transmitted in a circulative propagative manner by at least seven species of thrips (mainly, Frankliniella occidentalis). Tomato spotted wilt orthotospovirus (TSWV) (Figure 3a), Impatiens necrotic spot orthotospovirus (INSV), Groundnut ringspot orthotospovirus (GRSV), Tomato chlorotic spot orthotospovirus (TCSV), Watermelon silver mottle orthotospovirus (WSMoV), Capsicum chlorosis orthotospovirus (CaCV), Groundnut bud necrosis orthotospovirus (GBNV), Pepper necrotic spot orthotospovirus (PNSV), Pepper chlorotic spot orthotospovirus (PCSV) were reported to infect Capsicum species [127]. Among them, TSWV and INSV are worldwide distributed and represent the only two orthotospoviruses occurring in pepper cultivations of Mediterranean area whereas, CaCV, GRSV, and TCSV have emerged as serious pathogens of these crops in India, Australia, Greece (CaCV), Florida (GRSV, TCSV) and South America (TCSV), in more recent years [128,129,130,131,132].
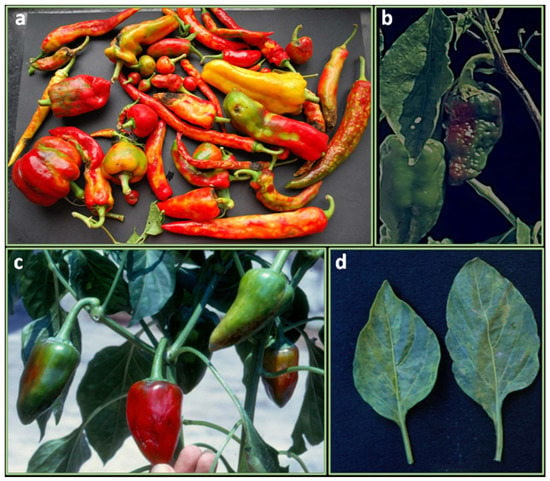
Figure 3.
Damages caused by viral diseases in leaves, plants, and fruits: (a) extensive spots on fruits caused by Tomato spotted wilt orthotospovirus (TSWV) in pepper landraces; (b) effect of Potato virus Y (PVY) on fruit and stem; (c) symptoms on mature fruit caused by Cucumber mosaic virus (CMV); (d) Tomato mosaic virus (ToMV) on leaves.
Heritable resistance to TSWV based on a hypersensitive response has been identified in several accessions of C. chinense, among them PI152225, PI159234 and PI159236 have been the most adopted in breeding programs [133].
The resistance is due to a single dominant gene (Tsw) or a tightly linked group of genes in several C. chinense accessions (PI159236, PI152225, CNPH-275 and 7204) [134,135]. The Tsw gene has been mapped in the distal portion of chromosome 10 [136]. A CAPS marker (SCAC 568) tightly linked 0.9 cM away to the Tsw locus has been identified in a segregant F2 population C. chinense (PI152225) × C. frutescens (PI195301) [137]. The resistance conferred by the Tsw gene is overcome by high temperatures (28–33°C) and early plant virus inoculations (two- to four-true-leaf stages) [138]. Recently, the position of Tsw has been more precisely assessed in a 295-kb candidate region on chromosome 10 in which NLR genes were clustered [139].
A new resistance inherited as a single dominant gene and indicated either to present a single allele at the Tsw locus or to be controlled by a different gene tightly linked to Tsw, was found in AC09-207, which is a C. chinense accession very similar to PI152225 [140].
Several other Capsicum species (C. frutescens, C. chacoense, C. pubescens, C. galapagoense, C. baccatum var pendulum and var baccatum), carrying the Tsw gene, are known as a good source of resistance too [141,142]. No extensive efforts have been instead carried out to discover the genetic basis of resistance for the other Orthotospovirus in pepper. Nowadays the Tsw gene is widely used in most commercial pepper hybrids as the unique source of TSWV resistance [143], however, its extensive adoption has triggered the rapid emergence of resistant-breaking (RB) isolates soon after their introduction. To date, reports on Tsw-resistance breakdown are from Italy, Spain, Australia, Hungary, Turkey, Argentina, and more recently in China and California [144,145,146].
Mixed infections with RB-TSWV and wild-type TSWV (WT-TSWV) isolates are very frequent in pepper cultivations. The co-infection by WT and RB isolates induces synergism effects with the appearance of necrosis on the apical leaves of TSWV-resistant genotypes [147].
For these reasons, the search for sources of resistance and/or tolerance (reduction of severity symptoms) to RB-TSWV strains in wild or exotic germplasm is essential to develop new varieties. Studies on Capsicum germplasm resistant to RB-TSWV are currently in progress in different countries [148,149,150]. A good level of tolerance to WT- and RB-TSWV isolates in the C. baccatum accession PIM26-1 [150].
Regarding CaCV, sources of resistance were found in C. chinense PI90972 [151]. A transcriptome analysis and expression profiling of CaCV evidenced about 2500 genes differentially expressed between susceptible and resistant genotypes with different functions (pathogenesis, cell death, and hormone-mediated signaling pathways and enzymes for defense-related pathways) [152]. Genes involved in localized cell death, cell signaling, synthesis of antimicrobial compounds and PR proteins were found highly upregulated. Moreover, two resistance NB-LRR candidates were putatively involved in a CaCV-resistant breeding line carrying introgressions from C. chinense.
4.2. Aphid-Transmitted Viruses
4.2.1. Potyviruses
Potyviruses likely represent the most spread viruses infecting peppers involving aphids as vectors of transmission [153] (Figure 3b). Seed transmission could occur, although, this has not been conclusively demonstrated in Capsicum [127,129]. Pepper can be infected by at least eleven different potyvirus species [127]. Among them, Potato virus Y (PVY) is worldwide distributed and is the only one severely affecting pepper crops in Europe [154]. PVY exists as three pathotypes (PVY-0, PVY-1, and PVY-1,2) according to the expressed virulence [153]. The other potyviruses infecting pepper have a narrowed geographical distribution. Therefore, many of them such as TEV, PepMoV, Pepper severe mosaic virus (PepSMV), Pepper yellow mosaic virus (PepYMV), Perù tomato mosaic virus (PTV), and the tentative species, Ecuadorian rocoto virus (EcRV) are distributed in South America and have been detected sporadically in other continents, while, Chilli veinal mottle virus (ChiVMV), Chilli ringspot virus (ChiRSV) and recently PepMoV are present in Asia, while Pepper veinal mottle virus (PVMV) is confined in Africa [127,154].
Several resistance genes to potyvirus are reported in pepper. The pvr1 locus, showing various alleles with different resistance levels to TEV (pvr1, pvr12), PVY-0 (pvr1, pvr11, pvr12) and PepMoV (pvr1), was identified in C. chinense PI159236 and PI152225 [155,156], while the pvr11 was detected in C. annuum cv. Avelar, Yolo Y, CM334, PI264281, and C. frutescens I5491 [141,156]. The pvr2 resistance alleles (pvr21, pvr22, pvr23) are effective against PVY-0, PVY-1, TEV (common strain), and are found in the C. annuum accessions Yolo Y, PI264281, SC46252, Florida VR2135. The allele pvr21 (Yolo Y) is effective only against PVY-0, while pvr22 (Florida VR2) is effective against PVY-0, PVY-0,1, and TEV. The allele pvr23 (Perennial) confers partial resistance to PVY. Mapping results showed that these genes were organized in a cluster of recessive genes on Chr 4.
Based on the co-segregation analysis, pvr2 was found to corresponds to the eukaryotic translation initiation factor 4E (eIF4E) [157]. The recessive resistance was probably related to the incompatibility between the potyvirus genome-linked protein (VPg) and eIF4E which occurred in resistant genotypes. Based on homology to eIF4E and allelism tests between pvr1 and pvr22 (both mapping in the same genetic locus of Chr 3) it has been suggested a nomenclature re-designation of pvr21 and pvr22 in pvr11 and pvr12, respectively [158]. Mutations in the eIF4E and eIF(iso)4E genes in pepper were identified through a cDNA eco-tilling platform within 233 cultivated accessions of Capsicum [159]. The authors reported five new eIF4E variants (named as pvr210, pvr211, pvr212, pvr213, and pvr214) related to PVY-resistance responses which represent an excellent allele reserve against the changing nature of viruses, to use in breeding programs.
The pvr2 alleles, pvr21 and pvr22, have been used extensively to breed potyvirus resistant pepper cultivars for more than 50 years. Both alleles confer efficient resistance toward PVY, while the only pvr22 is effective against TEV. The resistance of pvr22 proved extremely durable against PVY. To date, some pvr21 and pvr22-breaking isolates have been described [160]. However, they are not very prevalent so the cultivars carrying the pvr21 and pvr22 resistance continue to be used in breeding programs.
The pvr3 gene was reported in C. annuum cv. Avelar and confers resistance to PepMoV141. The pvr4 gene derived from C. annuum CM334 confers resistance to PVY-0, PVY-1,2, and PepMoV142. Other sources of this gene were found in C. chinense, C. frutescens, C. baccatum var. pendulum, C. praetermissum and C. galapagoense accessions using the CAPS marker named CSO [141,161].
The recessive loci pvr5 and pvr8 from C. annuum CM334 provide resistance to PVY-0 and PVY-1 isolate P-62-81, respectively [162].
Pepper plants expressing the pvr6 gene from C. annuum cv. Perennial, mapped on Ch 3, are resistant to ChiVMV145. The dominant gene Pvr7 from C. chinense PI159236 confers resistance to PepMoV Florida (V1182) strain and is tightly linked to Pvr4146. Pvr4 and Pvr7 are mapped on Chr 10 tightly linked to Tsw, which confers resistance to TSWV116. Therefore, this chromosome is considered a main cluster of dominant resistance genes in pepper. Venkatesh and collaborators [163], demonstrated that the dominant PepMoV resistance in C. annuum cv. 9093 could be derived from C. annuum CM334, and that Pvr4 and Pvr7 loci should be considered as the same locus.
Moreover, dominant allele Pvr4 confers a wide range of resistance against several potyviruses (PVY, PepMoV, PTV, PepSMV, and PepYMV) [154].
QTLs involved in the complete and partial resistance to some PVY isolates (To72 and Son41) were identified in eleven chromosomal regions, near pvr2 and pvr6 (Table 2) [164]. These QTLs reduce PVY symptom intensity and improve greatly the durability of the major-effect gene pvr23, which alone can be rapidly broken down [165]. Four additional major QTLs explaining over 70% of the variation with additive and epistatic interaction were identified [89]. The authors showed how the resistance breakdown frequency for pvr23 was under the control of three main QTLs, suggesting a pleiotropic effect on the durability of the major resistance gene.
Different markers have been developed for resistance-assisted breeding to potyviruses. A CAPS marker tightly linked to Pvr4 was developed by BSA-AFLP [161]. A SCAR marker (SCUBC191423) linked to Pvr4 was instead developed by BSA-RAPD [166]. Both markers were mapped on Chr 10 at distance variable from 5 to 10 cM and can be used for routine selection of PVY resistant lines.
Three allele-specific CAPS markers able to detect three recessive viral resistance alleles pvr1, pvr11, and pvr12 and a functional SNP marker at the pvr2-eIF4E locus, have been developed [156,167]. The use of the four primers in a single PCR experiment, allow differentiating alleles in homozygous and heterozygous genotypes. Through KASP-PCR, it was possible to develop a marker in the coding region for the cloned pvr1 resistance gene [35]. The KASP_pvr1 was validated using a C. chinense F2 population derived from Habanero (pvr1+/pvr1+) x PI159234 (pvr1/pvr1) [158]. The genetic factors underlying the number of PVY particles entering the plant and the accumulation at the systemic level have been studied using a genome-wide association study (GWAS) approach in a collection of ~260 C. annuum accessions [168]. Among the over 10 thousand SNPs identified through GBS, seven were highly associated with the resistance being located on chromosomes 4, 6, 9 and 12. Two of them on Chr 4 were closely linked to pvr2 in the region encoding the eIF4E, whereas, the SNPs detected on Chr 6 and 12 colocalized with previously reported QTLs.
Investigations toward the dissection of the genetic basis of ChiVMV has also been carried out reporting novel codominant markers for ChiVMV [169]. One CAPS marker tightly linked to the ChiVMV resistance locus and two high resolution melting (HRM) markers were developed through BSA-AFLP and mapped on Chr 6. Next-generation sequencing (NGS) has been also used to generate molecular markers tightly linked to Pvr4. Over 5000 single nucleotides variances in the NB-LRR gene regions were identified and converted into PCR-based markers [170]. More recently, the Cvr1 gene has been mapped to the short arm of Chr 6 of the resistant variety CV3 [171]. The region was reported to cluster several other NLR genes involved in resistance mechanisms. Furthermore, the authors identified SNP markers useful for assisted breeding of ChiVMV and for the fine mapping of resistance genes.
4.2.2. Cucumoviruses
Cucumber mosaic virus (CMV), is the main representative of Cucumovirus and is transmitted mainly by Myzus persicae and Aphis gossypii. CMV reduces quality and fruit yields (Figure 3c), especially in the early infections; yield losses greatly can reach 80% [172]. CMV can occur in nature in mixed infection with other viruses with synergistic effects, i.e., CMV and PepMoV [173]. Furthermore, the coinfection with CMV can reduce plant resistance against other viruses as in PepMoV and ChiVMV resistant pepper plants [174].
CMV isolates are classified in subgroups I (clade A and B) and II. Isolates of subgroup I, clade IA, and subgroup II are distributed worldwide while most of the isolates of clade IB are from East Asia. Pepper is more frequently affected by CMV isolates of subgroup I.
A single dominant resistance gene against CMV (Cmr1), identified from the C. annuum cv Bukang, was located in the centromeric region of pepper Chr 2. It inhibits the systemic movement of CMV isolates of subgroup IA [175]. A new isolate of CMV belonging to subgroup IB and designated as CMV-P1, has emerged in Korea and is able to break down the resistance conferred by Cmr1 [176]. Recently, a new single recessive gene, cmr2, able to confer resistance to CMV-P1 has been identified using a combining BSA and allelism tests [177]. BSA allowed detecting a single AFLP marker located at 16 cM from cmr2. The analysis has been corroborated by inheritance and allelism tests in segregating populations developed using as a source of resistance Lam32 (an Indian C. annuum cultivar carrying the cmr2 gene). This novel gene provides a broad spectrum of resistance to several CMV strains including the common CMVKorean and CMVFNY.
Almost all the CMV resistance sources identified in Capsicum spp. (Table 3) display a partial resistance controlled by multiple genes [177]. The resistance reported in C. annuum Perennial is due to various mechanisms [178]: partial resistance to initial virus infection [88], inhibition of virus multiplication [179], and inhibition of long-distance movement of the virus [180]. The resistance in C. frutescens BG2814-6 is instead expressed at the level of replication and cell-to-cell movement [181]. Several of these are ontogenetic depending on the pepper developmental stage [182]. These resistance mechanisms restrict only partially the virus translocation within plants but confer a good level of protection in the field, particularly when different sources were combined into a cultivar [179].

Table 3.
Sources of resistance/tolerance to virus diseases in Capsicum spp.
Three chromosomal regions on Chr 3, 11 and 12 with additive or epistatic effects involved in resistance to the CMV systemic movement and explaining 57% of the phenotypic variation (Table 2) were reported [164]. In addition, four QTLs significantly associated with resistance to CMV and a major QTL with digenic interaction on Chr 11 associated with genes conferring resistance to TMV were identified [90]. This QTL was confirmed by Caranta and colleagues [91], which reported the existence of four additive and two epistatic QTLs, as well as of a major QTL on Chr 12 (cmv 12.1) explaining between the 45% and 63.6% of the phenotypic variation [91]. Two major QTLs on Chr 5 and 11 explaining a total of 55% of the total phenotypic variation associated with the tolerance to CMVHB-jz strain were further identified [92].
Recently, NGS has been used to identify novel genomic regions underlying CMV resistance. By means of GBS, two novel major QTLs responsible for the resistance to CMV-P1 were identified [93]. The two QTLs were positioned on the Chr 5 (52.7–58.1 cM) and 10 (21.9-32.5 cM) and explained about 20% of the phenotypic variation, respectively. Using SLAF-seq a single gene located on Chr 2 (CA02g19570) was reported to be the candidate for the QTL qCmr2.1 conferring resistance to CMVFNY [94]. Furthermore, a major QTL on Chr 11 was identified. By means of the same genomic strategy, it was possible to detect three additional QTLs for resistance to the CMVHB-jz strain [95]. The major QTL, explaining about 20% of the phenotypic variation, was identified on Chr 11 confirming the importance of this chromosomal region for resistance to CMV. Besides the identification of QTLs, SLAF-seq has allowed the development of functional markers linked to CMV-resistant to be used for MAS in pepper.
4.3. Whitefly-Transmitted Viruses
Viruses belonging to the genera Begomovirus and Crinivirus are transmitted by different species of whiteflies, representing a danger for the cultivation of pepper in different World regions.
4.3.1. Begomoviruses
The genus Begomovirus contains viruses transmitted by the whitefly Bemisia tabaci persistently. At least 37 ratified and 6 candidate species have been described as naturally infecting pepper. Many of them cause serious diseases in pepper crops in Asia and America [127]. The diseases caused by Begomoviruses are easily recognized by their distinctive symptoms ascribed to three types: a) vein yellowing; b) yellow mosaic and c) leaf curl.
Among Begomoviruses, Chilli leaf curl virus (ChiLCV) is one of the most destructive disease for chilli pepper. The virus is distributed in almost all equatorial regions of the World [183]. Pepper golden mosaic virus (PepGMV) (previously named Serrano golden mosaic begomovirus and Texas pepper begomovirus) and Pepper huastego yellow vein virus (PHYVV) represent a new threat for pepper production in Central America. Pepper leaf curl virus (PepLCV) has been reported in India, United States, Nigeria and several other countries such as Pakistan, Bangladesh, and Indonesia [184]. Tomato yellow leaf curl virus (TYLCV) is one of the most devastating plant viruses of tomato whereas in other crops such as cucurbits and peppers is asymptomatic [185]. The virus has been reported on pepper crops in some areas of the Mediterranean basin [186,187].
The begomovirus, Tomato leaf curl New Delhi virus (ToLCNDV), represents an important constraint to tomato production, in the Indian sub-continent. In recent years the virus has been rapidly spreading into several countries of the Mediterranean basin causing significant economic losses on cucurbit and tomato [188]. Recently, it has been recovered in Italy in pepper plants showing yellowing and leaf curling [189].
Synergistic interactions between different begomoviruses infecting pepper can cause the breakdown of natural resistance in the host plant [190].
Despite the increasingly devastating effect of begomoviruses of pepper in many areas of Asia, Central America, and West Africa, breeding programs have not yet produced resistant commercial varieties due to the genetic nature of resistance, which is governed by major recessive genes [191]. The extent in the identification of resistant germplasm and of markers linked to minor genes were done for the ChiLCV-VNS (Varanasi isolate) strain [191].
With respect to PepLCV, an inheritance study of resistance using the partially compatible interspecific cross (PBC-535 X Bhut Jolokia), revealed the monogenic recessive nature [192]. Transcriptomic analysis evidenced 234 unique genes up-regulated in resistant genotype BS-35 respect the susceptible IVPBC535 indicating that gene expression in the resistant genotype responded strongly to PepLCV [193].
Recently, the analysis of 100 Capsicum spp. accessions in two locations of Thailand, allowed to identify the accession PP99 as the main source of resistance [194]. The other four genotypes (PP1037-7644-1, PBC148, PBC149, PBC502, PBC518, and PBC601) were classified as highly resistant at both locations. In any case, no accession was identified as being immune to the disease.
There are several reports of resistance sources to PHYVV in Capsicum. Trujillo-Aguirre and Díaz-Plaza [195], found genetic resistance to PHYVV and PepGMV in wild populations of C. chinense from Southeast Mexico. Hernández-Verdugo and colleagues [196], found genetic resistance to PHYVV in wild populations of Capsicum from Northwest Mexico. More recently, Retes-Manjarrez and collaborators [197], reported the UAS12 line (C. annuum) as the most promising genetic resource for its high resistance conferred by at least two genes.
Resistance to PepGMV in BG-3821 accession (C. chinense) is probably controlled by two genes with either additive or duplicate recessive epistatic action [198]. Moreover, the author indicated that the resistance is associated with reduced virus replication and movement, and the induction of genes associated with systemic acquired resistance (SAR).
4.3.2. Crinivirus
Tomato chlorosis virus (ToCV) is emerging as a problem worldwide resulting in severe damage, especially to tomato crops [199]. This virus is transmitted in a semipersistent manner by the whitefly species Bemisia tabaci, Trialeurodes abutiloneus and T. vaporariorum [200]. Although tomato is the main crop affected by this crinivirus, the virus has been also reported on sweet pepper plants in greenhouses of southern Spain, Brazil, Costa Rica, Tunisia, and Saudi Arabia [200]. Stunting accompanied by curling, interveinal yellowing and abnormal elongation of leaves, reduced fruit number and size are characteristic of ToCV infections in pepper. No information on sources of resistance to ToCV has been reported in Capsicum germplasm, to date.
4.4. Viruses Transmitted by Contact
Tobamoviruses
Tobamoviruses are mechanically transmitted and represent the most damaging viruses for pepper in protected cultivations [127]. The most prevalent in pepper are Tobacco mosaic virus (TMV), Tomato mosaic virus (ToMV) (Figure 3d), Bell pepper mottle virus (BPeMV), Pepper mild mottle virus (PMMoV), Paprika mild mottle virus (PaMMV), Obuda pepper virus (OBPV), Tobacco mild green mosaic virus (TMGMV)109. These viruses are particularly stable and for this reason, they remain infectious in contaminated plant residues, compost, soil, and irrigation water. They are easily transmitted by contact and seeds. Seeds can be externally or more rarely internally (endosperm) infected [211]. Tobamoviruses infecting Capsicum plants are classified into four pathotypes, P0 (TMV and ToMV), P1 (PaMMV), P1.2 and P1.2.3 (PMMoV), based on the reaction of pepper cultivars carrying different L resistance genes (L1, L2, L3, and L4) [212]. L1 confers resistance to P0 strains; L2 confers resistance to P0 and P1, L3 confers resistance to P0, P1 and P1,2, L4 confers resistance to all strains (P0, P1, P1,2 and P1,2,3) [213]. Studies have identified the viral coat proteins (CPs) as elicitors of L genes-mediated resistance [214,215] and amino acid changes responsible for overcoming L3 and L4-gene-mediated resistance in the CP [215,216,217]. The L locus was mapped to the sub-telomeric region of pepper Chr 11, 4.0 cM apart from the RFLP marker TG36 [218]. This region was syntenic to the tomato Chr 11 which carries the I2 resistance genes for F. oxysporum [219]. L1 was mapped in C. annuum to Chr 11 through an integrated molecular linkage map of cultivated pepper (C. annuum) obtained from the alignment of three DH (double haploids) maps [218]. L4 from C. chacoense was mapped by Matsunaga and collaborators [220] and confers resistance to the most aggressive and common tobamovirus pathotypes P1.2.3 [221]. Good sources of resistance to pathotypes P1.2.3 were recently found in several accession of C. baccatum var. pendulum and in germplasm belonging to C. pubescens, C. frutescens, C. chinense and C. praetermissum using the dominant marker 060I2END linked to the L4 locus [141].
In addition to these classical L genes, another Tobamovirus resistance gene, L1a, has been identified [210]. The authors demonstrated that in contrast to L1, the gene L1a mediates resistance to P0 pathotype (TMV and ToMV), independently by the temperature, and to P1 (PaMMV) at 24 °C. A year later, the same research group, identified a single incompletely dominant gene different from the L gene designated as Hk, which confers resistance to P1 pathotype (PaMMV) at 30 °C but not at 24 °C. The source of resistance C. annuum cv Nanbu–Ohnaga, although resistant to PaMMV was ineffective against any of the other Tobamovirus pathotypes (TMV P0 and P1, and PaMMV P1,2) [210]. The P1.2.3.4 pathotype of PMMoV, which differs from P1,2 for two amino acids in the coat protein, can break the L4 resistance, indicating the need to identify R genes effective against this virus strain [217]. Efforts to develop molecular markers linked to L genes are reported, such as the SCAR marker WA31-1500S linked 1.5 cM to L4 and able to distinguish resistant from susceptible accessions [220].
The L3 resistance gene of C. chinense was positioned in a 400-kb region of pepper Chr 11 containing clusters of R-like genes and highly repetitive sequences, confirming, the presence of many repetitive sequences of the L locus [222,223]. Several tightly linked markers, including the 189D23M located within 0.1 cM of the L3 gene, were identified. However, inconsistencies in the genetic distances of these markers from the L3 locus [222], suggested linkage disequilibrium in the underlying region containing the L3 locus. Via comparative analysis, Yang and collaborators [221], developed L-linked markers using the BAC sequence information corresponding to the syntenic tomato I2 (conferring resistance to F. oxysporum f. sp. lycopersici) and potato R3 (conferring resistance to P. infestans) loci, three of which (087H3T7, 060I2END and 158K24) were found to be in linkage to the L3 and L4 loci. Further mapping analysis demonstrated a different linkage of the previously identified 189D23M to L4 respect L3, suggesting the possible existence of different genes closely linked instead that different alleles at the same locus. Three years later, the same research group [224], developed a marker (L4segF&R) located within 0.3 cM from L4 using diverse segregating populations and breeding lines. Given its not complete co-segregation with the L4 gene, the marker is considered as a candidate of resistance not- L4 related. Furthermore, several allele-specific markers for the L locus were developed using the LRR-encoding domain of the NBS-LRR disease resistance gene candidate for the different L alleles.
Functional studies reported different transcription factors involved in the infection of Tobamovirus. The CaWRKYb gene of the WRKY family was reported to be rapidly induced during TMV (pathotype P0) infection in hot pepper [225]. A CaWRKYb-knockdown evidenced a reduced resistance level in plants as a result of minor hypersensitive response upon TMV-P0 infection. The compromised resistance to TMV-P0 was due to major TMV accumulation through decreased expression of pathogenesis-related genes of C. annuum (CaPR-1, CaPR-5 and CaPR-10). The results suggested that CaWRKYb plays as a positive role in defense-related signal transduction pathways in hot pepper [225]. The gene CaWRKYd, isolated from microarray analysis in TMV-P0-inoculated hot pepper (C. annuum) plants is a new transcription factor that belongs with a subgroup (IIa) of the WRKY family [226]. CaWRKYd transcripts were reported to be induced by P0 inoculation and hormone treatments [226]. The silencing of this gene affected TMV-P0-mediated HR cell death and the accumulation of TMV-P0 coat protein in local and systemic leaves. Moreover, a reduction of expression of some pathogenesis-related (PR) and HR (hypersensitivity response)-related genes was evidenced after silencing, confirming that this gene modulates HR cell death by regulating downstream gene expression. The same year, Huh and collaborators [227], analyzed the function of C. annuum basic transcription factor 3 (CaBtf3) of the NAC family through VIGS and found its involvement in HR cell death related to TMV-P0 infection.
4.5. Pollen Transmitted Viruses
Ilarviruses
Ilarviruses are transmitted mechanically by thrips feeding on pollen grains containing the virus or by carrying pollen grains contaminated by the virus. Tobacco streak virus (TSV), is the main species including a wide host range, with at least 200 susceptible species. TSV was reported causing systemic necrosis, dark streaks on stems and petioles and tip necrosis on pepper in Argentina and in India [228,229].
Parietaria mottle virus (PMoV), was identified on bell pepper in Southeast Spain [230], and on pepper ecotypes and commercial hybrids in Southern Italy [231]. Infected plants showed rings, mosaic and necrotic patches of the leaves, necrotic stems, and brown patches and corky rings on fruits [231]. No source of genetic resistance has been investigated in Capsicum spp., to date.
5. Arthropods and Nematode Pests
In plants, insects and arthropods exert their activity destroying tissues, causing energy stresses and competing for nutrients. Furthermore, insects are key vectors of several pathogens. In pepper, more than 21 insect and non-insect pests cause heavy yield losses worldwide [232]. A strategy to reduce pest damages and minimize the use of insecticide applications is the adoption of pest-resistant genotypes. Unfortunately, studies on plant genotypic variation in resistance to arthropods and pests in the genus Capsicum are still scarce to date and resistant commercial varieties (or rootstocks) are available only for root–knot nematodes.
5.1. Thrips
Thrips (Thysanoptera: Thripidae) cause damages directly by feeding on leaves, fruits or flowers, and indirectly by transferring viruses, especially TSWV in pepper worldwide. There are at least 16 species of thrips that attack Capsicum [233]. Among them, F. occidentalis (Figure 4a), is the major species found on pepper in Europe [234], and in Asia as well [235]. Several pepper accessions have been found to carry resistance to thrips which may be exploited further to breed resistant varieties increasing the effectiveness of thrips control and delay or reduce the transmission of viruses [236,237]. Six C. annuum and C. baccatum accessions (Table 4) were identified as good sources for resistance against Thrips parvispinus and F. occidentalis [96]. These studies also confirmed the good level of resistance of two accessions: Keystone Resistant Giant and CPRO-1 [237,238]. The latter showed a reducing of thrips reproduction. Moreover, the leaf-based resistance to F. occidentalis and T. tabaci have been demonstrated species-specific, being not correlated [239].

Figure 4.
Arthropods and nematodes pests: (a) stripes on fruits caused by thrips feeding (Frankliniella occidentalis); (b) adult stages of whiteflies (Bemisia Tabaci) on the underside of leaves; (c) damages on fruit caused by of cotton bollworm larvae (Helicoverpa armigera); (d) distorted leaves and damages on inflorescences caused by broad mite feeding (Polyphagotarsonemus latus); (e) galls or “knots” on pepper roots caused by nematode (Meloydogyne spp.) feeding.

Table 4.
Sources of resistance to arthropod and nematode pests in Capsicum spp.
An attempt to identify chromosomal regions responsible for resistance has been reported [96]. The authors developed a genetic map in an F2 population derived from the cross between C. annuum AC 1979 (female parent, susceptible) x C. chinense 4661 (male parent, resistant). A single QTL explaining about 50% of the genetic variation was detected for three traits of resistance (damage caused by larvae and the survival of first and second instar larval stages), all co-localized near the same marker on Chr 6. Resistance parameters and trichomes density were not correlated suggesting that the latter don’t exert major effects on resistance mechanisms to thrips.
5.2. Tobacco Whitefly
Bemisia tabaci (Hemiptera: Aleyrodidae) (Figure 4b) has become a serious threat to crop production not only by causing direct feeding damages but also being a vector capable of transmitting efficiently more than 200 plant viruses, 90% of them Begomoviruses [240]. The frequent use of pesticides leads to resistant whiteflies so that the use of resistant varieties, biological control or a combination of them is strongly recommended [241]. Sources of resistance were found surveying 44 Capsicum accessions under both screen-house (Wageningen, Holland) and in-field test conditions (tropical area in Indonesia) [242] (Table 4). A strong antixenotic and antibiosis effect against B. tabaci was found in P2, P4, ACC1 and ACC12 accessions of spp. (Table 3) [243]. An antibiosis mechanism was also suggested for the whitefly-resistant accessions IAC-1544 (C. frutescens), IAC-1545 (C. chinense), 1579 (C. annuum) [244].
5.3. Aphids
The cotton aphid (CA), Aphis gossypii and the green peach aphid (GPA), Myzus persicae, (Hemiptera: Aphididae), are the main species [245]. As direct pest, GPA causes chlorosis, leaf defoliation, flower, and fruit abortion and reduces photosynthesys. Moreover, GPA represents an efficient vector for many pepper destroying viruses including PepSMV, PepMoV and PepYMV [127].
Only a few studies to identify sources of resistance to GPA in Capsicum spp. have been published. Bosland and Ellington [246], found one C. pubescens accession showing antixenosis rather than antibiosis resistance to the GPA. However, no information has so far been reported on the use of this germplasm in C. annuum breeding for aphid resistance.
Sun and colleagues [247], screened 74 pepper accessions, belonging to C. annuum, C. frutescens, C. chinense and C. baccatum, for resistance to GPA. The authors identified three C. baccatum accessions with high (PB2013071) or intermediate resistance (PB2013062 and PB2012022) and elucidated possible mechanisms of aphid resistance. The highly resistant genotype resulted in a severely reduced uptake of phloem, a significant callose deposition due to feeding of GPA, and in the accumulation of ROS (reactive oxygen species) [247].
Very recently, two major QTLs for resistance were detected and validated on pepper Chr 2 [248]. The analysis was carried out in an F2 population derived from the intraspecific cross between the highly resistant C. baccatum PB2013071 and the susceptible PB2013046. The identified QTLs Rmprp-1 and Rmpas-1 inhibited the reproduction and affected GPA survival, respectively. Moreover, Rmprp-1 was located in a genomic region of 96 kb which is predicted to encode four analogs of resistance genes of the receptor-like kinase family containing a leucine-rich repeat domain (LRR-RLKs). Regarding CA, sources of resistance were found in C. annuum germplasm by the choice and non-choice tests [249]. The resistant accession IPB C20 made the shortest longevity and reproduction time of melon aphid compared to the other genotypes tested. Moreover, the same genotype IPB C20 was able to suppress aphid progenies.
5.4. Lepidopterous and Leaf Miner Pests
Cotton bollworm (Helicoverpa armigera) (Lepidoptera: Noctuidae) is the main moth that causes pepper damage. In Europe, the pest is of economic importance in Portugal and Spain and of lesser importance in other countries where it is also established. In 2003, H. armigera was a serious problem on pepper crops in Southern Italy (Metaponto area). Thirty percent of the pepper fruits and 70%–80% of the pepper plants were damaged. The larvae fed on leaves, flowers and fruits, with fruits recording the most serious damages [250] (Figure 4c).
The analysis of thirty-three genotypes with different levels of damages caused by cotton bollworm under field conditions allowed to identify seven pepper genotypes (SL-37, Arka Lohith, Purired, Devarhippargi, TC-1, Button and H.C.-28) as resistant [251].
American serpentine leaf miner (Liriomyza trifolii) (Diptera: Agromyzidae) is a well-known pest with a broad host range among and leaf and fruit vegetable crops, attacking over 120 plant species. After hatching from the eggs, feed on the mesophyll tissues in the leaves and form serpentine mines, which can reduce significantly the photosynthetic activity of the plant. Resistance mechanisms against this pest were found in some inbred lines of C. chinense (G84, G110, and G37) [252]. Another source of resistance was detected in cv. Sakigake 2-go (C. annuum var. angulosum) [253].
5.5. Broad Mites
The broad mite, Polyphagotarsonemus latus (Acari: Tarsonemidae), is a polyphagous pest that attacks several important crops worldwide. This pest damages the outer cells of leaves as they feed on the plant sap. Leaves become distorted, bronze-colored, stiff, and rolled; flowers become distorted and fail to open normally; fruits are distorted and loss of yield is observed (Figure 4d). In extreme cases, plants are killed by the infestation. Resistant genotypes (Jwala, RHRC, Errect, and AGC-77) to P. latus were found [257] (Table 4); moreover, sources of resistance (Pant C-1; LCA-304 and LCA-312) both to P. latus and thrips Scirtothrips dorsalis, were identified [255]. More recently, Latha and Hunumanthraya [256], screening thirty-one chilli genotypes for thrips (S. dorsalis) and mite resistance under field condition, identified four Capsicum spp. accessions (DCC-3, DCC-185, DCC-109, and DCC-89) as moderately resistant to both pests. The authors highlighted that some morphological and biochemical characters (trichome density, chlorophyll, and phenol content) were negatively correlated with the population of thrips, mites and Leaf Curl Index.
5.6. Root-knot Nematodes
Root-knot nematodes (RKN) (Nematoda: Meloidogyne) belong to the genus Meloidogyne which includes 90 species, the most important of which in terms of damages and diffusion are M. incognita, M. arenaria and M. javanica (Figure 4e). Nematodes disease occurs in both open field and greenhouses, and prediction of the crop losses that a certain population density of nematode may cause is of importance to decide whether to cultivate pepper or not [272]. Another species that has gained importance recently is M. enterolobii for which, the sources of resistance against the major species of Meloidogyne are ineffective on its control [273]. Two genotypes, named UFGFR 05 (C. frutescens) and UFGCH 24 (C. chinense) are recently identified as resistant to M. enterolobii [269].
Resistance mechanisms to M. arenaria (races 1 and 2), M. incognita and M. javanica identified in C. annuum, C. chinense, C. chacoense and C. frutescens are conditioned by a single dominant gene designated N gene [2]. In C. annuum, resistance to RKN is also associated with several dominant genes (Me genes) that act independently in gene-for-gene interactions [262,268]. Six Me genes have previously been shown to be stable at high temperatures in three highly resistant and genetically distant accessions, PI322719, PI201234, and CM334. Some genes (Me4, Mech1 and Mech2) are specific to certain Meloidogyne species or populations, whereas others (Me1, Me3, and Me7) are effective against a wide range of species, including M. arenaria, M. javanica, and M. incognita.
However, the high genomic plasticity and genetic diversity exhibited by RKNs confer them a high potential to adapt to the host and an ability to develop virulent populations that break down the pepper plant resistance [274,275]. Nonetheless, a fitness cost associated with virulence has been observed and the joint management of diversified resistance sources together with adapted cultivation practices may well provide effective and sustainable control [276]. Particularly, two major R genes that differ in their mechanisms (Me1 and Me3) into a single cultivar, seems the most secure and durable strategy after three years of experimentation [277].
Me3 and Me4 were found to be linked 10 cM each through BSA in a segregant population derived from the cross-Yolo Wonder (susceptible) X PM687 (resistant) [277]. These genes, along with Mech1, Mech2, Me1 and Me7 made the main cluster of 28 cM on Chr 9 [262]. Comparative mapping evidenced a colinearity with Chr 12 in both tomato and potato demonstrating the existence of orthologous regions for nematode resistance in Solanaceae. Crossing lines homozygous for N (Carolina Wonder and Charleston Belle) to lines homozygous for Me3 (HDA 149 and PM 687) and employing allelism test, showed that the two genes were distinct [278]. A subsequent study found co-localization of N-gene in the Me genes cluster on the Chr 9, reporting the N gene allelic to Me7 and located 7 cM apart from Me1, and 2 cM from Me3 [261].
A genetic mapping study using F2:3 families derived from the cross Yolo Wonder × Doux Longd es Landes, allowed to identify a cluster on Chr 1 including three tightly linked QTLs with broad mechanisms of resistance against M. incognita, M. arenaria, M. javanica, respectively. A fourth QTL, providing specific resistance to M. javanica was mapped on Chr 9 [97].
Although several genes against root-knot have been identified, none of them has been cloned. Chen and collaborators [263], reported the first cloning study of CaMi, a candidate root-knot nematode resistance gene isolated from the resistant pepper line PR205. Transgenic tomato plants carrying the full coding genomic region of CaMi evidenced improved resistance against the root-knot nematodes compared to untransformed susceptible plants although not heritable. CaMi gene exerted a hypersensitive response (HR) as well as many necrotic cells around nematodes. Mao and colleagues [279], isolated and cloned from the resistant line HDA149, CaRKNR, an NBS-LRR gene showing homology to the tomato root–knot resistant gene Mi-1.2. The gene was mapped on Chr 6 and did not belong to Me family genes. In cloned plants, the expression level of CaRKNR increased up to four times while the silencing of the gene in HDA149 reduced the resistance to nematodes.
Different studies targeted at the development of markers linked to nematode-resistant gene for assisted breeding have been performed. Fazari and colleagues [261], developed PCR based markers tightly linked to Me1, Me3, Me7 and N genes. A codominant CAPS marker located 1.13 cM away from the Me1 gene, and a set of microsatellites tightly linked 0.8 cM away from the N gene have been also reported [280,281]. Finally, Wang and collaborators [282], fine mapped the region surrounding Me1, developing different PCR based markers closely linked. All these markers are useful for the marker-assisted breeding of nematode resistance in pepper.
6. Impact of Genomics and Future Challenges in Plant Disease Research
In recent years, a rapid increase in genomics has enabled the implementation of novel approaches toward the understanding of the molecular mechanisms underpinning resistance to pathogens. Next-generation sequencing technologies can be applied to provide whole-genome sequencing of pathogens and to develop high-throughput molecular markers for QTL mapping and gene discovery. Breeding programs are benefiting from these signs of progress in terms of precision and speediness to achieve results. Indeed, conventional molecular approaches are laborious and time-consuming. The QTL studies performed up to the early 2000s led to the development of genetic maps consisting of a few hundred markers to use for gene discovery and markers assisted selection. QTLs were often positioned in large intervals with difficulty in transferring them due to linkage drag. Moreover, markers were not always reliable due to recombination mechanisms leading to linkage breaking. As a result, many available molecular markers are not applicable in breeding for resistance. NGS-based genotyping produces instead thousands of single nucleotide polymorphism which allow detecting loci involved in the resistance to pathogens narrowing down the regions underlying genes of interest. Reduced representation sequencing method for genotyping such GBS (genotyping by sequencing) or RADseq (restriction site-associated DNA sequencing) are paramount, allowing high throughput genome scans at relatively low cost [93,168,283]. These NGS technologies, therefore, can be used to generate diagnostic markers able to detect the allelic variation within resistance genes. In addition to genomic-based breeding, NGS can be applied to unravel the diversity of the genome sequences of pathogen strains to identify specific virulence genes. Moreover, it can be applied to generate a large dataset of sequenced transcriptomes associated with pathogen virulence or to investigate the expression of effector proteins during the early stages of infection [284]. The future of NGS is shifting toward whole-genome sequencing, allowing to resolve key questions related to the function of virulence genes, the mechanism of resistance and the evolution of pathogens. The major accessibility to platforms as well the easiest analysis and management of data make the use of these technologies affordable to pathologists, geneticists, and breeders, covering different branches of research with the final target of better management and control of diseases.
Author Contributions
M.P., D.A., P.T. conceptualized the work and jointly wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by EU-Horizon 2020 research and innovation program G2P-SOL project under grant agreement number. 677379.
Acknowledgments
We are thankful to Jundae Lee (Chonbuk National University), Bruno Parisi (CREA Research Centre for Cereal and Industrial Crops), Catello Pane (CREA Research Centre for Vegetable and Ornamental Crops), Antonio Ragozzino (University of Naples) and Astolfo Zoina (University of Naples) for providing valuable phots of fungal bacterial, and viral diseases.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| QTL | Quantitative Trait Locus |
| NBS-LRR | Nucleotide-binding site leucine-rich repeat |
| SCAR | Sequence Characterized Amplified Region |
| SNP | Single Nucleotide Polymorphysms |
| MAS | Marker-assisted selection |
| GBS | Genotyping by sequencing |
| RAPD | Random amplification of Polymorphic DNA |
| RFLP | Restriction Fragment Length Polymorphism |
| cM | Centimorgans |
| RIL | Recombinant inbred lines |
| AFLP | Amplified Fragment Length Polymorphism |
| BSA | Bulked Segregant Analysis |
| Xs. | Xanthomonas |
| KASP | Kompetitive Allele-Specific PCR |
| BW | Bacterial wilt |
| TSWV | Tomato spotted wilt orthotospovirus |
| CaCV | Capsicum chlorosis orthotospovirus |
| PVY | Potyviruses |
| TEV | Tobacco etch virus |
| PepMoV | Pepper mottle virus |
| TMV | Tobacco mosaic virus |
| ToMV | Tomato mosaic virus |
| CMV | Cucumber mosaic virus |
References
- Faostat 2018. Available online: http://www.fao.org/ (accessed on 12 March 2020).
- Sarath Babu, B.; Pandravada, S.R.; Pasada Rao, R.D.V.J.; Anitha, K.; Chakrabarty, S.K.; Varaprasad, K.S. Global sources of pepper genetic resources against arthropods, nematodes and pathogens. Crop Prot. 2011, 30, 389–400. [Google Scholar] [CrossRef]
- Pitrat, M. Vegetable crops in the Mediterranean Basin with an overview of virus resistance. Adv. Virus Res. 2012, 84, 1–29. [Google Scholar]
- Djian-Caporalino, C.; Palloix, A.; Fazari, A.; Marteu, N.; Barbary, A.; Abad, P.; Sage-Palloix, A.M.; Mateille, T.; Risso, S.; Lanza, R.; et al. Pyramiding, alternating or mixing: Comparative performances of deployment strategies of nematode resistance genes to promote plant resistance efficiency and durability. BMC Plant Biol. 2014, 14, 53. [Google Scholar] [CrossRef]
- Ros, C.; Lacasa, C.M.; Martínez, V.; Bielza, P.; Lacasa, A. Response of pepper rootstocks to co-infection of Meloidogyne incognita and Phytophthora spp. Eur. J. Hortic. Sci. 2014, 79, 22–28. [Google Scholar]
- Özkaynak, E.; Devran, Z.; Kahveci, E.; Doğanlar, S.; Başköylü, B.; Doğan, F.; Yüksel, M. Pyramiding multiple genes for resistance to PVY, TSWV and PMMoV in pepper using molecular markers. Eur. J. Hortic. Sci. 2014, 79, 233–239. [Google Scholar]
- Tan, M.Y.; Hutten, R.C.; Visser, R.G.; van Eck, H.J. The effect of pyramiding Phytophthora infestans resistance genes R Pi-mcd1 and R Pi-ber in potato. Theor. Appl. Genet. 2010, 121, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Pilet-Nayel, M.L.; Moury, B.; Caffier, V.; Montarry, J.; Kerlan, M.C.; Fournet, S.; Durel, C.E.; Delourme, R. Quantitative Resistance to Plant Pathogens in Pyramiding Strategies for Durable Crop Protection. Front. Plant Sci. 2017, 8, 1838. [Google Scholar] [CrossRef] [PubMed]
- Chile Database. Resistance Sources from Chile Variety Database. 2018. Available online: www.g6csy.net/chile/database.html (accessed on 12 March 2020).
- Peppers. Resistance Sources From USDA-ARS Database. 2018. Available online: http://www.ars-grin.gov/npgs/acc/ (accessed on 12 March 2020).
- AVGRIS. The Asian Vegetable Center (ARVDC) (Taiwan). 2018. Available online: http://www.avrdc.org (accessed on 12 March 2020).
- CGN. The Centre for Genetic Resources, the Netherlands (CGN) of Wageningen University. 2018. Available online: https://www.wur.nl/en/Research-Results/Statutory-research-tasks/Centre-for-Genetic-Resources-the-Netherlands-1/Expertise-areas/Plant-Genetic-Resources.htm (accessed on 12 March 2020).
- PGR Portal. National Bureau of Plant India. 2018. Available online: www.nbpgr.ernet.in/pgrportal (accessed on 12 March 2020).
- Anand, N.; Deshpande, A.A.; Sridhar, T.S. Resistance to powdery mildew in an accession of Capsicum frutescens and its inheritance pattern. Capsicum Eggplant Newsl 1987, 6, 77–78. [Google Scholar]
- De Souza, V.L.; Café-Filho, A.C. Resistance to Leveillula taurica in the genus Capsicum. Plant Pathol. 2003, 52, 613–619. [Google Scholar] [CrossRef]
- Lee, O.H.; Hwang, H.S.; Kim, J.Y.; Han, J.H.; Yoo, Y.S.; Kim, B.S. A search for sources of resistance to powdery mildew (Leveillula taurica (Lev.) Arn.) in pepper (Capsicum spp.). Korean J. Hort. Sci. Tech. 2001, 19, 7e11. [Google Scholar]
- Daubèze, A.M.; Hennart, J.W.; Palloix, A. Resistance to Leveillula taurica in pepper (Capsicum annuum) is oligogenically controlled and stable in Mediterranean regions. Plant Breed. 1995, 114, 327–332. [Google Scholar] [CrossRef]
- Lefebvre, V.; Daubèze, A.M.; Rouppe van der Voort, J.; Peleman, J.; Bardin, M.; Palloix, A. QTLs for resistance to powdery mildew in pepper under natural and artificial infections. Theor. Appl. Genet. 2003, 107, 661–666. [Google Scholar] [CrossRef] [PubMed]
- Jo, J.; Venkatesh, J.; Han, K.; Lee, H.Y.; Choi, G.J.; Lee, H.J.; Choi, D.; Kang, B.C. Molecular mapping of PMR1, a novel locus conferring resistance to powdery mildew in pepper (Capsicum annuum). Front. Plant Sci. 2017, 8, 2090. [Google Scholar] [CrossRef] [PubMed]
- Gabor, B.K.; Just, B.J.; Huang, C.; Jones, C.M.; Vreugdenhil, D.; Kniskern, J.M.; Quijada, P.A.; Berke, T.G.; Allersma, A.P.; Xiang, W. Methods and Compositions for Producing Capsicum Plants with Powdery Mildew Resistance. U.S. Patent No 9,689,045., 2017. [Google Scholar]
- Black, L.L. Studies on Phytophthora blight in pepper. In AVRDC Report 1998; Talekar, N.S., Ed.; Asian Vegetable Research and Development Center: Shanhua, Taiwan, 1999; pp. 25–27. [Google Scholar]
- McGregor, C.; Waters, V.; Nambeesan, S.; MacLean, D.; Candole, B.L.; Conner, P. Genotypic and phenotypic variation among pepper accessions resistant to Phytophthora capsici. Hortsci. 2011, 46, 1235–1240. [Google Scholar] [CrossRef]
- Guerrero-Moreno, A.; Laborde, J.A. Current status of pepper breeding for resistance to Phytophthora capsici in Mexico. In Proceedings of the 4th Eucarpia Meeting on Genetics and Breeding on Capsicum & Eggplant, Wageningen (Países Bajos), The Netherlands, 14–16 October 2004; pp. 52–56. [Google Scholar]
- Smith, P.G.; Kinble, K.A.; Grogan, R.G.; Millett, A.H. Inheritance of resistance in pepper to Phytophthora root rot. Phytopathology 1967, 57, 377–379. [Google Scholar]
- Gurung, S.; Short, D.P.G.; Hu, X.; Sandoya, G.V.; Hayes, R.J.; Subbarao, K.V. Screening of wild and cultivated Capsicum germplasm reveals new sources of Verticillium wilt resistance. Plant Dis. 2015, 10, 1404–1409. [Google Scholar] [CrossRef]
- Anaya-López, J.L.; González-Chavira, M.M.; Villordo-Pineda, E.; Rodríguez-Guerra, R.; Rodríguez-Martínez, R.; Guevara-González, R.G.; Guevara-Olvera, L.; Montero-Tavera, V.; Torres-Pacheco, I. Selection of chili pepper genotypes resistant to pathogenic wilt disease complex. Rev. Mex. Cienc. Agric. 2011, 2, 373–383. [Google Scholar]
- Montri, P.; Taylor, P.W.J.; Mongkolporn, O. Pathotypes of Colletotrichum capsici, the causal agent of chili anthracnose, in Thailand. Plant Dis 2009, 93, 17–20. [Google Scholar] [CrossRef]
- Mongkolporn, O.; Montri, P.; Supakaew, T.T.; Taylor, P.W.J. Differential reactions on mature green and ripe chili fruit infected by three Colletotrichum species. Plant Dis. 2010, 94, 306–310. [Google Scholar] [CrossRef]
- Voorrips, R.E.; Finkers, R.; Sanjaya, L.; Groenwold, R. QTL mapping of anthracnose (Colletotrichum spp.) resistance in a cross between Capsicum annuum and C. chinense. Theor. Appl. Genet. 2004, 109, 1275–1282. [Google Scholar] [CrossRef]
- Pakdeevaraporn, P.; Wasee, S.; Taylor, P.W.J.; Mongkolporn, O. Inheritance of resistance to anthracnose caused by Colletotrichum capsici in Capsicum. Plant Breed. 2005, 124, 206–208. [Google Scholar] [CrossRef]
- Silva, S.A.M.; Rodrigues, R.; Goncalves, L.S.A.; Sudre, C.P.; Bento, C.S.; Carmo, M.G.F.; Medeiros, A.M. Resistance in Capsicum spp. to anthracnose affected by different stages of fruit development during pre- and postharvest. Trop. Plant Pathol. 2014, 39, 335–341. [Google Scholar] [CrossRef]
- Mishra, R.; Rout, E.; Kumar Joshi, R. Identification of resistant sources against anthracnose disease caused by Colletotrichum truncatum and Colletotrichum gloeosporioides in Capsicum annuum L. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2018. [Google Scholar] [CrossRef]
- Maruti, T.B.; Tembhurne, B.V.; Chavan, R.L.; Amaresh, Y.S. Reaction of chilli (Capsicum annuum L.) genotypes and hybrids against Fusarium wilt (Fusarium solani). J. Spices Aromat. Crops 2014, 23, 186–191. [Google Scholar]
- Singh, A.; Singh, A.K.; Singh, A. Screening of chilli germplasms against Fusarium wilt. Crop Res. 1998, 15, 132–133. [Google Scholar]
- Holdsworth, W.L.; Mazourek, M. Development of user-friendly markers for the pvr1 and Bs3 disease resistance genes in pepper. Mol. Breed. 2015, 35, 28. [Google Scholar] [CrossRef]
- Kim, D.S.; Hwang, B.K. The pepper MLO gene, CaMLO2, is involved in the susceptibility cell death response and bacterial and oomycete proliferation. PlantJ. 2012, 72, 843–855. [Google Scholar]
- Muhyi, R.; Bosland, P.W. Evaluation of Capsicum germplasm for sources of resistance to Rhizoctonia solani. HortScience 1995, 30, 341–342. [Google Scholar] [CrossRef]
- Jones, J.B.; Lacy, G.H.; Bouzar, H.; Stall, R.E.; Schaad, N.W. Reclassification of the xanthomonads associated with bacterial spot disease of tomato and pepper. Syst. Appl. Microbiol. 2004, 27, 755–762. [Google Scholar] [CrossRef]
- Wai, K.P.; Siddique, M.I.; Mo, H.S.; Yoo, H.J.; Byeon, S.E.; Jegal, Y.; Mekuriaw, A.A.; Kim, B.S. Pathotypes of bacterial spot pathogen infecting Capsicum peppers in Korea. Plant Pathol. J. 2015, 31, 428–432. [Google Scholar] [CrossRef]
- Csillery, G.; Szarka, E.; Sardi, E.; Mityko, J.; Kapitany, J.; Nagy, B.; Szarka, J. The unity of plant defense: Genetics, breeding and physiology. In Proceedings of the 12th Eucarpia Meeting on Genetics and Breeding of Capsicum and Egg-Plant, Noordwijkerhout, The Netherlands, 17–19 May 2004; pp. 147–153. [Google Scholar]
- Cook, A.A.; Stall, R.E. Inheritance of resistance in pepper to bacterial spot. Phytopathology 1963, 53, 1060–1062. [Google Scholar]
- Cook, A.A.; Guevara, Y.G. Hypersensitivity in Capsicum chacoense to race 1 of pepper. Plant Dis. 1984, 68, 329–330. [Google Scholar] [CrossRef]
- Kim, B.S.; Hartmann, R.W. Inheritance of a gene (Bs3) conferring hypersensitive resistance to Xanthomonas campestris pv. vesicatoria in pepper (Capsicum annuum). Plant Dis. 1985, 69, 233–235. [Google Scholar]
- Hibberd, A.M.; Bassett, M.J.; Stall, R.E. Allelism tests of three dominant genes for hypersensitive resistance to bacterial spot of pepper. Phytopathology 1987, 77, 1304–1307. [Google Scholar] [CrossRef]
- Lafortune, D.; Béramis, M.; Daubèze, A.M.; Boissot, N.; Palloix, A. Partial resistance of pepper to bacterial wilt is oligogenic and stable under tropical conditions. Plant Dis. 2005, 89, 501–506. [Google Scholar] [CrossRef]
- Kang, Y.J.; Ahn, Y.K.; Kim, T.K.; Jun, T.H. Resequencing of Capsicum annuum parental lines (YCM334 and Taean) for the genetic analysis of bacterial wilt resistance. BMC Plant Biol. 2016, 16, 235. [Google Scholar] [CrossRef]
- Lebeau, A.; Daunay, M.C.; Frary, A.; Palloix, A.; Wang, J.F.; Dintinger, J.; Chiroleu, F.; Wicker, E.; Prior, P. Bacterial wilt resistance in tomato, pepper, and eggplant: Genetic resources respond to diverse strains in the Ralstonia solanacearum species complex. Phytopathology 2011, 101, 154–165. [Google Scholar] [CrossRef]
- Matos, F.S.A.; Lopes, C.A.; Takatsu, A. Identification of sources of resistance to Pseudomonas solanacearum in Capsicum spp. Hort. Bras. 1990, 8, 22–23. [Google Scholar]
- Eggink, P.M.; D’hoop, B.B.; Brouwer, M.; Deniau, A.X. Resistance against Leveillula taurica in pepper. U.S. Patent No 9,351,451, 2016. [Google Scholar]
- Zheng, Z.; Nonomura, T.; Appiano, M.; Pavan, S.; Matsuda, Y.; Toyoda, H.; Wolters, A.A.; Visser, R.G.F. Loss of function in MLO orthologs reduces susceptibility of pepper and tomato to powdery mildew disease caused by Leveillula taurica. Plos One 2013, 8, e70723. [Google Scholar] [CrossRef]
- Quirin, E.A.; Ogundiwin, E.A.; Prince, J.P.; Mazourek, M.; Briggs, M.O.; Chlada, T.S.; Kim, K.T.; Falise, M.; Kang, B.C.; Jahn, M.M. Development of sequence characterized amplified region (SCAR) primers for the detection of Phyto.5.2, a major QTL for resistance to Phytophthora capsici Leon in pepper. Theor. Appl. Genet. 2005, 110, 605–612. [Google Scholar] [CrossRef]
- Walker, S.J.; Bosland, P.W. Inheritance of Phytophthora root rot and foliar blight resistance in pepper. J. Am. Soc. Hortic. Sci. 1999, 124, 14–18. [Google Scholar] [CrossRef]
- Sy, O.; Steiner, R.; Bosland, P.W. Inheritance of Phytophthora stem blight resistance as compared to Phytophthora root rot and Phytophthora foliar blight resistance in Capsicum annuum L. J. Am. Hortic. Soc. 2005, 130, 75–78. [Google Scholar] [CrossRef]
- Sy, O.; Steiner, R.; Bosland, P.W. Recombinant inbred line differential identifies race-specific resistance to Phytophthora root rot in Capsicum annuum. Phytopathology 2008, 98, 867–870. [Google Scholar] [CrossRef] [PubMed]
- Monroy-Barbosa, A.; Bosland, P.W. Genetic analysis of Phytophthora root rot race-specific resistance in chile pepper. J. Am. Hortic. Sci. 2008, 133, 825–829. [Google Scholar] [CrossRef]
- Barchenger, D.W.; Lamour, K.H.; Bosland, P.W. Challenges and strategies for breeding resistance in Capsicum annuum to the multifarious pathogen, Phytophthora capsici. Front. Plant Sci. 2018, 9, 628. [Google Scholar] [CrossRef]
- Candole, B.L.; Conner, P.J.; McGregor, C.; Waters, V.; Ji, P. The disease reactions of heirloom bell pepper ‘California Wonder’ to Phytophthora Capsici. Agric. Sci. 2012, 3, 417–424. [Google Scholar]
- Barchenger, D.W.; Sheu, Z.M.; Kumar, S.; Lin, S.W.; Burlakoti, R.R.; Bosland, P.W. Race characterization of Phytophthora capsici as a basis for global anticipatory resistance breeding in Capsicum. Phytopathology 2018, 108, 964–971. [Google Scholar] [CrossRef]
- Kim, B.; Hur, J. Inheritance of resistance to bacterial spot and to Phytophthora blight in peppers. J. Korean Soc. Hort. Sci. 1990, 31, 350–357. [Google Scholar]
- Lefebvre, V.; Palloix, A. Both epistatic and additive effects of QTLs are involved in polygenic induced resistance to disease: A case study, the interaction pepper-Phytophthora capsici Leonian. Theor. Appl. Genet. 1996, 93, 503–511. [Google Scholar] [CrossRef]
- Thabuis, A.; Palloix, A.; Pflieger, S.; Daubeze, A.M.; Caranta, C.; Lefebvre, V. Comparative mapping of Phytophthora resistance loci in pepper germplasm: Evidence for conserved resistance loci across Solanaceae and for a large genetic diversity. Theor. Appl. Genet. 2003, 106, 1473–1485. [Google Scholar] [CrossRef]
- Ogundiwin, E.A.; Berke, T.F.; Massoudi, M.; Black, L.L.; Huestis, G.; Choi, D.; Lee, S.; Prince, J.P. Construction of 2 intraspecific linkage maps and identification of resistance QTLs for Phytophthora capsici root-rot and foliar-blight diseases of pepper (Capsicum annuum L.). Genome 2005, 48, 698–711. [Google Scholar] [CrossRef]
- Minamiyama, Y.; Tsuro, M.; Kubo, T.; Hirai, M. QTL analysis for resistance to Phytophthora capsici in pepper using a high density SSR-based map. Breed. Sci. 2007, 57, 129–134. [Google Scholar] [CrossRef]
- Kim, H.J.; Nahm, S.H.; Lee, H.R.; Yoon, G.B.; Kim, K.T.; Kang, B.C.; Choi, D.; Kweon, O.Y.; Cho, M.C.; Kwon, J.K.; et al. BAC-derived markers converted from RFLP linked to Phytophthora capsici resistance in pepper (Capsicum annuum L.). Theor. Appl. Genet. 2008, 118, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Sugita, T.; Yamaguchi, K.; Kinoshita, T.; Yuji, K.; Sugimura, Y.; Nagata, R.; Kawasaki, S.; Todoroki, A. QTL analysis for resistance to Phytophthora blight (Phytophthora capsici Leon.) using an intraspecific doubled-haploid population of Capsicum annuum. Breed. Sci. 2006, 56, 137–145. [Google Scholar] [CrossRef]
- Truong, H.T.H.; Kim, K.T.; Kim, D.W.; Kim, S.; Chae, Y.; Park, J.H.; Oh, D.G.; Cho, M.C. Identification of isolate-specific resistance QTLs to Phytophthora root rot using an intraspecific recombinant inbred line population of pepper (Capsicum annuum). Plant Pathol. 2012, 61, 48–56. [Google Scholar] [CrossRef]
- Lu, F.H.; Kwoon, S.H.; Yoon, M.I.; Kim, K.T.; Cho, M.C.; Yoon, M.K.; Park, Y.J. SNP marker integration and QTL analysis of 12 agronomic and morphological traits in F8 RILs of pepper (Capsicum annuum L.). Mol. Cells 2012, 34, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.Y.; Kang, J.H.; Jeong, H.S.; Choi, H.J.; Yang, H.B.; Kim, K.T.; Choi, D.; Choi, G.J.; Jahn, M.; Kang, B.C. Combined used of bulk-segregated analysis of microarrays reveals SNP markers pinpointing a major QTL for resistance to Phytophthora capsici in pepper. Theor. Appl. Genet. 2014, 127, 2503–2513. [Google Scholar] [CrossRef]
- Mallard, S.; Cantet, M.; Massire, A.; Bachellez, A.; Ewert, S.; Lefebvre, V. A key QTL cluster is conserved among accessions and exhibits broad-spectrum resistance to Phytophthora capsici: A valuable locus for pepper breeding. Mol. Breed. 2013, 32, 349–364. [Google Scholar] [CrossRef]
- Naegele, R.P.; Ashrafi, H.; Hill, T.A.; Chin-Wo, S.R.; van Deynze, A.E.; Hausbeck, M.K. QTL mapping of fruit rot resistance to the plant pathogen Phytophthora capsici in a recombinant inbred line Capsicum annuum population. Phytopathology 2014, 104, 479–483. [Google Scholar] [CrossRef]
- Rehrig, W.Z.; Ashrafi, H.; Hill, T.; Prince, J.; Van Deynze, A. CaDMR1 cosegregates with QTL Pc5.1 for resistance to Phytophthora capsici in pepper (Capsicum annuum). Plant Genome 2014, 7, 1–12. [Google Scholar] [CrossRef]
- Xu, X.; Chao, J.; Cheng, X.; Wang, R.; Sun, B.; Wang, H.; Luo, S.; Xu, X.; Wu, T.; Li, Y. Mapping of a Novel Race Specific Resistance Gene to Phytophthora Root Rot of Pepper (Capsicum annuum) Using Bulked Segregant Analysis Combined with Specific Length Amplified Fragment Sequencing Strategy. PLoS ONE 2016, 11, e0151401. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.K.; Baek, K.H.; Seong, E.S.; Joung, Y.H.; Choi, G.J.; Park, J.M.; Cho, H.S.; Kim, E.A.; Lee, S.; Choi, D. MsrB2, pepper methionine sulfoxide reductase B2, is a novel defense regulator against oxidative stress and pathogen attack. Plant Physiol. 2010, 154, 245–261. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.L.; Jia, Q.L.; Li, D.W.; Wang, J.E.; Yin, Y.X.; Gong, Z.H. Characteristic of the pepper CaRGA2 gene in defense responses against Phytophthora capsici Leonian. Int. J. Mol. Sci. 2013, 14, 8985–9004. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wang, L.; Guo, J.; Yang, W.; Shen, H. Molecular mapping of a gene conferring resistance to Phytophthora capsici Leonian race 2 in pepper line PI201234 (Capsicum annuum L.). Mol Breed. 2016, 36, 66. [Google Scholar] [CrossRef]
- Ridzuan, R.; Rafii, M.Y.; Ismail, S.I.; Mohammad Yusoff, M.; Miah, G.; Usman, M. Breeding for anthracnose disease resistance in chili: Progress and prospects. Int. J. Mol. Sci. 2018, 19, 3122. [Google Scholar] [CrossRef] [PubMed]
- Mongkolporn, O.; Taylor, P.W.J. Chili anthracnose: Colletotrichum taxonomy and pathogenicity. Plant Pathol. 2018, 67, 1255–1263. [Google Scholar] [CrossRef]
- Suwor, P.; Thummabenjapone, P.; Sanitchon, J.; Kumar, S.; Techawongstien, S. Phenotypic and genotypic responses of chili (Capsicum annuum L.) progressive lines with different resistant genes against anthracnose pathogen (Colletotrichum spp.). Eur. J. Plant Pathol. 2015, 143, 725–736. [Google Scholar] [CrossRef]
- Kim, S.; Kim, K.T.; Kim, D.H.; Yang, E.Y.; Cho, M.C.; Jamal, A.; Chae, Y.; Pae, D.H.; Oh, D.G.; Hwang, J.K. Identification of quantitative trait loci associated with anthracnose resistance in chili pepper (Capsicum spp.) Korean, J. Hort. Sci. Technol. 2010, 28, 1014–1024. [Google Scholar]
- Sun, C.Y. Resistances to anthracnose (Colletotrichum acutatum) of Capsicum mature green and ripe fruit are controlled by a major dominant cluster of QTLs on chromosome P5. Sci Hortic. 2015, 181, 81–88. [Google Scholar] [CrossRef]
- AVRDC Report; AVRDC-The World Vegetable Center: Shanhua, Taiwan, 1998; pp. 54–57.
- Yoon, J.B.; Yang, D.C.; Do, J.W.; Park, H.G. Overcoming two post-fertilization genetic barriers in interspecific hybridization between Capsicum annuum and C. baccatum for introgression of anthracnose resistance. Breed. Sci. 2006, 56, 31–38. [Google Scholar] [CrossRef]
- Cremona, S.; Iovene, M.; Festa, G.; Conicella, C.; Parisi, M. Production of embryo rescued hybrids between the landrace ‘‘Friariello’’ (Capsicum annuum var annuum) and C baccatum var pendulum, phenotypic and cytological characterization. Euphytica 2018, 214, 129. [Google Scholar] [CrossRef]
- Mahasuk, P.; Taylor, P.W.J.; Mongkolporn, O. Identification of two new genes conferring resistance to Colletotrichum acutatum in Capsicum baccatum. Phytopathology 2009, 99, 1100–1104. [Google Scholar] [CrossRef] [PubMed]
- Mahasuk, P.; Struss, D.; Mongkolporn, O. QTLs for resistance to anthracnose identified in two Capsicum sources. Mol Breed. 2016, 36, 10. [Google Scholar] [CrossRef]
- Mimura, Y.; Kageyama, T.; Minamiyama, Y.; Masashi, H. QTL analysis for resistance to Ralstonia solanacearum in Capsicum accession ‘LS2341’. J. Japan. Soc. Hort. Sci. 2009, 78, 307–313. [Google Scholar] [CrossRef]
- Mahbou-Somo-Toukam, G. Diversité de Ralstonia Solanacearum au Cameroun et bases génétiques de la résistance chez le piment (Capsicum Annuum) et les Solanacées. Amélioration des plantes. AgroParisTech. 2010. Available online: https://pastel.archives-ouvertes.fr/pastel-00607879 (accessed on 12 March 2020).
- Caranta, C.; Lefebvre, V.; Palloix, A. Polygenic resistance of pepper to potyviruses consists of a combination of isolate-specific and broad-spectrum quantitative trait loci. Mol. Plant Microbe Int. 1997, 10, 872–878. [Google Scholar] [CrossRef]
- Quenouille, J.; Paulhiac, E.; Moury, B.; Palloix, A. Quantitative trait loci from the host genetic background modulate the durability of a resistance gene: A rational basis for sustainable resistance breeding in plants. Heredity 2014, 112, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Ben Chaim, A.; Grube, R.C.; Lapidot, M.; Jahn, M.; Paran, I. Identification of quantitative trait loci associated with resistance to Cucumber mosaic virus in Capsicum Annum. Theor. Appl. Genet. 2001, 102, 1213–1220. [Google Scholar] [CrossRef]
- Caranta, C.; Pflieger, S.; Lefebvre, V.; Daubèze, A.M.; Thabuis, A.; Palloix, A. QTLs involved in the restriction of Cucumber mosaic virus (CMV) long-distance movement in pepper. Theor. Appl. Genet. 2002, 104, 586–591. [Google Scholar] [CrossRef]
- Yao, M.; Li, N.; Wang, F.; Ye, Z. Genetic analysis and identification of QTLs for resistance to Cucumber mosaic virus in chili pepper (Capsicum annuum L.). Euphytica 2013, 193, 135–145. [Google Scholar] [CrossRef]
- Eun, M.H.; Han, J.; Yoon, J.B.; Lee, J. QTL mapping of resistance to the Cucumber mosaic virus P1 strain in pepper using a genotyping-by-sequencing analysis. Hortic. Env. Biotechnol. 2016, 57, 589–597. [Google Scholar] [CrossRef]
- Guo, G.J.; Wang, S.B.; Liu, J.B.; Pan, B.G.; Diao, W.P.; Ge, W.; Gao, C.Z.; Snyder, J.C. Rapid identification of QTLs underlying resistance to Cucumber mosaic virus in pepper (Capsicum frutescens). Theor. Appl. Genet. 2017, 130, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Yin, Y.; Wang, F.; Yao, M. Construction of a high-density genetic map and identification of QTLs for Cucumber mosaic virus resistance in pepper (Capsicum annuum L.) using specific length amplified fragment sequencing (SLAF-seq). Breed. Sci. 2018, 68, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Maharijaya, A.; Vosman, B.; Steenhuis-Broers, G.; Pelgrom, K.; Purwito, A.; Visser, R.G.; Voorrips, R.E. QTL mapping of thrips resistance in pepper. Theor. Appl. Genet. 2015, 128, 1945–1956. [Google Scholar] [CrossRef] [PubMed]
- Barbary, A.; Djian-Caporalino, C.; Marteu, N.; Fazari, A.; Caromel, B.; Castagnone-Sereno, P.; Palloix, A. Plant genetic background increasing the efficiency and durability of major resistance genes to root-knot nematodes can be resolved into a few resistance QTLs. Front. Plant Sci. 2016, 7, 632. [Google Scholar] [CrossRef] [PubMed]
- Goicoechea, N. Verticillium-induced wilt in pepper: Physiological disorders and perspectives for controlling the disease. Plant Pathol. J. 2006, 5, 258–265. [Google Scholar]
- González-Salán, M.; Bosland, P. Sources of resistance to Verticillium wilt in Capsicum. Euphytica 1992, 59, 49–53. [Google Scholar]
- Barchenger, D.W.; Rodriguez, K.; Jiang, L.; Hanson, S.F.; Bosland, P.W. Allele-specific CAPS marker in a Ve1 homolog of Capsicum annuum for improved selection of Verticillium dahliae resistance. Mol. Breed. 2017, 37, 134. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, Z.; Seidl, M.F.; Majer, A.; Jakse, J.; Javornik, B.; Thomma, B.P. Broad taxonomic characterization of Verticillium wilt resistance genes reveals an ancient origin of the tomato Ve1 immune receptor. Mol. Plant Pathol. 2017, 18, 195–209. [Google Scholar] [CrossRef]
- Loganathan, M.; Venkataravanappa, V.; Saha, S.; Sharma, B.K.; Tirupathi, S.; Verma, M.K. Morphological, cultural and molecular characterizations of Fusarium wilt infecting tomato and chilli. Paper presented at National Symposium on Abiotic and Biotic Stress Management in Vegetable Crops. Indian Society of Vegetable Science, IIVR. 2013; 12–14. [Google Scholar]
- Lomas-Cano, T.; Palmero-Llamas, D.; de Cara, M.; García Rodríguez, C.; Boix-Ruiz, A.; Camacho-Ferre, F.; Tello-Marquina, T.C. First report of Fusarium oxysporum on sweet pepper seedlings in Almería, Spain. Plant Dis. 2014, 98, 1435. [Google Scholar] [CrossRef]
- Miller, S.A.; Rowe, R.C.; Riedel, R.M. Fusarium and Verticillium wilts of tomato, potato, pepper and eggplant. Ohio State Univ. Exten. Fact. Hyg. 1996, 96, 3122. [Google Scholar]
- Rahin, A.A.; Sharif, F.M. A study of pepper wilt in northern Iraq. In Ecology and Management of Soilborne Plant Pathogens; Parker, C.A., Rovira, A.D., Moore, K.J., Wong, P.T.W., Kollmorgen, J.F., Eds.; American Phytopathological Society: Saint Paul, MN, USA, 1985; pp. 59–62. [Google Scholar]
- Rivelli, V.C. A wilt of pepper incited by Fusarium oxysporum f. sp. capsici forma specialis nova. M. Sc. Dissertation, Louisiana State University, Baton Rouge, LA, USA, 1989. [Google Scholar]
- Naik, M.K.; Devika Rani, G.S.; Madhukar, H.M. Identification of resistance sources against wilt of chilli (Capsicum annuum L.) resistance caused by Fusarium solani (Mart.) Sacc. J. Mycopathol. Res. 2008, 46, 93–96. [Google Scholar]
- Shobha, F.; Tembhurne, B.V.; Naik, M.K.; Khan, H.; Patil, B.V. Screening of M4 mutants of chilli (Capsicum annuum L.) against Fusarium wilt (Fusarium solani) resistance. Int. J. Curr. Microbiol. App. Sci. 2018, 7, 879–884. [Google Scholar]
- Manu, D.G.; Tembhurne, B.V.; Kisan, B.; Aswathnarayana, D.S.; Diwan, J.R. Inheritance of Fusarium wilt and qualitative and quantitative characters in chilli (Capsicum annuum L). J. Agr. Env. Sci. 2014, 3, 433–444. [Google Scholar]
- Nayeema, J.; Ahmed, N.; Tanki, M.I.; Das, G.M. Screening of hot pepper germplasm for resistance to Fusarium wilt [F. pallidoroseum (Cook) Sacc.]. Capsicum Egg. Plant Newslett. 1995, 14, 68–71. [Google Scholar]
- Ahmed, N.; Tanki, M.I.; Mir, N.M. Screening of advance breeding lines of chilli and sweet and hot pepper cultivars against Fusarium wilt. Plant Dis.Res. 1994, 9, 153–154. [Google Scholar]
- Mannai, S.; Jabnoun-Khiareddine, H.; Nasraoui, B.; Daami-Remadi, M. Rhizoctonia root rot of pepper (Capsicum annuum): Comparative pathogenicity of causal agent and biocontrol attempt using fungal and bacterial agents. J. Plant Pathol. Microbiol. 2018, 9, 431. [Google Scholar]
- Sahin, F.; Miller, S.A. Resistance in Capsicum pubescens to Xanthomonas campestris pv. vesicatoria pepper race 6. Plant Dis. 1998, 82, 794–799. [Google Scholar] [CrossRef] [PubMed]
- Tai, T.; Dahlbeck, D.; Stall, R.E.; Peleman, J.; Staskawicz, B.J. High-resolution genetic and physical mapping of the region containing the Bs2 resistance gene of pepper. Theor. Appl. Genet. 1999, 99, 1201–1206. [Google Scholar] [CrossRef]
- Pierre, M.; Noel, L.; Lahaye, T.; Ballvora, A.; Veuskens, J.; Ganal, M.; Bonas, U. High-resolution genetic mapping of the pepper resistance locus Bs3 governing recognition of the Xanthomonas campestris pv vesicatoria AvrBs3 protein. Theor. Appl. Genet. 2000, 101, 255–263. [Google Scholar] [CrossRef]
- Romer, P.; Jordan, T.; Lahaye, T. Identification and application of a DNA-based marker that is diagnostic for the pepper (Capsicum annuum) bacterial spot resistance gene Bs3. Plant Breed. 2010, 129, 737–740. [Google Scholar] [CrossRef]
- Vallejos, C.E.; Jones, V.; Stall, R.E.; Jones, J.B.; Minsavage, G.V.; Schultz, D.C.; Rodrigues, R.; Olsen, L.E.; Mazourek, M. Characterization of two recessive genes controlling resistance to all races of bacterial spot in peppers. Theor. Appl. Genet. 2010, 121, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.W.; Kim, Y.J.; Lee, S.C.; Hong, J.K.; Hwang, B.K. Hydrogen peroxide generation by the pepper extracellular peroxidase CaPO2 activates local and systemic cell death and defense response to bacterial pathogens. Plant Physiol. 2007, 145, 890–904. [Google Scholar] [CrossRef] [PubMed]
- Elphinstone, J.G.; Allen, C.; Prior, P.; Hayward, A.C. The current bacterial wilt situation: A global overview. In Bacterial Wilt Disease and the Ralstonia Solanacearum Species Complex; American Phytopathological Society: St Paul, MN, USA, 2005; pp. 9–28. [Google Scholar]
- Grimault, V.; Prior, P. Invasiveness of Pseudomonas solanacearum in tomato, eggplant and pepper: A comparative study. Eur. J. Plant Pathol. 1994, 100, 259–267. [Google Scholar] [CrossRef]
- Safni, I.; Cleenwerck, I.; De Vos, P.; Fegan, M.; Sly, L.; Kappler, U. Polyphasic taxonomic revision of the Ralstonia solanacearum species complex: Proposal to emend the descriptions of Ralstonia solanacearum and Ralstonia syzygii and reclassify current R. syzygii strains as Ralstonia syzygii subsp. syzygii subsp. nov., R. solanacearum phylotype IV strains as Ralstonia syzygii subsp. indonesiensis subsp. nov., banana blood disease bacterium strains as Ralstonia syzygii subsp. celebesensis subsp. nov. and R. solanacearum phylotype I and III strains as Ralstonia pseudosolanacearum sp. nov. Int. J. Syst. Evol. Micr. 2014, 64, 3087–3103. [Google Scholar]
- Genin, S.; Denny, T.P. Pathogenomics of the Ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 2012, 50, 67–89. [Google Scholar] [CrossRef]
- Buddenhagen, I.; Sequeira, L.; Kelman, A. Designation of races in Pseudomonas solanacearum. Phytopathology 1962, 52, 726. [Google Scholar]
- Hayward, A.C. Encyclopedia of Microbiology, 2nd ed.; Lederberg, J., Alexander, M., Bloom, B.R., Eds.; Academic Press: Cambridge, MA, USA, 2000; Volume 4. [Google Scholar]
- Hashimoto, N.; Matsumoto, S.; Yoshikawa, M.; Horita, M.; Tsuchiya, K. Varietal resistance among red pepper and sweet pepper cultivars to Ralstonia solanacearum isolated in Kyoto Prefecture. Jpn. J. Phytopathol. 2001, 67, 201–202. [Google Scholar]
- Mou, S.L.; Liu, Z.Q.; Gao, F.; Yang, S.; Su, M.X.; Shen, L.; Wu, Y.; He, S.L. CaHDZ27, a Homeodomain-Leucine Zipper I (HD-Zip I) protein, positively regulates the resistance to Ralstonia solanacearum Infection in pepper. Mol. Plant-Microbe Interact. 2017, 30, 960–973. [Google Scholar] [CrossRef]
- Kenyon, L.; Kumar, S.; Tsai, W.S.; Hughes, J.d.A. Virus Diseases of Peppers (Capsicum spp.) and Their Control. Adv. Virus Res. 2014, 90, 297–354. [Google Scholar]
- Pappu, H.R.; Jones, R.A.C.; Jain, R.K. Global status of tospovirus epidemics in diverse cropping systems: Successes achieved and challenges ahead. Virus Res. 2009, 141, 219–236. [Google Scholar] [CrossRef]
- Moury, B.; Verdin, E. Viruses of pepper crops in the Mediterranean basin: A remarkable stasis. Adv. Virus Res. 2012, 84, 127–162. [Google Scholar] [PubMed]
- Krishnareddy, M.; Usha Rani, R.; Anil Kumar, K.S.; Madhavi Reddy, K.; Pappu, H.R. Capsicum chlorosis virus (Genus Tospovirus) infecting chili pepper (Capsicum annuum) in India. Plant Dis. 2008, 92, 1469. [Google Scholar] [CrossRef] [PubMed]
- Orfanidou, C.G.; Boutsika, A.; Tsiolakis, G.; Winter, S.; Katis, N.I.; Maliogka, V.I. Capsicum Chlorosis Virus: A new viral pathogen of pepper in Greece. Plant Dis. 2019, 103, 379. [Google Scholar] [CrossRef]
- Webster, C.G.; Frantz, G.; Reitz, S.R.; Funderburk, J.E.; Mellinger, H.C.; McAvoy, E.; Turechek, W.W.; Marshall, S.H.; Tantiwanich, Y.; McGrath, M.T.; et al. Emergence of Groundnut ringspot virus and Tomato chlorotic spot virus in vegetables in Florida and the southeastern United States. Phytopathology 2015, 105, 388–398. [Google Scholar] [CrossRef]
- Boiteux, L.S.; De Avila, A.C. Inheritance of a resistance specific to Tomato spotted wilt tospovirus in Capsicum chinense ‘PI 159236’. Euphytica 1994, 75, 139–142. [Google Scholar] [CrossRef]
- Boiteux, L.S. Allelic relationships between genes for resistance to Tomato spotted wilt tospovirus in Capsicum chinense. Theor. Appl. Genet. 1995, 90, 146–149. [Google Scholar] [CrossRef]
- Moury, B.; Palloix, A.; Selassie, K.G.; Marchoux, G. Hypersensitive resistance to Tomato spotted wilt virus in three Capsicum chinense accessions is controlled by a single gene and is overcome by virulent strains. Euphytica 1997, 94, 45–52. [Google Scholar] [CrossRef]
- Jahn, M.; Paran, I.; Hoffmann, K.; Radwanski, E.R.; Livingstone, K.D.; Grube, R.C.; Aftergoot, E.; Lapidot, M.; Moyer, J. Genetic mapping of the Tsw locus for resistance to the Tospovirus tomato spotted wilt virus in Capsicum spp. and its relationship to the Sw5 gene for resistance to the same pathogen in tomato. Mol. Plant Micr. Interact. 2000, 13, 673–682. [Google Scholar] [CrossRef]
- Moury, B.; Pflieger, S.; Blattes, A.; Lefebvre, V.; Palloix, A. A CAPS marker to assist selection of Tomato spotted wilt virus (TSWV) resistance in pepper. Genome 2000, 43, 137–142. [Google Scholar] [CrossRef]
- Moury, B.; Selassie, K.G.; Marchoux, G.; Daubeze, A.M.; Palloix, A. High temperature effects on hypersensitive resistance to Tomato spotted wilt Tospovirus (TSWV) in pepper (Capsicum chinense Jacq.). Eur. J. Plant Pathol. 1998, 104, 489–498. [Google Scholar] [CrossRef]
- Kim, S.B.; Kang, W.H.; Huy, H.N.; Yeom, S.I.; An, J.T.; Kim, S.; Kang, M.Y.; Kim, H.J.; Jo, Y.D.; Ha, Y.; et al. Divergent evolution of multiple virus-resistance genes from a progenitor in Capsicum spp. New Phytol. 2017, 213, 886–899. [Google Scholar] [CrossRef] [PubMed]
- Hoang, N.; Yang, H.B.; Kang, B.C. Identification and inheritance of a new source of resistance against Tomato spotted wilt virus (TSWV) in Capsicum. Sci. Hortic. 2013, 161, 8–14. [Google Scholar] [CrossRef]
- Di Dato, F.; Parisi, M.; Cardi, T.; Tripodi, P. Genetic diversity and assessment of markers linked to resistance and pungency genes in Capsicum germplasm. Euphytica. 2015, 204, 103–119. [Google Scholar] [CrossRef]
- Cebolla-Cornejo, J.; Soler, S.; Gomar, B.; Soria, M.D.; Nuez, F. Screening Capsicum germplasm for resistance to Tomato spotted wilt virus (TSWV). Ann. Appl. Biol. 2003, 143, 143–152. [Google Scholar] [CrossRef]
- Turina, M.; Tavella, L.; Ciuffo, M. Advances in Virus Research; Chapter 12; Loebenstein, G., Lecoq, H., Eds.; Academic Press: San Diego, CA, USA, 2012; Volume 84. [Google Scholar]
- Turina, M.; Kormelink, R.; Resende, R.O. Resistance to Tospoviruses in Vegetable Crops: Epidemiological and molecular aspects. Annu. Rev. Phytopathol. 2016, 54, 347–371. [Google Scholar] [CrossRef]
- Jiang, L.; Huang, Y.; Sun, L.; Wang, B.; Zhu, M.; Li, J.; Huang, C.; Liu, Y.; Li, F.; Liu, Y. Occurrence and diversity of Tomato spotted wilt virus isolates breaking the Tsw resistance gene of Capsicum chinense in Yunnan, Southwest China. Plant Pathol. 2016, 6, 980–989. [Google Scholar] [CrossRef]
- Macedo, M.A.; Rojas, M.R.; Gilbertson, R.L. First report of a resistance-breaking strain of Tomato spotted wilt orthotospovirus infecting sweet pepper with the Tsw resistance gene in California, U.S.A. Plant Dis. 2019, 103, 1048. [Google Scholar] [CrossRef]
- Aramburu, J.; Galipienso, L.; Soler, S.; Rubio, L.; López, C. A severe symptom phenotype in pepper cultivars carrying the Tsw resistance gene is caused by a mixed infection between resistance-breaking and non-resistance-breaking isolates of Tomato spotted wilt virus. Phytoparasitica 2015, 43, 597–605. [Google Scholar] [CrossRef][Green Version]
- Parisi, M.; Di Dato, F.; Minutolo, M.; Festa, G.; Alioto, D. Screening Capsicum spp. for tolerance to a resistance-breaking strain of Tomato spotted wilt virus by artificial inoculation. Plant Pathol. 2015, 2015 (Suppl. 97), S57. [Google Scholar]
- Almási, A.; Csilléry, G.; Salánki, K.; Nemes, K.; Palkovics, L.; Tóbiás, I. Comparison of wild type and resistance-breaking isolates of Tomato spotted wilt virus and searching for resistance on pepper. In Proceedings of the 15th EUCARPIA Capsicum and Eggplant Working Group Meeting, Kecskemét, Hungary, 12–14 September 2016; pp. 574–578. [Google Scholar]
- Soler, S.; Debreczeni, D.E.; Vidal, E.; Aramburu, J.; López, C.; Galipienso, L.; Rubio, L. A new Capsicum baccatum accession shows tolerance to wild-type and resistance-breaking isolates of Tomato spotted wilt virus. Ann. Appl. Biol. 2015, 167, 343–353. [Google Scholar] [CrossRef]
- Persley, D.M.; McGrath, D.; Sharman, M.; Walker, I.O. Breeding for tospovirus resistance in a package of disease resistances for Capsicum and tomato. J. Insect Sci. 2009, 141, 36–37. [Google Scholar]
- Widana Gamage, S.M.; McGrath, D.J.; Persley, D.M.; Dietzgen, R.G. Transcriptome analysis of Capsicum chlorosis virus-induced hypersensitive resistance response in bell Capsicum. PLoS ONE 2016, 11, e0159085. [Google Scholar] [CrossRef]
- Kim, S.B.; Lee, H.Y.; Seo, S.; Lee, J.H.; Choi, D. RNA-Dependent RNA Polymerase (NIb) of the Potyviruses is an avirulence factor for the broad-spectrum resistance gene Pvr4 in Capsicum annuum cv. CM334. PLoS ONE 2015, 10, e0119639. [Google Scholar]
- Janzac, B.; Fabre, M.F.; Palloix, A.; Moury, B. Characterization of a new potyvirus infecting pepper crops in Ecuador. Arch. Virol. 2008, 153, 1543–1548. [Google Scholar] [CrossRef] [PubMed]
- Kyle, M.M.; Palloix, A. Proposed revision of nomenclature for potyvirus resistance genes in Capsicum. Euphytica 1997, 97, 183–188. [Google Scholar] [CrossRef]
- Yeam, I.; Kang, B.C.; Lindeman, W.; Frantz, J.D.; Faber, N.; Jahn, M.M. Allele-specific CAPS markers based on point mutations in resistance alleles at the pvr1 locus encoding eIF4E in Capsicum. Theor. Appl. Genet. 2005, 112, 178–186. [Google Scholar] [CrossRef]
- Ruffel, S.; Dussault, M.H.; Palloix, A.; Moury, B.; Bendahmane, A.; Robaglia, C.; Caranta, C. A natural recessive resistance gene against Potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant J. 2002, 32, 1067–1075. [Google Scholar] [CrossRef]
- Kang, B.C.; Yeam, I.; Frantz, J.D.; Murphy, J.F.; Jahn, M.M. The pvr1 locus in Capsicum encodes a translation initiation factor eIF4E that interacts with Tobacco etch virus VPg. Plant J. 2005, 42, 392–405. [Google Scholar] [CrossRef]
- Ibiza, V.P.; Cañizares, J.; Nuez, F. EcoTILLING in Capsicum species: Searching for new virus resistances. Bmc Genom. 2010, 11, 631. [Google Scholar] [CrossRef]
- Luis-Arteaga, M.; Gil-Ortega, R. Biological characterization of PVY as isolated from pepper in Spain. In Proceedings of the VI Meeting on Capsicum and Eggplant, Zaragoza, Spain, 21–24 October 1986; pp. 183–188. [Google Scholar]
- Caranta, C.; Thabuis, A.; Palloix, A. Development of a CAPS marker for the Pvr4 locus: A tool for pyramiding potyvirus resistance genes in pepper. Genome 1999, 42, 1111–1116. [Google Scholar] [CrossRef]
- Andrés, A.; Luis Arteaga, M.; Gil Ortega, R. New genes related to PVY resistance in C. annuum L. ‘Serrano Criollo de Morelos-334’. In Proceedings of the XIIth EUCARPIA Meeting on genetics and breeding of Capsicum and eggplant, Noordwijkerhout, The Netherlands, 17–19 May 2004; Voorrips, R.E., Ed.; Pubblisher Plant Research International: Wageningen UR, The Netherlands; pp. 134–138. [Google Scholar]
- Venkatesh, J.; An, J.; Kang, W.-H.; Jahn, M.; Kang, B.C. Fine mapping of the dominant potyvirus resistance gene Pvr7 reveals a relationship with Pvr4 in Capsicum annuum. Phytopathology 2018, 108, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Caranta, C.; Palloix, A.; Lefebvre, V.; Daubéze, A.M. QTLs for a component of partial resistance to Cucumber mosaic virus in pepper: Restriction of virus installation in host-cells. Theor. Appl. Genet. 1997, 94, 431–438. [Google Scholar] [CrossRef]
- Palloix, A.; Ayme, V.; Moury, B. Durability of plant major resistance genes to pathogens depends on the genetic background, experimental evidence and consequences for breeding strategies. New Phytol. 2009, 183, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Arnedo-Andrés, M.; Gil-Ortega, R.; Luis-Arteaga, M.; Hormaza, I. Development of RAPD and SCAR markers linked to the Pvr4 locus for resistance to PVY in pepper (Capsicum annuum L.). Theor. Appl. Genet. 2002, 105, 1067–1074. [Google Scholar]
- Rubio, M.; Caranta, C.; Palloix, A. Functional markers for selection of potyvirus resistance alleles at the pvr2-eIF4E locus in pepper using tetra-primer ARMS-PCR. Genome 2008, 51, 767–777. [Google Scholar] [CrossRef]
- Tamisier, L.; Szadkowski, M.; Nemouchi, G.; Lefebvre, V.; Szadkowski, E.; Duboscq, R.; Santoni, S.; Sarah, G.; Sauvage, C.; Palloix, A.; et al. Genome-wide association mapping of QTLs implied in potato virus Y population sizes in pepper: Evidence for widespread resistance QTL pyramiding. Mol. Plant Pathol. 2020, 21, 3–16. [Google Scholar] [CrossRef]
- Lee, H.R.; An, H.J.; You, Y.G.; Lee, J.; Kim, H.J.; Kang, B.C.; Harn, C.H. Development of a novel codominant molecular marker for Chili veinal mottle virus resistance in Capsicum annuum L. Euphytica 2013, 193, 197–205. [Google Scholar] [CrossRef]
- Devran, Z.; Kahveci, E.; Özkaynak, E.; Studholme, D.J.; Tör, M. Development of molecular markers tightly linked to Pvr4 gene in pepper using next-generation sequencing. Mol. Breed. 2015, 35, 101. [Google Scholar] [CrossRef]
- Lee, J.H.; An, J.T.; Siddique, M.I.; Han, K.; Choi, S.; Kwon, J.K.; Kang, B.C. Identification and molecular genetic mapping of Chili veinal mottle virus (ChiVMV) resistance genes in pepper (Capsicum annuum). Mol. Breed. 2017, 37, 121. [Google Scholar] [CrossRef]
- Avilla, C.; Collar, J.L.; Duque, M.; Pérez, P.; Fereres, A. Impact of floating rowcovers on bell pepper yield and virus incidence. Hortsci. 1997, 32, 882–883. [Google Scholar] [CrossRef]
- Murphy, J.F.; Bowen, K.L. Synergistic disease in pepper caused by the mixed infection of Cucumber mosaic virus and Pepper Mottle Virus. Phytopathol. 2006, 96, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Guerini, M.N.; Murphy, J.F. Resistance of Capsicum annuum ‘Avelar’ to Pepper mottle potyvirus and alleviation of this resistance by co-infection with Cucumber mosaic cucumovirus are associated with virus movement. J. Gen. Virol. 1999, 80, 2785–2792. [Google Scholar] [CrossRef] [PubMed]
- Kang, W.-H.; Hoang, N.; Yang, H.B.; Kwon, J.K.; Jo, S.H.; Seo, J.K.; Kim, K.H.; Choi, D.; Kang, B.C. Molecular mapping and characterization of a single dominant gene controlling CMV resistance in peppers (Capsicum annuum L.). Theor. Appl. Genet. 2010, 120, 1587–1596. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.Y.; Lee, J.H.; Ahn, H.I.; Yoon, J.Y.; Her, N.H.; Choi, J.K.; Choi, G.S.; Kim, D.-S.; Ryu, G.-H. Identification and sequence analysis of RNA3 of a resistance-breaking Cucumber mosaic virus isolate on Capsicum annuum. Plant Pathol. J. 2006, 22, 265–270. [Google Scholar] [CrossRef]
- Choi, S.; Lee, J.H.; Kang, W.H.; Kim, J.; Huy, H.N.; Park, S.W.; Son, E.H.; Kwon, J.K.; Kang, B.C. Identification of Cucumber mosaic resistance 2 (cmr2) that confers resistance to a new Cucumber mosaic virus isolate P1 (CMV-P1) in pepper (Capsicum spp.). Front. Plant Sci. 2018, 9, 1106. [Google Scholar] [CrossRef]
- Suzuki, K.; Kuroda, T.; Miura, Y.; Murai, J. Screening and field trials of virus resistant sources in Capsicum spp. Plant Dis. 2003, 87, 779–783. [Google Scholar] [CrossRef]
- Nono-Womdim, R.; Gèbre-Selassie, K.; Palloix, A.; Pochard, E.; Marchoux, G. Study of multiplication of Cucumber mosaic virus in susceptible and resistant Capsicum annuum lines. Ann. Appl. Biol. 1993, 122, 49–56. [Google Scholar] [CrossRef]
- Nono-Womdim, R.; Palloix, A.; Gèbre-Selassie, K.; Marchoux, G. Partial resistance of bell pepper to Cucumber mosaic virus movement within plants: Field evaluation of its efficiency in southern France. J. Phytopathol. 1993, 37, 125–132. [Google Scholar] [CrossRef]
- Grube, R.C.; Zhang, Y.; Murphy, J.F.; Loaiza-Figueroa, F.; Lackney, V.K.; Provvidenti, R.; Jahn, M.K. New source of resistance to Cucumber mosaic virus in Capsicum frutescens. Plant Dis. 2000, 84, 885–891. [Google Scholar] [CrossRef]
- Garcia-Ruiz, H.; Murphy, J.F. Age-related resistance in bell pepper to Cucumber mosaic virus. Ann. Appl. Biol. 2001, 139, 307–317. [Google Scholar] [CrossRef]
- Thakur, H.; Jindal, S.K.; Sharma, A.; Dhaliwal, M.S. Chilli leaf curl virus disease: A serious threat for chilli cultivation. J. Plant Dis. Prot. 2018, 125, 239–249. [Google Scholar] [CrossRef]
- Kumar, S.; Kumar, S.; Singh, M.; Singh, A.K.; Rai, M. Identification of host plant resistance to Pepper leaf curl virus in chilli (Capsicum species). Sci. Hortic. 2006, 110, 359–361. [Google Scholar] [CrossRef]
- Kil, E.-J.; Byun, H.-S.; Kim, S.; Kim, J.; Park, J.; Cho, S.; Yang, D.C.; Lee, K.Y.; Choi, H.S.; Kim, J.K. Sweet pepper confirmed as a reservoir host for Tomato yellow leaf curl virus by both agro-inoculation and whitefly-mediated inoculation. Arch. Virol. 2014, 159, 2387–2395. [Google Scholar] [CrossRef] [PubMed]
- Gharsallah Chouchane, S.; Gorsane, F.; Nakhla, M.K.; Maxwell, D.P.; Marrakchi, M.; Fakhfakh, H. First report of Tomato yellow leaf curl virus-Israel species infecting tomato, pepper and bean in Tunisia. J. Phytopathol. 2007, 155, 236–240. [Google Scholar] [CrossRef]
- Comes, S.; Fanigliulo, A.; Pacella, R. & Crescenzi, A. Pepper leaf curl disease caused by Tomato yellow leaf curl Sardinia virus on pepper in Southern Italy. J. Plant Pathol. 2009, 91, S42. [Google Scholar]
- Juárez, M.; Rabadán, M.P.; Martínez, L.D.; Tayahi, M.; Grande-Pérez, A.; Gómez, P. Natural hosts and genetic diversity of the emerging Tomato Leaf Curl New Delhi Virus in Spain. Front. Microbiol. 2019, 10, 140. [Google Scholar] [CrossRef]
- Luigi, M.; Bertin, S.; Manglii, A.; Troiano, E.; Davino, S.; Tomassoli, L.; Parrella, G. First report of Tomato Leaf Curl New Delhi Virus causing Yellow Leaf Curl of pepper in Europe. Plant Dis. 2019, 103, 2970. [Google Scholar] [CrossRef]
- Singh, A.K.; Kushwaha, N.; Chakraborty, S. Synergistic interaction among Begomoviruses leads to the suppression of host defense-related gene expression and breakdown of resistance in chilli. Appl. Microbiol. Biotechnol. 2016, 100, 4035–4049. [Google Scholar] [CrossRef]
- Rai, V.P.; Rai, A.C.; Kumar, S.; Kumar, R.; Kumar, S.; Singh, M.; Rai, A.B.; Singh, S.P. Emergence of new variant of Chilli leaf curl virus in North India. Veg. Sci. 2010, 37, 124–128. [Google Scholar]
- Rai, V.P.; Kumar, R.; Singh, S.P.; Kumar, S.; Kumar, S.; Singh, M.; Rai, M. Monogenic recessive resistance to Pepper leaf curl virus in an interspecific cross of Capsicum. Sci. Hortic. 2014, 172, 34–38. [Google Scholar] [CrossRef]
- Rai, V.P.; Rai, A.; Kumar, R.; Kumar, S.; Kumar, S.; Singh, M.; Singh, S.P. Microarray analyses for identifying genes conferring resistance to Pepper leaf curl virus in chilli pepper (Capsicum spp.). Genom. Data 2016, 9, 140–142. [Google Scholar] [CrossRef] [PubMed]
- Barchenger, D.; Jeeatid, N.; Yule, S.; Lin, S.W.; Wang, Y.W.; Kenyon, L. Novel sources of resistance to Pepper leaf curl virus disease (Begomovirus). In Proceedings of the 17th Eucarpia Meeting on Genetics and Breeding of Capsicum and Eggplant, Avignon, France, 11–13 September 2019; Lefebvre, V., Editor Daunay, M.C., Eds.; Institut National de la Recherche Agronomique (INRA): Avignon, France, 2019. RS-P/14. pp. 78–79. [Google Scholar]
- Trujillo-Aguirre, J.; Díaz-Plaza, R. Obtención de cultivares de chile habanero con resistencia a virosis transmitida por mosca blanca. IV.; Zamorano, Honduras: Taller latino americano sobre moscas blancas y gemini virus. 1995. [Google Scholar]
- Hernández-Verdugo, S.; Guevara-González, R.G.; Rivera-Bustamante, R.F.; Oyama, K. Screening wild plants of Capsicum annuum for resistance to Pepper huasteco virus (PHV): Presence of viral DNA and differentiation among populations. Euphytica 2001, 122, 31–36. [Google Scholar] [CrossRef]
- Retes-Manjarrez, J.E.; Hernández-Verdugo, S.; Pariaud, B.; Hernández-Espinal, L.A.; Parra-Terraza, S.; Trejo-Saavedra, D.L.; Rivera-Bustamante, R.F.; Garzón-Tiznado, J.A. Resistance to Pepper huasteco yellow vein virus and its heritability in wild genotypes of Capsicum annuum. Bot. Sci. 2018, 96, 52–62. [Google Scholar] [CrossRef]
- Garcìa-Neria, M.A.; Rivera-Bustamante, R.F. Characterization of geminivirus resistance in an accession of Capsicum chinense Jacq. Mol. Plant Microbe. 2011, 24, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Velasco, L.; Simon, B.; Janssen, D.; Cenis, J.L. Incidences and progression of Tomato chlorosis virus disease and Tomato yellow leaf curl virus disease in tomato under different greenhouse covers in southeast Spain. Ann. Appl. Biol. 2008, 153, 335–344. [Google Scholar] [CrossRef]
- Wintermantel, W.M.; Wisler, G.C. Vector specificity, host range, and genetic diversity of Tomato Chlorosis Virus. Plant Dis. 2006, 90, 814–819. [Google Scholar] [CrossRef]
- Black, L.L.; Hobbs, H.A.; Gatti, J.M. Tomato spotted wilt virus resistance in Capsicum chinense PI152225 and 159236. Plant Dis. 1991, 75, 863. [Google Scholar] [CrossRef]
- Green, S.K.; Kim, J.S. Source of resistance to viruses of pepper (Capsicum spp.): A catalog. Tech. Bull. Avrdc. 1994, 20, 5–64. [Google Scholar]
- Parrella, G.; Ruffel, S.; Moretti, A.; Morel, C.; Palloix, A.; Caranta, C. Recessive resistance genes against potyviruses are localized in colinear genomic regions of the tomato (Lycopersicon spp.) and pepper (Capsicum spp.) genomes. Theor. Appl. Genet. 2002, 105, 855–861. [Google Scholar] [CrossRef]
- Bento, C.S.; Rodrigues, R.; Gonçalves, L.S.; Oliveira, H.S.; Santos, M.H.; Pontes, M.C.; Sudré, C.P. Inheritance of resistance to Pepper yellow mosaic virus in Capsicum baccatum var. pendulum. Genet. Mol. Res. 2013, 12, 1074–1082. [Google Scholar] [CrossRef]
- Caranta, C.; Palloix, A.; Gebre-Selassie, G.; Lefebvre, V.; Moury, B.; Daubeze, A.M. A complementation of two genes originating from susceptible Capsicum annuum lines confers a new and complete resistance to Pepper veinal mottle virus. Phytopathology 1996, 86, 739–743. [Google Scholar] [CrossRef]
- Dogimont, C.; Palloix, A.; Daubèze, A.M.; Marchoux, G.; Gèbre-Selassie, K.; Pochard, E. Genetic analysis of broad spectrum resistance to potyviruses using doubled haploid lines of pepper (Capsicum annuum L.). Euphytica 1996, 88, 231–239. [Google Scholar] [CrossRef]
- Grube, R.C.; Blauth, J.R.; Arnedo Andrés, M.S.; Caranta, C.; Jahn, M.K. Identification and comparative mapping of a dominant potyvirus resistance gene cluster in Capsicum. Theor. Genet. 2000, 101, 852–859. [Google Scholar] [CrossRef]
- Srivastava, A.; Mangal, M.; Saritha, R.K.; Kalia, P. Screening of chilli pepper (Capsicum spp.) lines for resistance to the begomoviruses chilli leaf curl disease in India. Crop Prot. 2017, 100, 177–185. [Google Scholar] [CrossRef]
- Tomita, R.; Sekine, K.T.; Mizumoto, H.; Sakamoto, M.; Murai, J.; Kiba, A.; Hikichi, Y.; Suzuki, K.; Kobayashi, K. Genetic basis for the hierarchical interaction between Tobamovirus spp. and L resistance gene alleles from different pepper species. Mol. Plant Micr. Interact. 2011, 24, 108–111. [Google Scholar] [CrossRef]
- Sawada, H.; Takeuchi, S.; Hamada, H.; Kiba, A.; Matsumoto, M.; Hikichi, Y. A new tobamovirus-resistance gene, L-1a, of sweet pepper (Capsicum annuum L.). J. Jpn. Soc. Hortic. Sci. 2004, 73, 552–557. [Google Scholar] [CrossRef]
- Genda, Y.; Sato, K.; Nunomura, O.; Hirabayashi, T.; Tsuda, S. Immunolocalization of Pepper mild mottle virus in developing seeds and seedlings of Capsicum Annu. J. Gen. Plant Pathol. 2011, 77, 201–208. [Google Scholar] [CrossRef]
- Rast, A.T.B. Pepper tobamoviruses and pathotypes used in resistance breeding. Capsicum Newsl. 1988, 7, 20–23. [Google Scholar]
- Boukema, I.W. Allelism of genes controlling resistance to TMV in Capsicum, L. Euphytica 1980, 29, 433–439. [Google Scholar] [CrossRef]
- Matsumoto, K.; Sawada, H.; Matsumoto, K.; Hamada, H.; Yoshimoto, E.; Ito, T.; Takeuchi, S.; Tsuda, S.; Suzuki, K.; Kobayashi, K.; et al. The coat protein gene of tobamovirus P (0) pathotype is a determinant for activation of temperature-insensitive L (1a)-gene-mediated resistance in Capsicum plants. Arch. Virol. 2008, 153, 645–650. [Google Scholar] [CrossRef]
- Hamada, H.; Takeuchi, S.; Morita, Y.; Sawada, H.; Kiba, A.; Hikichi, Y. Amino acid changes in Pepper mild mottle virus coat protein that affect L3 gene-mediated resistance in pepper. J. Gen. Plant Pathol. 2002, 68, 155–162. [Google Scholar] [CrossRef]
- Berzal-Herranz, A.; De La Cruz, A.; Tenllado, F.; Diaz-Ruiz, J.R.; Lopez, L.; Sanz, A.I.; Vaquero, C.; Serra, M.T.; Garcia-Luque, I. The Capsicum L3 gene-mediated resistance against the tobamoviruses is elicited by the coat protein. Virology 1995, 209, 498–505. [Google Scholar] [CrossRef] [PubMed]
- Genda, Y.; Kanda, A.; Hamada, H.; Sato, K.; Ohnishi, J.; Tsuda, S. Two amino acid substitutions in the coat protein of Pepper mild mottle virus are responsible for overcoming the L4 gene mediated resistance in Capsicum spp. Phytopathology 2007, 97, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, V.; Pflieger, S.; Thabuis, A.; Caranta, C.; Blattes, A.; Chauvet, J.C.; Daubeze, A.M.; Palloix, A. Towards the saturation of the pepper linkage map by alignment of three intraspecific maps including known-function genes. Genome 2002, 45, 839–854. [Google Scholar] [CrossRef]
- Livingstone, K.D.; Lackney, V.K.; Blauth, J.R.; Van Wijk, R.; Jahn, M.K. Genome mapping in Capsicum and the evolution of genome structure in the solanaceae. Genetics 1999, 152, 1183–1202. [Google Scholar] [PubMed]
- Matsunaga, H.; Saito, T.; Hirai, M.; Nunome, T.; Yoshida, T. DNA markers linked to Pepper mild mottle virus (PMMoV) resistant locus (L4) in Capsicum. J. Jpn. Soc. Hortic. Sci. 2003, 72, 218–220. [Google Scholar] [CrossRef]
- Yang, H.; Liu, W.Y.; Kang, W.; Jahn, M.; Kang, B.C. Development of SNP markers linked to the L locus in Capsicum spp. by a comparative genetic analysis. Mol. Breed. 2009, 24, 433. [Google Scholar] [CrossRef]
- Tomita, R.; Murai, J.; Miura, Y.; Ishihara, H.; Liu, S.; Kubotera, Y.; Honda, A.; Hatta, R.; Kuroda, T.; Hamada, H.; et al. Fine mapping and DNA fiber FISH analysis locates the tobamovirus resistance gene L3 of Capsicum chinense in a 400-kb region of R-like genes cluster embedded in highly repetitive sequences. Appl. Genet. 2008, 117, 1107–1118. [Google Scholar] [CrossRef]
- Yoo, E.Y.; Kim, S.; Kim, Y.H.; Lee, C.J.; Kim, B.D. Construction of a deep coverage BAC library from Capsicum annuum, ‘CM334’. Theor. Appl. Genet. 2003, 107, 540–543. [Google Scholar] [CrossRef]
- Yang, H.-B.; Liu, W.-Y.; Kang, W.-H.; Kim, J.-H.; Cho, H.; Yoo, J.-H. Development and validation of L allele-specific markers in Capsicum. Mol. Breed. 2012, 30, 819–829. [Google Scholar] [CrossRef]
- Lim, J.H.; Park, C.-J.; Huh, S.U.; Choi, L.M.; Lee, G.L.; Kim, Y.J.; Paek, K.-H. Capsicum annuum WRKYb transcription factor that binds to the CaPR-10 promoter functions as a positive regulator in innate immunity upon TMV infection. Biochem. Biophys. Res. Commun. 2011, 411, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Huh, S.H.; Choi, L.M.; Lee, G.J.; Kim, J.Y.; Paek, K.-H. Capsicum annuum WRKY transcription factor d (CaWRKYd) regulates hypersensitive response and defense response upon Tobacco mosaic virus infection. Plant Sci. 2012, 197, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Huh, S.H.; Kim, K.-J.; Paek, K.-H. Capsicum annuum basic transcription factor 3 (CaBtf3) regulates transcription of pathogenesis-related genes during hypersensitive response upon Tobacco mosaic virus infection. Biochem. Biophys. Res. Commun. 2012, 417, 910–917. [Google Scholar] [CrossRef] [PubMed]
- Gracia, O.; Feldman, J.M. Tobacco streak virus in pepper. J. Phytopathol. 1974, 80, 313–323. [Google Scholar] [CrossRef]
- Jain, R.K.; Bag, S.; Awasthi, L.P. First report of natural infection of Capsicum annuum by Tobacco streak virus in India. Plant Pathol. 2005, 54, 257. [Google Scholar] [CrossRef]
- Janssen, D.; Sáez, E.; Segundo, E.; Martín, G.; Gil, F.; Cuadrado, I.M. Capsicum annuum - a new host of Parietaria mottle vírus in Spain. Plant Pathol. 2005, 54, 567. [Google Scholar] [CrossRef]
- Parrella, G.; Greco, B.; Troiano, E. Severe symptoms of mosaic and necrosis in bell pepper associated with Parietaria mottle virus in Italy. Plant Dis. 2016, 100, 151. [Google Scholar] [CrossRef]
- Dey, P.K.; Sarkar, P.K.; Somchoudhury, A.K. Efficacy of different treatment schedules of profenofos against major pests of chilli. Pestol. 2001, 25, 26–29. [Google Scholar]
- Capinera, J.L. Order Thysanoptera-thrips. In Handbook of Vegetable Pests, 1st ed.; Capinera, J.L., Ed.; Academic Press: San Diego, CA, USA, 2007; pp. 535–550. [Google Scholar]
- Tommasini, M.; Maini, S. Frankliniella occidentalis and other thrips harmful to vegetable and ornamental crops in Europe. In Biological Control of Thrips Pests, 1st ed.; Van Lenteren, J., Loomans, A.J.M., Tommasini, M.G., Maini, S., Ruidavets, J., Eds.; Wageningen University Papers: Wageningen, The Netherland, 1995; Volume 95, pp. 1–42. [Google Scholar]
- Zhang, Z.J.; Wu, Q.; Li, X.F.; Zhang, Y.J.; Xu, B.Y.; Zhu, G.R. Life history of western flower thrips, Frankliniella occidentalis (Thysanoptera, Thripidae), on five different vegetable leaves. J. Appl. Entomol. 2007, 131, 347–354. [Google Scholar] [CrossRef]
- Maris, P.C.; Joosten, N.N.; Goldbach, R.W.; Peters, D. Restricted spread of Tomato spotted wilt virus in thrips-resistant pepper. Phytopathology 2003, 93, 1223–1227. [Google Scholar] [CrossRef]
- Fery, R.L.; Schalk, J.M. Resistance in pepper (Capsicum annuum L.) to western flower thrips [Frankliniella occidentalis (Pergande)]. HortScience 1991, 26, 1073–1074. [Google Scholar] [CrossRef]
- Maris, P.C.; Joosten, N.N.; Peters, D.; Goldbach, R.W. Thrips resistance in pepper and its consequences for the acquisition and inoculation of Tomato spotted wilt virus by the western flower thrips. Phytopathology 2002, 93, 96–101. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Visschers, I.G.S.; Peters, J.L.; van de Vondervoort, J.A.H.; Hoogveld, R.H.M.; van Dam, N.M. Thrips resistance screening is coming of age: Leaf position and ontogeny are important determinants of leaf-based resistance in pepper. Front. Plant Sci. 2019, 10, 510. [Google Scholar] [CrossRef] [PubMed]
- Morales, F.J. Tropical whitefly IPM project. Adv. Virus Res. 2007, 69, 249–311. [Google Scholar] [PubMed]
- Roditakis, E.; Grispou, M.; Morou, E.; Kristoffersen, J.B.; Roditakis, N.; Nauen, R. Current status of insecticide resistance in Q biotype Bemisia tabaci populations from Crete. Pest Manag. Sci. 2009, 65, 313–322. [Google Scholar] [CrossRef]
- Firdaus, S.; Van Heusden, A.; Harpenas, A.; Supena, E.D.J.; Visser, G.F.; Vosman, B. Identification of silverleaf whitefly resistance in pepper. Plant Breed. 2011, 130, 708–714. [Google Scholar] [CrossRef]
- Jeevanandham, N.; Marimuthu, M.; Natesan, S.; Gandhi, K.; Appachi, S. Plant resistance in chillies Capsicum spp. against whitefly, Bemisia tabaci under field and greenhouse condition. J. Entomol. Zool. Stud. 2018, 6, 1904–1914. [Google Scholar]
- Pantoja, K.F.C.; Rocha, K.C.G.; Melo, A.M.; Marubayashi, J.M.; Baldin, E.L.L.; Bentivenha, J.P.F.; Gioria, R.; Kobori, R.F.; Pavan, M.A.; Krause-Sakate, R. Identification of Capsicum accessions tolerant to Tomato severe rugose virus and resistant to Bemisia tabaci Middle East-Asia Minor 1 (MEAM1). Trop. Plant Pathol. 2018, 43, 138. [Google Scholar] [CrossRef]
- Weintraub, P.G. Integrated control of pests in tropical and subtropical sweet pepper production. Pest Manag. Sci. 2007, 63, 753–760. [Google Scholar] [CrossRef] [PubMed]
- Bosland, P.W.; Ellington, J.J. Comparison of Capsicum annuum and C. pubescens for antixenosis as a means of aphid resistance. HortScience 1996, 31, 1017–1018. [Google Scholar] [CrossRef]
- Sun, M.; Voorrips, R.E.; Steenhuis-Broers, G.; Van’t Westende, W.; Vosman, B. Reduced phloem uptake of Myzus persicae on an aphid resistant pepper accession. BMC Plant Biol. 2018, 18, 138. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Voorrips, R.E.; Vosman, B. Aphid populations showing differential levels of virulence on Capsicum Access. Insect Sci. 2019, 27, 336–348. [Google Scholar] [CrossRef] [PubMed]
- Daryanto, A.; Syukur, M.; Hidayat, P.; Maharijaya, A. Antixenosis and antibiosis-base resistance of chili pepper to melon aphid. J. Appl. Hort. 2017, 19, 147–151. [Google Scholar]
- Sannino, L.; Espinosa, B.; Caponero, A. Helicoverpa armigera (Hübner) harmful to pepper crops in Italy. Inf. Fitopatol. 2004, 54, 23–25. [Google Scholar]
- Shivaramu, K.; Kulkarni, K.A. Screening of chilli germplasm for resistance to Helicoverpa armigera (Hübner) in chilli. Pest Manag. Hort. Ecosyst. 2008, 14, 59–66. [Google Scholar]
- Berny-Mier y Teran, J.C.; Abdala-Roberts, L.; Duran-Yanez, A.; Tut-Pech, F. Variation in insect pest and virus resistance among Habanero peppers (Capsicum chinense Jacq.) in Yucatán, México. Agrociencia 2013, 47, 471–482. [Google Scholar]
- Kashiwagi, T.; Horibata, Y.; Mekuria, D.B.; Tebayashi, S.-I.; Kim, C.-S. Ovipositional deterrent in the sweet pepper, Capsicum annuum, at the mature stage against Liriomyza trifolii (Burgess). Biosci. Biotechnol. Biochem. 2005, 69, 1831–1835. [Google Scholar] [CrossRef]
- Maharijaya, A.; Vosman, B.; Steenhuis-Broers, G.; Harpenas, A.; Purwito, A.; Visser, R.G.F.; Voorrips, R.E. Screening of pepper accessions for resistance against two thrips species (Frankliniella occidentalis and Thrips parvispinus). Euphytica 2011, 177, 401–410. [Google Scholar] [CrossRef]
- Tatagar, M.H.; Prabhu, S.T.; Jagadeesha, R.C. Screening chilli genotypes for resistance to thrips, Scirtothrips dorsalis (Hood) and mite, Polyphagotarsonemus latus (Banks). Pest Manag. Hort. Ecosyst. 2001, 7, 113–116. [Google Scholar]
- Latha, S.; Hunumanthraya, L. Screening of chilli genotypes against chilli thrips (Scirtothrips dorsalis Hood) and yellow mite [Polyphagotarsonemus latus (Banks)]. J. Entomol. Zool. Stud. 2018, 6, 2739–2744. [Google Scholar]
- Desai, H.R.; Bandhania, K.A.; Patel, A.J.; Patel, M.B.; Rai, A.B. Screening of chilli varieties/germplasms for resistance to yellow mite, Polyphagotarsonemus latus (Banks) in South Gujarat. Pest Manag. Hort. Ecosyst. 2006, 12, 55–62. [Google Scholar]
- Di Vito, M.; Saccardo, F.; Zaccheo, G. Response of lines of Capsicum spp. to italian populations of four species of Meloidogyne. Nematol. Mediterr. 1991, 19, 1. [Google Scholar]
- Di Vito, M.; Saccardo, F.; Errico, A.; Zema, V.; Zaccheo, G. Genetics of resistance to root-knot nematodes (Meloidogyne spp.) in Capsicum chacoense, C. chinense and C. frutescens. J. Gen. Breed. 1993, 47, 23–26. [Google Scholar]
- Thies, J.A.; Mueller, J.D.; Fery, R.L. Effectiveness of resistance to southern root-knot nematode in ‘Carolina Cayenne’ pepper (Capsicum annuum L.) in greenhouse, microplot, and field tests. J. Am. Soc. Hort. Sci. 1997, 122, 200–204. [Google Scholar] [CrossRef]
- Fazari, A.; Palloix, A.; Wang, L.; Yan Hua, M.; Sage-Palloix, A.M.; Zhang, B.X.; Djian-Caporalino, C. The root-knot nematode resistance N-gene co-localizes in the Me-genes cluster on the pepper (Capsicum annuum L.) P9 chromosome. Plant Breed. 2012, 131, 665–673. [Google Scholar] [CrossRef]
- Djian-Caporalino, C.; Fazari, A.; Arguel, M.J.; Vernie, T.; Van de Casteele, C.; Faure, I.; Brunoud, G.; Pijarowski, L.; Palloix, A.; Lefebvre, V.; et al. Root-knot nematode (Meloidogyne spp.) Me resistance genes in pepper (Capsicum annuum L.) are clustered on the P9 chromosome. Theor. Appl. Genet. 2007, 114, 473–476. [Google Scholar] [CrossRef]
- Chen, R.; Li, H.; Zhang, L.; Zhang, J.; Xiao, J.; Ye, Z. CaMi, a root-knot nematode resistance gene from hot pepper (Capsicum annuum L.) confers nematode resistance in tomato. Plant Cell Rep. 2007, 26, 895–905. [Google Scholar] [CrossRef]
- Khan, A.A.; Khan, M.W. Suitability of some cultivars of pepper as hosts for Meloidogyne javanica and races of m. incognita. Nematol. Mediterr. 1991, 19, 1. [Google Scholar]
- Fery, R.L.; Thies, J.A. Genetic analysis of resistance to the southern root-knot nematode in Capsicum chinense Jacq. J. Amer. Soc. Hort. Sci. 1998, 126, 1008–1011. [Google Scholar] [CrossRef]
- Hendy, H.; Pochard, E.; Dalmasso, A. Transmission héréditaire de la résistance aux nématodes Meloidogyne chitwood (Tylenchida) portée par 2 lignées de Capsicum annuum L: Étude de descendances homozygotes issues d’androgenèse. Agronomie 1985, 5, 93–100. [Google Scholar] [CrossRef]
- Thies, J.A.; Fery, R.L. Characterization of Capsicum chinense cultigens for resistance to Meloidogyne arenaria, M. hapla, and M. Javanica. Plant Dis. 2001, 85, 267–270. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Barbary, A.; Djian-Caporalino, C.; Palloix, A.; Castagnone-Sereno, P. Host genetic resistance to root-knot nematodes, Meloidogyne spp.; in Solanaceae: From genes to the field. Pest. Manag. Sci. 2015, 71, 1591–1598. [Google Scholar] [CrossRef] [PubMed]
- Marques, M.L.S.; Chadud, J.V.G.; Oliveira, M.F.; Nascimento, A.R.; Rocha, M.R. Identification of chili pepper genotypes (Capsicum spp.) resistant to Meloidogyne Enterolobii. J. Agric. Sci. 2019, 11, 165–175. [Google Scholar] [CrossRef]
- Gonçalves, L.S.A. Resistance to root-knot nematode (Meloidogyne enterolobii) in Capsicum spp. accessions. Braz. J. Agr. Sci. 2014, 9, 1. [Google Scholar] [CrossRef]
- Thies, J.A.; Fery, R.L. Heat stability ofresistance to southern root-knot nematode inbell pepper genotypes homozygous and hetero-zygous for the N gene. J. Amer. Soc. Hort. Sci. 2002, 127, 371–375. [Google Scholar] [CrossRef]
- Moosavi, M.R. Damage of the root-knot nematode Meloidogyne javanica to bell pepper, Capsicum Annu. J. Plant Dis. Prot. 2015, 122, 244–249. [Google Scholar] [CrossRef]
- Pinheiro, J.B.; Boiteux, L.S.; Almeida, M.R.A.; Pereira, R.B.; Galhardo, L.C.S.; Carneiro, R.M.D.G. First report of Meloidogyne enterolobii in Capsicum rootstocks carrying the Me1 and Me3/Me7 genes in Central Brazil. Nematropica 2015, 45, 184–188. [Google Scholar]
- Castagnone-Sereno, P.; Danchin, E.G.J.; Perfus-Barbeoch, L.; Abad, P. Diversity and evolution of root-knot nematodes, genus Meloidogyne: New insights from the genomic era. Annu. Rev. Phytopathol. 2013, 51, 203–220. [Google Scholar] [CrossRef]
- Ros-Ibáñez, C.; Robertson, L.; Martínez-Lluch, M.C.; Cano-García, A.; Lacasa-Plasencia, A. Development of virulence to Meloidogyne incognita on resistant pepper rootstocks. Span. J. Agric. Res. 2014, 12, 225–232. [Google Scholar] [CrossRef]
- Castagnone-Sereno, P.; Bongiovanni, M.; Wajnberg, E. Selection and parasite evolution: A reproductive fitness cost associated with virulence in the parthenogenetic nematode Meloidogyne Incogn. Evol. Ecol. 2007, 21, 259–270. [Google Scholar] [CrossRef]
- Djian-Caporalino, C.; Pijarowski, L.; Fazari, A.; Samson, M.; Gaveau, L.; O’Byrne, C.; Lefebvre, V.; Caranta, C.; Palloix, A.; Abad, P. High-resolution genetic mapping of the pepper (Capsicum annuum L.) resistance loci Me3 and Me4 conferring heat-stable resistance to root-knot nematodes (Meloidogyne spp.). Theor. Appl. Genet. 2001, 103, 592–600. [Google Scholar] [CrossRef]
- Thies, J.A.; Ariss, J.J. Comparison between the N and Me3 genes conferring resistance to the root-knot nematode (Meloidogyne incognita) in genetically different pepper lines (Capsicum annuum). Eur. J. Plant Pathol. 2009, 125, 545. [Google Scholar] [CrossRef]
- Mao, Z.; Zhu, P.; Liu, F.; Huang, Y.; Ling, J.; Chen, G.; Yang, Y.; Feng, D.; Xie, B. Cloning and functional analyses of pepper CaRKNR involved in Meloidogyne incognita resistance. Euphytica 2015, 205, 903–913. [Google Scholar] [CrossRef]
- Uncu, A.T.; Celik, I.; Devran, Z.; Özkaynak, E.; Frary, A.; Frary, A.; Doganlar, S. Development of a SNP-based CAPS assay for the Me1 gene conferring resistance to root knot nematode in pepper. Euphytica 2015, 206, 393–399. [Google Scholar] [CrossRef]
- Celik, I.; Sogut, M.A.; Özkaynak, E.; Doganlar, S.; Frary, A. Physical mapping of NBS-coding resistance genes to the Me-gene cluster on chromosome P9 reveals markers tightly linked to the N gene for root-knot nematode resistance in pepper. Mol. Breed. 2016, 36, 137. [Google Scholar] [CrossRef]
- Wang, X.; Fazari, A.; Cao, Y.; Zhang, Z.; Palloix, A.; Mao, S.; Zhang, B.; Caporalino, C.; Wang, L. Fine mapping of the root-knot nematode resistance gene Me1 in pepper (Capsicum annuum L.) and development of markers tightly linked to Me1. Mol. Breed. 2018, 38, 39. [Google Scholar] [CrossRef]
- Jo, J.; Purushotham, P.M.; Han, K.; Lee, H.R.; Nah, G.; Kang, B.C. Development of a genetic map for onion (Allium cepa L.) using reference-free genotyping-by-sequencing and SNP assays. Front. Plant Sci. 2017, 14, 1606. [Google Scholar] [CrossRef]
- Kiran, K.; Rawal, H.C.; Dubey, H.; Jaswal, R.; Bhardwaj, S.C.; Prasad, P.; Pal, D.; Devanna, B.N.; Sharma, T.T. Dissection of genomic features and variations of three pathotypes of Puccinia striiformis through whole genome sequencing. Sci. Rep. 2017, 7, 42419. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).