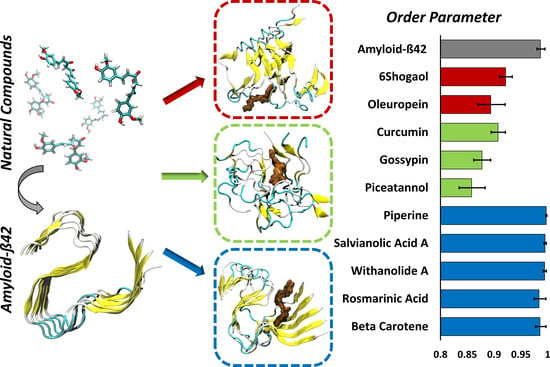
The Impact of Natural Compounds on S-Shaped Aβ42 Fibril: From Molecular Docking to Biophysical Characterization
Abstract
1. Introduction
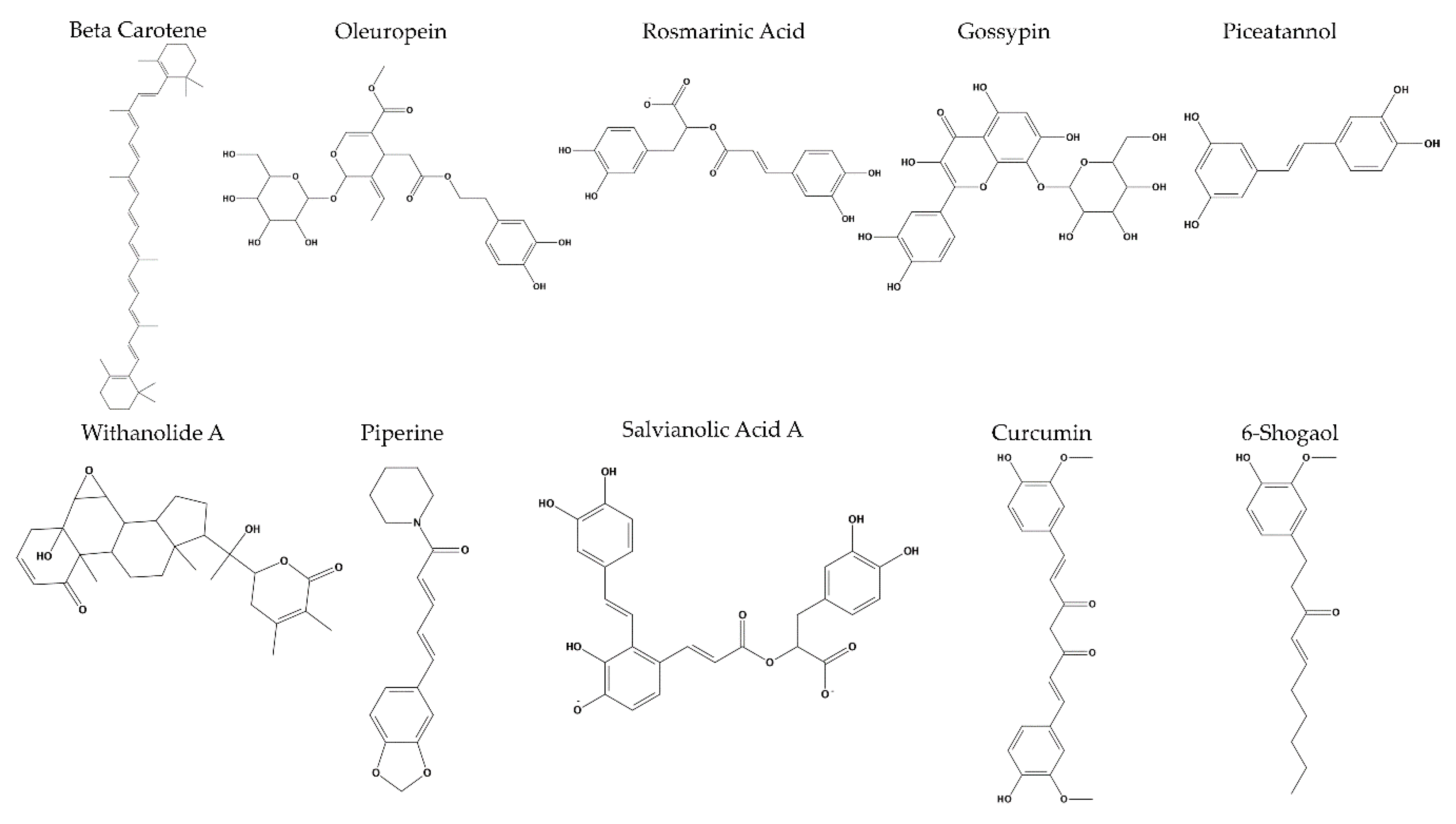
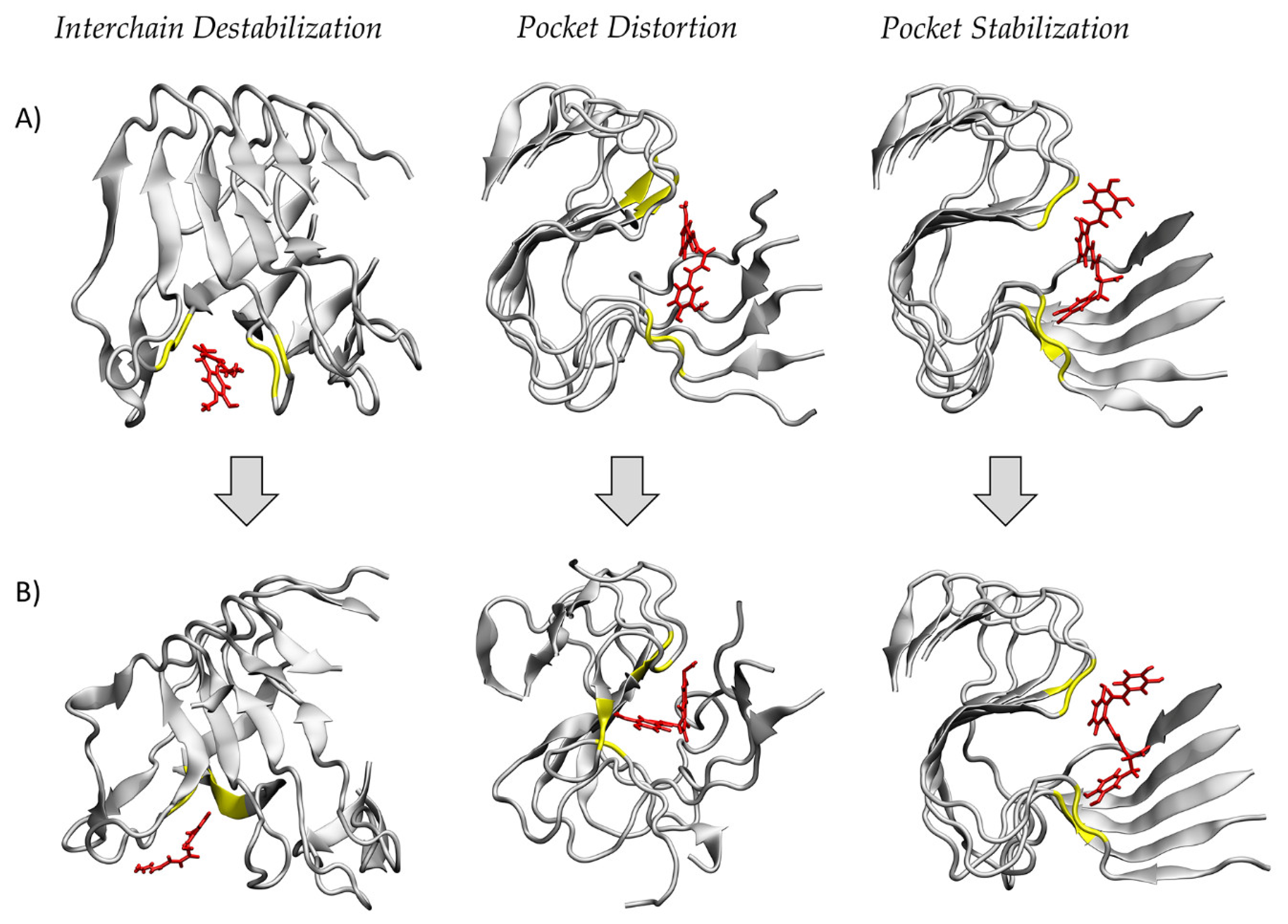
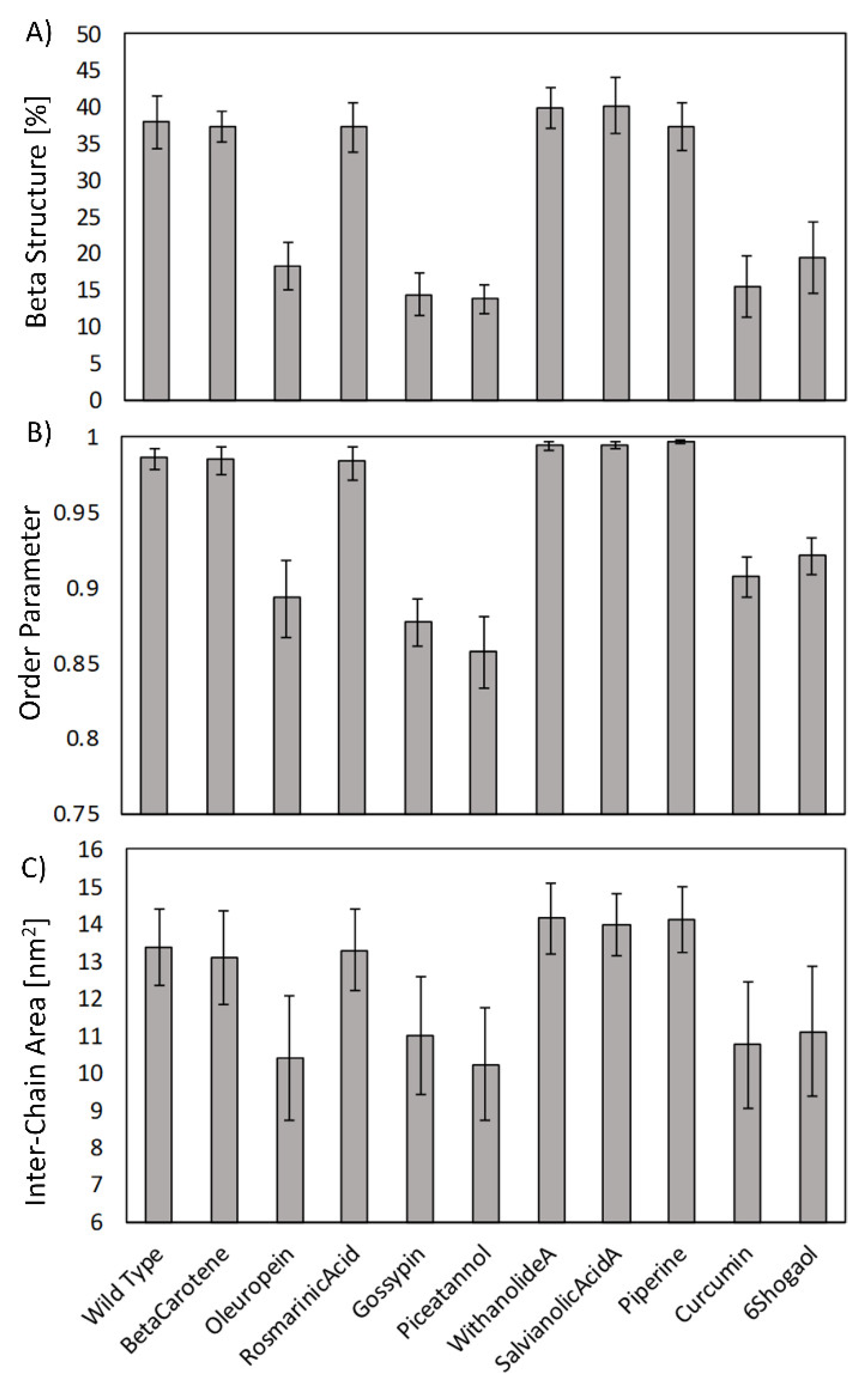
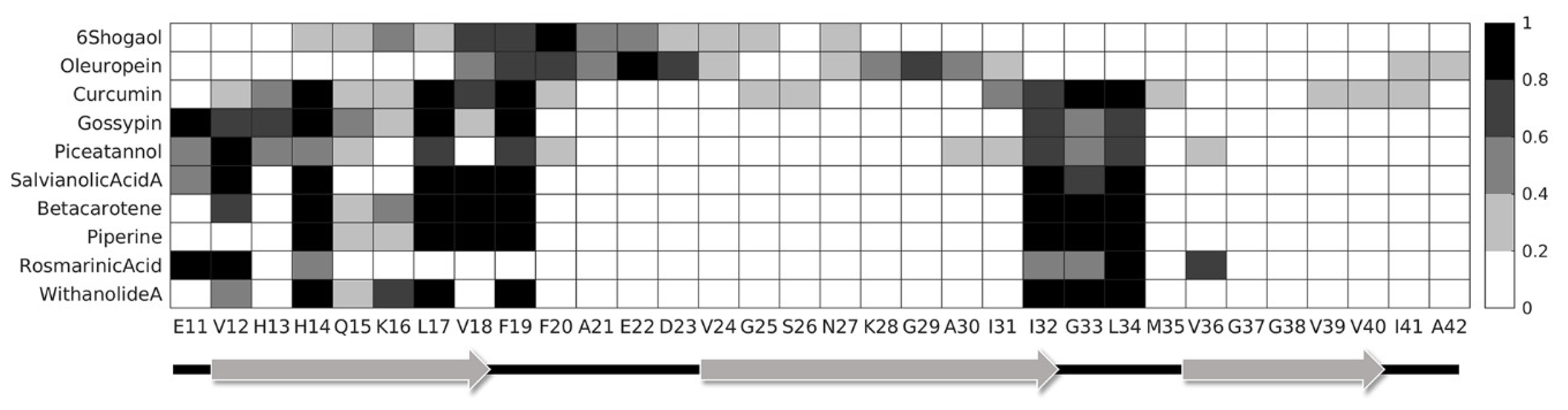
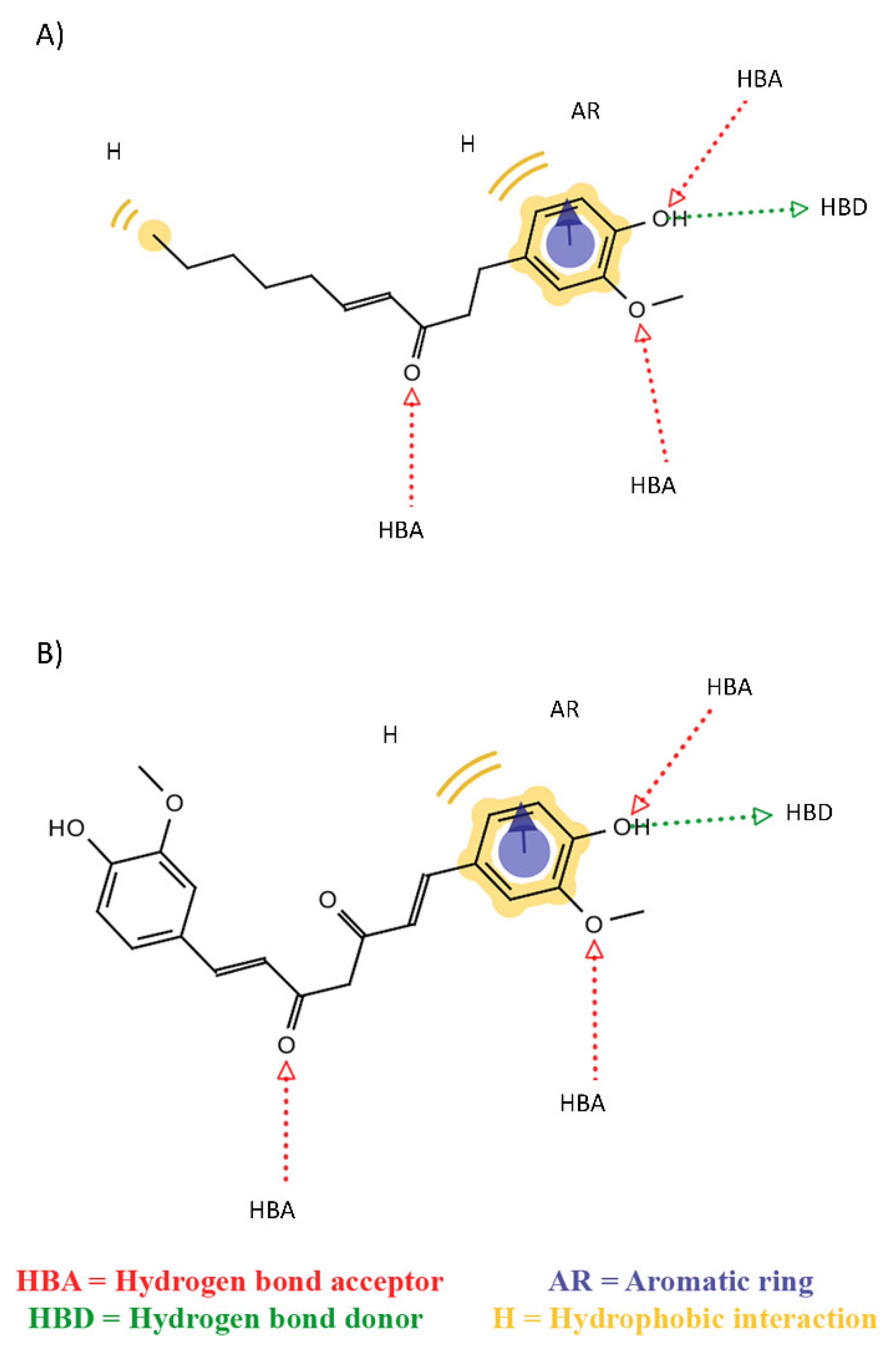
2. Results
3. Discussion
4. Materials and Methods
4.1. Molecular Dynamics Setup
4.2. Molecular Docking Protocol
4.3. Binding Energy Estimation and Protein-Compounds Conformational Dynamics
4.4. Order Parameter
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s Disease |
| BBB | Blood Brain Barrier |
| APP | Amyloid Precursor Protein |
| MM–GBSA | Molecular Mechanics–Generalized Born Surface Area |
| VDW | Van der Waals |
| PME | Particle Mesh Ewald |
| GAFF | General Amber Force Field |
| RMSD | Root Mean Square Deviations |
| ordP | Order Parameter |
References
- Selkoe, D.J. The molecular pathology of Alzheimer’s disease. Neuron 1991, 6, 487–498. [Google Scholar] [CrossRef]
- Cummings, J.L. Alzheimer’s Disease. N. Engl. J. Med. 2004, 351, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Andrade, S.; Ramalho, M.J.; Loureiro, J.A.; Do Carmo Pereira, M. Natural compounds for alzheimer’s disease therapy: A systematic review of preclinical and clinical studies. Int. J. Mol. Sci. 2019, 20, 2313. [Google Scholar] [CrossRef]
- Selkoe, D.J.; Hardy, J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 2016, 8, 595–608. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.I.A.; Linse, S.; Luheshi, L.M.; Hellstrand, E.; White, D.A.; Rajah, L.; Otzen, D.E.; Vendruscolo, M.; Dobson, C.M.; Knowles, T.P.J. Proliferation of amyloid- 42 aggregates occurs through a secondary nucleation mechanism. Proc. Natl. Acad. Sci. USA 2013, 110, 9758–9763. [Google Scholar] [CrossRef] [PubMed]
- Schütz, A.K.; Vagt, T.; Huber, M.; Ovchinnikova, O.Y.; Cadalbert, R.; Wall, J.; Güntert, P.; Böckmann, A.; Glockshuber, R.; Meier, B.H. Atomic-Resolution Three-Dimensional Structure of Amyloid $β$ Fibrils Bearing the Osaka Mutation. Angew. Chemie Int. Ed. 2015, 54, 331–335. [Google Scholar] [CrossRef]
- Guo, S.; Akhremitchev, B.B. Packing density and structural heterogeneity of insulin amyloid fibrils measured by AFM nanoindentation. Biomacromolecules 2006, 7, 1630–1636. [Google Scholar] [CrossRef]
- VandenAkker, C.C.; Engel, M.F.M.; Velikov, K.P.; Bonn, M.; Koenderink, G.H. Morphology and Persistence Length of Amyloid Fibrils Are Correlated to Peptide Molecular Structure. J. Am. Chem. Soc. 2011, 133, 18030–18033. [Google Scholar] [CrossRef]
- Palhano, F.L.; Lee, J.; Grimster, N.P.; Kelly, J.W. Toward the Molecular Mechanism(s) by Which EGCG Treatment Remodels Mature Amyloid Fibrils. J. Am. Chem. Soc. 2013, 135, 7503–7510. [Google Scholar] [CrossRef]
- Wang, T.; Jo, H.; DeGrado, W.F.; Hong, M. Water Distribution, Dynamics, and Interactions with Alzheimer’s β-Amyloid Fibrils Investigated by Solid-State NMR. J. Am. Chem. Soc. 2017, 139, 6242–6252. [Google Scholar] [CrossRef]
- Grasso, G.; Rebella, M.; Muscat, S.; Morbiducci, U.; Tuszynski, J.; Danani, A.; Deriu, M. Conformational Dynamics and Stability of U-Shaped and S-Shaped Amyloid β Assemblies. Int. J. Mol. Sci. 2018, 19, 571. [Google Scholar] [CrossRef] [PubMed]
- Grasso, G.; Rebella, M.; Morbiducci, U.; Tuszynski, J.A.; Danani, A.; Deriu, M.A. The Role of Structural Polymorphism in Driving the Mechanical Performance of the Alzheimer’s Beta Amyloid Fibrils. Front. Bioeng. Biotechnol. 2019, 7, 83. [Google Scholar] [CrossRef] [PubMed]
- Miceli, M.; Muscat, S.; Morbiducci, U.; Cavaglià, M.; Deriu, M.A. Ultrasonic waves effect on S-shaped β-amyloids conformational dynamics by non-equilibrium molecular dynamics. J. Mol. Graph. Model. 2020, 96, 107518. [Google Scholar] [CrossRef] [PubMed]
- Muscat, S.; Stojceski, F.; Danani, A. Elucidating the Effect of Static Electric Field on Amyloid Beta 1–42 Supramolecular Assembly. J. Mol. Graph. Model. 2020, 96, 107535. [Google Scholar] [CrossRef]
- Lambracht-Washington, D.; Rosenberg, R.N. Advances in the development of vaccines for Alzheimer’s disease. Discov. Med. 2013, 15, 319–326. [Google Scholar]
- Wang, C.Y.; Wang, P.-N.; Chiu, M.-J.; Finstad, C.L.; Lin, F.; Lynn, S.; Tai, Y.-H.; De Fang, X.; Zhao, K.; Hung, C.-H.; et al. UB-311, a novel UBITh ® amyloid β peptide vaccine for mild Alzheimer’s disease. Alzheimer’s Dement. Transl. Res. Clin. Interv. 2017, 3, 262–272. [Google Scholar] [CrossRef]
- Gold, M. Phase II clinical trials of anti–amyloid β antibodies: When is enough, enough? Alzheimer’s Dement. Transl. Res. Clin. Interv. 2017, 3, 402–409. [Google Scholar] [CrossRef]
- van Dyck, C.H. Anti-Amyloid-β Monoclonal Antibodies for Alzheimer’s Disease: Pitfalls and Promise. Biol. Psychiatry 2018, 83, 311–319. [Google Scholar] [CrossRef]
- Rajasekhar, K.; Madhu, C.; Govindaraju, T. Natural Tripeptide-Based Inhibitor of Multifaceted Amyloid β Toxicity. ACS Chem. Neurosci. 2016, 7, 1300–1310. [Google Scholar] [CrossRef]
- Viet, M.H.; Ngo, S.T.; Lam, N.S.; Li, M.S. Inhibition of Aggregation of Amyloid Peptides by Beta-Sheet Breaker Peptides and Their Binding Affinity. J. Phys. Chem. B 2011, 115, 7433–7446. [Google Scholar] [CrossRef]
- Lin, D.; Qi, R.; Li, S.; He, R.; Li, P.; Wei, G.; Yang, X. Interaction Dynamics in Inhibiting the Aggregation of Aβ Peptides by SWCNTs: A Combined Experimental and Coarse-Grained Molecular Dynamic Simulation Study. ACS Chem. Neurosci. 2016, 7, 1232–1240. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wang, W.; Sang, J.; Jia, L.; Lu, F. Hydroxylated Single-Walled Carbon Nanotubes Inhibit Aβ 42 Fibrillogenesis, Disaggregate Mature Fibrils, and Protect against Aβ 42 -Induced Cytotoxicity. ACS Chem. Neurosci. 2019, 10, 588–598. [Google Scholar] [CrossRef] [PubMed]
- Xiong, N.; Dong, X.Y.; Zheng, J.; Liu, F.F.; Sun, Y. Design of LVFFARK and LVFFARK-Functionalized Nanoparticles for Inhibiting Amyloid β-Protein Fibrillation and Cytotoxicity. ACS Appl. Mater. Interfaces 2015, 7, 5650–5662. [Google Scholar] [CrossRef] [PubMed]
- MacLeod, R.; Hillert, E.-K.; Cameron, R.T.; Baillie, G.S. The role and therapeutic targeting of α-, β- and γ-secretase in Alzheimer’s disease. Futur. Sci. OA 2015, 1, fso.15.9. [Google Scholar] [CrossRef]
- Cui, J.; Wang, X.; Li, X.; Wang, X.; Zhang, C.; Li, W.; Zhang, Y.; Gu, H.; Xie, X.; Nan, F.; et al. Targeting the γ-/β-secretase interaction reduces β-amyloid generation and ameliorates Alzheimer’s disease-related pathogenesis. Cell Discov. 2015, 1, 15021. [Google Scholar] [CrossRef]
- Nie, Q.; Du, X.G.; Geng, M.Y. Small molecule inhibitors of amyloid β peptide aggregation as a potential therapeutic strategy for Alzheimer’s disease. Acta Pharmacol. Sin. 2011, 32, 545–551. [Google Scholar] [CrossRef]
- Zhu, M.; De Simone, A.; Schenk, D.; Toth, G.; Dobson, C.M.; Vendruscolo, M. Identification of small-molecule binding pockets in the soluble monomeric form of the Aβ42 peptide. J. Chem. Phys. 2013, 139. [Google Scholar] [CrossRef]
- Doig, A.J.; Derreumaux, P. Inhibition of protein aggregation and amyloid formation by small molecules. Curr. Opin. Struct. Biol. 2015, 30, 50–56. [Google Scholar] [CrossRef]
- Habchi, J.; Chia, S.; Limbocker, R.; Mannini, B.; Ahn, M.; Perni, M.; Hansson, O.; Arosio, P.; Kumita, J.R.; Challa, P.K.; et al. Systematic development of small molecules to inhibit specific microscopic steps of Aβ42 aggregation in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2017, 114, E200–E208. [Google Scholar] [CrossRef]
- Liu, F.; Ma, Z.; Sang, J.; Lu, F. Edaravone inhibits the conformational transition of amyloid-β42: Insights from molecular dynamics simulations. J. Biomol. Struct. Dyn. 2019, 1–12. [Google Scholar] [CrossRef]
- Liang, C.; Savinov, S.N.; Fejzo, J.; Eyles, S.J.; Chen, J. Modulation of Amyloid-β42 Conformation by Small Molecules Through Nonspecific Binding. J. Chem. Theory Comput. 2019, 15, 5169–5174. [Google Scholar] [CrossRef] [PubMed]
- Orgogozo, J.M.; Gilman, S.; Dartigues, J.F.; Laurent, B.; Puel, M.; Kirby, L.C.; Jouanny, P.; Dubois, B.; Eisner, L.; Flitman, S.; et al. Subacute meningoencephalitis in a subset of patients with AD after Aβ42 immunization. Neurology 2003, 61, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Frid, P.; Anisimov, S.V.; Popovic, N. Congo red and protein aggregation in neurodegenerative diseases. Brain Res. Rev. 2007, 53, 135–160. [Google Scholar] [CrossRef] [PubMed]
- Zenaro, E.; Piacentino, G.; Constantin, G. The blood-brain barrier in Alzheimer’s disease. Neurobiol. Dis. 2017, 107, 41–56. [Google Scholar] [CrossRef] [PubMed]
- Bui, T.T.; Nguyen, T.H. Natural product for the treatment of Alzheimer’s disease. J. Basic Clin. Physiol. Pharmacol. 2017, 28, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.S.; Robertson, A.A.B.; Cooper, M.A. Natural product and natural product derived drugs in clinical trials. Nat. Prod. Rep. 2014, 31, 1612–1661. [Google Scholar] [CrossRef]
- Andrade, S.; Ramalho, M.J.; Loureiro, J.A.; Pereira, M.C. Interaction of natural compounds with biomembrane models: A biophysical approach for the Alzheimer’s disease therapy. Colloids Surfaces B Biointerfaces 2019, 180, 83–92. [Google Scholar] [CrossRef]
- Rasool, M.; Malik, A.; Qureshi, M.S.; Manan, A.; Pushparaj, P.N.; Asif, M.; Qazi, M.H.; Qazi, A.M.; Kamal, M.A.; Gan, S.H.; et al. Recent Updates in the Treatment of Neurodegenerative Disorders Using Natural Compounds. Evidence-Based Complement. Altern. Med. 2014, 2014, 1–7. [Google Scholar] [CrossRef]
- Hiremathad, A. A Review: Natural Compounds as Anti-Alzheimer’s Disease Agents. Curr. Nutr. Food Sci. 2017, 13. [Google Scholar] [CrossRef]
- Deb, S.; Mazumder, M.K.; Dutta, A.; Phukan, B.C.; Bhattacharya, P.; Paul, R.; Borah, A. Therapeutic implications of anti-inflammatory natural products in Alzheimer’s disease. In Discovery and Development of Anti-Inflammatory Agents from Natural Products; Elsevier: Amsterdam, The Netherlands, 2019; pp. 241–258. ISBN 9780128169926. [Google Scholar]
- Mourtas, S.; Canovi, M.; Zona, C.; Aurilia, D.; Niarakis, A.; La Ferla, B.; Salmona, M.; Nicotra, F.; Gobbi, M.; Antimisiaris, S.G. Curcumin-decorated nanoliposomes with very high affinity for amyloid-β1-42 peptide. Biomaterials 2011, 32, 1635–1645. [Google Scholar] [CrossRef]
- Ringman, J.; Frautschy, S.; Cole, G.; Masterman, D.; Cummings, J. A Potential Role of the Curry Spice Curcumin in Alzheimers Disease. Curr. Alzheimer Res. 2005, 2, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Knowles, T.P.; Fitzpatrick, A.W.; Meehan, S.; Mott, H.R.; Vendruscolo, M.; Dobson, C.M.; Welland, M.E. Role of intermolecular forces in defining material properties of protein nanofibrils. Science 2007, 318, 1900–1903. [Google Scholar] [CrossRef] [PubMed]
- Bidone, T.C.; Kim, T.; Deriu, M.A.; Morbiducci, U.; Kamm, R.D. Multiscale impact of nucleotides and cations on the conformational equilibrium, elasticity and rheology of actin filaments and crosslinked networks. Biomech. Model. Mechanobiol. 2015, 14, 1143–1155. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.-M.; Xu, Q.; Wei, D.-Q. Recent Studies on Mechanisms of New Drug Candidates for Alzheimer’s Disease Interacting with Amyloid-β Protofibrils Using Molecular Dynamics Simulations. In Translational Bioinformatics and Its Application; Wei, D.-Q., Ma, Y., Cho, W.C., Xu, Q., Zhou, F., Eds.; Springer: Dordrecht, The Netherlands, 2017; pp. 135–151. [Google Scholar]
- Tang, M.; Wang, Z.; Zhou, Y.; Xu, W.; Li, S.; Wang, L.; Wei, D.; Qiao, Z. A Novel Drug Candidate for Alzheimer’s Disease Treatment: Gx-50 Derived from Zanthoxylum Bungeanum. J. Alzheimer’s Dis. 2013, 34, 203–213. [Google Scholar] [CrossRef]
- Hou, S.; Gu, R.-X.; Wei, D.-Q. Inhibition of β-Amyloid Channels with a Drug Candidate wgx-50 Revealed by Molecular Dynamics Simulations. J. Chem. Inf. Model. 2017, 57, 2811–2821. [Google Scholar] [CrossRef]
- Fan, H.-M.; Gu, R.-X.; Wang, Y.-J.; Pi, Y.-L.; Zhang, Y.-H.; Xu, Q.; Wei, D.-Q. Destabilization of Alzheimer’s Aβ42 Protofibrils with a Novel Drug Candidate wgx-50 by Molecular Dynamics Simulations. J. Phys. Chem. B 2015, 119, 11196–11202. [Google Scholar] [CrossRef]
- Kanchi, P.K.; Dasmahapatra, A.K. Polyproline chains destabilize the Alzheimer’s amyloid-β protofibrils: A molecular dynamics simulation study. J. Mol. Graph. Model. 2019, 93, 107456. [Google Scholar] [CrossRef]
- Sharma, B.; Kalita, S.; Paul, A.; Mandal, B.; Paul, S. The role of caffeine as an inhibitor in the aggregation of amyloid forming peptides: A unified molecular dynamics simulation and experimental study. RSC Adv. 2016, 6, 78548–78558. [Google Scholar] [CrossRef]
- Battisti, A.; Palumbo Piccionello, A.; Sgarbossa, A.; Vilasi, S.; Ricci, C.; Ghetti, F.; Spinozzi, F.; Marino Gammazza, A.; Giacalone, V.; Martorana, A.; et al. Curcumin-like compounds designed to modify amyloid beta peptide aggregation patterns. RSC Adv. 2017, 7, 31714–31724. [Google Scholar] [CrossRef]
- Masuda, Y.; Fukuchi, M.; Yatagawa, T.; Tada, M.; Takeda, K.; Irie, K.; Akagi, K.; Monobe, Y.; Imazawa, T.; Takegoshi, K. Solid-state NMR analysis of interaction sites of curcumin and 42-residue amyloid β-protein fibrils. Bioorg. Med. Chem. 2011, 19, 5967–5974. [Google Scholar] [CrossRef]
- Rao, P.P.N.; Mohamed, T.; Teckwani, K.; Tin, G. Curcumin Binding to Beta Amyloid: A Computational Study. Chem. Biol. Drug Des. 2015, 86, 813–820. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Ma, B.; McElheny, D.; Parthasarathy, S.; Long, F.; Hoshi, M.; Nussinov, R.; Ishii, Y. Aβ(1–42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat. Struct. Mol. Biol. 2015, 22, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Fändrich, M.; Nyström, S.; Nilsson, K.P.R.; Böckmann, A.; LeVine, H.; Hammarström, P. Amyloid fibril polymorphism: A challenge for molecular imaging and therapy. J. Intern. Med. 2018, 283, 218–237. [Google Scholar] [CrossRef] [PubMed]
- Acosta, D.M.Á.V.; Vega, B.C.; Basurto, J.C.; Morales, L.G.F.; Rosales Hernández, M.C. Recent Advances by In Silico and In Vitro Studies of Amyloid-β 1-42 Fibril Depicted a S-Shape Conformation. Int. J. Mol. Sci. 2018, 19, 2415. [Google Scholar] [CrossRef] [PubMed]
- Su, P.C.; Tsai, C.C.; Mehboob, S.; Hevener, K.E.; Johnson, M.E. Comparison of radii sets, entropy, QM methods, and sampling on MM-PBSA, MM-GBSA, and QM/MM-GBSA ligand binding energies of F. tularensis enoyl-ACP reductase (FabI). J. Comput. Chem. 2015, 36, 1859–1873. [Google Scholar] [CrossRef] [PubMed]
- Wolber, G.; Langer, T. LigandScout: 3-D Pharmacophores Derived from Protein-Bound Ligands and Their Use as Virtual Screening Filters. J. Chem. Inf. Model. 2005, 45, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Singh, A. Ekavali A review on Alzheimer’s disease pathophysiology and its management: An update. Pharmacol. Rep. 2015, 67, 195–203. [Google Scholar] [CrossRef]
- Auld, D.S.; Kornecook, T.J.; Bastianetto, S.; Quirion, R. Alzheimer’s disease and the basal forebrain cholinergic system: Relations to β-amyloid peptides, cognition, and treatment strategies. Prog. Neurobiol. 2002, 68, 209–245. [Google Scholar] [CrossRef]
- Farlow, M.R.; Miller, M.L.; Pejovic, V. Treatment Options in Alzheimer’s Disease: Maximizing Benefit, Managing Expectations. Dement. Geriatr. Cogn. Disord. 2008, 25, 408–422. [Google Scholar] [CrossRef]
- Du, W.J.; Guo, J.J.; Gao, M.T.; Hu, S.Q.; Dong, X.Y.; Han, Y.F.; Liu, F.F.; Jiang, S.; Sun, Y. Brazilin inhibits amyloid β-protein fibrillogenesis, remodels amyloid fibrils and reduces amyloid cytotoxicity. Sci. Rep. 2015, 5, 1–10. [Google Scholar] [CrossRef]
- Tavanti, F.; Pedone, A.; Menziani, M. Computational Insight into the Effect of Natural Compounds on the Destabilization of Preformed Amyloid-β(1–40) Fibrils. Molecules 2018, 23, 1320. [Google Scholar] [CrossRef] [PubMed]
- Pourkhodadad, S.; Alirezaei, M.; Moghaddasi, M.; Ahmadvand, H.; Karami, M.; Delfan, B.; Khanipour, Z. Neuroprotective effects of oleuropein against cognitive dysfunction induced by colchicine in hippocampal CA1 area in rats. J. Physiol. Sci. 2016, 66, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Luccarini, I.; Ed Dami, T.; Grossi, C.; Rigacci, S.; Stefani, M.; Casamenti, F. Oleuropein aglycone counteracts Aβ42 toxicity in the rat brain. Neurosci. Lett. 2014, 558, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Ferrándiz, M.L.; Alcaraz, M.J. Anti-inflammatory activity and inhibition of arachidonic acid metabolism by flavonoids. Agents Actions 1991, 32, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Thamizhiniyan, V.; Vijayaraghavan, K.; Subramanian, S.P. Gossypin, a flavonol glucoside protects pancreatic beta-cells from glucotoxicity in streptozotocin-induced experimental diabetes in rats. Biomed. Prev. Nutr. 2012, 2, 239–245. [Google Scholar] [CrossRef]
- Ono, K.; Hasegawa, K.; Naiki, H.; Yamada, M. Curcumin Has Potent Anti-Amyloidogenic Effects for Alzheimer’s β-Amyloid Fibrils In Vitro. J. Neurosci. Res. 2004. [Google Scholar] [CrossRef]
- Yang, F.; Lim, G.P.; Begum, A.N.; Ubeda, O.J.; Simmons, M.R.; Ambegaokar, S.S.; Chen, P.; Kayed, R.; Glabe, C.G.; Frautschy, S.A.; et al. Curcumin inhibits formation of amyloid β oligomers and fibrils, binds plaques, and reduces amyloid in vivo. J. Biol. Chem. 2005. [Google Scholar] [CrossRef]
- Diomede, L.; Rigacci, S.; Romeo, M.; Stefani, M.; Salmona, M. Oleuropein Aglycone Protects Transgenic C. elegans Strains Expressing Aβ42 by Reducing Plaque Load and Motor Deficit. PLoS ONE 2013, 8, e58893. [Google Scholar] [CrossRef]
- Rigacci, S.; Guidotti, V.; Bucciantini, M.; Nichino, D.; Relini, A.; Berti, A.; Stefani, M. Aβ(1-42) aggregates into non-toxic amyloid assemblies in the presence of the natural polyphenol oleuropein aglycon. Curr. Alzheimer Res. 2011, 8, 841–852. [Google Scholar] [CrossRef]
- Pantano, D.; Luccarini, I.; Nardiello, P.; Servili, M.; Stefani, M.; Casamenti, F. Oleuropein aglycone and polyphenols from olive mill waste water ameliorate cognitive deficits and neuropathology. Br. J. Clin. Pharmacol. 2017, 83, 54–62. [Google Scholar] [CrossRef]
- Kumar, J.; Namsechi, R.; Sim, V.L. Structure-based peptide design to modulate amyloid beta aggregation and reduce cytotoxicity. PLoS ONE 2015, 10, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Leri, M.; Natalello, A.; Bruzzone, E.; Stefani, M.; Bucciantini, M. Oleuropein aglycone and hydroxytyrosol interfere differently with toxic Aβ 1-42 aggregation. Food Chem. Toxicol. 2019, 129, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 2010, 78, 1950–1958. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926. [Google Scholar] [CrossRef]
- Hess, B.; Hess, B.; Bekker, H.; Berendsen, H.J.C.C.; Fraaije, J.G.E.M.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 14101. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.J.C.; Postma, J.P.M.; Van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 27–28, 33–38. [Google Scholar] [CrossRef]
- Grasso, G.; Muscat, S.; Rebella, M.; Morbiducci, U.; Audenino, A.; Danani, A.; Deriu, M.A. Cell penetrating peptide modulation of membrane biomechanics by Molecular dynamics. J. Biomech. 2018, 73, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2019, 47, D1102–D1109. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006, 25, 247–260. [Google Scholar] [CrossRef]
- Jakalian, A.; Jack, D.B.; Bayly, C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002, 23, 1623–1641. [Google Scholar] [CrossRef]
- Klejborowska, G.; Urbaniak, A.; Preto, J.; Maj, E.; Moshari, M.; Wietrzyk, J.; Tuszynski, J.A.; Chambers, T.C.; Huczyński, A. Synthesis, biological evaluation and molecular docking studies of new amides of 4-bromothiocolchicine as anticancer agents. Bioorg. Med. Chem. 2019, 76, 115144. [Google Scholar] [CrossRef]
- Sahakyan, H.; Abelyan, N.; Arakelov, V.; Arakelov, G.; Nazaryan, K. In silico study of colchicine resistance molecular mechanisms caused by tubulin structural polymorphism. PLoS ONE 2019, 14, e0221532. [Google Scholar] [CrossRef]
- Kumbhar, B.V.; Borogaon, A.; Panda, D.; Kunwar, A. Exploring the Origin of Differential Binding Affinities of Human Tubulin Isotypes αβII, αβIII and αβIV for DAMA-Colchicine Using Homology Modelling, Molecular Docking and Molecular Dynamics Simulations. PLoS ONE 2016, 11, e0156048. [Google Scholar] [CrossRef]
- Gajewski, M.M.; Tuszynski, J.A.; Barakat, K.; Huzil, J.T.; Klobukowski, M. Interactions of laulimalide, peloruside, and their derivatives with the isoforms of β-tubulin. Can. J. Chem. 2013, 91, 511–517. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Sun, H.; Duan, L.; Chen, F.; Liu, H.; Wang, Z.; Pan, P.; Zhu, F.; Zhang, J.Z.H.; Hou, T. Assessing the performance of MM/PBSA and MM/GBSA methods. 7. Entropy effects on the performance of end-point binding free energy calculation approaches. Phys. Chem. Chem. Phys. 2018, 20, 14450–14460. [Google Scholar] [CrossRef]
- Hou, T.; Wang, J.; Li, Y.; Wang, W. Assessing the performance of the molecular mechanics/Poisson Boltzmann surface area and molecular mechanics/generalized Born surface area methods. II. The accuracy of ranking poses generated from docking. J. Comput. Chem. 2011, 32, 866–877. [Google Scholar] [CrossRef]
- Grasso, G.; Leanza, L.; Morbiducci, U.; Danani, A.; Deriu, M.A. Aminoacid Substitutions in the Glycine Zipper Affect the Conformational Stability of Amyloid Beta Fibrils. J. Biomol. Struct. Dyn. 2019, 1–13. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muscat, S.; Pallante, L.; Stojceski, F.; Danani, A.; Grasso, G.; Deriu, M.A. The Impact of Natural Compounds on S-Shaped Aβ42 Fibril: From Molecular Docking to Biophysical Characterization. Int. J. Mol. Sci. 2020, 21, 2017. https://doi.org/10.3390/ijms21062017
Muscat S, Pallante L, Stojceski F, Danani A, Grasso G, Deriu MA. The Impact of Natural Compounds on S-Shaped Aβ42 Fibril: From Molecular Docking to Biophysical Characterization. International Journal of Molecular Sciences. 2020; 21(6):2017. https://doi.org/10.3390/ijms21062017
Chicago/Turabian StyleMuscat, Stefano, Lorenzo Pallante, Filip Stojceski, Andrea Danani, Gianvito Grasso, and Marco Agostino Deriu. 2020. "The Impact of Natural Compounds on S-Shaped Aβ42 Fibril: From Molecular Docking to Biophysical Characterization" International Journal of Molecular Sciences 21, no. 6: 2017. https://doi.org/10.3390/ijms21062017
APA StyleMuscat, S., Pallante, L., Stojceski, F., Danani, A., Grasso, G., & Deriu, M. A. (2020). The Impact of Natural Compounds on S-Shaped Aβ42 Fibril: From Molecular Docking to Biophysical Characterization. International Journal of Molecular Sciences, 21(6), 2017. https://doi.org/10.3390/ijms21062017