The Major Cat Allergen Fel d 1 Binds Steroid and Fatty Acid Semiochemicals: A Combined In Silico and In Vitro Study
Abstract
1. Introduction
2. Results
2.1. In Silico Molecular Docking of Fel d 1 with Putative Ligands
2.2. Fluorescence Binding Studies
2.3. Visualization of Molecular Interactions
3. Discussion
4. Materials and Methods
4.1. System Configuration
4.2. Collection and Structure Conversion of Ligands
4.3. Physio-Chemical Properties Analysis
4.4. Molecular Docking Analysis
4.4.1. Ligand Optimization
4.4.2. Protein Grid Parameters
4.4.3. Semi-Empirical Calculations
4.4.4. Molecular Visualization
4.5. Fluorescence Measurement and Binding Assays
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van Milligen, F.J.; Vroom, T.M.; Aalberse, R.C. Presence of Felis domesticus Allergen I in the Cat’s Salivary and Lacrimal Glands. Int. Arch. Allergy Appl. Immunol. 1990, 92, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Charpin, C.; Mata, P.; Charpin, D.; Lavaut, M.N.; Allasia, C.; Vervloet, D. Fel d I allergen distribution in cat fur and skin. J. Allergy Clin. Immunol. 1991, 88, 77–82. [Google Scholar] [CrossRef]
- De Andrade, A.D.; Birnbaum, J.; Magalon, C.; Magnol, J.P.; Lanteaume, A.; Charpin, D.; Vervloet, D. Fel d I levels in cat anal glands. Clin. Exp. Allergy 1996, 26, 178–180. [Google Scholar] [CrossRef]
- Bonnet, B.; Messaoudi, K.; Jacomet, F.; Michaud, E.; Fauquert, J.L.; Caillaud, D.; Evrard, B. An update on molecular cat allergens: Fel d 1 and what else? Chapter 1: Fel d 1, the major cat allergen. Allergy Asthma Clin. Immunol. 2018, 14, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zielonka, T.M.; Charpin, D.; Berbis, P.; Lucciani, P.; Casanova, D.; Vervloet, D. Effects of castration and testosterone on Fel dI production by sebaceous glands of male cats: I--Immunological assessment. Clin. Exp. Allergy 1994, 24, 1169–1173. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, L.; Grönlund, H.; Sandalova, T.; Ljunggren, H.G.; van Hage-Hamsten, M.; Achour, A.; Schneider, G. The crystal structure of the major cat allergen Fel d 1, a member of the secretoglobin family. J. Biol. Chem. 2003, 278, 37730–37735. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, L.; Velickovic, T.C.; Badia-Martinez, D.; Adedoyin, J.; Thunberg, S.; Hallen, D.; Berndt, K.; Grönlund, H.; Gafvelin, G.; Van Hage, M.; et al. Structural characterization of the tetrameric form of the major cat allergen Fel d 1. J. Mol. Biol. 2007, 370, 714–727. [Google Scholar] [CrossRef]
- Kristensen, A.K.; Schou, C.; Roepstorff, P. Determination of isoforms, N-linked glycan structure and disulfide bond linkages of the major cat allergen Fel d1 by a mass spectrometric approach. Biol. Chem. 1997, 378, 899–908. [Google Scholar] [CrossRef]
- Bienboire-Frosini, C.; Lebrun, R.; Vervloet, D.; Pageat, P.; Ronin, C. Distribution of core fragments from the major cat allergen Fel d 1 is maintained among the main anatomical sites of production. Int. Arch. Allergy Immunol. 2010, 152, 197–206. [Google Scholar] [CrossRef]
- Bienboire-Frosini, C.; Lebrun, R.; Vervloet, D.; Pageat, P.; Ronin, C. Variable content of Fel d 1 variants in house dust and cat extracts may have an impact on allergen measurement. J. Investig. Allergol. Clin. Immunol. 2012, 22, 270–279. [Google Scholar]
- Liccardi, G.; D’Amato, G.; Russo, M.; Canonica, G.W.; D’Amato, L.; De Martino, M.; Passalacqua, G. Focus on cat allergen (Fel d 1): Immunological and aerodynamic characteristics, modality of airway sensitization and avoidance strategies. Int. Arch. Allergy Immunol. 2003, 132, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zahradnik, E.; Raulf, M. Animal allergens and their presence in the environment. Front. Immunol. 2014, 5, 76. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.B.; Zhang, Z.; Chilton, B.S. Uteroglobin: A steroid-inducible immunomodulatory protein that founded the Secretoglobin superfamily. Endocr. Rev. 2007, 28, 707–725. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, S.; Cai, Y.; Murata, M.; Tomita, T.; Yoneda, M.; Xu, L.; Pilon, A.L.; Cachau, R.E.; Kimura, S. A novel pathway of LPS uptake through syndecan-1 leading to pyroptotic cell death. Elife 2018, 7, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Chiba, Y.; Kurotani, R.; Kusakabe, T.; Miura, T.; Link, B.W.; Misawa, M.; Kimura, S. Uteroglobin-related protein 1 expression suppresses allergic airway inflammation in mice. Am. J. Respir. Crit. Care Med. 2006, 173, 958–964. [Google Scholar] [CrossRef]
- Maccioni, M.; Riera, C.M.; Rivero, V.E. Identification of rat prostatic steroid binding protein (PSBP) as an immunosuppressive factor. J. Reprod. Immunol. 2001, 50, 133–149. [Google Scholar] [CrossRef]
- Karn, R.C. Evolution of Rodent Pheromones: A Review of the ABPs with Comparison to the ESPs and the MUPs. Int. J. Biochem. Res. Rev. 2013, 3, 328–363. [Google Scholar] [CrossRef]
- Chung, A.G.; Belone, P.M.; Bímová, B.V.; Karn, R.C.; Laukaitis, C.M. Studies of an Androgen-Binding Protein Knockout Corroborate a Role for Salivary ABP in Mouse Communication. Genetics 2017, 205, 1517–1527. [Google Scholar] [CrossRef]
- Austin, C.J.; Emberson, L.; Nicholls, P. Purification and characterization of pheromaxein, the porcine steroid-binding protein. A member of the secretoglobin superfamily. Eur. J. Biochem. 2004, 271, 2593–2606. [Google Scholar] [CrossRef]
- Carayol, N.; Birnbaum, J.; Magnan, A.; Ramadour, M.; Lanteaume, A.; Vervloet, D.; Tessier, Y.; Pageat, P. Fel d 1 production in the cat skin varies according to anatomical sites. Allergy 2000, 55, 570–573. [Google Scholar] [CrossRef]
- Pageat, P.; Gaultier, E. Current research in canine and feline pheromones. Vet. Clin. N. Am. Small Anim. Pract. 2003, 33, 187–211. [Google Scholar] [CrossRef]
- Bienboire-Frosini, C.; Cozzi, A.; Lafont-Lecuelle, C.; Vervloet, D.; Ronin, C.; Pageat, P. Immunological differences in the global release of the major cat allergen Fel d 1 are influenced by sex and behaviour. Vet. J. 2012, 193, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Ligabue-Braun, R.; Sachett, L.G.; Pol-Fachin, L.; Verli, H. The Calcium Goes Meow: Effects of Ions and Glycosylation on Fel d 1, the Major Cat Allergen. PLoS ONE 2015, 10, e0132311. [Google Scholar] [CrossRef] [PubMed]
- Karn, R.C. The mouse salivary androgen-binding protein (ABP) alpha subunit closely resembles chain 1 of the cat allergen Fel dI. Biochem. Genet. 1994, 32, 271–277. [Google Scholar] [CrossRef]
- Durairaj, R.; Pageat, P.; Bienboire-Frosini, C. Another cat and mouse game: Deciphering the evolution of the SCGB superfamily and exploring the molecular similarity of major cat allergen Fel d 1 and mouse ABP using computational approaches. PLoS ONE 2018, 13, e0197618. [Google Scholar] [CrossRef]
- Karn, R.C.; Clements, M.A. A comparison of the structures of the alpha:beta and alpha:gamma dimers of mouse salivary androgen-binding protein (ABP) and their differential steroid binding. Biochem. Genet. 1999, 37, 187–199. [Google Scholar] [CrossRef]
- Chapman, M.D.; Smith, A.M.; Vailes, L.D.; Arruda, L.K.; Dhanaraj, V.; Pomés, A. Recombinant allergens for diagnosis and therapy of allergic disease. J. Allergy Clin. Immunol. 2000, 106, 409–418. [Google Scholar] [CrossRef]
- Vailes, L.D.; Sun, A.W.; Ichikawa, K.; Wu, Z.; Sulahian, T.H.; Chapman, M.D.; Guyre, P.M. High-level expression of immunoreactive recombinant cat allergen (Fel d 1): Targeting to antigen-presenting cells. J. Allergy Clin. Immunol. 2002, 110, 757–762. [Google Scholar] [CrossRef][Green Version]
- Chen, X.; Zaro, J.L.; Shen, W.C. Fusion protein linkers: Property, design and functionality. Adv. Drug Deliv. Rev. 2013, 65, 1357–1369. [Google Scholar] [CrossRef]
- Wuenschmann, S.; Vailes, L.D.; King, E.; Aalberse, R.C.; Chapman, M.D. Expression of a Deglycosylated Recombinant Fel d 1 in Pichia pastoris. J. Allergy Clin. Immunol. 2008, 121, S214. [Google Scholar] [CrossRef]
- Vitale Shreve, K.R.; Udell, M.A.R. Stress, security, and scent: The influence of chemical signals on the social lives of domestic cats and implications for applied settings. Appl. Anim. Behav. Sci. 2017, 187, 69–76. [Google Scholar] [CrossRef]
- Mills, D.S.; White, J.C. Long-term follow up of the effect of a pheromone therapy on feline spraying behaviour. Vet. Rec. 2000, 147, 746–747. [Google Scholar] [PubMed]
- Mills, D.S.; Mills, C. Evaluation of a novel method for delivering a synthetic analogue of feline facial pheromone to control urine spraying by cats. Vet. Rec. 2001, 149, 197–199. [Google Scholar] [CrossRef]
- Kronen, P.W.; Ludders, J.W.; Erb, H.N.; Moon, P.F.; Gleed, R.D.; Koski, S. A synthetic fraction of feline facial pheromones calms but does not reduce struggling in cats before venous catheterization. Vet. Anaesth. Analg. 2006, 33, 258–265. [Google Scholar] [CrossRef]
- Pereira, J.S.; Fragoso, S.; Beck, A.; Lavigne, S.; Varejão, A.S.; da Graça Pereira, G. Improving the feline veterinary consultation: The usefulness of Feliway spray in reducing cats’ stress. J. Feline Med. Surg. 2016, 18, 959–964. [Google Scholar] [CrossRef]
- Cozzi, A.; Monneret, P.; Lafont-Lecuelle, C.; Bougrat, L.; Gaultier, E.; Pageat, P. The maternal cat appeasing pheromone: Exploratory study of the effects on aggressive and affiliative interactions in cats. J. Vet. Behav. Clin. Appl. Res. 2010, 5, 37–38. [Google Scholar] [CrossRef]
- DePorter, T.L.; Bledsoe, D.L.; Beck, A.; Ollivier, E. Evaluation of the efficacy of an appeasing pheromone diffuser product vs placebo for management of feline aggression in multi-cat households: A pilot study. J. Feline Med. Surg. 2019, 21, 293–305. [Google Scholar] [CrossRef]
- Fridlansky, F.; Milgrom, E. Interaction of uteroglobin with progesterone, 5αpregnane-3, 20-dione and estrogens. Endocrinology 1976, 99, 1244–1251. [Google Scholar] [CrossRef]
- Karn, R.C. Steroid binding by mouse salivary proteins. Biochem. Genet. 1998, 36, 105–117. [Google Scholar] [CrossRef]
- Chen, C.; Schilling, K.; Hiipakka, R.A.; Huang, I.Y.; Liao, S. Prostate α-protein. Isolation and characterization of the polypeptide components and cholesterol binding. J. Biol. Chem. 1982, 257, 116–121. [Google Scholar]
- Taylor, R.D.; Jewsbury, P.J.; Essex, J.W. A review of protein-small molecule docking methods. J. Comput. Aided Mol. Des. 2002, 16, 151–166. [Google Scholar] [CrossRef]
- Pelosi, P.; Zhu, J.; Knoll, W. From radioactive ligands to biosensors: Binding methods with olfactory proteins. Appl. Microbiol. Biotechnol. 2018, 102, 8213–8227. [Google Scholar] [CrossRef]
- Dorries, K.M.; Adkins-Regan, E.; Halpern, B.P. Sensitivity and behavioral responses to the pheromone androstenone are not mediated by the vomeronasal organ in domestic pigs. Brain Behav. Evol. 1997, 49, 53–62. [Google Scholar] [CrossRef]
- Guiraudie-Capraz, G.; Pageat, P.; Cain, A.H.; Madec, I.; Nagnan-Le Meillour, P. Functional characterization of olfactory binding proteins for appeasing compounds and molecular cloning in the vomeronasal organ of pre-pubertal pigs. Chem. Senses 2003, 28, 609–619. [Google Scholar] [CrossRef]
- Nagnan-Le Meillour, P.; Joly, A.; Le Danvic, C.; Marie, A.; Zirah, S.; Cornard, J.P. Binding specificity of native odorant-binding protein isoforms is driven by phosphorylation and O-N-acetylglucosaminylation in the pig Sus scrofa. Front. Endocrinol. 2019, 9, 816. [Google Scholar] [CrossRef]
- Pelosi, P. The role of perireceptor events in vertebrate olfaction. Cell. Mol. Life Sci. 2001, 58, 503–509. [Google Scholar] [CrossRef]
- Stopkova, R.; Klempt, P.; Kuntova, B.; Stopka, P. On the tear proteome of the house mouse (Mus musculus musculus) in relation to chemical signalling. PeerJ 2017, 5, e3541. [Google Scholar] [CrossRef]
- Morgenstern, J.P.; Griffith, I.J.; Brauer, A.W.; Rogers, B.L.; Bond, J.F.; Chapman, M.D.; Kuo, M.-C. Amino acid sequence of Fel dI, the major allergen of the domestic cat: Protein sequence analysis and cDNA cloning. Proc. Natl. Acad. Sci. USA 1991, 88, 9690–9694. [Google Scholar] [CrossRef]
- Emes, R.D.; Riley, M.C.; Laukaitis, C.M.; Goodstadt, L.; Karn, R.C.; Ponting, C.P. Comparative evolutionary genomics of androgen-binding protein genes. Genome Res. 2004, 14, 1516–1529. [Google Scholar] [CrossRef]
- Jackson, B.C.; Thompson, D.C.; Wright, M.W.; McAndrews, M.; Bernard, A.; Nebert, D.W.; Vasiliou, V. Update of the human secretoglobin (SCGB) gene superfamily and an example of “evolutionary bloom” of androgen-binding protein genes within the mouse Scgb gene superfamily. Hum. Genomics 2011, 5, 691–702. [Google Scholar] [CrossRef]
- Golebiowski, J.; Topin, J.; Charlier, L.; Briand, L. Interaction between odorants and proteins involved in the perception of smell: The case of odorant-binding proteins probed by molecular modelling and biophysical data. Flavour Fragr. J. 2012, 27, 445–453. [Google Scholar] [CrossRef]
- Grönlund, H.; Bergman, T.; Sandström, K.; Alvelius, G.; Reininger, R.; Verdino, P.; Hauswirth, A.; Liderot, K.; Valent, P.; Spitzauer, S.; et al. Formation of disulfide bonds and homodimers of the major cat allergen Fel d 1 equivalent to the natural allergen by expression in Escherichia coli. J. Biol. Chem. 2003, 278, 40144–40151. [Google Scholar] [CrossRef] [PubMed]
- Le Danvic, C.; Guiraudie-Capraz, G.; Abderrahmani, D.; Zanetta, J.P.; Nagnan-Le Meillour, P. Natural ligands of porcine olfactory binding proteins. J. Chem. Ecol. 2009, 35, 741–751. [Google Scholar] [CrossRef] [PubMed]
- Brimau, F.; Cornard, J.P.; Le Danvic, C.; Lagant, P.; Vergoten, G.; Grebert, D.; Pajot, E.; Nagnan-Le Meillour, P. Binding specificity of recombinant odorant-binding protein isoforms is driven by phosphorylation. J. Chem. Ecol. 2010, 36, 801–813. [Google Scholar] [CrossRef] [PubMed]
- Burger, B.V. Mammalian semiochemicals. In The Chemistry of Pheromones and Other Semiochemicals II; Springer: Berlin/Heidelberg, Germany, 2005; pp. 231–278. [Google Scholar]
- Beynon, R.J.; Armstrong, S.D.; Gómez-Baena, G.; Lee, V.; Simpson, D.; Unsworth, J.; Hurst, J.L. The complexity of protein semiochemistry in mammals. Biochem. Soc. Trans. 2014, 42, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, J.W.S.; Cameron-Beaumont, C. The signalling repertoire of the domestic cat and its undomesticated relatives. In The Domestic Cat, The Biology of Its Behaviour; Turner, D.C., Bateson, P., Eds.; Cambridge University Press: Cambridge, UK, 2000; pp. 68–93. [Google Scholar]
- Natoli, E.; De Vito, E. Agonistic behaviour, dominance rank and copulatory success in a large multi-male feral cat, Felis catus L., colony in central Rome. Anim. Behav. 1991, 42, 227–241. [Google Scholar] [CrossRef]
- Natoli, E. Behavioural Responses of Urban Feral Cats to Different Types of Urine Marks. Behaviour 1985, 94, 234–243. [Google Scholar] [CrossRef]
- Smith, J.L.D.; McDougal, C.; Miquelle, D. Scent marking in free-ranging tigers, Panthera tigris. Anim. Behav. 1989, 37, 1–10. [Google Scholar] [CrossRef]
- De Groot, H.; van Swieten, P.; Aalberse, R.C. Evidence for a Fel d I-like molecule in the “big cats” (Felidae species). J. Allergy Clin. Immunol. 1990, 86, 107–116. [Google Scholar] [CrossRef]
- Burgos, T.; Virgós, E.; Valero, E.S.; Arenas-Rojas, R.; Rodríguez-Siles, J.; Recio, M.R. Prey density determines the faecal-marking behaviour of a solitary predator, the Iberian lynx (Lynx pardinus). Ethol. Ecol. Evol. 2019, 31, 219–230. [Google Scholar] [CrossRef]
- Darden, S.K.; Steffensen, L.K.; Dabelsteen, T. Information transfer among widely spaced individuals: Latrines as a basis for communication networks in the swift fox? Anim. Behav. 2008, 75, 425–432. [Google Scholar] [CrossRef]
- Tegoni, M.; Pelosi, P.; Vincent, F.; Spinelli, S.; Grolli, S.; Ramoni, R.; Cambillau, C. Mammalian odorant binding proteins. Biochim. Biophys. Acta 2000, 1482, 229–240. [Google Scholar] [CrossRef]
- Papes, F.; Logan, D.W.; Stowers, L. The Vomeronasal Organ Mediates Interspecies Defensive Behaviors through Detection of Protein Pheromone Homologs. Cell 2010, 141, 692–703. [Google Scholar] [CrossRef]
- Callebaut, I.; Poupon, A.; Bally, R.; Demaret, J.P.; Housset, D.; Delettre, J.; Hossenlopp, P.; Mornon, J.P. The uteroglobin fold. Ann. N. Y. Acad. Sci. 2000, 923, 90–112. [Google Scholar] [CrossRef]
- Herre, J.; Grönlund, H.; Brooks, H.; Hopkins, L.; Waggoner, L.; Murton, B.; Gangloff, M.; Opaleye, O.; Chilvers, E.R.; Fitzgerald, K.; et al. Allergens as immunomodulatory proteins: The cat dander protein Fel d 1 enhances TLR activation by lipid ligands. J. Immunol. 2013, 191, 1529–1535. [Google Scholar] [CrossRef]
- Bublin, M.; Eiwegger, T.; Breiteneder, H. Do lipids influence the allergic sensitization process? J. Allergy Clin. Immunol. 2014, 134, 521–529. [Google Scholar] [CrossRef]
- Satyaraj, E.; Wedner, H.J.; Bousquet, J. Keep the cat, change the care pathway: A transformational approach to managing Fel d 1, the major cat allergen. Allergy Eur. J. Allergy Clin. Immunol. 2019, 74, 5–17. [Google Scholar] [CrossRef]
- Satyaraj, E.; Gardner, C.; Filipi, I.; Cramer, K.; Sherrill, S. Reduction of active Fel d1 from cats using an antiFel d1 egg IgY antibody. Immun. Inflamm. Dis. 2019, 7, 68–73. [Google Scholar] [CrossRef]
- Thoms, F.; Jennings, G.T.; Maudrich, M.; Vogel, M.; Haas, S.; Zeltins, A.; Hofmann-Lehmann, R.; Riond, B.; Grossmann, J.; Hunziker, P.; et al. Immunization of cats to induce neutralizing antibodies against Fel d 1, the major feline allergen in human subjects. J. Allergy Clin. Immunol. 2019, 144, 193–203. [Google Scholar] [CrossRef]
- Jayaram, B.; Singh, T.; Mukherjee, G.; Mathur, A.; Shekhar, S.; Shekhar, V. Sanjeevini: A freely accessible web-server for target directed lead molecule discovery. BMC Bioinform. 2012, 13, S7. [Google Scholar] [CrossRef]
- Bikadi, Z.; Hazai, E. Application of the PM6 semi-empirical method to modeling proteins enhances docking accuracy of AutoDock. J. Cheminform. 2009, 1, 15. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Solis, F.J.; Wets, R.J.-B. Minimization by Random Search Techniques. Math. Oper. Res. 1981, 6, 19–30. [Google Scholar] [CrossRef]
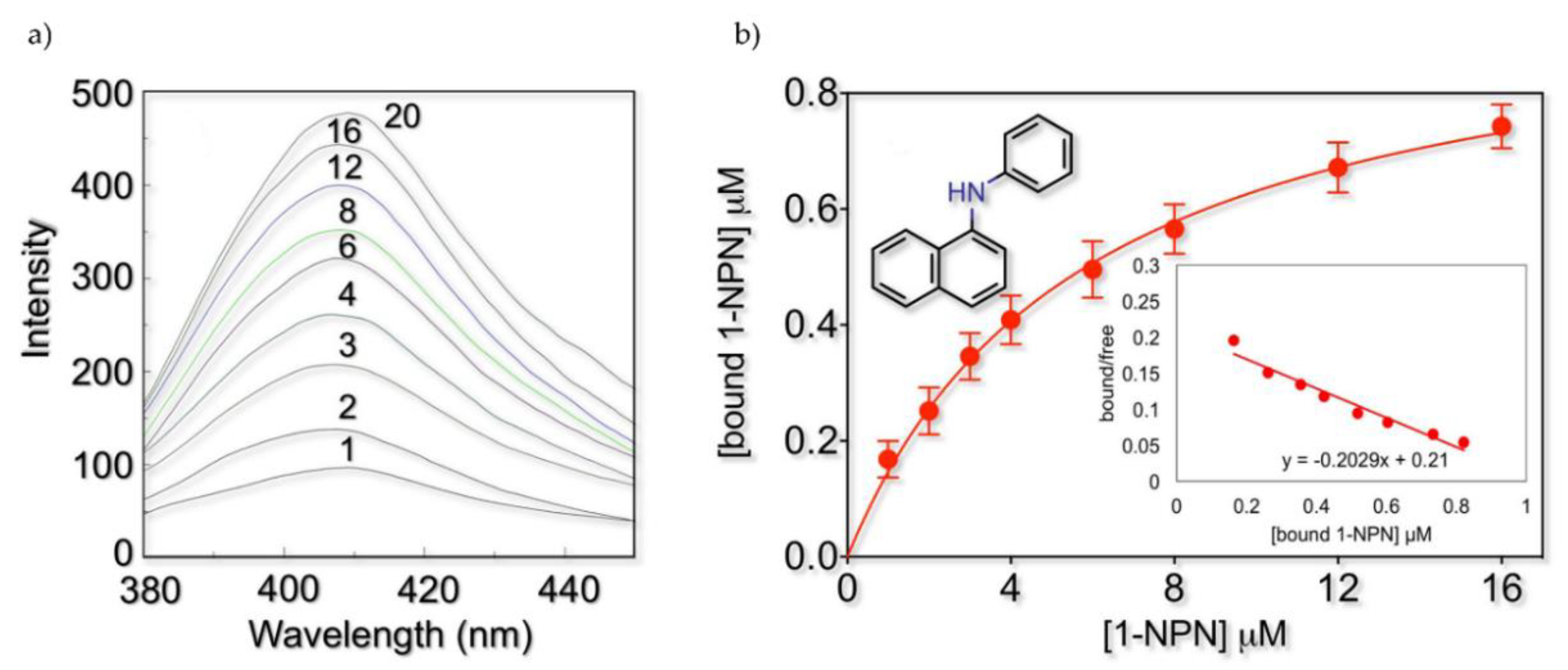
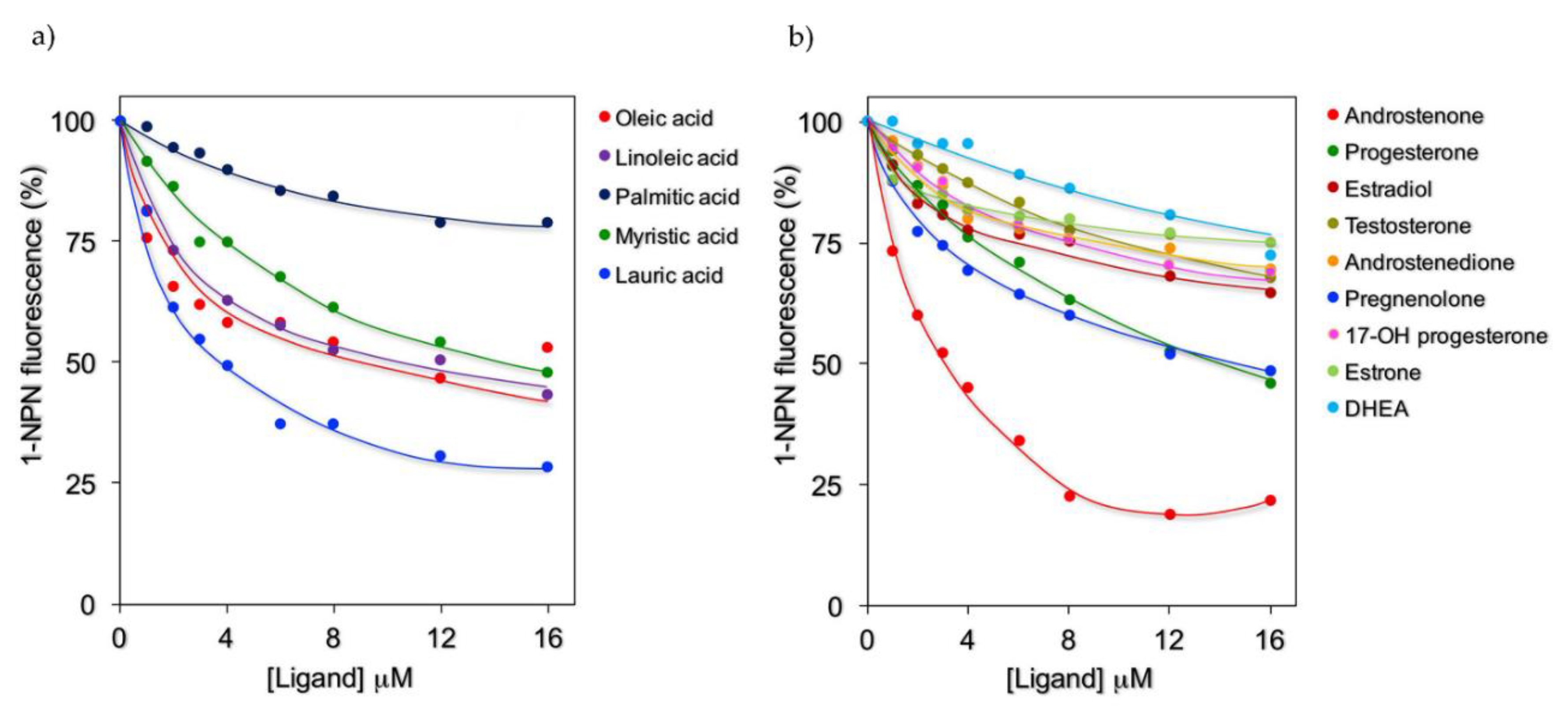
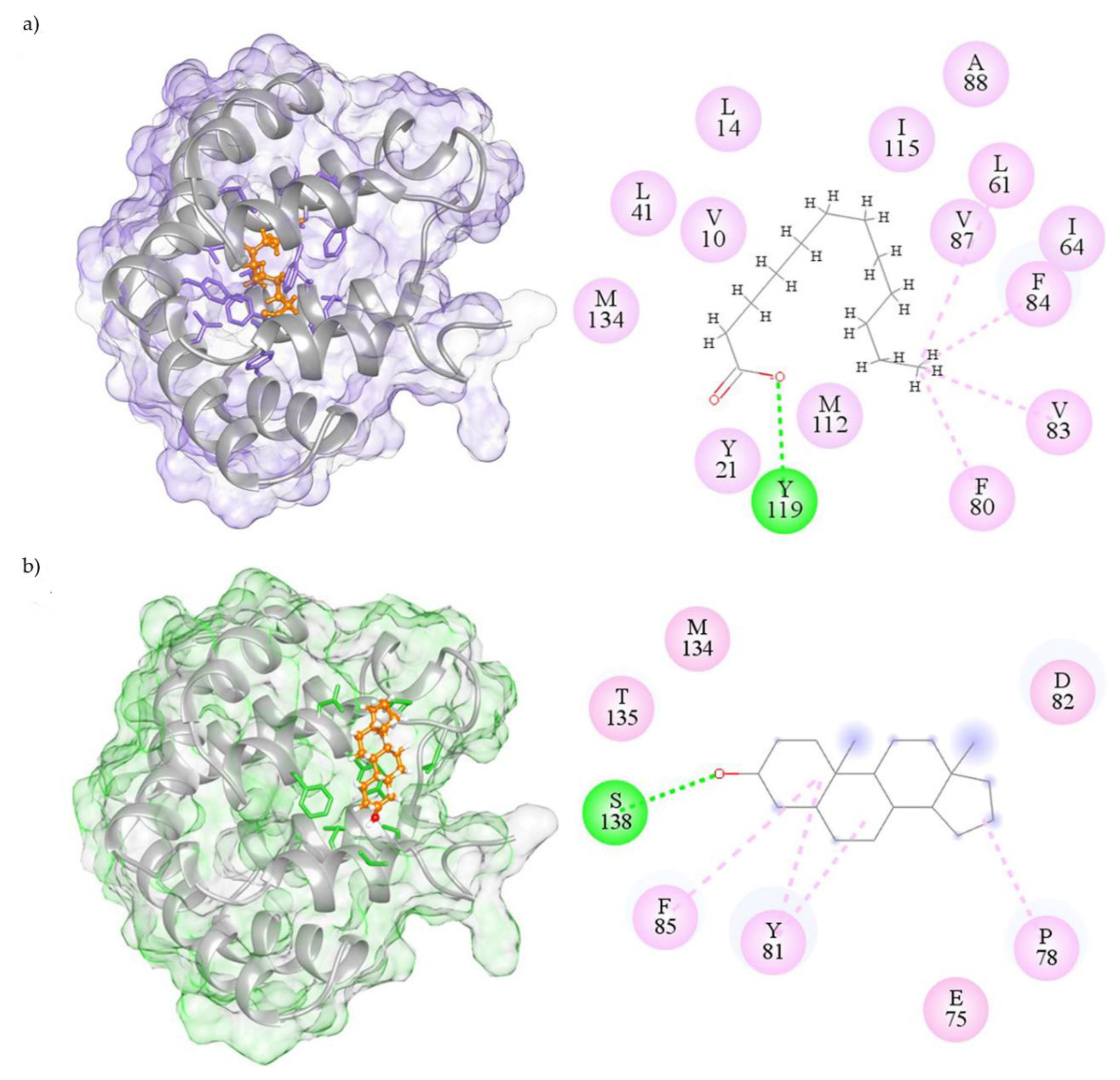
| n° | Compound Names | Estimated Free Energy of Binding (kcal/mol) | Total Intermolecular Energy (kcal/mol) | Frequency | H-Bond Residue | Hydrophobic Residue (Alkyl/Pi-Alkyl/Pi-Sigma) | In Silico Screening a | 1-NPN Displacement Screening |
|---|---|---|---|---|---|---|---|---|
| Fatty Acids and Other Derivatives | ||||||||
| 1 | Isobutyric acid | −2.96 | −3.26 | 27% | A88, Y119, L129 | No | ND | |
| 2 | Capric Acid | −5.16 | −7.4 | 23% | L61, F80, V83, F84 | No | No | |
| 3 | Lauric Acid | −5.84 | −8.58 | 60% | Y119 | L61, F80, V83, F84 | Yes | Yes |
| 4 | Myristic Acid | −3.35 | −7.02 | 36% | F84 | F13, V133, M134, I137 | Yes | Yes |
| 5 | Palmitic Acid | −2.33 | −5.88 | 16% | F84 | V133, M134 | Yes | Yes |
| 6 | Oleic Acid | −2.82 | −7.05 | 50% | M134 | I64, F80, V83 | Yes | Yes |
| 7 | Linoleic Acid | −2.95 | −6.88 | 40% | P78 | A88, Y119, L129 | Yes | Yes |
| 8 | Dodecanal | −4.88 | −7.42 | 2% | F84, M134 | No | No | |
| 9 | Dodecanol | −3.93 | −7.02 | 2% | F84, V133, M134 | No | No | |
| 10 | Tetradecanol | −3.97 | −7.89 | 6% | P78, Y81 | No | No | |
| 11 | Ethyl Laurate | −4.7 | −8.02 | 12% | F84 | L61, I64, F80, V83, V133, M134, I137 | Yes | No |
| 12 | Methyl Palmitate | −2.53 | −6.67 | 20% | T76 | Yes | No | |
| 13 | Nonanamide | −4.53 | −6.51 | 4% | L61, I64, V83, F80 | No | ND | |
| 14 | Hexadecanamide | −2.84 | −6.3 | 18% | T135 | Y81 | Yes | ND |
| 15 | Octadecanamide | −2.81 | −6.96 | 6% | G131 | Y81, F85 | Yes | ND |
| Steroids | ||||||||
| 1 | Androstenone | −5.84 | −5.84 | 65% | S138 | P78, Y81, F85 | Yes | Yes |
| 2 | Androstenedione | −5.83 | −5.83 | 44% | Y81, F85 | Yes | Yes | |
| 3 | Androstenol | −5.06 | −5.36 | 22% | Y81, F85 | No | No | |
| 4 | Progesterone | −5.74 | −6.04 | 62% | T76 | Y81, F85 | Yes | Yes |
| 5 | Hydroxyprogesterone | −5.14 | −5.54 | 39% | Y81 | F85 | Yes | Yes |
| 6 | Pregnenolone | −5.59 | −6.17 | 58% | T76 | Y81, F85 | Yes | Yes |
| 7 | Estradiol | −4.94 | −5.54 | 26% | T76 | Y81, F85 | Yes | Yes |
| 8 | Testosterone | −5.6 | −5.9 | 35% | T76 | Y81, F85 | Yes | Yes |
| 9 | Dihydrotestosterone | −5.06 | −5.35 | 12% | Y81, F85 | No | No | |
| 10 | Estrone | −3.56 | −3.86 | 10% | D82, G131 | F85 | Yes | Yes |
| 11 | Dehydroepiandrosterone (DHEA) | −4.64 | −4.94 | 30% | Y81, F85 | Yes | Yes | |
| 12 | Corticosterone | −5.35 | −6.38 | 30% | T76, N89 | Y81, F85 | Yes | No |
| 13 | Deoxycorticosterone | −4.99 | −5.29 | 12% | Y81, F85 | No | No | |
| Fluorescent Probe | ||||||||
| 1 | 1-NPN (Central) | −6.7 | −7.41 | 50% | Y119 | L14, L61, M112 | Yes | / |
| 2 | 1-NPN (Surface) | −4.74 | −5.45 | 30% | Y81, F85 | Yes | / | |
| Ligand | (IC50) (µM) | Kd (µM) |
|---|---|---|
| Lauric acid | 3.3 | 2.6 |
| Oleic acid | 10.0 | 7.7 |
| Linoleic acid | 10.1 | 7.8 |
| Myristic acid | 14.4 | 11.1 |
| Androstenone | 3.1 | 2.4 |
| Pregnenolone | 13.1 | 10.1 |
| Progesterone | 13.6 | 10.5 |
| n° | Compounds | PubChem Compound ID (CID) | Chemical Formula | Molecular Weight (g/mol) | H-Bond Donor | H-Bond Acceptor | Topological Polar Surface Area (Ų) | Lipinski Rule of Five (RO5) |
|---|---|---|---|---|---|---|---|---|
| Fatty Acids and Their Derivatives | ||||||||
| 1 | Isobutyric acid | CID_6590 | C4H8O2 | 88.106 | 1 | 2 | 37.3 | 0 |
| 2 | Capric acid | CID_2969 | C10H20O2 | 172.268 | 1 | 2 | 37.3 | 0 |
| 3 | Lauric acid | CID_3893 | C12H24O2 | 200.322 | 1 | 2 | 37.3 | 0 |
| 4 | Myristic acid | CID_11005 | C14H28O2 | 228.376 | 1 | 2 | 37.3 | 0 |
| 5 | Palmitic acid | CID_985 | C16H32O2 | 256.43 | 1 | 2 | 37.3 | 1 |
| 6 | Oleic acid | CID_445639 | C18H34O2 | 282.468 | 1 | 2 | 37.3 | 1 |
| 7 | Linoleic acid | CID_5280450 | C18H32O2 | 280.442 | 1 | 2 | 37.3 | 1 |
| 8 | Dodecanal | CID_8194 | C12H24O | 184.323 | 0 | 1 | 17.1 | 0 |
| 9 | Dodecanol | CID_8193 | C12H26O | 186.339 | 1 | 1 | 20.2 | 0 |
| 10 | Tetradecanol | CID_8209 | C14H30O | 214.393 | 1 | 1 | 20.2 | 0 |
| 11 | Ethyl Laurate | CID_7800 | C14H28O2 | 228.376 | 0 | 2 | 26.3 | 0 |
| 12 | Methyl palmitate | CID_8181 | C17H34O2 | 270.457 | 0 | 2 | 26.3 | 1 |
| 13 | Nonanamide | CID_70709 | C9H19NO | 157.257 | 1 | 1 | 43.1 | 0 |
| 14 | Hexadecanamide | CID_69421 | C16H33NO | 255.446 | 1 | 1 | 43.1 | 0 |
| 15 | Octadecanamide | CID_31292 | C18H37NO | 283.5 | 1 | 1 | 43.1 | 1 |
| Steroids | ||||||||
| 1 | Androstenone | CID_6852393 | C19H28O | 272.432 | 0 | 1 | 17.1 | 1 |
| 2 | Androstenedione | CID_6128 | C19H26O2 | 286.415 | 0 | 2 | 34.1 | 0 |
| 3 | Androstenol | CID_101989 | C19H30O | 274.448 | 1 | 1 | 20.2 | 1 |
| 4 | Progesterone | CID_5994 | C21H30O2 | 314.469 | 0 | 2 | 34.1 | 0 |
| 5 | Hydroxyprogesterone | CID_6238 | C21H30O3 | 330.468 | 1 | 3 | 54.4 | 0 |
| 6 | Pregnenolone | CID_8955 | C21H32O2 | 316.485 | 1 | 2 | 37.3 | 0 |
| 7 | Estradiol | CID_5757 | C18H24O2 | 272.388 | 2 | 2 | 40.5 | 0 |
| 8 | Testosterone | CID_6013 | C19H28O2 | 288.431 | 1 | 2 | 37.3 | 0 |
| 9 | Dihydrotestosterone | CID_10635 | C19H30O2 | 290.447 | 1 | 2 | 37.3 | 0 |
| 10 | Estrone | CID_5870 | C18H22O2 | 270.372 | 1 | 2 | 37.3 | 0 |
| 11 | Dehydroepiandrosterone (DHEA) | CID_5881 | C19H28O2 | 288.431 | 1 | 2 | 37.3 | 0 |
| 12 | Corticosterone | CID_5753 | C21H30O4 | 346.467 | 2 | 4 | 74.6 | 0 |
| 13 | Deoxycorticosterone | CID_6166 | C21H30O3 | 330.468 | 1 | 3 | 54.4 | 0 |
| Fluorescent Probe | ||||||||
| 1 | N-phenyl-1-naphthylamine (1-NPN) | CID_7013 | C16H13N | 219.287 | 1 | 1 | 12 | 0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bienboire-Frosini, C.; Durairaj, R.; Pelosi, P.; Pageat, P. The Major Cat Allergen Fel d 1 Binds Steroid and Fatty Acid Semiochemicals: A Combined In Silico and In Vitro Study. Int. J. Mol. Sci. 2020, 21, 1365. https://doi.org/10.3390/ijms21041365
Bienboire-Frosini C, Durairaj R, Pelosi P, Pageat P. The Major Cat Allergen Fel d 1 Binds Steroid and Fatty Acid Semiochemicals: A Combined In Silico and In Vitro Study. International Journal of Molecular Sciences. 2020; 21(4):1365. https://doi.org/10.3390/ijms21041365
Chicago/Turabian StyleBienboire-Frosini, Cécile, Rajesh Durairaj, Paolo Pelosi, and Patrick Pageat. 2020. "The Major Cat Allergen Fel d 1 Binds Steroid and Fatty Acid Semiochemicals: A Combined In Silico and In Vitro Study" International Journal of Molecular Sciences 21, no. 4: 1365. https://doi.org/10.3390/ijms21041365
APA StyleBienboire-Frosini, C., Durairaj, R., Pelosi, P., & Pageat, P. (2020). The Major Cat Allergen Fel d 1 Binds Steroid and Fatty Acid Semiochemicals: A Combined In Silico and In Vitro Study. International Journal of Molecular Sciences, 21(4), 1365. https://doi.org/10.3390/ijms21041365







