Effect of Pepper-Containing Diets on the Diversity and Composition of Gut Microbiome of Drosophila melanogaster
Abstract
1. Introduction
2. Results
2.1. Phytochemical Content
2.2. Sequencing Data
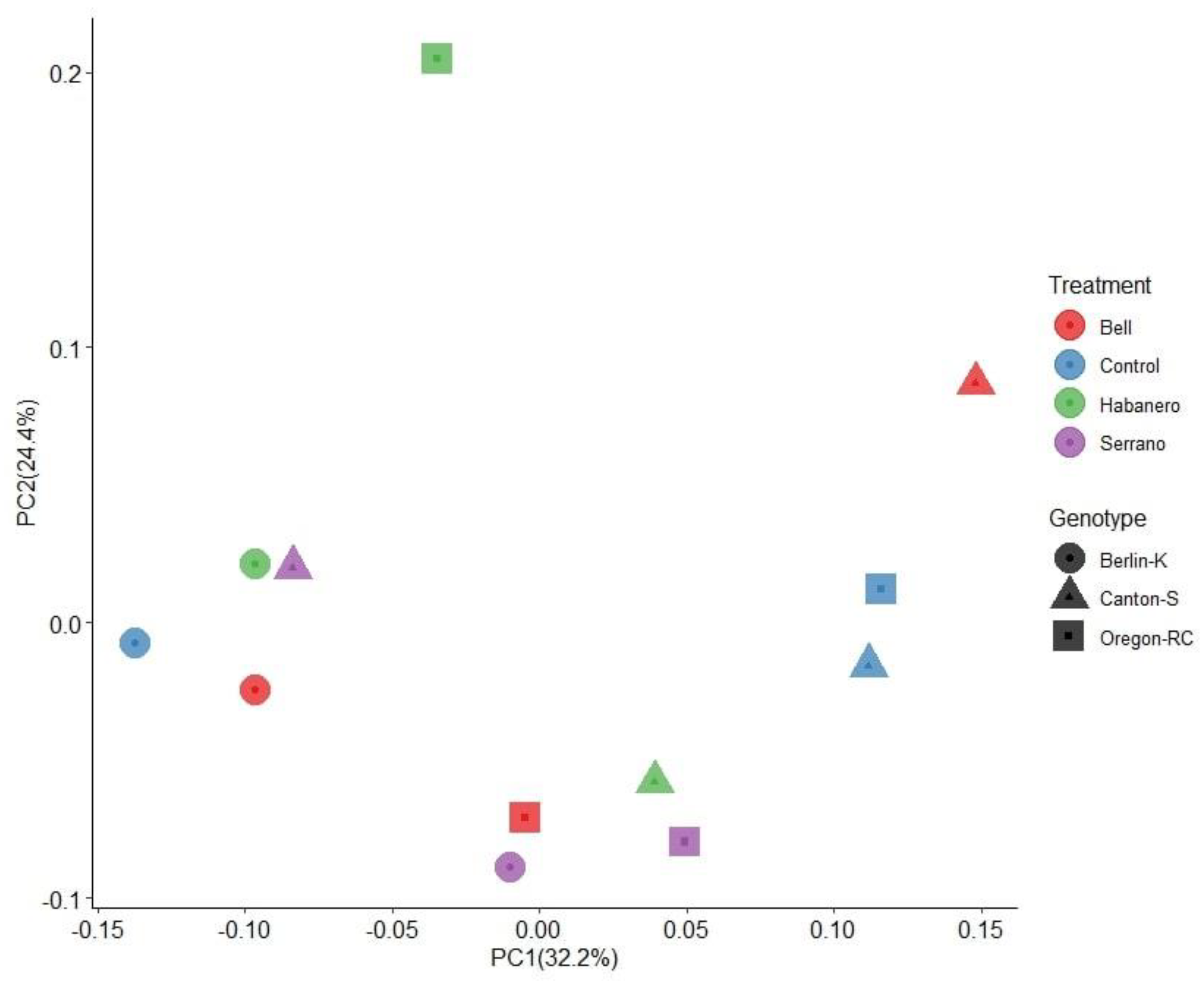
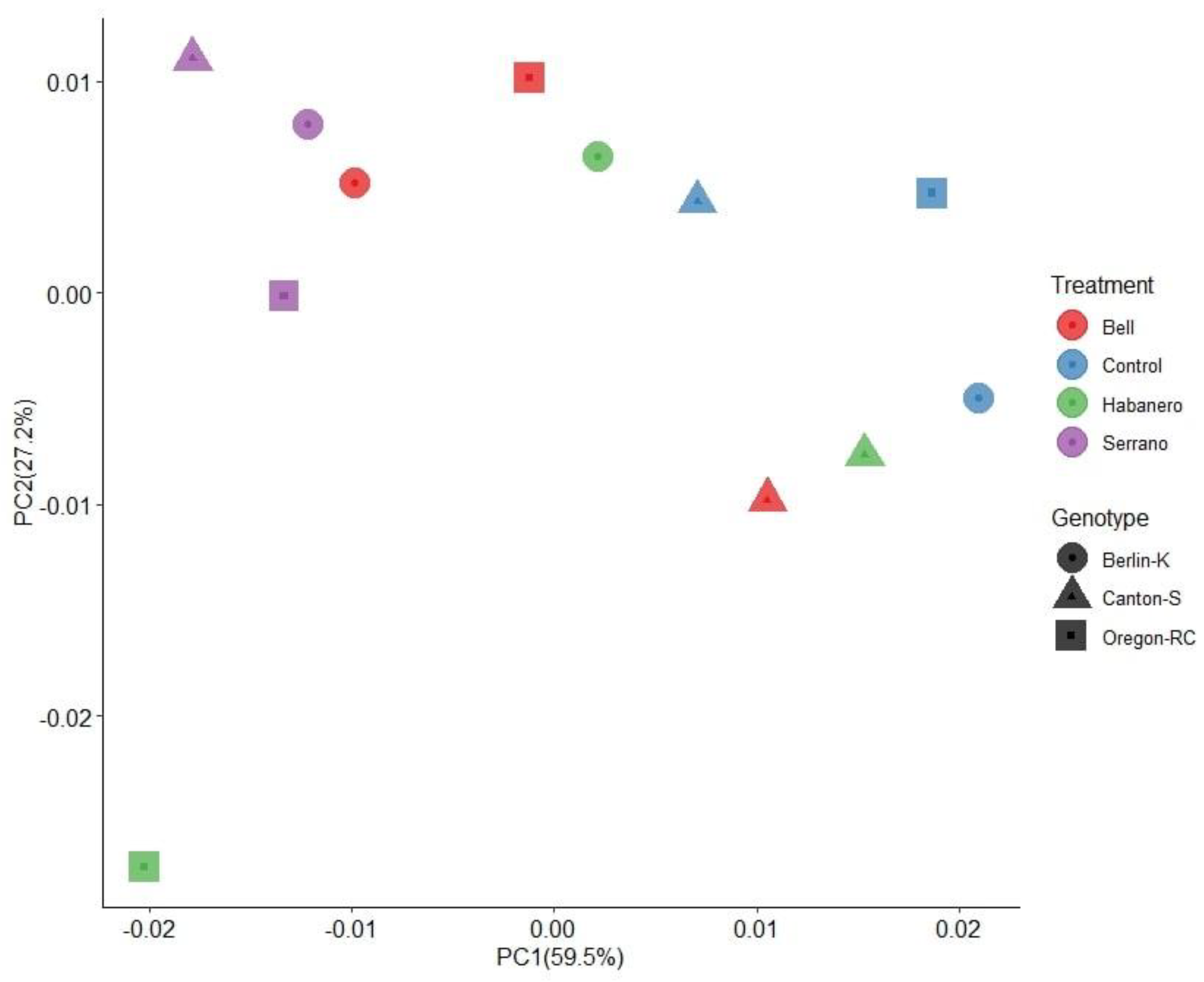
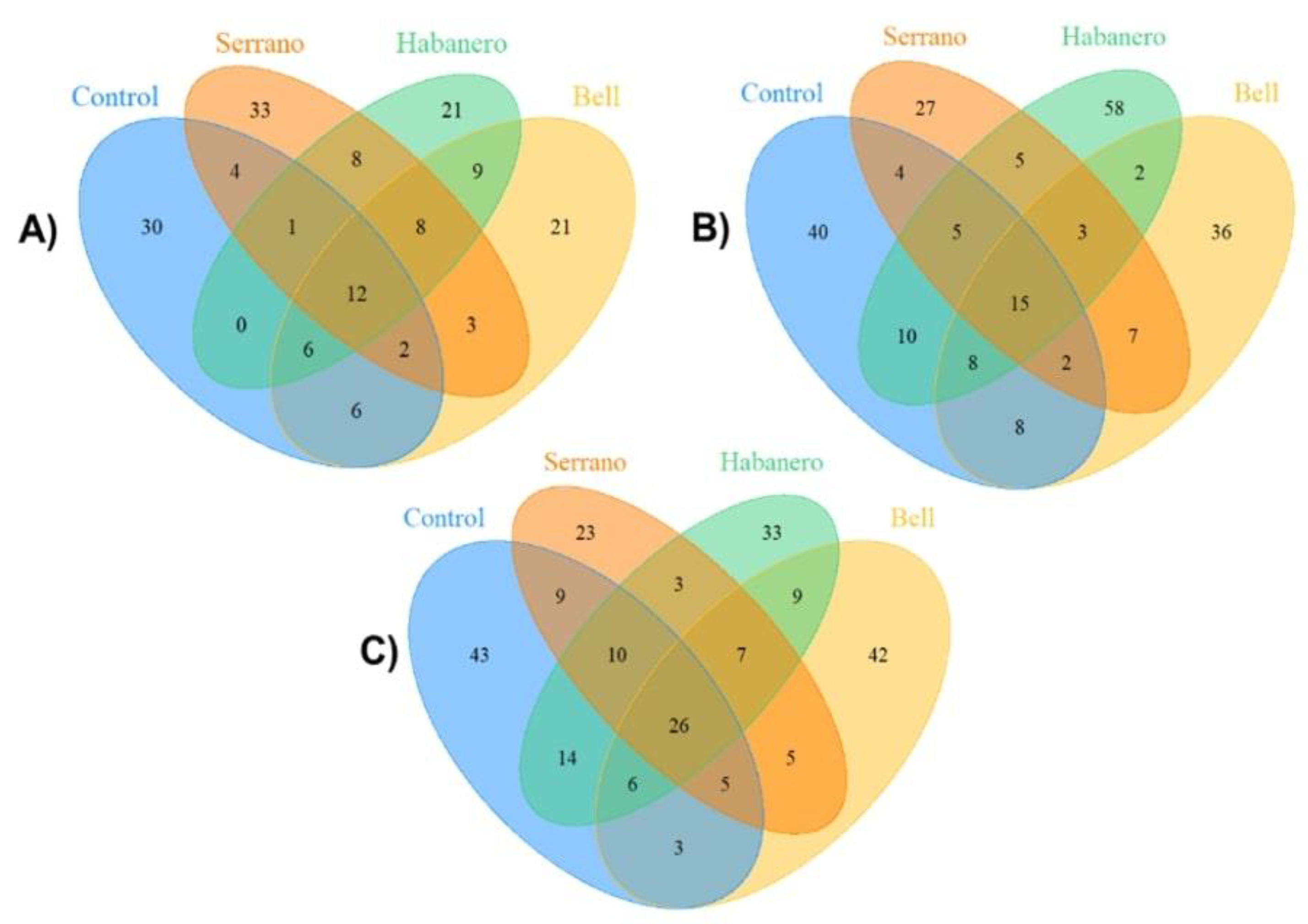
2.3. Gut Microbiome Structure of D. melanogaster Strains Reared on the Different Diets
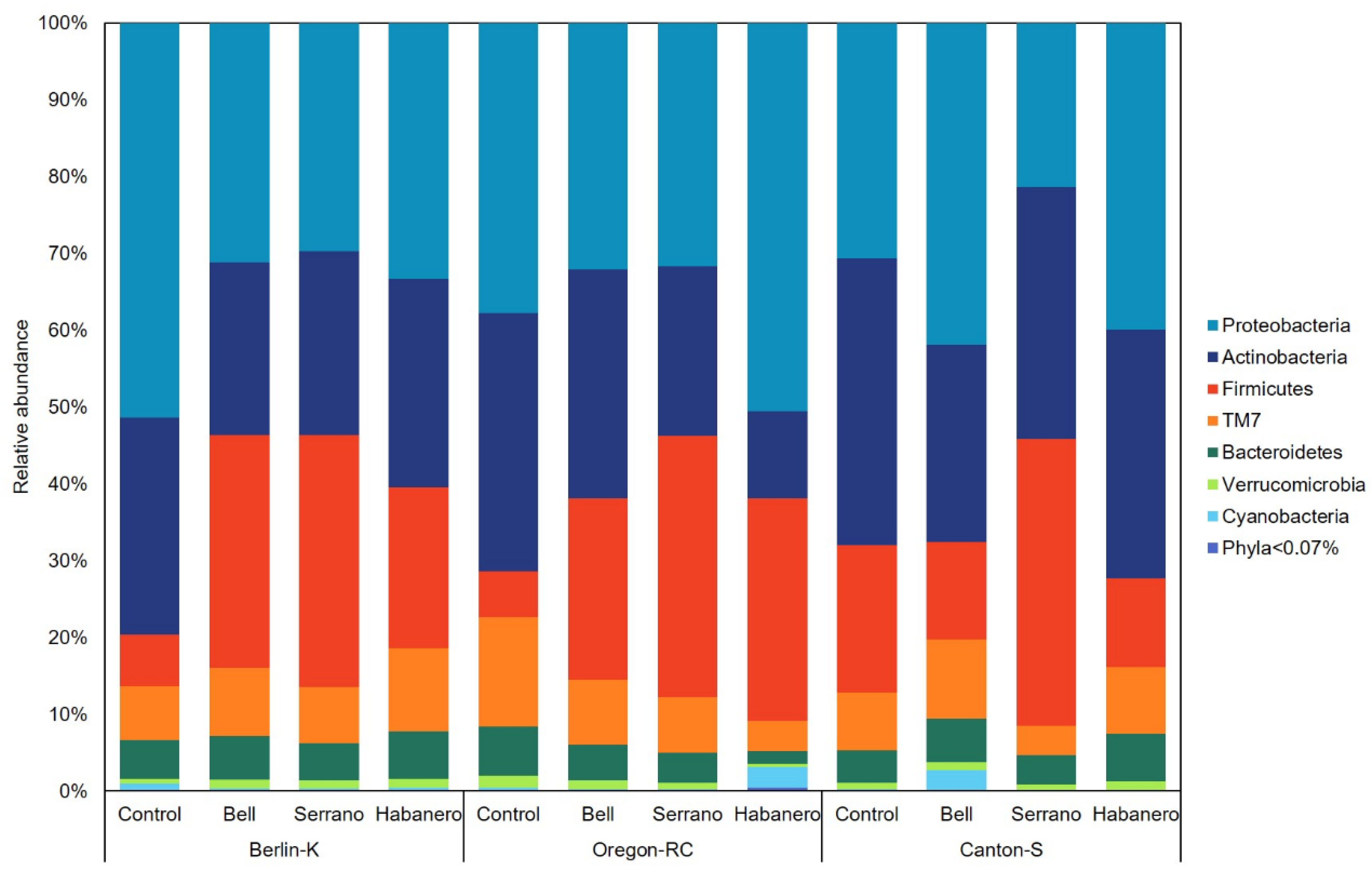
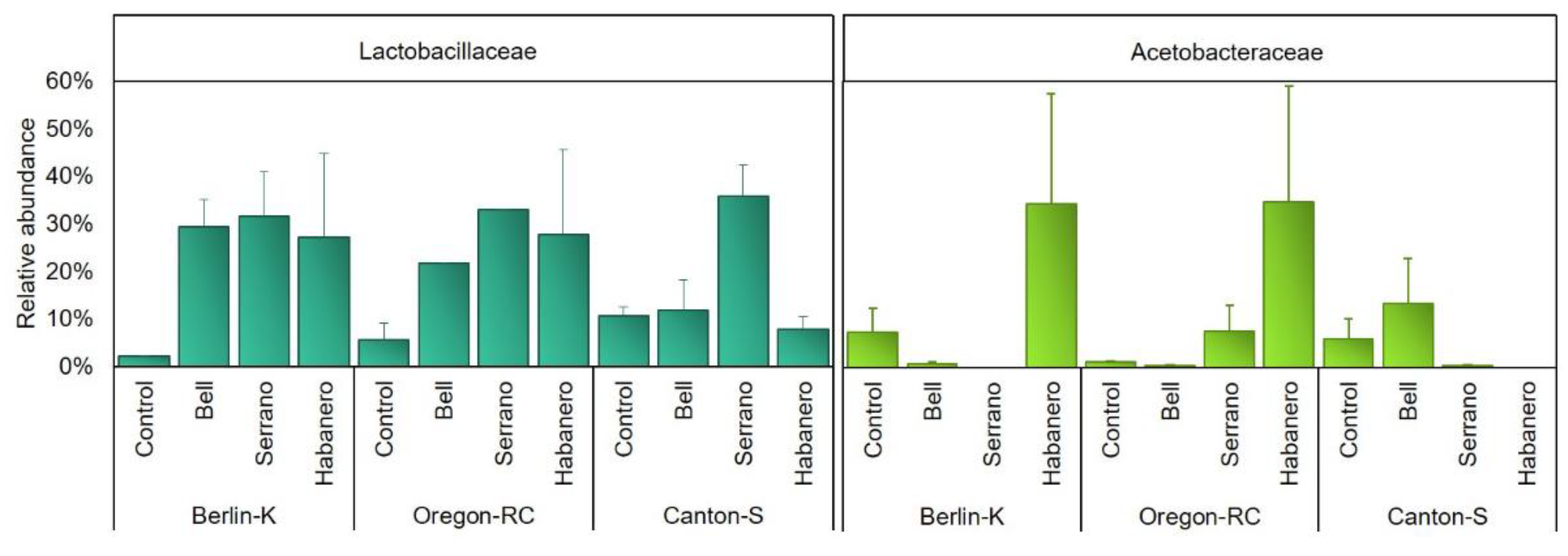
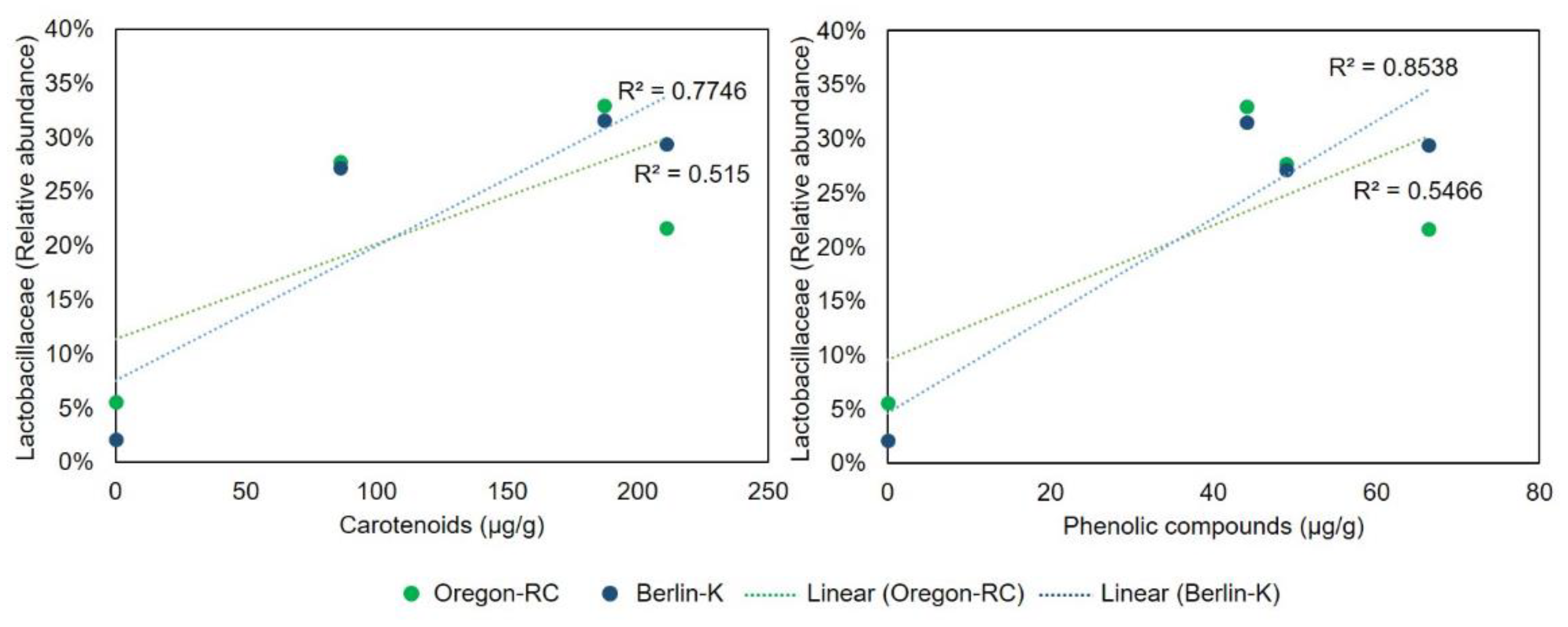
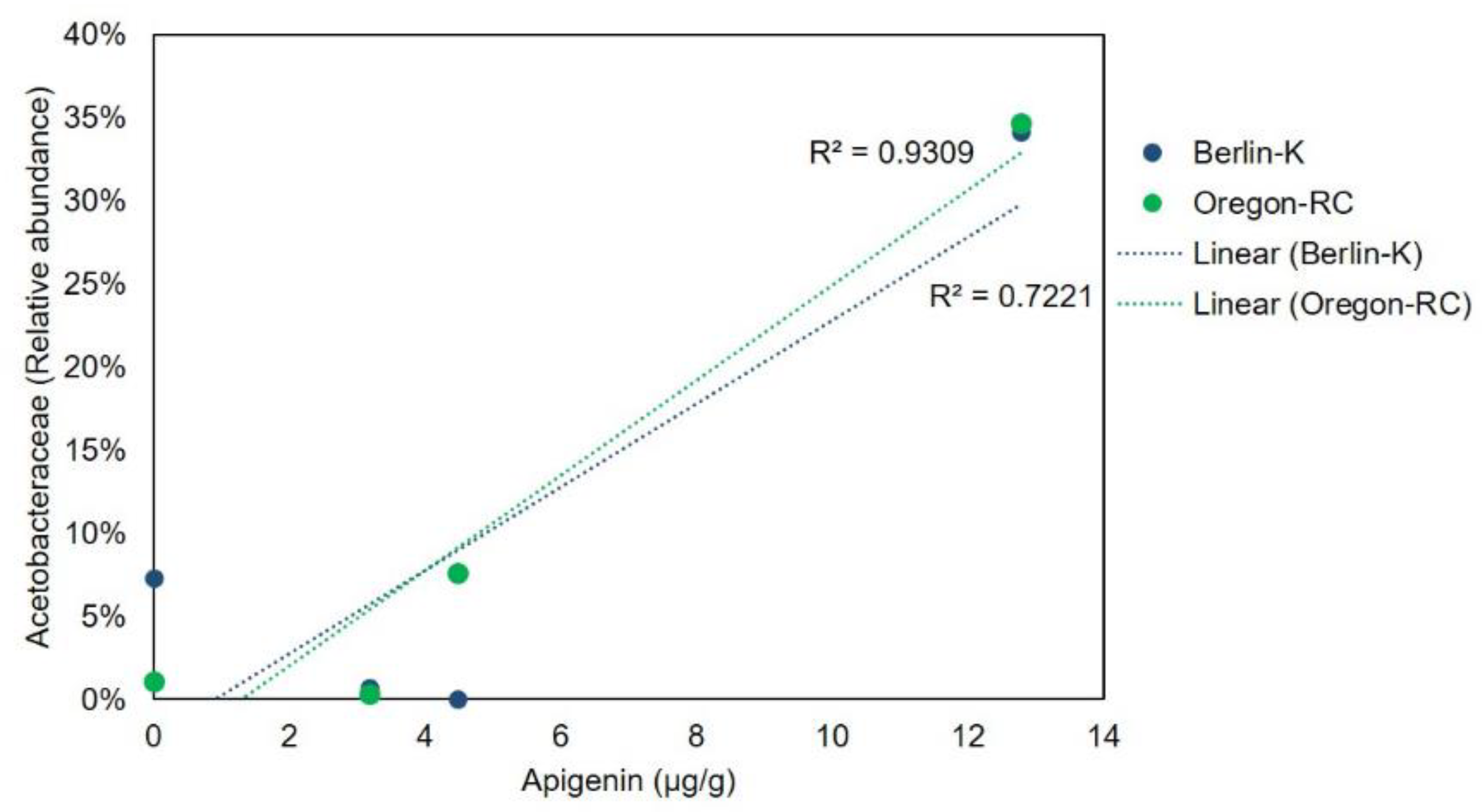
2.4. Gut Microbiota Composition of Drosophila Genetic Backgrounds Reared on the Different Diets
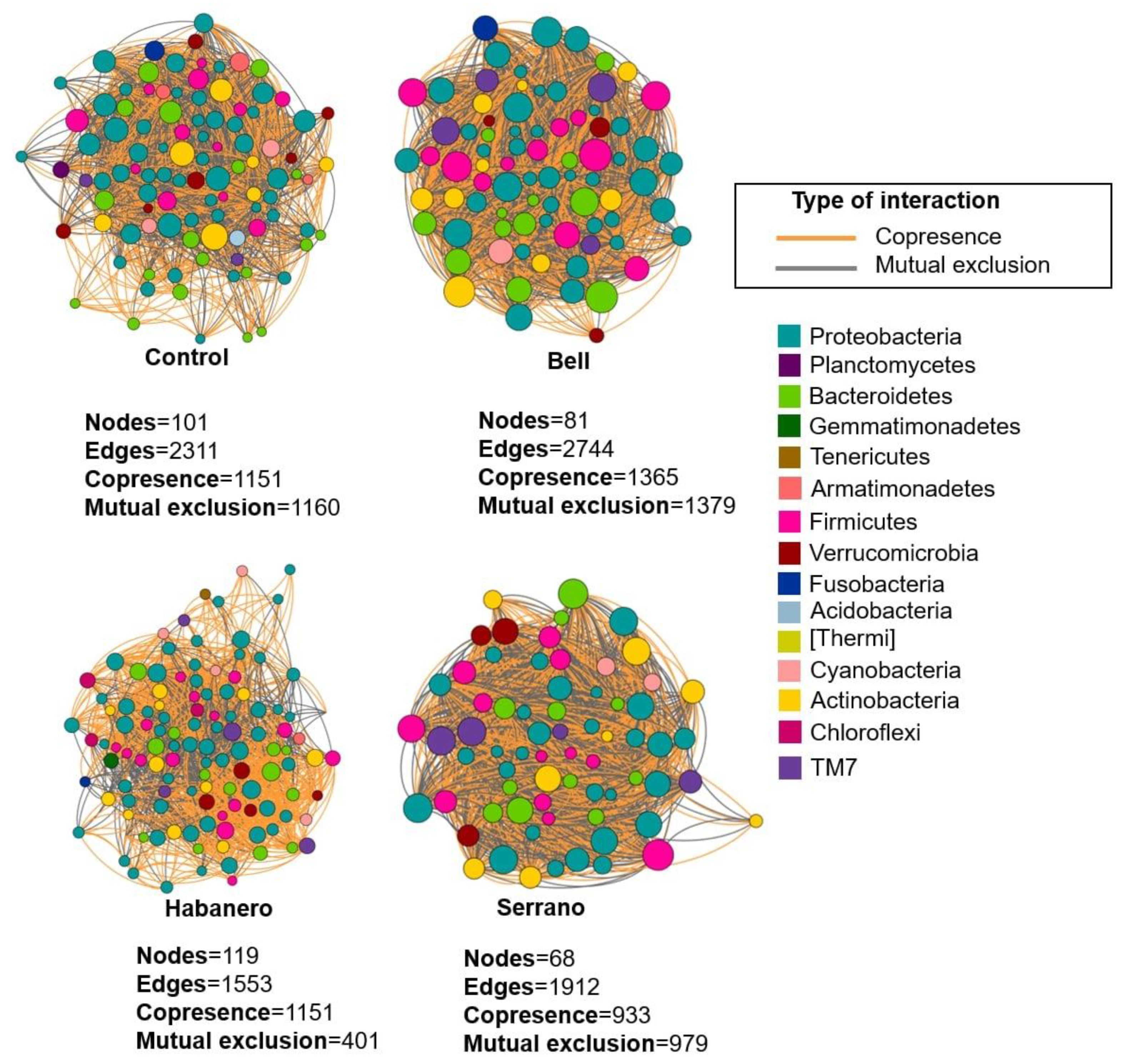
2.5. Microbiome Interaction Networks among Drosophila Populations Reared on Different Diets
3. Discussion
4. Materials and Methods
4.1. Drosophila Stocks and Cultures
4.2. Maintenance of Drosophila Strains on Control and Pepper Containing Diets
4.3. Gut Dissection and DNA Extraction
4.4. PCR Conditions and Product Purification
4.5. Library Preparation and Sequencing
4.6. Data Analysis
4.7. Diversity Metrics
4.8. Network Analysis
4.9. Quantification of Phytochemicals
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hillman, E.T.; Lu, H.; Yao, T.; Nakatsu, C.H. Microbial Ecology along the Gastrointestinal Tract. Microbes. Env. 2017, 32, 300–313. [Google Scholar] [CrossRef]
- Xu, Z.; Knight, R. Dietary effects on human gut microbiome diversity. Br. J. Nutr. 2015, 113, S1–S5. [Google Scholar] [CrossRef]
- Duda-Chodak, A.; Tarko, T.; Satora, P.; Sroka, P. Interaction of dietary compounds, especially polyphenols, with the intestinal microbiota: A review. Eur. J. Nutr. 2015, 54, 325–341. [Google Scholar] [CrossRef] [PubMed]
- Baenas, N.; Belovic, M.; Ilic, N.; Moreno, D.A.; Garcia-Viguera, C. Industrial use of pepper (Capsicum annum L.) derived products: Technological benefits and biological advantages. Food Chem. 2019, 274, 872–885. [Google Scholar] [CrossRef]
- Fiedor, J.; Burda, K. Potential role of carotenoids as antioxidants in human health and disease. Nutrients 2014, 6, 466–488. [Google Scholar] [CrossRef] [PubMed]
- Jeon, G.; Choi, Y.; LEE, S.M.; Kim, Y.; Oh, M.; JEONG, H.S.; Lee, J. Antioxidant and antiproliferative properties of hot pepper (Capsicum annuum L.) seeds. J. Food Biochem. 2012, 36, 595–603. [Google Scholar] [CrossRef]
- Navarro, J.M.; Flores, P.; Garrido, C.; Martinez, V. Changes in the contents of antioxidant compounds in pepper fruits at different ripening stages, as affected by salinity. Food Chem. 2006, 96, 66–73. [Google Scholar] [CrossRef]
- Pandey, K.B.; Rizvi, S.I. Plant polyphenols as dietary antioxidants in human health and disease. Oxid Med. Cell Longev. 2009, 2, 270–278. [Google Scholar] [CrossRef]
- Jehrke, L.; Stewart, F.A.; Droste, A.; Beller, M. The impact of genome variation and diet on the metabolic phenotype and microbiome composition of Drosophila melanogaster. Sci. Rep. 2018, 8, 6215. [Google Scholar] [CrossRef]
- Wong, A.C.; Vanhove, A.S.; Watnick, P.I. The interplay between intestinal bacteria and host metabolism in health and disease: Lessons from Drosophila melanogaster. Dis. Model. Mech. 2016, 9, 271–281. [Google Scholar] [CrossRef]
- Broderick, N.A.; Buchon, N.; Lemaitre, B. Microbiota-induced changes in Drosophila melanogaster host gene expression and gut morphology. MBio 2014, 5, e01117-14. [Google Scholar] [CrossRef] [PubMed]
- Broderick, N.A.; Lemaitre, B. Gut-associated microbes of Drosophila melanogaster. Gut Microbes 2012, 3, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.A.; Lang, J.M.; Bhatnagar, S.; Eisen, J.A.; Kopp, A. Bacterial communities of diverse Drosophila species: Ecological context of a host–microbe model system. PLoS Genet. 2011, 7, e1002272. [Google Scholar] [CrossRef] [PubMed]
- Staubach, F.; Baines, J.F.; Künzel, S.; Bik, E.M.; Petrov, D.A. Host species and environmental effects on bacterial communities associated with Drosophila in the laboratory and in the natural environment. PLoS ONE 2013, 8, e70749. [Google Scholar] [CrossRef] [PubMed]
- Early, A.M.; Shanmugarajah, N.; Buchon, N.; Clark, A.G. Drosophila genotype influences commensal bacterial levels. PLoS ONE 2017, 12, e0170332. [Google Scholar] [CrossRef] [PubMed]
- Leftwich, P.T.; Clarke, N.V.; Hutchings, M.I.; Chapman, T. Gut microbiomes and reproductive isolation in Drosophila. Proc. Natl. Acad. Sci. USA 2017, 114, 12767–12772. [Google Scholar] [CrossRef]
- Chaston, J.M.; Dobson, A.J.; Newell, P.D.; Douglas, A.E. Host Genetic Control of the Microbiota Mediates the Drosophila Nutritional Phenotype. Appl. Env. Microbiol. 2016, 82, 671–679. [Google Scholar] [CrossRef]
- Cox, C.R.; Gilmore, M.S. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect. Immun. 2007, 75, 1565–1576. [Google Scholar] [CrossRef]
- Erkosar, B.; Storelli, G.; Defaye, A.; Leulier, F. Host-intestinal microbiota mutualism: “learning on the fly”. Cell Host Microbe 2013, 13, 8–14. [Google Scholar] [CrossRef]
- Blum, J.E.; Fischer, C.N.; Miles, J.; Handelsman, J. Frequent replenishment sustains the beneficial microbiome of Drosophila melanogaster. MBio 2013, 4, e00860-13. [Google Scholar] [CrossRef]
- Ren, C.; Webster, P.; Finkel, S.E.; Tower, J. Increased internal and external bacterial load during Drosophila aging without life-span trade-off. Cell Metab 2007, 6, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.C.; Chaston, J.M.; Douglas, A.E. The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis. ISME J. 2013, 7, 1922–1932. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A quality Control Tool for High throughput Sequence Data; Babraham Bioinformatics, Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. QIIME 2: Reproducible, Interactive, Scalable, and Extensible Microbiome Data Science; 2167-9843. PeerJ Preprints, 2018. Available online: https://peerj.com/preprints/27295/ (accessed on 10 November 2018).
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Boutros, P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 2001, 26, 32–46. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; O’hara, R.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community ecology package. R package version 1.17-4. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 11 March 2019).
- Martinez, P. pairwiseAdonis: Pairwise multilevel comparison using adonis. R Package Version 0.3 2019. Available online: https://github.com/pmartinezarbizu/pairwiseAdonis (accessed on March 2019).
- Faust, K.; Sathirapongsasuti, J.F.; Izard, J.; Segata, N.; Gevers, D.; Raes, J.; Huttenhower, C. Microbial co-occurrence relationships in the human microbiome. PLoS Comput. Biol. 2012, 8, e1002606. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Assenov, Y.; Ramirez, F.; Schelhorn, S.E.; Lengauer, T.; Albrecht, M. Computing topological parameters of biological networks. Bioinformatics 2008, 24, 282–284. [Google Scholar] [CrossRef] [PubMed]
- Bae, H.; Jayaprakasha, G.; Jifon, J.; Patil, B.S. Extraction efficiency and validation of an HPLC method for flavonoid analysis in peppers. Food Chem. 2012, 130, 751–758. [Google Scholar] [CrossRef]
- Chebrolu, K.K.; Jayaprakasha, G.; Jifon, J.; Patil, B.S. Production system and storage temperature influence grapefruit vitamin C, limonoids, and carotenoids. J. Agric. Food Chem. 2012, 60, 7096–7103. [Google Scholar] [CrossRef] [PubMed]
- Yoo, K.S.; Bang, H.; Lee, E.J.; Crosby, K.; Patil, B.S. Variation of carotenoid, sugar, and ascorbic acid concentrations in watermelon genetic backgrounds and genetic analysis. Hortic. Environ. Biotechnol. 2012, 53, 552–560. [Google Scholar] [CrossRef]
- Nimmakayala, P.; Abburi, V.L.; Abburi, L.; Alaparthi, S.B.; Cantrell, R.; Park, M.; Choi, D.; Hankins, G.; Malkaram, S.; Reddy, U.K. Linkage disequilibrium and population-structure analysis among Capsicum annuum L. cultivars for use in association mapping. Mol. Genet. Genom. 2014, 289, 513–521. [Google Scholar] [CrossRef]
| Bell | Serrano | Habanero | |
|---|---|---|---|
| Carotenoids (µg/g) | |||
| Capsanthin | 52.001 | 37.676 | 11.145 |
| α-carotene | 12.88 | 23.435 | 1.781 |
| β-carotene | 129.153 | 115.670 | 64.470 |
| β-cryptoxanthin | 16.845 | 10.267 | 8.816 |
| Phenolic compounds (µg/g) | |||
| Myricetin | 9.672 | 8.864 | 4.979 |
| Quercetin | 41.355 | 26.028 | 21.532 |
| Kaempferol | ND | ND | 0.801 |
| Apigenin | 3.177 | 4.483 | 12.782 |
| Luteolin | 12.140 | 4.672 | 8.764 |
| Capsaicinoids (µg/g) | |||
| Capsaicin | ND | 2529.117 | 2478.723 |
| Dihydrocapsaicin | ND | 1524.960 | 746.127 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garcia-Lozano, M.; Haynes, J.; Lopez-Ortiz, C.; Natarajan, P.; Peña-Garcia, Y.; Nimmakayala, P.; Stommel, J.; Alaparthi, S.B.; Sirbu, C.; Balagurusamy, N.; et al. Effect of Pepper-Containing Diets on the Diversity and Composition of Gut Microbiome of Drosophila melanogaster. Int. J. Mol. Sci. 2020, 21, 945. https://doi.org/10.3390/ijms21030945
Garcia-Lozano M, Haynes J, Lopez-Ortiz C, Natarajan P, Peña-Garcia Y, Nimmakayala P, Stommel J, Alaparthi SB, Sirbu C, Balagurusamy N, et al. Effect of Pepper-Containing Diets on the Diversity and Composition of Gut Microbiome of Drosophila melanogaster. International Journal of Molecular Sciences. 2020; 21(3):945. https://doi.org/10.3390/ijms21030945
Chicago/Turabian StyleGarcia-Lozano, Marleny, Joshua Haynes, Carlos Lopez-Ortiz, Purushothaman Natarajan, Yadira Peña-Garcia, Padma Nimmakayala, John Stommel, Suresh B. Alaparthi, Cristian Sirbu, Nagamani Balagurusamy, and et al. 2020. "Effect of Pepper-Containing Diets on the Diversity and Composition of Gut Microbiome of Drosophila melanogaster" International Journal of Molecular Sciences 21, no. 3: 945. https://doi.org/10.3390/ijms21030945
APA StyleGarcia-Lozano, M., Haynes, J., Lopez-Ortiz, C., Natarajan, P., Peña-Garcia, Y., Nimmakayala, P., Stommel, J., Alaparthi, S. B., Sirbu, C., Balagurusamy, N., & Reddy, U. K. (2020). Effect of Pepper-Containing Diets on the Diversity and Composition of Gut Microbiome of Drosophila melanogaster. International Journal of Molecular Sciences, 21(3), 945. https://doi.org/10.3390/ijms21030945







