Constitutive Activation of Guanylate Cyclase by the G86R GCAP1 Variant Is Due to “Locking” Cation-π Interactions that Impair the Activator-to-Inhibitor Structural Transition
Abstract
1. Introduction
2. Results and Discussion
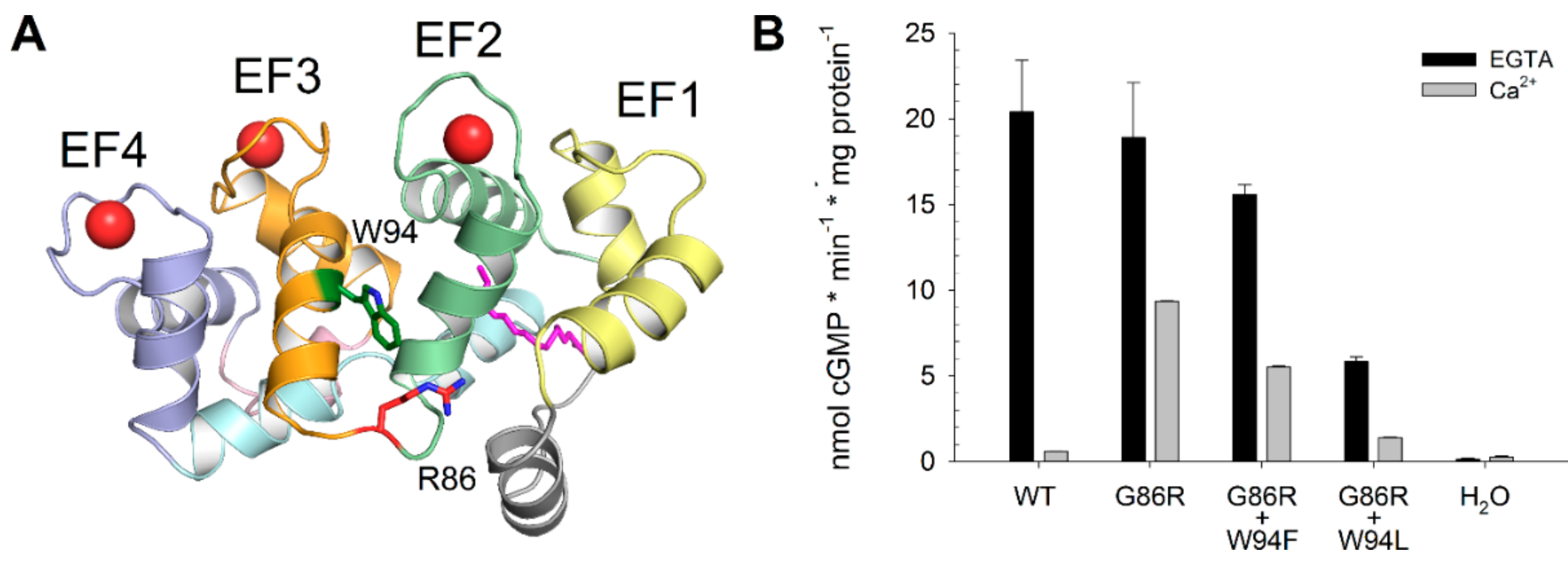
2.1. GC Regulation by GCAP1 Variants
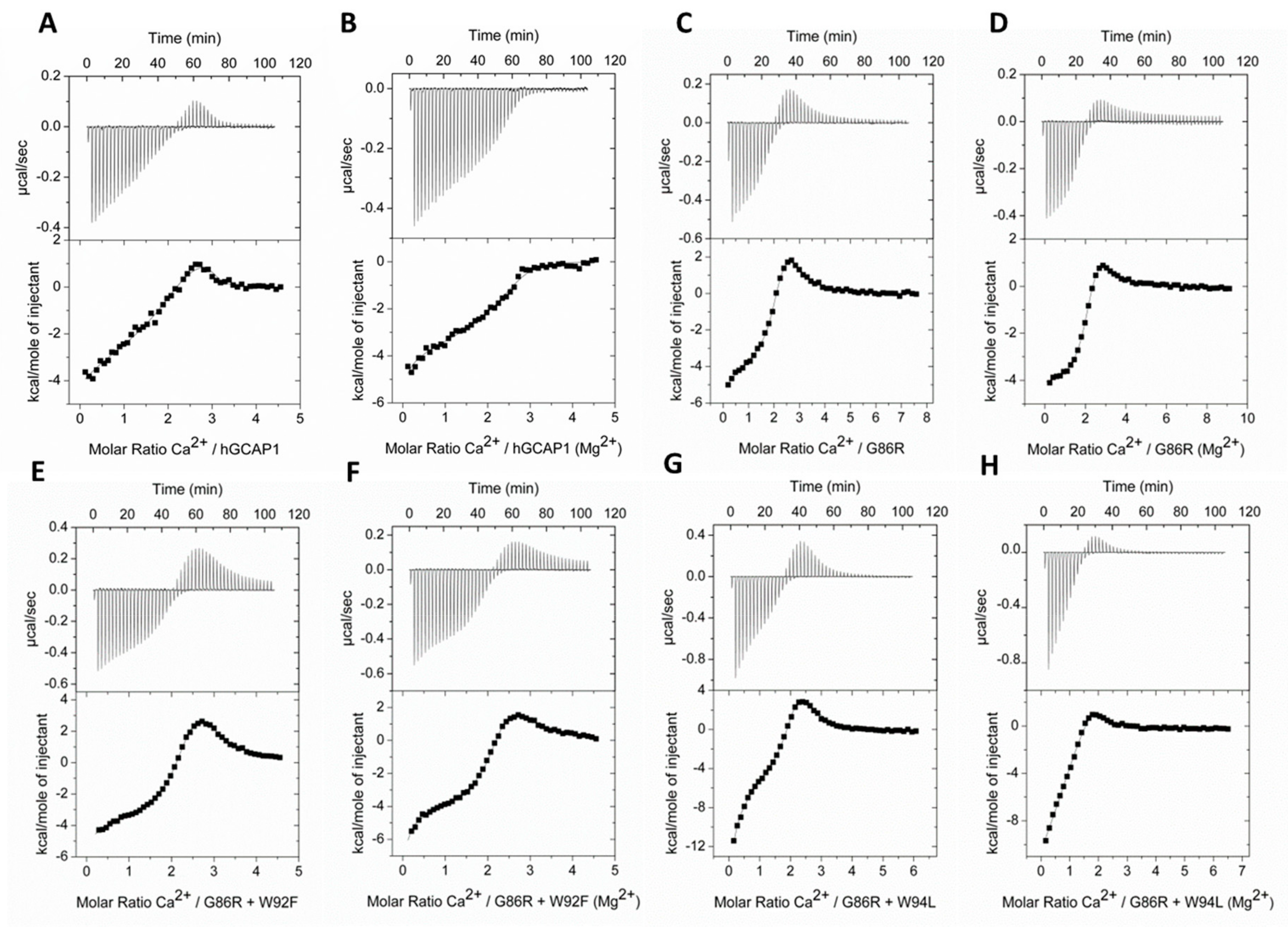
2.2. Mg2+/Ca2+ Affinity of GCAP1 G86R+W94F/L
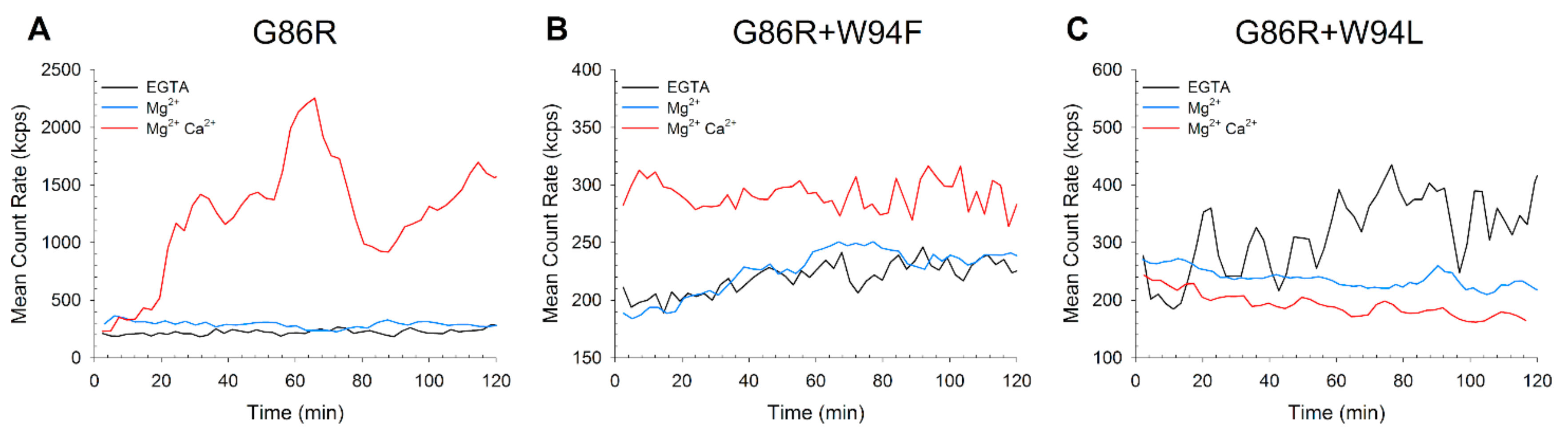
2.3. Quaternary Structure and Aggregation Propensity of GCAP1 Variants
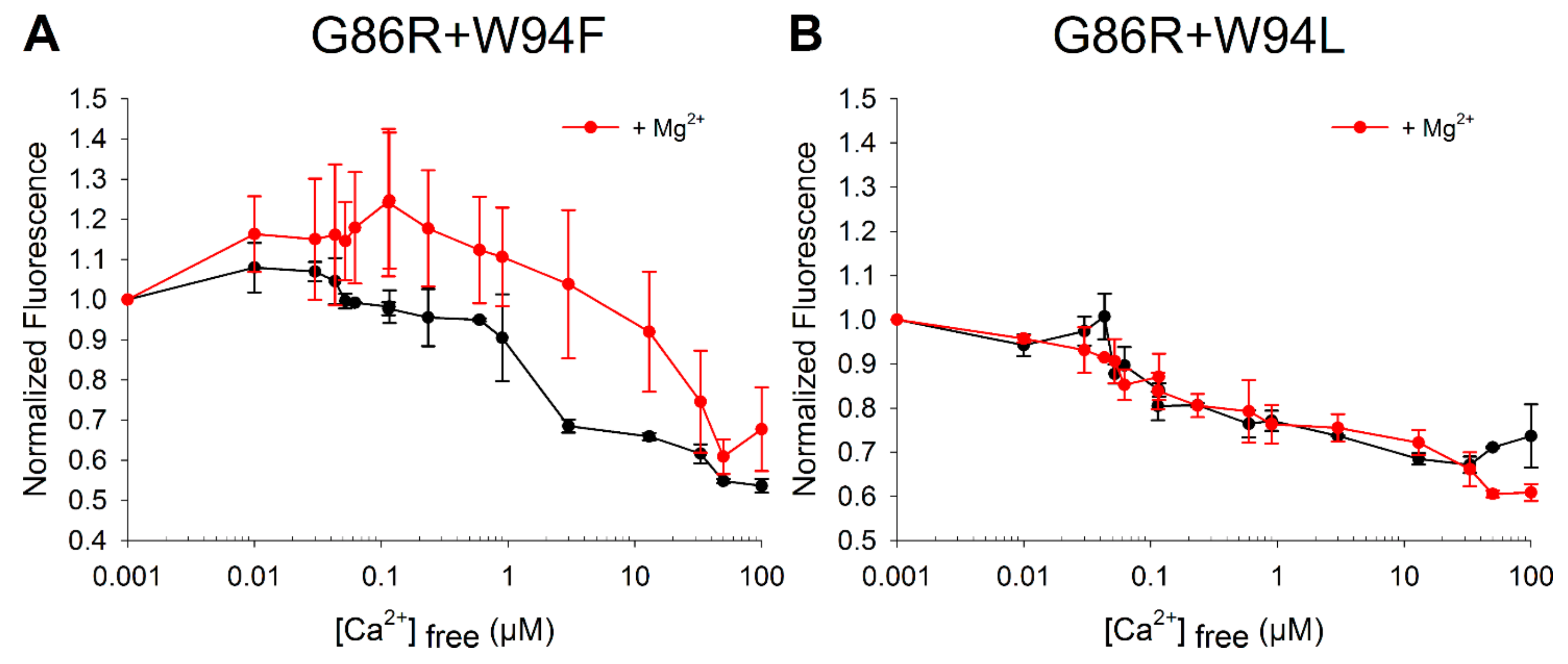
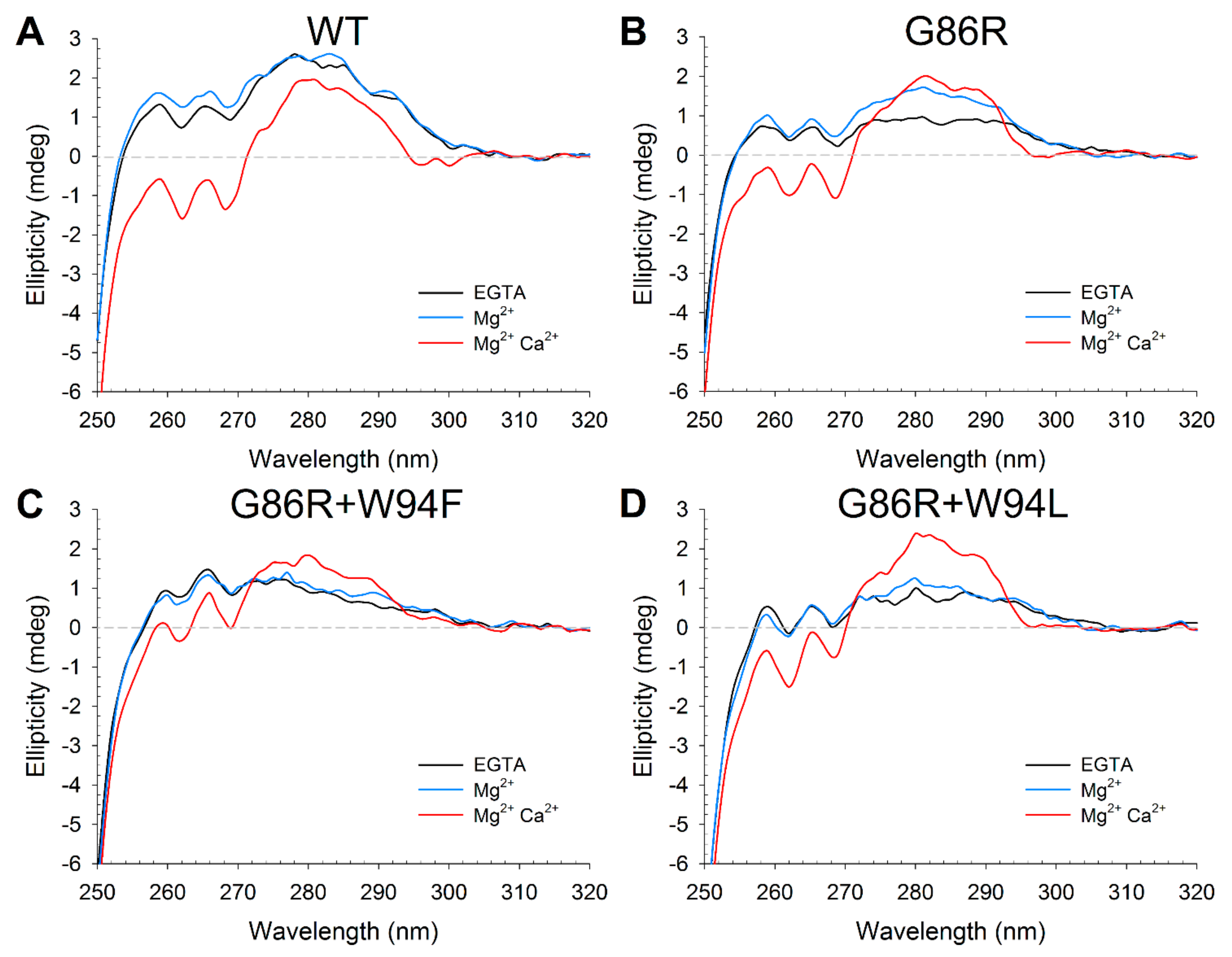
2.4. Effects of Ca2+ Binding on the Intrinsic Trp Fluorescence of GCAP1 Mutants
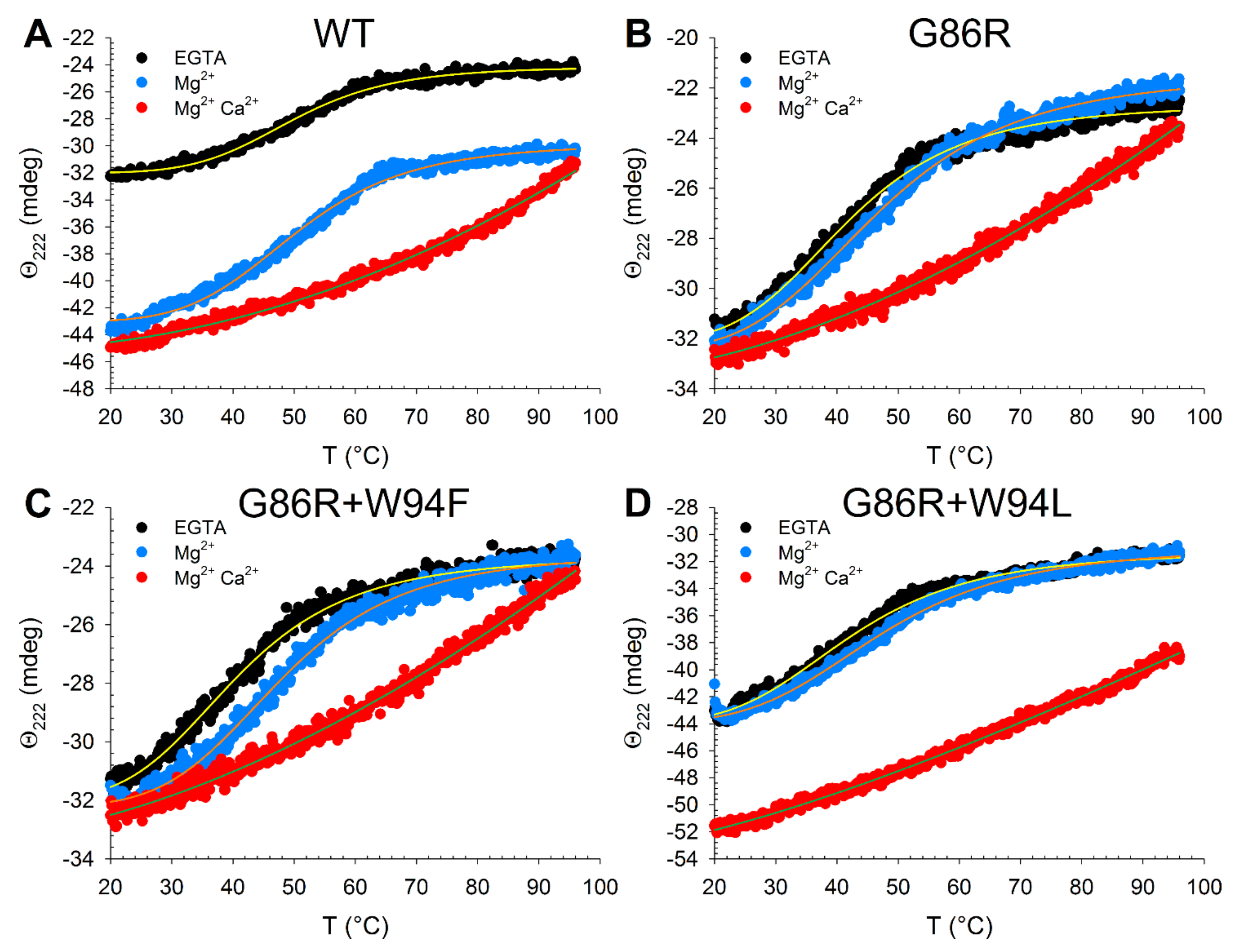
2.5. Effects of Ion Binding on the Conformation and Thermal Stability of GCAP1 Mutants
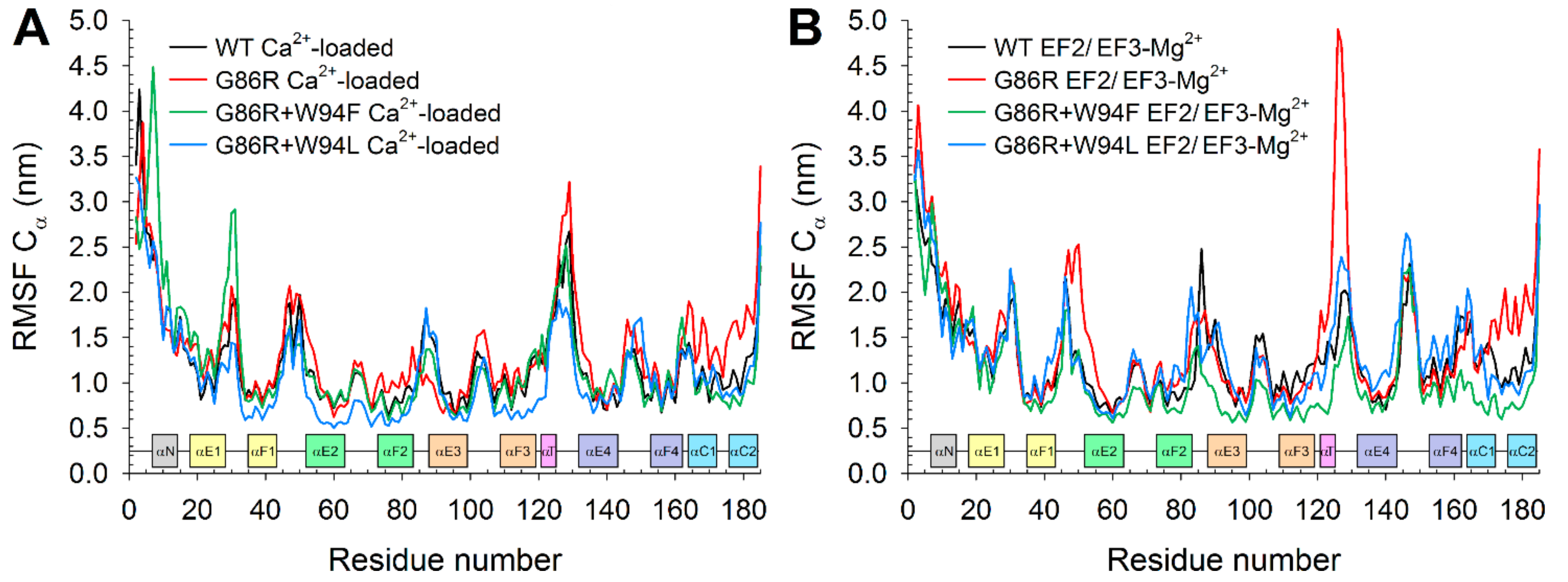
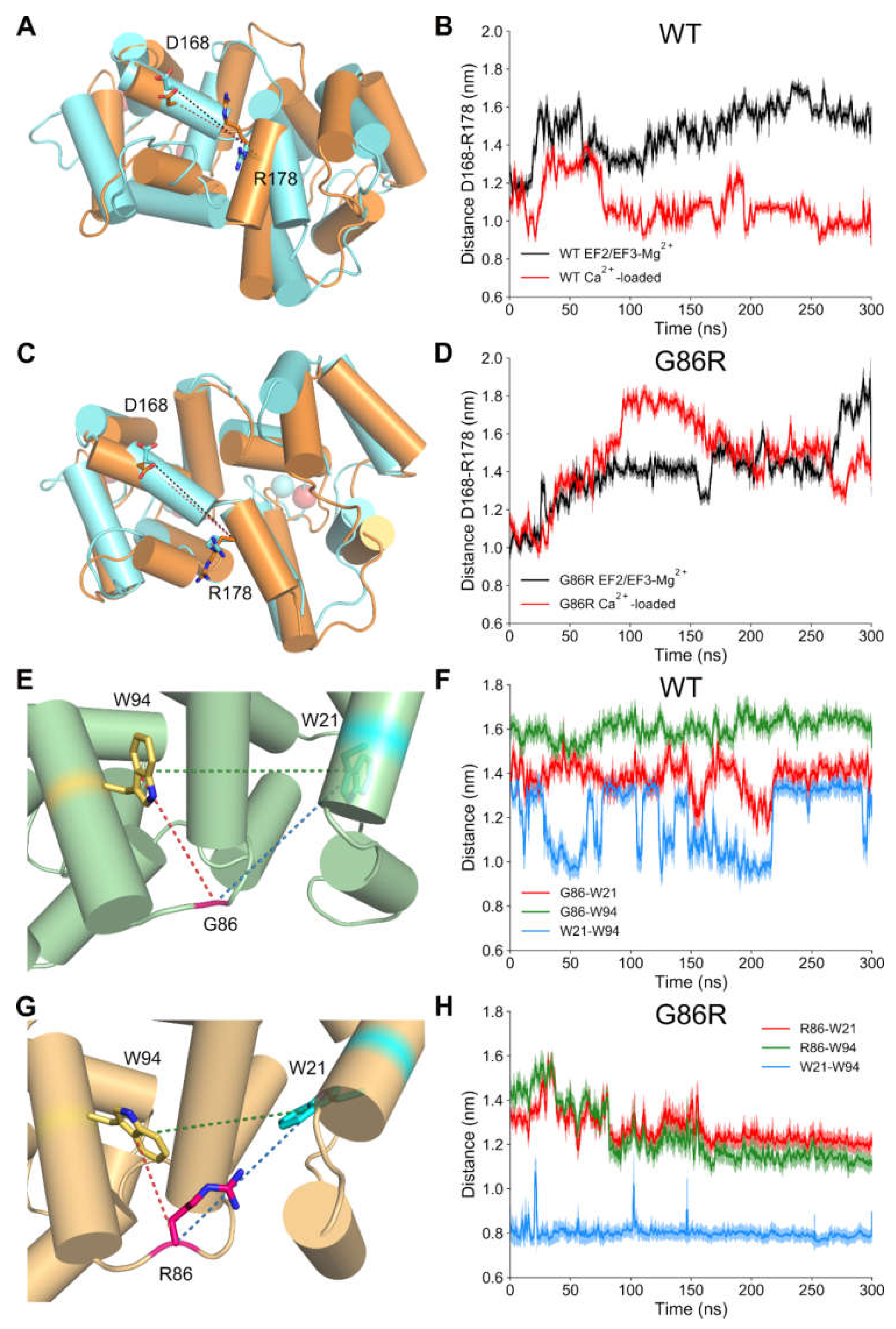
2.6. Dynamic Structural Insights on the Interaction between G/R86 and W/F/L94 Provided by Molecular Dynamics Simulations
3. Materials and Methods
3.1. Cloning, Protein Expression and Purification
3.2. Tryptophan Fluorescence Measurements
3.3. Guanylate Cyclase Assay
3.4. Isothermal Titration Calorimetry
3.5. Analytical Size Exclusion Chromatography
3.6. Gel Shift Assay
3.7. Circular Dichroism Spectroscopy and Thermal Denaturation Profiles
3.8. Dynamic Light Scattering
3.9. Molecular Dynamics Simulations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| GCAP1 | Guanylate Cyclase activating protein 1 |
| GC | Guanylate Cyclase 1 |
| NCS | Neuronal Calcium Sensor |
| EGTA | ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid |
| ITC | Isothermal Titration Calorimetry |
| Tris | tris(hydroxymethyl)aminomethane |
| DTT | Dithiothreitol |
| HEPES | 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid |
| DLS | Dynamic Light Scattering |
| SEC | Size Exclusion Chromatography |
| CD | Circular Dichroism |
| UV | Ultraviolet |
| MD | Molecular Dynamics |
| RMSF | Root-Mean Square Fluctuation |
| MOPS | 3-(N-morpholino)propanesulfonic acid |
| SDS-PAGE | Sodium dodecyl sulfate–polyacrylamide gel electrophoresis |
References
- Koch, K.W.; Dell’Orco, D. Protein and Signaling Networks in Vertebrate Photoreceptor Cells. Front. Mol. Neurosci. 2015, 8, 67. [Google Scholar] [CrossRef] [PubMed]
- Koch, K.W.; Dell’Orco, D. A calcium-relay mechanism in vertebrate phototransduction. ACS Chem. Neurosci. 2013, 4, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Korenbrot, J.I. Speed, sensitivity, and stability of the light response in rod and cone photoreceptors: Facts and models. Prog. Retin. Eye Res. 2012, 31, 442–466. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Dizhoor, A.M.; Ames, J.B. Structural diversity of neuronal calcium sensor proteins and insights for activation of retinal guanylyl cyclase by GCAP1. Front. Mol. Neurosci. 2014, 7, 19. [Google Scholar] [CrossRef] [PubMed]
- Dizhoor, A.M.; Olshevskaya, E.V.; Peshenko, I.V. Mg2+/Ca2+ cation binding cycle of guanylyl cyclase activating proteins (GCAPs): Role in regulation of photoreceptor guanylyl cyclase. Mol. Cell. Biochem. 2010, 334, 117–124. [Google Scholar] [CrossRef]
- Koch, K.W.; Duda, T.; Sharma, R.K. Ca(2+)-modulated vision-linked ROS-GC guanylate cyclase transduction machinery. Mol. Cell. Biochem. 2010, 334, 105–115. [Google Scholar] [CrossRef]
- Peshenko, I.V.; Dizhoor, A.M. Activation and inhibition of photoreceptor guanylyl cyclase by guanylyl cyclase activating protein 1 (GCAP-1): The functional role of Mg2+/Ca2+ exchange in EF-hand domains. J. Biol. Chem. 2007, 282, 21645–21652. [Google Scholar] [CrossRef]
- Marino, V.; Sulmann, S.; Koch, K.W.; Dell’Orco, D. Structural effects of Mg2+ on the regulatory states of three neuronal calcium sensors operating in vertebrate phototransduction. Biochim. Biophys. Acta 2015, 1853, 2055–2065. [Google Scholar] [CrossRef]
- Peshenko, I.V.; Olshevskaya, E.V.; Lim, S.; Ames, J.B.; Dizhoor, A.M. Calcium-myristoyl Tug is a new mechanism for intramolecular tuning of calcium sensitivity and target enzyme interaction for guanylyl cyclase-activating protein 1: Dynamic connection between N-fatty acyl group and EF-hand controls calcium sensitivity. J. Biol. Chem. 2012, 287, 13972–13984. [Google Scholar] [CrossRef]
- Marino, V.; Dell’Orco, D. Allosteric communication pathways routed by Ca(2+)/Mg(2+) exchange in GCAP1 selectively switch target regulation modes. Sci. Rep. 2016, 6, 34277. [Google Scholar] [CrossRef]
- Hamel, C.P. Cone rod dystrophies. Orphanet J. Rare Dis. 2007, 2, 7. [Google Scholar] [CrossRef] [PubMed]
- Michaelides, M.; Hardcastle, A.J.; Hunt, D.M.; Moore, A.T. Progressive cone and cone-rod dystrophies: Phenotypes and underlying molecular genetic basis. Surv. Ophthalmol. 2006, 51, 232–258. [Google Scholar] [CrossRef] [PubMed]
- Roosing, S.; Thiadens, A.A.; Hoyng, C.B.; Klaver, C.C.; den Hollander, A.I.; Cremers, F.P. Causes and consequences of inherited cone disorders. Prog. Retin. Eye Res. 2014, 42, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Sheng, X.; Zhuang, W.; Sun, X.; Liu, G.; Shi, X.; Huang, G.; Mei, Y.; Li, Y.; Pan, X.; et al. GUCA1A mutation causes maculopathy in a five-generation family with a wide spectrum of severity. Genet. Med. 2017, 19, 945–954. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Li, S.; Xiao, X.; Jia, X.; Sun, W.; Gao, Y.; Li, L.; Wang, P.; Guo, X.; Zhang, Q. Novel GUCA1A mutation identified in a Chinese family with cone-rod dystrophy. Neurosci. Lett. 2013, 541, 179–183. [Google Scholar] [CrossRef]
- Jiang, L.; Katz, B.J.; Yang, Z.; Zhao, Y.; Faulkner, N.; Hu, J.; Baird, J.; Baehr, W.; Creel, D.J.; Zhang, K. Autosomal dominant cone dystrophy caused by a novel mutation in the GCAP1 gene (GUCA1A). Mol. Vis. 2005, 11, 143–151. [Google Scholar]
- Jiang, L.; Wheaton, D.; Bereta, G.; Zhang, K.; Palczewski, K.; Birch, D.G.; Baehr, W. A novel GCAP1(N104K) mutation in EF-hand 3 (EF3) linked to autosomal dominant cone dystrophy. Vis. Res. 2008, 48, 2425–2432. [Google Scholar] [CrossRef][Green Version]
- Kamenarova, K.; Corton, M.; Garcia-Sandoval, B.; Fernandez-San Jose, P.; Panchev, V.; Avila-Fernandez, A.; Lopez-Molina, M.I.; Chakarova, C.; Ayuso, C.; Bhattacharya, S.S. Novel GUCA1A mutations suggesting possible mechanisms of pathogenesis in cone, cone-rod, and macular dystrophy patients. Biomed. Res. Int. 2013, 2013, 517570. [Google Scholar] [CrossRef]
- Manes, G.; Mamouni, S.; Herald, E.; Richard, A.C.; Senechal, A.; Aouad, K.; Bocquet, B.; Meunier, I.; Hamel, C.P. Cone dystrophy or macular dystrophy associated with novel autosomal dominant GUCA1A mutations. Mol. Vis. 2017, 23, 198–209. [Google Scholar]
- Marino, V.; Dal Cortivo, G.; Oppici, E.; Maltese, P.E.; D’Esposito, F.; Manara, E.; Ziccardi, L.; Falsini, B.; Magli, A.; Bertelli, M.; et al. A novel p.(Glu111Val) missense mutation in GUCA1A associated with cone-rod dystrophy leads to impaired calcium sensing and perturbed second messenger homeostasis in photoreceptors. Hum. Mol. Genet. 2018, 27, 4204–4217. [Google Scholar] [CrossRef]
- Nishiguchi, K.M.; Sokal, I.; Yang, L.; Roychowdhury, N.; Palczewski, K.; Berson, E.L.; Dryja, T.P.; Baehr, W. A novel mutation (I143NT) in guanylate cyclase-activating protein 1 (GCAP1) associated with autosomal dominant cone degeneration. Investig. Ophthalmol. Vis. Sci. 2004, 45, 3863–3870. [Google Scholar] [CrossRef] [PubMed]
- Nong, E.; Lee, W.; Merriam, J.E.; Allikmets, R.; Tsang, S.H. Disease progression in autosomal dominant cone-rod dystrophy caused by a novel mutation (D100G) in the GUCA1A gene. Doc. Ophthalmol. 2014, 128, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Payne, A.M.; Downes, S.M.; Bessant, D.A.; Taylor, R.; Holder, G.E.; Warren, M.J.; Bird, A.C.; Bhattacharya, S.S. A mutation in guanylate cyclase activator 1A (GUCA1A) in an autosomal dominant cone dystrophy pedigree mapping to a new locus on chromosome 6p21.1. Hum. Mol. Genet. 1998, 7, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Vocke, F.; Weisschuh, N.; Marino, V.; Malfatti, S.; Jacobson, S.G.; Reiff, C.M.; Dell’Orco, D.; Koch, K.W. Dysfunction of cGMP signalling in photoreceptors by a macular dystrophy-related mutation in the calcium sensor GCAP1. Hum. Mol. Genet. 2017, 26, 133–144. [Google Scholar] [CrossRef]
- Wilkie, S.E.; Li, Y.; Deery, E.C.; Newbold, R.J.; Garibaldi, D.; Bateman, J.B.; Zhang, H.; Lin, W.; Zack, D.J.; Bhattacharya, S.S.; et al. Identification and functional consequences of a new mutation (E155G) in the gene for GCAP1 that causes autosomal dominant cone dystrophy. Am. J. Hum. Genet. 2001, 69, 471–480. [Google Scholar] [CrossRef]
- Peshenko, I.V.; Cideciyan, A.V.; Sumaroka, A.; Olshevskaya, E.V.; Scholten, A.; Abbas, S.; Koch, K.W.; Jacobson, S.G.; Dizhoor, A.M. A G86R mutation in the calcium-sensor protein GCAP1 alters regulation of retinal guanylyl cyclase and causes dominant cone-rod degeneration. J. Biol. Chem. 2019, 294, 3476–3488. [Google Scholar] [CrossRef]
- Dell’Orco, D.; Behnen, P.; Linse, S.; Koch, K.W. Calcium binding, structural stability and guanylate cyclase activation in GCAP1 variants associated with human cone dystrophy. Cell. Mol. Life Sci. 2010, 67, 973–984. [Google Scholar] [CrossRef]
- Dizhoor, A.M.; Boikov, S.G.; Olshevskaya, E.V. Constitutive activation of photoreceptor guanylate cyclase by Y99C mutant of GCAP-1. Possible role in causing human autosomal dominant cone degeneration. J. Biol. Chem. 1998, 273, 17311–17314. [Google Scholar] [CrossRef]
- Kitiratschky, V.B.; Behnen, P.; Kellner, U.; Heckenlively, J.R.; Zrenner, E.; Jagle, H.; Kohl, S.; Wissinger, B.; Koch, K.W. Mutations in the GUCA1A gene involved in hereditary cone dystrophies impair calcium-mediated regulation of guanylate cyclase. Hum. Mutat. 2009, 30, E782–E796. [Google Scholar] [CrossRef]
- Olshevskaya, E.V.; Calvert, P.D.; Woodruff, M.L.; Peshenko, I.V.; Savchenko, A.B.; Makino, C.L.; Ho, Y.S.; Fain, G.L.; Dizhoor, A.M. The Y99C mutation in guanylyl cyclase-activating protein 1 increases intracellular Ca2+ and causes photoreceptor degeneration in transgenic mice. J. Neurosci. 2004, 24, 6078–6085. [Google Scholar] [CrossRef]
- Sokal, I.; Dupps, W.J.; Grassi, M.A.; Brown, J., Jr.; Affatigato, L.M.; Roychowdhury, N.; Yang, L.; Filipek, S.; Palczewski, K.; Stone, E.M.; et al. A novel GCAP1 missense mutation (L151F) in a large family with autosomal dominant cone-rod dystrophy (adCORD). Investig. Ophthalmol. Vis. Sci. 2005, 46, 1124–1132. [Google Scholar] [CrossRef] [PubMed]
- Sokal, I.; Li, N.; Surgucheva, I.; Warren, M.J.; Payne, A.M.; Bhattacharya, S.S.; Baehr, W.; Palczewski, K. GCAP1 (Y99C) mutant is constitutively active in autosomal dominant cone dystrophy. Mol. Cell 1998, 2, 129–133. [Google Scholar] [CrossRef]
- Marino, V.; Scholten, A.; Koch, K.W.; Dell’Orco, D. Two retinal dystrophy-associated missense mutations in GUCA1A with distinct molecular properties result in a similar aberrant regulation of the retinal guanylate cyclase. Hum. Mol. Genet. 2015, 24, 6653–6666. [Google Scholar] [CrossRef] [PubMed]
- Viviano, J.; Krishnan, A.; Wu, H.; Venkataraman, V. Electrophoretic mobility shift in native gels indicates calcium-dependent structural changes of neuronal calcium sensor proteins. Anal. Biochem. 2016, 494, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Dell’Orco, D.; Sulmann, S.; Linse, S.; Koch, K.W. Dynamics of conformational Ca2+-switches in signaling networks detected by a planar plasmonic device. Anal. Chem. 2012, 84, 2982–2989. [Google Scholar] [CrossRef] [PubMed]
- Ames, J.B. Dimerization of Neuronal Calcium Sensor Proteins. Front. Mol. Neurosci. 2018, 11, 397. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Roseman, G.; Peshenko, I.; Manchala, G.; Cudia, D.; Dizhoor, A.M.; Millhauser, G.; Ames, J.B. Retinal guanylyl cyclase activating protein 1 forms a functional dimer. PLoS ONE 2018, 13, e0193947. [Google Scholar] [CrossRef]
- Dal Cortivo, G.; Marino, V.; Iacobucci, C.; Vallone, R.; Arlt, C.; Rehkamp, A.; Sinz, A.; Dell’Orco, D. Oligomeric state, hydrodynamic properties and target recognition of human Calcium and Integrin Binding protein 2 (CIB2). Sci. Rep. 2019, 9, 15058. [Google Scholar] [CrossRef]
- Peshenko, I.V.; Dizhoor, A.M. Guanylyl cyclase-activating proteins (GCAPs) are Ca2+/Mg2+ sensors: Implications for photoreceptor guanylyl cyclase (RetGC) regulation in mammalian photoreceptors. J. Biol. Chem. 2004, 279, 16903–16906. [Google Scholar] [CrossRef]
- Peshenko, I.V.; Dizhoor, A.M. Ca2+ and Mg2+ binding properties of GCAP-1. Evidence that Mg2+-bound form is the physiological activator of photoreceptor guanylyl cyclase. J. Biol. Chem. 2006, 281, 23830–23841. [Google Scholar] [CrossRef]
- Stephen, R.; Bereta, G.; Golczak, M.; Palczewski, K.; Sousa, M.C. Stabilizing function for myristoyl group revealed by the crystal structure of a neuronal calcium sensor, guanylate cyclase-activating protein 1. Structure 2007, 15, 1392–1402. [Google Scholar] [CrossRef] [PubMed]
- Kelly, S.M.; Jess, T.J.; Price, N.C. How to study proteins by circular dichroism. Biochim. Biophys. Acta 2005, 1751, 119–139. [Google Scholar] [CrossRef] [PubMed]
- Marino, V.; Astegno, A.; Pedroni, M.; Piccinelli, F.; Dell’Orco, D. Nanodevice-induced conformational and functional changes in a prototypical calcium sensor protein. Nanoscale 2014, 6, 412–423. [Google Scholar] [CrossRef] [PubMed]
- Robin, J.; Brauer, J.; Sulmann, S.; Marino, V.; Dell’Orco, D.; Lienau, C.; Koch, K.W. Differential Nanosecond Protein Dynamics in Homologous Calcium Sensors. ACS Chem. Biol. 2015, 10, 2344–2352. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.Y.; Koch, K.W. Calcium- and myristoyl-dependent properties of guanylate cyclase-activating protein-1 and protein-2. Biochemistry 2002, 41, 13021–13028. [Google Scholar] [CrossRef]
- Koch, K.W.; Helten, A. Guanylate cyclase-based signaling in photoreceptors and retina. In Signal Transduction in the Retina; Taylor and Francis CRC Press: Boca Raton, FL, USA, 2008; pp. 121–143. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Hwang, J.Y.; Lange, C.; Helten, A.; Hoppner-Heitmann, D.; Duda, T.; Sharma, R.K.; Koch, K.W. Regulatory modes of rod outer segment membrane guanylate cyclase differ in catalytic efficiency and Ca(2+)-sensitivity. Eur. J. Biochem. 2003, 270, 3814–3821. [Google Scholar] [CrossRef]
- Zagel, P.; Dell’Orco, D.; Koch, K.W. The dimerization domain in outer segment guanylate cyclase is a Ca(2)(+)-sensitive control switch module. Biochemistry 2013, 52, 5065–5074. [Google Scholar] [CrossRef]
- Dell’Orco, D.; Sulmann, S.; Zagel, P.; Marino, V.; Koch, K.W. Impact of cone dystrophy-related mutations in GCAP1 on a kinetic model of phototransduction. Cell. Mol. Life Sci. 2014, 71, 3829–3840. [Google Scholar] [CrossRef]
- Vallone, R.; Dal Cortivo, G.; D’Onofrio, M.; Dell’Orco, D. Preferential binding of Mg2+ over Ca2+ to CIB2 triggers an allosteric switch impaired in Usher Syndrome type 1J. Front. Mol. Neurosci. 2018, 11, 274. [Google Scholar] [CrossRef]
- Laurent, T.C.; Killander, J. A theory of gel filtration and its exeperimental verification. J. Chromatogr. A 1964, 14, 317–330. [Google Scholar] [CrossRef]
- Astegno, A.; Maresi, E.; Marino, V.; Dominici, P.; Pedroni, M.; Piccinelli, F.; Dell’Orco, D. Structural plasticity of calmodulin on the surface of CaF2 nanoparticles preserves its biological function. Nanoscale 2014, 6, 15037–15047. [Google Scholar] [CrossRef] [PubMed]
- Marino, V.; Borsatto, A.; Vocke, F.; Koch, K.W.; Dell’Orco, D. CaF2 nanoparticles as surface carriers of GCAP1, a calcium sensor protein involved in retinal dystrophies. Nanoscale 2017, 9, 11773–11784. [Google Scholar] [CrossRef] [PubMed]
- Sulmann, S.; Dell’Orco, D.; Marino, V.; Behnen, P.; Koch, K.W. Conformational changes in calcium-sensor proteins under molecular crowding conditions. Chemistry 2014, 20, 6756–6762. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmuller, H.; MacKerell, A.D., Jr. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef]
- Marino, V.; Dell’Orco, D. Evolutionary-Conserved Allosteric Properties of Three Neuronal Calcium Sensor Proteins. Front. Mol. Neurosci. 2019, 12, 50. [Google Scholar] [CrossRef]
| GCAP1 Variant | GCAP1 Ca2+ Titration (Three Site Model) | |||||
|---|---|---|---|---|---|---|
| Apparent Dissociation Constant KD | Enthalpy Change ΔH (kcal/mol) | |||||
| KD1 (nM) | KD2 (nM) | KD3 (µM) | ΔH1 | ΔH2 | ΔH3 | |
| WT | 16.3 ± 1.2 | 36 ± 8 | 0.25 ± 0.05 | −4.2 ± 0.2 | −5 ± 4 | 1.20 ± 0.05 |
| G86R | 33 ± 2 | 78 ± 12 | 1.80 ± 0.05 | −5.2 ± 0.1 | −2.2 ± 0.1 | 3.20 ± 0.08 |
| G86R+W94L | 6 ± 4 | 7 ± 4 | 0.82 ± 0.13 | −12 ± 2 | 3.5 ± 0.5 | 2.6 ± 0.2 |
| G86R+W94F | 1.3 ± 0.8 (µM) | 101 ± 23 | 3.1 ± 0.3 | −7.4 ± 1.2 | −1.2 ± 0.2 | 5.4 ± 0.2 |
| +1 mM Mg2+ | ||||||
| WT | 77 ± 28 | 129 ± 17 | 0.8 ± 0.2 | −3.5 ± 0.4 | −1.2 ± 0.5 | 0.10 ± 0.01 |
| G86R | 62 ± 27 | 145 ± 12 | 1.6 ± 1.1 | −4.0 ± 0.1 | −3.7 ± 0.1 | 0.20 ± 0.02 |
| G86R+W94L | 138 ± 12 | 628 ± 30 | 358.0 ± 0.4 | −11.0 ± 0.1 | 3.8 ± 0.4 | −6 ± 3 |
| G86R+W94F | 1.9 ± 1.2 (µM) | 133 ± 34 | 4.6 ± 1.8 | −9.2 ± 2.0 | −2.6 ± 0.5 | 3.6 ± 0.3 |
| GCAP1 Variant | GCAP1 Mg2+ Titration (Two Site Model) | |||||
| KD1 (µM) | KD2 (µM) | ΔH1 | ΔH2 | |||
| G86R+W94L | 648 ± 144 | 15 ± 8 | 0.34 ± 0.08 | 21 ± 6 | ||
| G86R+W94F | 238 ± 123 | 112 ± 41 | 1.4 ± 0.5 | 12.3 ± 1.4 | ||
| GCAP1 Variant | Experimental Condition | θ222/θ208 | Δθ222/θ222 | Tm (°C) |
|---|---|---|---|---|
| WT | EGTA | 0.88 | - | 49.8 |
| Mg2+ | 0.90 | 0.08 | 50.3 | |
| Mg2+ Ca2+ | 0.93 | 0.15 | >96 | |
| G86R | EGTA | 0.88 | - | 41.6 |
| Mg2+ | 0.88 | 0.03 | 46.8 | |
| Mg2+ Ca2+ | 0.92 | 0.10 | >96 | |
| G86R+W94F | EGTA | 0.90 | - | 40.6 |
| Mg2+ | 0.92 | 0.02 | 47.8 | |
| Mg2+ Ca2+ | 0.92 | 0.08 | >96 | |
| G86R+W94L | EGTA | 0.88 | - | 42 |
| Mg2+ | 0.90 | 0.02 | 47 | |
| Mg2+ Ca2+ | 0.90 | 0.09 | >96 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abbas, S.; Marino, V.; Bielefeld, L.; Koch, K.-W.; Dell’Orco, D. Constitutive Activation of Guanylate Cyclase by the G86R GCAP1 Variant Is Due to “Locking” Cation-π Interactions that Impair the Activator-to-Inhibitor Structural Transition. Int. J. Mol. Sci. 2020, 21, 752. https://doi.org/10.3390/ijms21030752
Abbas S, Marino V, Bielefeld L, Koch K-W, Dell’Orco D. Constitutive Activation of Guanylate Cyclase by the G86R GCAP1 Variant Is Due to “Locking” Cation-π Interactions that Impair the Activator-to-Inhibitor Structural Transition. International Journal of Molecular Sciences. 2020; 21(3):752. https://doi.org/10.3390/ijms21030752
Chicago/Turabian StyleAbbas, Seher, Valerio Marino, Laura Bielefeld, Karl-Wilhelm Koch, and Daniele Dell’Orco. 2020. "Constitutive Activation of Guanylate Cyclase by the G86R GCAP1 Variant Is Due to “Locking” Cation-π Interactions that Impair the Activator-to-Inhibitor Structural Transition" International Journal of Molecular Sciences 21, no. 3: 752. https://doi.org/10.3390/ijms21030752
APA StyleAbbas, S., Marino, V., Bielefeld, L., Koch, K.-W., & Dell’Orco, D. (2020). Constitutive Activation of Guanylate Cyclase by the G86R GCAP1 Variant Is Due to “Locking” Cation-π Interactions that Impair the Activator-to-Inhibitor Structural Transition. International Journal of Molecular Sciences, 21(3), 752. https://doi.org/10.3390/ijms21030752






