Function and Regulation of the Pyruvate Transporter CstA in Escherichia coli †
Abstract
1. Introduction
2. Results
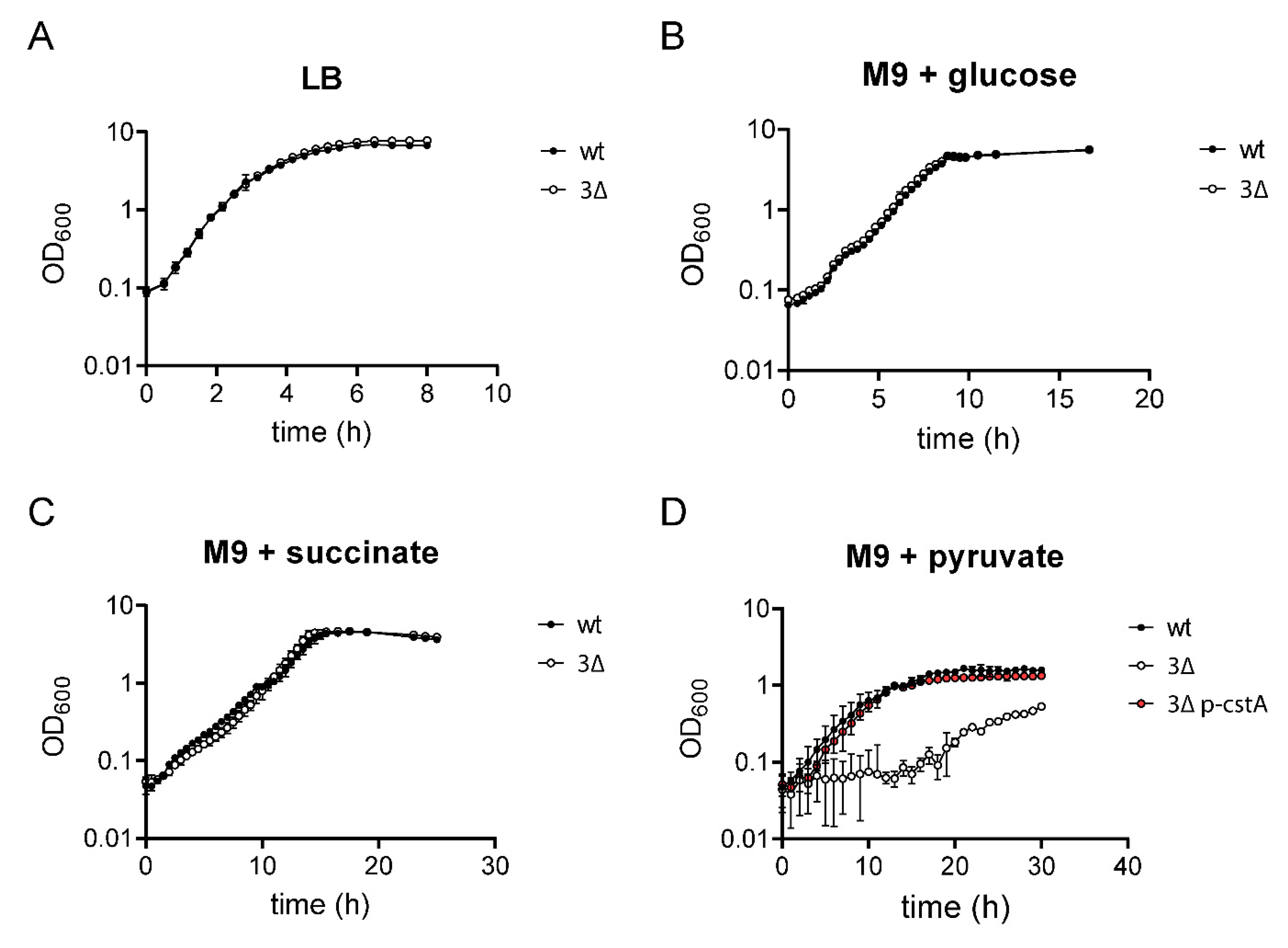
2.1. Construction of a Mutant that Is Unable to Grow on and Take up Pyruvate
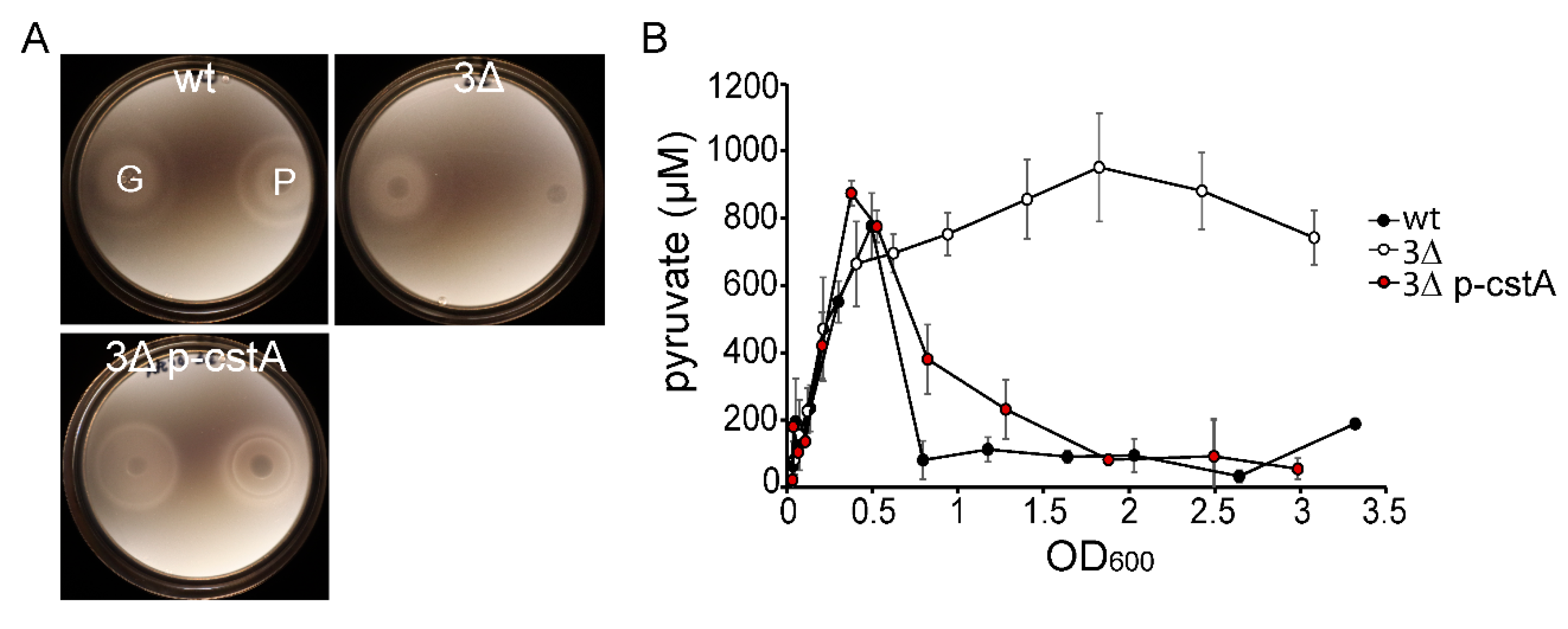
2.2. CstA Restores Growth and Chemotaxis towards Pyruvate
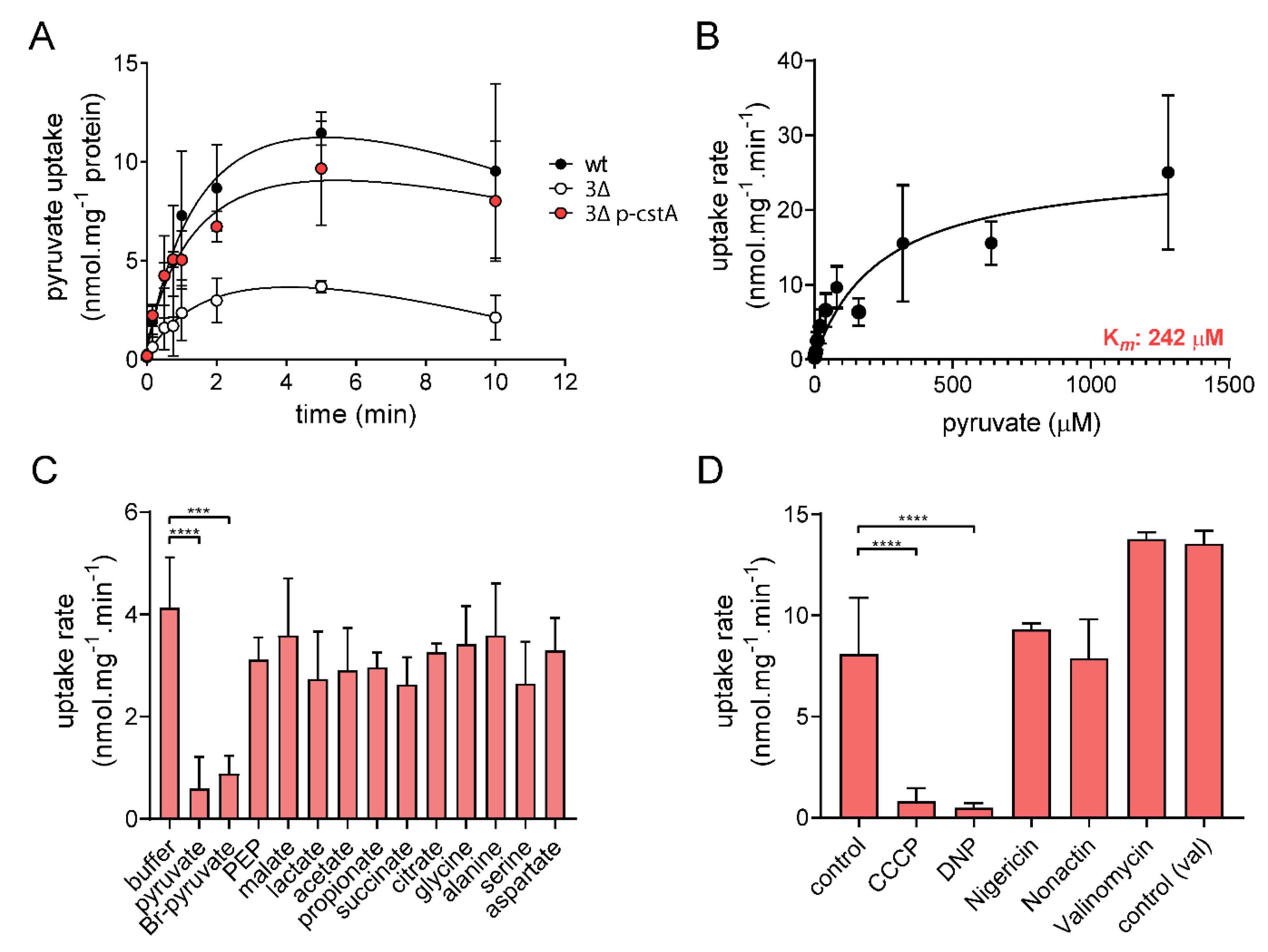
2.3. CstA Is a Specific Pyruvate Transporter with Moderate Affinity
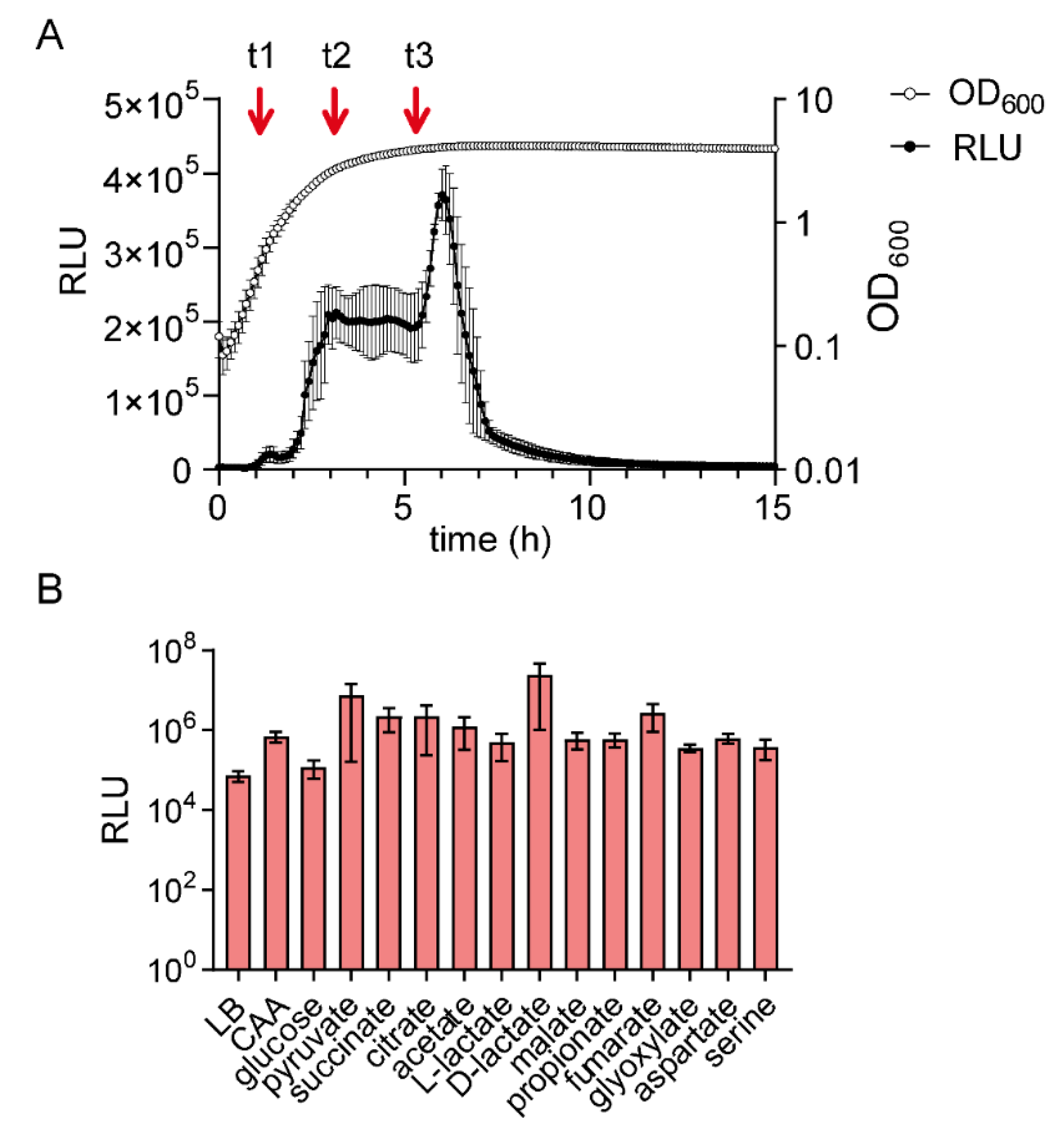
2.4. CstA Is Expressed in Late Exponential and Stationary Phase
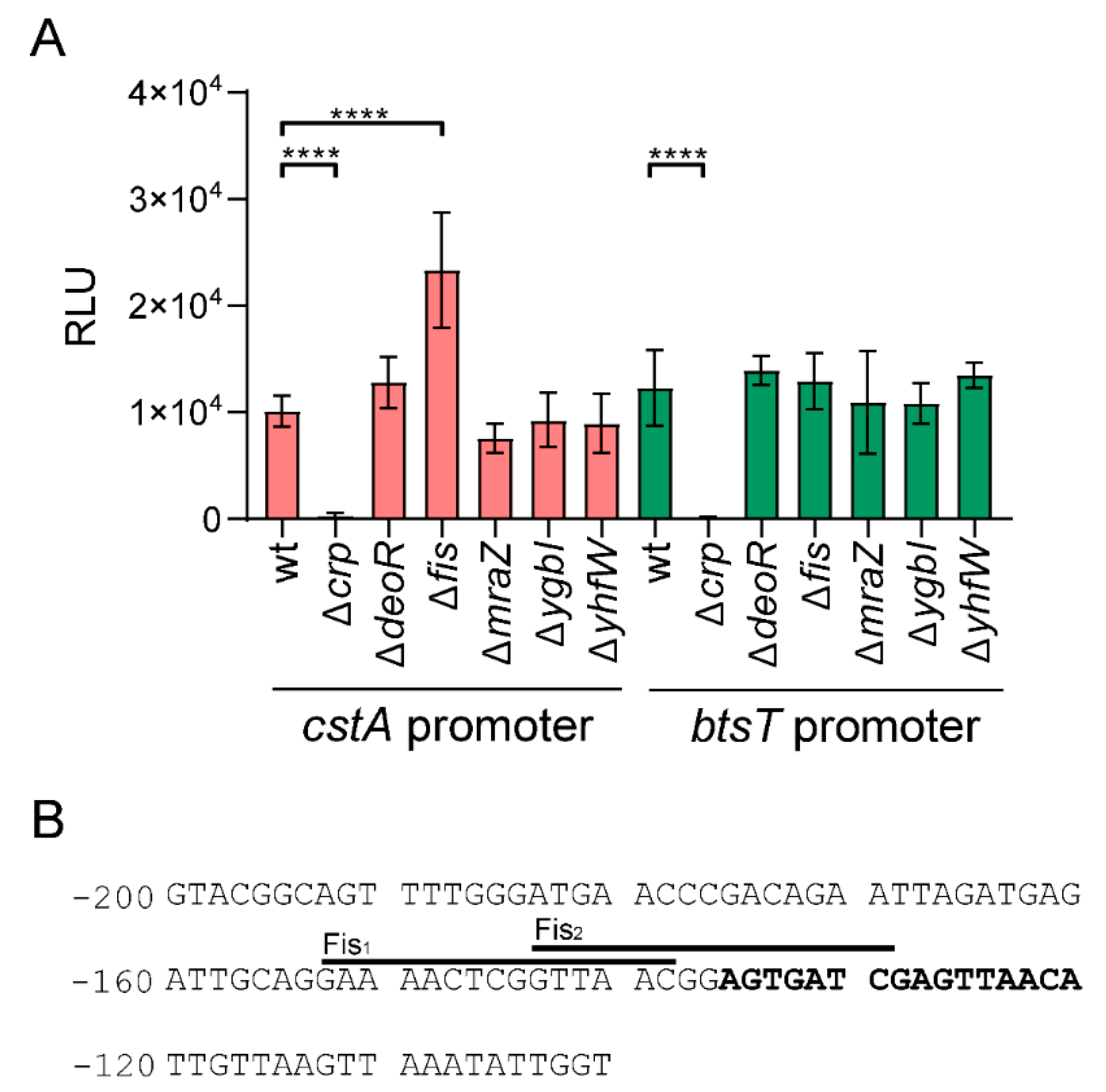
2.5. Identification of Fis as a Regulator of cstA
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Plasmids
4.2. Growth Conditions
4.3. Determination of the Extracellular Pyruvate Concentration
4.4. Chemotaxis Assay
4.5. Promoter Activity Assay
4.6. Transport Measurements with Intact Cells
4.7. DNA Affinity Capture Assay
4.8. Mass Spectrometry
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ahn, S.J.; Deep, K.; Turner, M.E.; Ishkov, I.; Waters, A.; Hagen, S.J.; Rice, K.C. Characterization of LrgAB as a stationary phase-specific pyruvate uptake system in Streptococcus mutans. BMC Microbiol. 2019, 19, 1–15. [Google Scholar] [CrossRef]
- Steiner, B.D.; Eberly, A.R.; Hurst, M.N.; Zhang, E.W.; Green, H.D.; Behr, S.; Jung, K.; Hadjifrangiskou, M. Evidence of cross-regulation in two closely related pyruvate-sensing systems in uropathogenic Escherichia coli. J. Membr. Biol. 2018, 251, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Tuntufye, H.N.; Lebeer, S.; Gwakisa, P.S.; Goddeeris, B.M. Identification of avian pathogenic Escherichia coli genes that are induced in vivo during infection in chickens. Appl. Environ. Microbiol. 2012, 78, 3343–3351. [Google Scholar] [CrossRef] [PubMed]
- Schär, J.; Stoll, R.; Schauer, K.; Loeffler, D.I.M.; Eylert, E.; Joseph, B.; Eisenreich, W.; Fuchs, T.M.; Goebel, W. Pyruvate carboxylase plays a crucial role in carbon metabolism of extra and intracellularly replicating Listeria monocytogenes. J. Bacteriol. 2010, 192, 1774–1784. [Google Scholar] [CrossRef] [PubMed]
- Harper, L.; Balasubramanian, D.; Ohneck, E.A.; Sause, W.E.; Chapman, J.; Mejia-Sosa, B.; Lhakhang, T.; Heguy, A.; Tsirigos, A.; Ueberheide, B.; et al. Staphylococcus aureus responds to the central metabolite pyruvate to regulate virulence. mBio 2018, 9, 1–17. [Google Scholar] [CrossRef]
- Bücker, R.; Heroven, A.K.; Becker, J.; Dersch, P.; Wittmann, C. The pyruvate-tricarboxylic acid cycle node: A focal point of virulence control in the enteric pathogen Yersinia pseudotuberculosis. J. Biol. Chem. 2014, 289, 30114–30132. [Google Scholar] [CrossRef]
- Behr, S.; Brameyer, S.; Witting, M.; Schmitt-Kopplin, P.; Jung, K. Comparative analysis of LytS/LytTR-type histidine kinase/response regulator systems in gamma-proteobacteria. PLoS ONE 2017, 12, 1–14. [Google Scholar] [CrossRef]
- Charbonnier, T.; Le Coq, D.; McGovern, S.; Calabre, M.; Delumeau, O.; Aymerich, S.; Jules, M. Molecular and physiological logics of the pyruvate-induced response of a novel transporter in Bacillus subtilis. mBio 2017, 8, 1–18. [Google Scholar] [CrossRef]
- Vilhena, C.; Kaganovitch, E.; Grünberger, A.; Motz, M.; Forné, I.; Kohlheyer, D.; Jung, K. Importance of pyruvate sensing and transport for the resuscitation of viable but nonculturable Escherichia coli K-12. J. Bacteriol. 2018, 201. [Google Scholar] [CrossRef]
- Vemuri, G.N.; Altman, E.; Sangurdekar, D.P.; Khodursky, A.B.; Eiteman, M.A. Overflow metabolism in Escherichia coli during steady-state growth: Transcriptional regulation and effect of the redox ratio. Appl. Environ. Microbiol. 2006, 72, 3653–3661. [Google Scholar] [CrossRef]
- Paczia, N.; Nilgen, A.; Lehmann, T.; Gätgens, J.; Wiechert, W.; Noack, S. Extensive exometabolome analysis reveals extended overflow metabolism in various microorganisms. Microb. Cell Fact. 2012, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Behr, S.; Fried, L.; Jung, K. Identification of a novel nutrient-sensing histidine kinase/response regulator network in Escherichia coli. J. Bacteriol. 2014, 196, 2023–2029. [Google Scholar] [CrossRef] [PubMed]
- Kraxenberger, T.; Fried, L.; Behr, S.; Jung, K. First insights into the unexplored two-component system YehU/YehT in Escherichia coli. J. Bacteriol. 2012, 194, 4272–4284. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fried, L.; Behr, S.; Jung, K. Identification of a target gene and activating stimulus for the YpdA/YpdB histidine kinase/response regulator system in Escherichia coli. J. Bacteriol. 2012, 195, 807–815. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ruby, E.G.; Nealson, K.H. Pyruvate production and excretion by the luminous marine bacteria. Appl. Environ. Microbiol. 1977, 34, 164–169. [Google Scholar] [CrossRef]
- Chubukov, V.; Gerosa, L.; Kochanowski, K.; Sauer, U. Coordination of microbial metabolism. Nat. Rev. Microbiol. 2014, 12, 327–340. [Google Scholar] [CrossRef]
- Kreth, J.; Lengeler, J.W.; Jahreis, K. Characterization of pyruvate uptake in Escherichia coli K-12. PLoS ONE 2013, 8, 6–12. [Google Scholar] [CrossRef]
- Miyake, Y.; Inaba, T.; Watanabe, H.; Teramoto, J.; Yamamoto, K.; Ishihama, A. Regulatory roles of pyruvate-sensing two-component system PyrSR (YpdAB) in Escherichia coli K-12. FEMS Microbiol. Lett. 2019, 366, 1–9. [Google Scholar] [CrossRef]
- Ogasawara, H.; Ishizuka, T.; Yamaji, K.; Kato, Y.; Shimada, T.; Ishihama, A. Regulatory role of pyruvate-sensing BtsSR in biofilm formation by Escherichia coli K-12. FEMS Microbiol. Lett. 2020, 366, 1–10. [Google Scholar] [CrossRef]
- Kristoficova, I.; Vilhena, C.; Behr, S.; Jung, K. BtsT, a novel and specific Pyruvate/H+ symporter in Escherichia coli. J. Bacteriol. 2018, 200, 1–11. [Google Scholar] [CrossRef]
- Hwang, S.; Choe, D.; Yoo, M.; Cho, S.; Kim, S.C.; Cho, S.; Cho, B.K. Peptide transporter CstA imports pyruvate in Escherichia coli K-12. J. Bacteriol. 2018, 200, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Saier, M.H.; Reddy, V.S.; Tamang, D.G.; Västermark, Å. The transporter classification database. Nucleic Acids Res. 2014, 42, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.E.; Matin, A. Molecular and functional characterization of a carbon starvation gene of Escherichia coli. J. Mol. Biol. 1991, 218, 129–140. [Google Scholar] [CrossRef]
- Rasmussen, J.J.; Vegge, C.S.; Frøkiær, H.; Howlett, R.M.; Krogfelt, K.A.; Kelly, D.J.; Ingmer, H. Campylobacter jejuni carbon starvation protein A (CstA) is involved in peptide utilization, motility and agglutination, and has a role in stimulation of dendritic cells. J. Med. Microbiol. 2013, 62, 1135–1143. [Google Scholar] [CrossRef]
- Garai, P.; Lahiri, A.; Ghosh, D.; Chatterjee, J.; Chakravortty, D. Peptide-utilizing carbon starvation gene yjiY is required for flagella-mediated infection caused by Salmonella. Microbiology 2016, 162, 100–116. [Google Scholar] [CrossRef]
- Dubey, A.K.; Baker, C.S.; Suzuki, K.; Jones, A.D.; Pandit, P.; Romeo, T.; Babitzke, P. CsrA regulates translation of the Escherichia coli carbon starvation gene, cstA, by blocking ribosome access to the cstA transcript. J. Bacteriol. 2003, 185, 4450–4460. [Google Scholar] [CrossRef]
- Gralla, J.D. Bacterial gene regulation from distant DNA sites. Cell 1989, 57, 193–195. [Google Scholar] [CrossRef]
- Somavanshi, R.; Ghosh, B.; Sourjik, V. Sugar influx sensing by the phosphotransferase system of Escherichia coli. PLoS Biol. 2016, 14, 1–19. [Google Scholar] [CrossRef]
- Schultz, J.E.; Latter, G.I.; Matin, A. Differential regulation by cyclic AMP of starvation protein synthesis in Escherichia coli. J. Bacteriol. 1988, 170, 3903–3909. [Google Scholar] [CrossRef]
- Gödeke, J.; Heun, M.; Bubendorfer, S.; Paul, K.; Thormann, K.M. Roles of two Shewanella oneidensis MR-1 extracellular endonucleases. Appl. Environ. Microbiol. 2011, 77, 5342–5351. [Google Scholar] [CrossRef]
- Koga, K.; Harada, T.; Shimizu, H.; Tanaka, K. Bacterial luciferase activity and the intracellular redox pool in Escherichia coli. Mol. Genet. Genom. 2005, 274, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Karp, M. Intracellular redox equilibrium and growth phase affect the performance of luciferase-based biosensors. J. Biotechnol. 2007, 127, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Park, S.S.; Ko, B.J.; Kim, B.G. Mass spectrometric screening of transcriptional regulators using DNA affinity capture assay. Anal. Biochem. 2005, 344, 152–154. [Google Scholar] [CrossRef] [PubMed]
- Kalinowski, J. Intracellular Signaling and Gene Target Analysis in Bacterial Signaling; Kramer, R., Jung, K., Eds.; Wiley-VCH Verlag GmbH & Co. KgaA: Weinheim, Germany, 2010; pp. 463–472. [Google Scholar]
- Shao, Y.; Feldman-Cohen, L.S.; Osuna, R. Functional characterization of the Escherichia coli Fis-DNA binding sequence. J. Mol. Biol. 2008, 376, 771–785. [Google Scholar] [CrossRef]
- Bradley, M.D.; Beach, M.B.; de Koning, A.P.J.; Pratt, T.S.; Osuna, R. Effects of Fis on Escherichia coli gene expression during different growth stages. Microbiology 2007, 153, 2922–2940. [Google Scholar] [CrossRef]
- Basan, M.; Hui, S.; Okano, H.; Zhang, Z.; Shen, Y.; Williamson, J.R.; Haw, T. Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature 2015, 528, 99–104. [Google Scholar] [CrossRef]
- Anderson, J.; Marks, V. Pyruvate in diabetes mellitus, concentrations in urine and blood. Lancet 1962, 1, 1159–1161. [Google Scholar] [CrossRef]
- Mathioudakis, D.; Engel, J.; Welters, I.D.; Dehne, M.G.; Matejec, R.; Harbach, H.; Henrich, M.; Schwandner, T.; Fuchs, M.; Weismüller, K.; et al. Pyruvate: Immunonutritional effects on neutrophil intracellular amino or alpha-keto acid profiles and reactive oxygen species production. Amino Acids 2011, 40, 1077–1090. [Google Scholar] [CrossRef]
- Mühling, J.; Fuchs, M.; Campos, M.E.; Gonter, J.; Engel, J.M.; Sablotzki, A.; Menges, T.; Weiss, S.; Dehne, M.G.; Krüll, M.; et al. Quantitative determination of free intracellular α-keto acids in neutrophils. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2003, 789, 383–392. [Google Scholar] [CrossRef]
- Hawver, L.A.; Giulietti, J.M.; Baleja, J.D.; Ng, W.L. Quorum sensing coordinates cooperative expression of pyruvate metabolism genes to maintain a sustainable environment for population stability. mBio 2016, 7, 1–11. [Google Scholar] [CrossRef]
- Jolkver, E.; Emer, D.; Ballan, S.; Krämer, R.; Eikmanns, B.J.; Marin, K. Identification and characterization of a bacterial transport system for the uptake of pyruvate, propionate, and acetate in Corynebacterium glutamicum. J. Bacteriol. 2009, 191, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Hosie, A.H.F.; Allaway, D.; Poole, P.S. A monocarboxylate permease of Rhizobium leguminosarum is the first member of a new subfamily of transporters. J. Bacteriol. 2002, 184, 5436–5448. [Google Scholar] [CrossRef] [PubMed]
- Behr, S.; Kristoficova, I.; Witting, M.; Breland, E.J.; Eberly, A.R.; Sachs, C.; Schmitt-Kopplin, P.; Hadjifrangiskou, M.; Jung, K. Identification of a high-affinity Pyruvate receptor in Escherichia coli. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Azam, T.A.; Iwata, A.; Nishimura, A.; Ueda, S.; Ishihama, A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999, 181, 6361–6370. [Google Scholar] [CrossRef] [PubMed]
- Ball, C.A.; Osuna, R.; Ferguson, K.C.; Johnson, R.C. Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J. Bacteriol. 1992, 174, 8043–8056. [Google Scholar] [CrossRef]
- Serra, D.O.; Klauck, G.; Hengge, R. Vertical stratification of matrix production is essential for physical integrity and architecture of macrocolony biofilms of Escherichia coli. Environ. Microbiol. 2015, 17, 5073–5088. [Google Scholar] [CrossRef]
- Ferenci, T. Regulation by nutrient limitation. Curr. Opin. Microbiol. 1999, 2, 208–213. [Google Scholar] [CrossRef]
- Ferenci, T. Hungry bacteria—Definition and properties of a nutritional state. Environ. Microbiol. 2001, 3, 605–611. [Google Scholar] [CrossRef]
- Blattner, F.R.; Plunkett, G.; Bloch, C.A.; Perna, N.T.; Burland, V.; Riley, M.; Collado-Vides, J.; Glasner, J.D.; Rode, C.K.; Mayhew, G.F.; et al. The complete genome sequence of Escherichia coli K-12. Science 1997, 277, 1453–1462. [Google Scholar] [CrossRef]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef]
- Guzman, L.M.; Belin, D.; Carson, M.J.; Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose P(BAD) promoter. J. Bacteriol. 1995, 177, 4121–4130. [Google Scholar] [CrossRef] [PubMed]
- Harwood, C.R.; Cutting, S.M. (Eds.) Chemically defined growth media and suplements. In Molecular Biological Methods for Bacillus; Wiley: London, UK, 1990; p. 548. [Google Scholar]
- O’donnell-Tormey, J.; Nathan, C.F.; Lanks, K.; Deboer, C.J.; De La Harpe, J. Secretion of pyruvate: An antioxidant defense of mammalian cells. J. Exp. Med. 1987, 165, 500–514. [Google Scholar] [CrossRef]
- Reyes-Darías, J.A.; Garcia-Fontana, C.; Corral-Lugo, A.; Rico-Jiménez, M.; Krell, T. Qualitative and quantitative assays for flagellum-mediated chemotaxis. Methods Mol. Biol. 2014, 1149, 87–97. [Google Scholar]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
| Time Point | Protein | UniProt Description | Fold Change |
|---|---|---|---|
| t1 | AtpE | ATP synthase subunit c | 2.47 |
| DeoR * | Regulator | 1.97 | |
| Fis * | Regulator | 3.53 | |
| HemY | Heme metabolic process | 1.82 | |
| MraZ * | Regulator | 1.85 | |
| RpmG | 50S ribosomal protein | 3.19 | |
| XseB | Exodeoxyribonuclease 7 small subunit | 2.10 | |
| YdjA | Putative NAD(P)H nitroreductase | 1.98 | |
| t2 | AceA | Isocitrate lyase | 2.06 |
| DeoR * | Regulator | 2.19 | |
| IhfA | Integration host factor | 2.22 | |
| Lpp | Major outer membrane lipoprotein | 2.00 | |
| Rph | Truncated inactive ribonuclease PH | 1.77 | |
| RplW | 30S ribosomal protein S5 | 1.94 | |
| RpmA | 50S ribosomal protein L27 | 2.85 | |
| RpsT | 30S ribosomal protein S20 | 1.95 | |
| YgbI * | Uncharacterized HTH-type transcriptional regulator | 2.08 | |
| YhfW * | Uncharacterized protein | 2.70 | |
| t3 | DeoR * | Regulator | 2.00 |
| JayE | Putative protein from lambdoid prophage | 3.34 | |
| Lpp | Major outer membrane lipoprotein | 2.01 | |
| RhlB | ATP-dependent RNA helicase RhlB | 2.13 | |
| RnpA | Ribonuclease P protein component | 2.68 | |
| YcaC | Probable hydrolase YcaC | 1.73 | |
| YgbI * | Uncharacterized HTH-type transcriptional regulator | 1.83 |
| Strains | ||
|---|---|---|
| E. coli MG1655 | [50] | |
| E. coli BW25113 | [51] | |
| JW5702 (E. coli BW25113 Δcrp) | [51] | |
| JW0824 (E. coli BW25113 ΔdeoR) | [51] | |
| JW3229 (E. coli BW25113 Δfis) | [51] | |
| JW0079 (E. coli BW25113 ΔmraZ) | [51] | |
| JW2705 (E. coli BW25113 ΔygbI) | [51] | |
| JW3343 (E. coli BW25113 ΔyhfW) | [51] | |
| E. coli MG1655 ΔbtsT ΔyhjX ΔcstA (3∆) | This study | |
| E. coli MG1655 ΔbtsT ΔcstA | This study | |
| E. coli MG1655 ΔbtsT ΔyhjX | This study | |
| E. coli MG1655 ΔcstA ΔyhjX | This study | |
| E. coli MG1655 ΔbtsT | [20] | |
| E. coli MG1655 ΔcstA | This study | |
| E. coli MG1655 ΔyhjX | [14] | |
| Plasmids | ||
| pBAD24 | [52] | |
| pBAD24-cstA6His (pBAD24-cstA6H) | This study | |
| pBBR1-cstAprom-lux | This study | |
| pBBR yjiY-lux (pBBR1-btsTprom-lux) | [13] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gasperotti, A.; Göing, S.; Fajardo-Ruiz, E.; Forné, I.; Jung, K. Function and Regulation of the Pyruvate Transporter CstA in Escherichia coli. Int. J. Mol. Sci. 2020, 21, 9068. https://doi.org/10.3390/ijms21239068
Gasperotti A, Göing S, Fajardo-Ruiz E, Forné I, Jung K. Function and Regulation of the Pyruvate Transporter CstA in Escherichia coli. International Journal of Molecular Sciences. 2020; 21(23):9068. https://doi.org/10.3390/ijms21239068
Chicago/Turabian StyleGasperotti, Ana, Stephanie Göing, Elena Fajardo-Ruiz, Ignasi Forné, and Kirsten Jung. 2020. "Function and Regulation of the Pyruvate Transporter CstA in Escherichia coli" International Journal of Molecular Sciences 21, no. 23: 9068. https://doi.org/10.3390/ijms21239068
APA StyleGasperotti, A., Göing, S., Fajardo-Ruiz, E., Forné, I., & Jung, K. (2020). Function and Regulation of the Pyruvate Transporter CstA in Escherichia coli. International Journal of Molecular Sciences, 21(23), 9068. https://doi.org/10.3390/ijms21239068





