Plant Single-Cell Metabolomics—Challenges and Perspectives
Abstract
1. Introduction
2. Technical Challenges
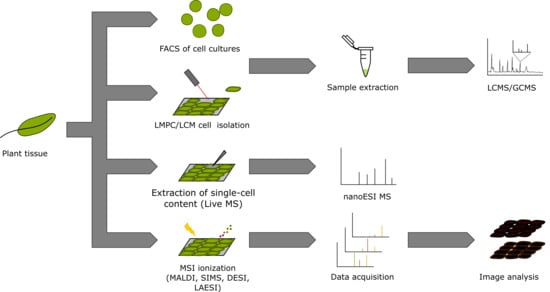
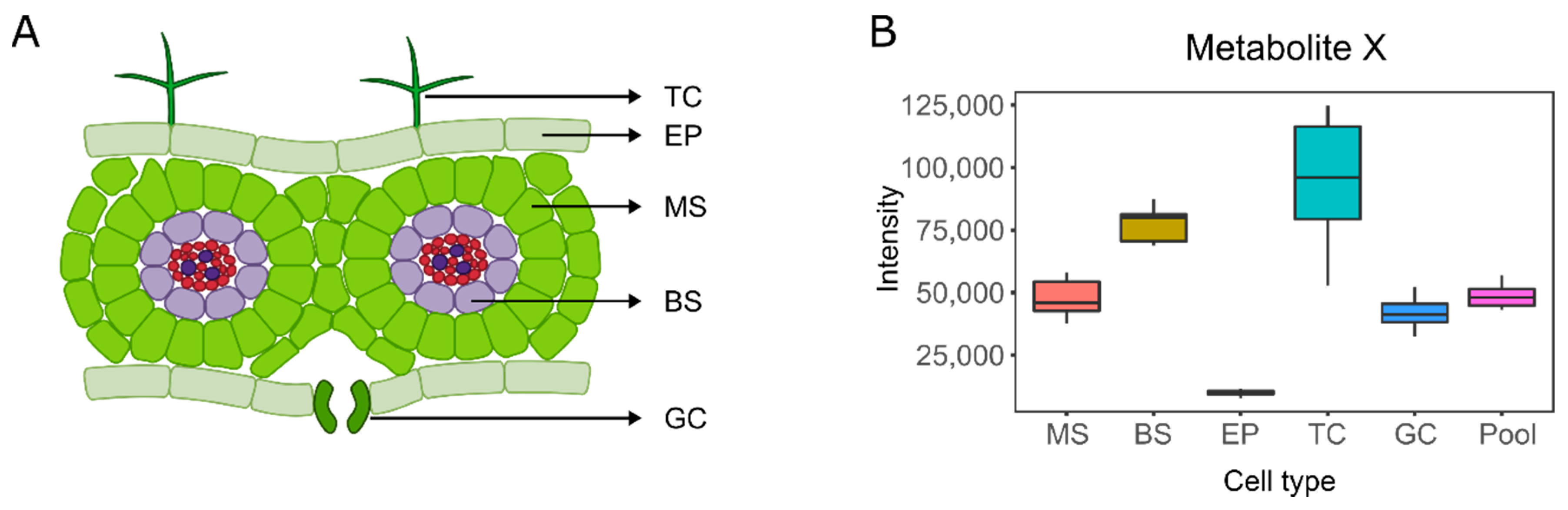
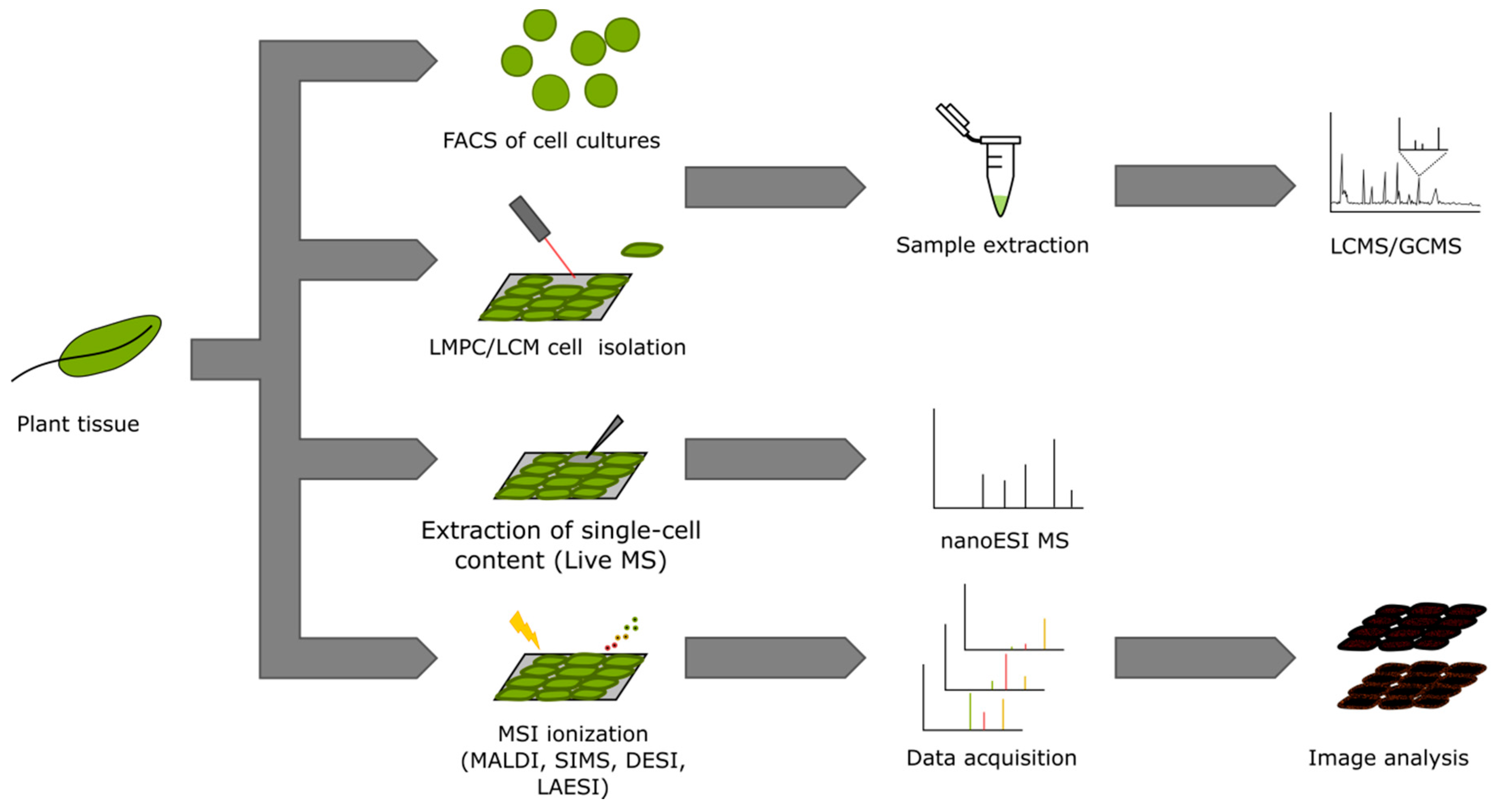
3. Single-Cell and Single-Cell-Type Metabolomics
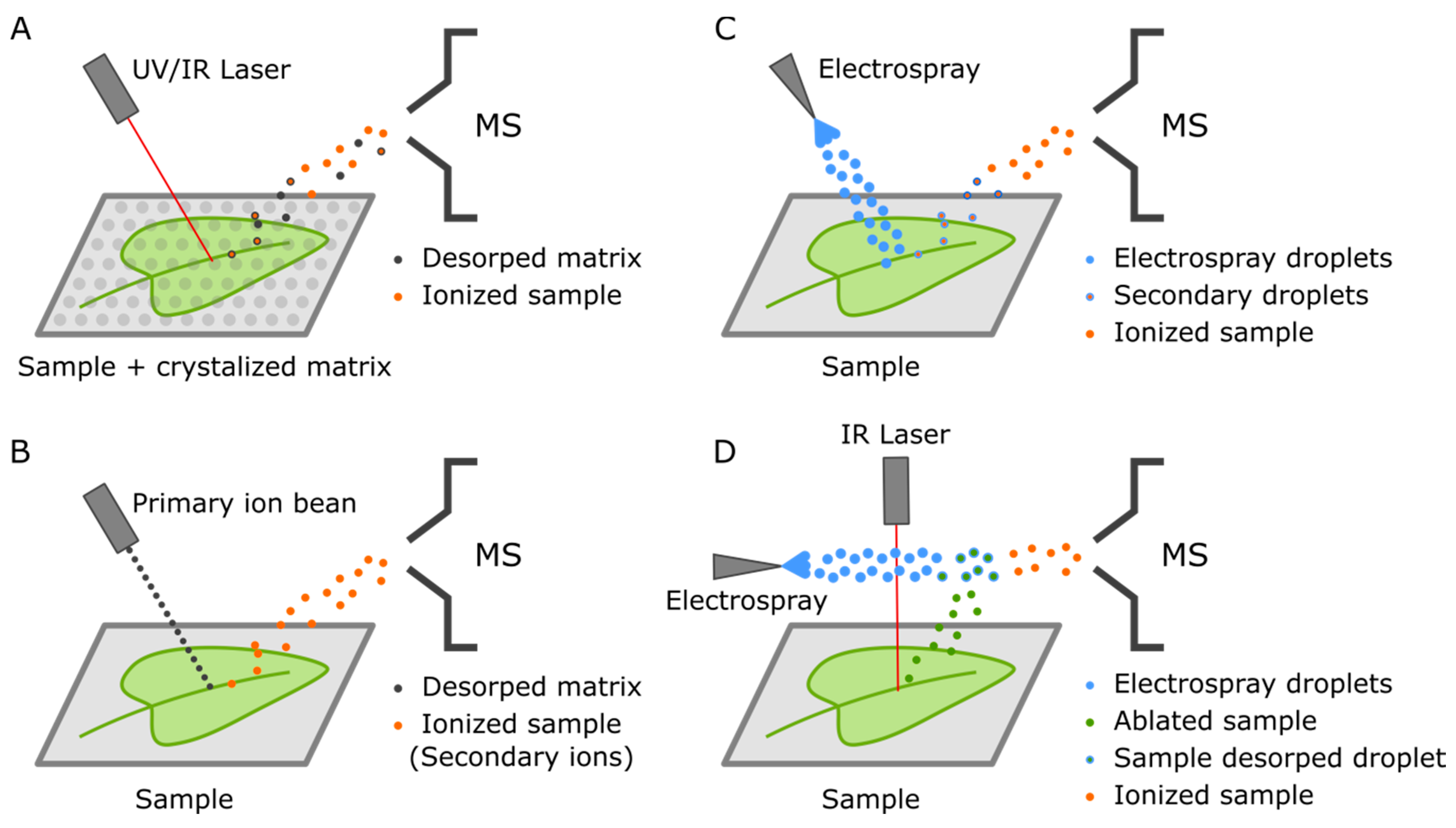
4. Mass Spectrometry Imaging (MSI)
5. Spatially Resolved Metabolomics in Plants: Current Status, Challenges, and Future Prospects
| Species | Technique | Cell-Type/Tissue | Compounds | Reference |
|---|---|---|---|---|
| Arabidopsis | FACS | Roots | Multiple | [27] |
| Arabidopsis | MALDI | Leaves | Glucosinolates | [60] |
| Catharanthus roseus | MALDI and Live-MS | Laticifers and idioblasts from leaves | TIA | [61] |
| Catharanthus roseus | MALDI and Live-MS | Laticifers, idioblast, parenchyma, and epidermal cells from stems | TIA | [62] |
| Viola cornuta | MALDI | Petals | Flavonoids | [64] |
| Rauvolfia tetraphylla | DESI | Stem, leaves, root, and fruits | Indole alkaloids | [69] |
| Hypericum perforatum | DESI | Petals and leaves | Hyperforin | [71] |
| Datura stramonium | DESI | Petals and leaves | Sugars, atropine, and scopolamine | [71] |
| Maize | MALDI | Roots | Amino acids | [76] |
| Maize | MALDI | Roots | Lipids, sugars, and benzoxazinoid | [77] |
| Glycyrrhiza glabra | MALDI | Roots | Flavonoids and triterpenoids | [78] |
| Camelina sativa | MALDI | Seed | Lipids | [80] |
| Camelina sativa | MALDI | Seed | Lipids | [81] |
| Camelina sativa | MALDI | Seed | Lipids | [82] |
| Brassica napus | MALDI | Seed | Lipids | [83] |
| Arabidopsis | MALDI | Seed | Lipids | [84] |
| Barley | MALDI | Germinating seeds | Multiple | [85] |
| Maize | MALDI | Germinating seeds | Multiple | [88] |
| Lycopodium clavatum | SIMS and MALDI | Polen | Sporopollenin | [90] |
| Poa alpina | MALDI | Polen | Multiple | [91] |
| Arabidopsis | MALDI | Leaves | Oxylipins | [108] |
| Rice | MALDI | Leaves | Multiple | [109] |
| Soybean | MALDI | Leaves | Multiple | [109] |
| Soybean | MALDI | Leaves | Isoflavones | [110] |
| Medicago truncatula | MALDI | Root nodules | Multiple | [111] |
| Medicago truncatula | MALDI | Root nodules | Multiple | [112] |
| Soybean | MALDI | Root nodules | Multiple | [113] |
| Soybean | LAESI | Root nodules | Multiple | [114] |
| Vicia faba | Live-MS | Leaves | Phytohormones | [117] |
6. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| DESI | Desorption electrospray ionization |
| FACS | Fluorescence-activated cell sorting |
| GC | Gas chromatography |
| LAESI | Laser-ablation electrospray ionization |
| LC | Liquid chromatography |
| LCM | Laser capture microdissection |
| LMD | Laser microdissection |
| LMPC | Laser microdissection and pressure catapulting |
| MALDI | Matrix-assisted laser desorption/ionization |
| MS | Mass spectrometry |
| MSI | Mass spectrometry imaging |
| QTOF | Quadrupole time-of-flight mass spectrometer |
| SIMS | Secondary ion mass spectrometry |
| TIA | Terpenoid indole alkaloid |
| UHPLC | Ultra-high-performance liquid chromatography |
| UV | Ultraviolet |
References
- Daloso, D.M.; Müller, K.; Obata, T.; Florian, A.; Tohge, T.; Bottcher, A.; Riondet, C.; Bariat, L.; Carrari, F.; Nunes-Nesi, A.; et al. Thioredoxin, a master regulator of the tricarboxylic acid cycle in plant mitochondria. Proc. Natl. Acad. Sci. USA 2015, 112, E1392–E1400. [Google Scholar] [CrossRef] [PubMed]
- Araújo, W.L.; Ishizaki, K.; Nunes-Nesi, A.; Larson, T.R.; Tohge, T.; Krahnert, I.; Witt, S.; Obata, T.; Schauer, N.; Graham, I.A.; et al. Identification of the 2-Hydroxyglutarate and Isovaleryl-CoA Dehydrogenases as Alternative Electron Donors Linking Lysine Catabolism to the Electron Transport Chain of Arabidopsis Mitochondria. Plant Cell 2010, 22, 1549–1563. [Google Scholar] [CrossRef] [PubMed]
- Tohge, T.; Wendenburg, R.; Ishihara, H.; Nakabayashi, R.; Watanabe, M.; Sulpice, R.; Hoefgen, R.; Takayama, H.; Saito, K.; Stitt, M.; et al. Characterization of a recently evolved flavonol-phenylacyltransferase gene provides signatures of natural light selection in Brassicaceae. Nat. Commun. 2016, 7, 12399. [Google Scholar] [CrossRef] [PubMed]
- Perez de Souza, L.; Garbowicz, K.; Brotman, Y.; Tohge, T.; Fernie, A.R. The Acetate Pathway Supports Flavonoid and Lipid Biosynthesis in Arabidopsis. Plant Physiol. 2020, 182, 857–869. [Google Scholar] [CrossRef] [PubMed]
- Cárdenas, P.D.; Sonawane, P.D.; Heinig, U.; Jozwiak, A.; Panda, S.; Abebie, B.; Kazachkova, Y.; Pliner, M.; Unger, T.; Wolf, D.; et al. Pathways to defense metabolites and evading fruit bitterness in genus Solanum evolved through 2-oxoglutarate-dependent dioxygenases. Nat. Commun. 2019, 10, 5169. [Google Scholar] [CrossRef] [PubMed]
- Altschuler, S.J.; Wu, L.F. Cellular Heterogeneity: Do Differences Make a Difference? Cell 2010, 141, 559–563. [Google Scholar] [CrossRef]
- Fleming, A. Metabolic aspects of organogenesis in the shoot apical meristem. J. Exp. Bot. 2006, 57, 1863–1870. [Google Scholar] [CrossRef]
- Medeiros, D.B.; Perez Souza, L.; Antunes, W.C.; Araújo, W.L.; Daloso, D.M.; Fernie, A.R. Sucrose breakdown within guard cells provides substrates for glycolysis and glutamine biosynthesis during light-induced stomatal opening. Plant J. 2018, 94, 583–594. [Google Scholar] [CrossRef]
- Wang, H.; Yan, S.; Xin, H.; Huang, W.; Zhang, H.; Teng, S.; Yu, Y.-C.; Fernie, A.R.; Lu, X.; Li, P.; et al. A Subsidiary Cell-Localized Glucose Transporter Promotes Stomatal Conductance and Photosynthesis. Plant Cell 2019, 31, 1328–1343. [Google Scholar] [CrossRef]
- Arrivault, S.; Obata, T.; Szecówka, M.; Mengin, V.; Guenther, M.; Hoehne, M.; Fernie, A.R.; Stitt, M. Metabolite pools and carbon flow during C4 photosynthesis in maize: 13CO2 labeling kinetics and cell type fractionation. J. Exp. Bot. 2016, 68, 283–298. [Google Scholar] [CrossRef]
- Islam, M.M.; Al-Siyabi, A.; Saha, R.; Obata, T. Dissecting metabolic flux in C4 plants: Experimental and theoretical approaches. Phytochem. Rev. 2018, 17, 1253–1274. [Google Scholar] [CrossRef]
- Weissmann, S.; Ma, F.; Furuyama, K.; Gierse, J.; Berg, H.; Shao, Y.; Taniguchi, M.; Allen, D.K.; Brutnell, T.P. Interactions of C4 Subtype Metabolic Activities and Transport in Maize Are Revealed through the Characterization of DCT2 Mutants. Plant Cell 2016, 28, 466–484. [Google Scholar] [CrossRef] [PubMed]
- Schenck, C.A.; Last, R.L. Location, location! cellular relocalization primes specialized metabolic diversification. FEBS J. 2020, 287, 1359–1368. [Google Scholar] [CrossRef]
- Luo, C.; Fernie, A.R.; Yan, J. Single-Cell Genomics and Epigenomics: Technologies and Applications in Plants. Trends Plant Sci. 2020, 25, 1030–1040. [Google Scholar] [CrossRef] [PubMed]
- Labib, M.; Kelley, S.O. Single-cell analysis targeting the proteome. Nat. Rev. Chem. 2020, 4, 143–158. [Google Scholar] [CrossRef]
- Marx, V. A dream of single-cell proteomics. Nat. Methods 2019, 16, 809–812. [Google Scholar] [CrossRef]
- Slavov, N. Unpicking the proteome in single cells. Science 2020, 367, 512–513. [Google Scholar] [CrossRef]
- Misra, B.B.; Assmann, S.M.; Chen, S. Plant single-cell and single-cell-type metabolomics. Trends Plant Sci. 2014, 19, 637–646. [Google Scholar] [CrossRef]
- Fujii, T.; Matsuda, S.; Tejedor, M.L.; Esaki, T.; Sakane, I.; Mizuno, H.; Tsuyama, N.; Masujima, T. Direct metabolomics for plant cells by live single-cell mass spectrometry. Nat. Protoc. 2015, 10, 1445–1456. [Google Scholar] [CrossRef]
- Yang, Y.; Huang, Y.; Wu, J.; Liu, N.; Deng, J.; Luan, T. Single-cell analysis by ambient mass spectrometry. TrAC Trends Anal. Chem. 2017, 90, 14–26. [Google Scholar] [CrossRef]
- Bjarnholt, N.; Li, B.; D’Alvise, J.; Janfelt, C. Mass spectrometry imaging of plant metabolites–principles and possibilities. Nat. Prod. Rep. 2014, 31, 818–837. [Google Scholar] [CrossRef] [PubMed]
- Hansen, R.L.; Lee, Y.J. High-Spatial Resolution Mass Spectrometry Imaging: Toward Single Cell Metabolomics in Plant Tissues. Chem. Rec. 2018, 18, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Masujima, T. Live Single-cell Mass Spectrometry. Anal. Sci. 2009, 25, 953–960. [Google Scholar] [CrossRef] [PubMed]
- Gross, A.; Schoendube, J.; Zimmermann, S.; Steeb, M.; Zengerle, R.; Koltay, P. Technologies for Single-Cell Isolation. Int. J. Mol. Sci. 2015, 16, 16897–16919. [Google Scholar] [CrossRef]
- Zhang, X.; Wei, X.; Wei, Y.; Chen, M.; Wang, J. The up-to-date strategies for the isolation and manipulation of single cells. Talanta 2020, 218, 121147. [Google Scholar] [CrossRef]
- Couvillion, S.P.; Zhu, Y.; Nagy, G.; Adkins, J.N.; Ansong, C.; Renslow, R.S.; Piehowski, P.D.; Ibrahim, Y.M.; Kelly, R.T.; Metz, T.O. New mass spectrometry technologies contributing towards comprehensive and high throughput omics analyses of single cells. Analyst 2019, 144, 794–807. [Google Scholar] [CrossRef]
- Moussaieff, A.; Rogachev, I.; Brodsky, L.; Malitsky, S.; Toal, T.W.; Belcher, H.; Yativ, M.; Brady, S.M.; Benfey, P.N.; Aharoni, A. High-resolution metabolic mapping of cell types in plant roots. Proc. Natl. Acad. Sci. USA 2013, 110, E1232–E1241. [Google Scholar] [CrossRef]
- Reichard, A.; Asosingh, K. Best Practices for Preparing a Single Cell Suspension from Solid Tissues for Flow Cytometry. Cytom. Part A 2019, 95, 219–226. [Google Scholar] [CrossRef]
- Arrivault, S.; Guenther, M.; Ivakov, A.; Feil, R.; Vosloh, D.; Van Dongen, J.T.; Sulpice, R.; Stitt, M. Use of reverse-phase liquid chromatography, linked to tandem mass spectrometry, to profile the Calvin cycle and other metabolic intermediates in Arabidopsis rosettes at different carbon dioxide concentrations. Plant J. 2009, 59, 826–839. [Google Scholar] [CrossRef]
- Brasko, C.; Smith, K.; Molnar, C.; Farago, N.; Hegedus, L.; Balind, A.; Balassa, T.; Szkalisity, A.; Sukosd, F.; Kocsis, K.; et al. Intelligent image-based in situ single-cell isolation. Nat. Commun. 2018, 9, 226. [Google Scholar] [CrossRef]
- Isozaki, A.; Mikami, H.; Hiramatsu, K.; Sakuma, S.; Kasai, Y.; Iino, T.; Yamano, T.; Yasumoto, A.; Oguchi, Y.; Suzuki, N.; et al. A practical guide to intelligent image-activated cell sorting. Nat. Protoc. 2019, 14, 2370–2415. [Google Scholar] [CrossRef] [PubMed]
- Karas, M.; Bahr, U. Matrix-Assisted Laser Desorption-Ionization (MALDI) Mass Spectrometry: Principles and Applications. In Selected Topics in Mass Spectrometry in the Biomolecular Sciences; Caprioli, R.M., Malorni, A., Sindona, G., Eds.; Springer Netherlands: Dordrecht, The Netherlands, 1997; pp. 33–53. [Google Scholar]
- Schwamborn, K.; Kriegsmann, M.; Weichert, W. MALDI imaging mass spectrometry—From bench to bedside. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2017, 1865, 776–783. [Google Scholar] [CrossRef]
- Walker, A.V. Secondary Ion Mass Spectrometry. In Encyclopedia of Spectroscopy and Spectrometry, 3rd ed.; Lindon, J.C., Tranter, G.E., Koppenaal, D.W., Eds.; Academic Press: Oxford, UK, 2017; pp. 44–49. [Google Scholar]
- Hollerbach, A.; Ayrton, S.; Jarmusch, A.; Graham Cooks, R. Desorption Electrospray Ionization: Methodology and Applications. In Encyclopedia of Spectroscopy and Spectrometry, 3rd ed.; Lindon, J.C., Tranter, G.E., Koppenaal, D.W., Eds.; Academic Press: Oxford, UK, 2017; pp. 401–408. [Google Scholar]
- Cooks, R.G.; Ouyang, Z.; Takats, Z.; Wiseman, J.M. Ambient Mass Spectrometry. Science 2006, 311, 1566–1570. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Dill, A.L.; Eberlin, L.S.; Cooks, R.G.; Ifa, D.R. Mass spectrometry imaging under ambient conditions. Mass Spectrom. Rev. 2013, 32, 218–243. [Google Scholar] [CrossRef] [PubMed]
- Boughton, B.A.; Hamilton, B. Spatial Metabolite Profiling by Matrix-Assisted Laser Desorption Ionization Mass Spectrometry Imaging. In Metabolomics: From Fundamentals to Clinical Applications; Sussulini, A., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 291–321. [Google Scholar]
- Sturtevant, D.; Lee, Y.-J.; Chapman, K.D. Matrix assisted laser desorption/ionization-mass spectrometry imaging (MALDI-MSI) for direct visualization of plant metabolites in situ. Curr. Opin. Biotechnol. 2016, 37, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Korte, A.R.; Yandeau-Nelson, M.D.; Nikolau, B.J.; Lee, Y.J. Subcellular-level resolution MALDI-MS imaging of maize leaf metabolites by MALDI-linear ion trap-Orbitrap mass spectrometer. Anal. Bioanal. Chem. 2015, 407, 2301–2309. [Google Scholar] [CrossRef]
- Gemperline, E.; Rawson, S.; Li, L. Optimization and Comparison of Multiple MALDI Matrix Application Methods for Small Molecule Mass Spectrometric Imaging. Anal. Chem. 2014, 86, 10030–10035. [Google Scholar] [CrossRef]
- Li, S.; Zhang, Y.; Liu, J.a.; Han, J.; Guan, M.; Yang, H.; Lin, Y.; Xiong, S.; Zhao, Z. Electrospray deposition device used to precisely control the matrix crystal to improve the performance of MALDI MSI. Sci. Rep. 2016, 6, 37903. [Google Scholar] [CrossRef]
- Calvano, C.D.; Monopoli, A.; Cataldi, T.R.I.; Palmisano, F. MALDI matrices for low molecular weight compounds: An endless story? Anal. Bioanal. Chem. 2018, 410, 4015–4038. [Google Scholar] [CrossRef]
- Korte, A.R.; Lee, Y.J. MALDI-MS analysis and imaging of small molecule metabolites with 1,5-diaminonaphthalene (DAN). J. Mass Spectrom. 2014, 49, 737–741. [Google Scholar] [CrossRef]
- Dueñas, M.E.; Larson, E.A.; Lee, Y.J. Toward Mass Spectrometry Imaging in the Metabolomics Scale: Increasing Metabolic Coverage Through Multiple On-Tissue Chemical Modifications. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Soltwisch, J.; Kettling, H.; Vens-Cappell, S.; Wiegelmann, M.; Müthing, J.; Dreisewerd, K. Mass spectrometry imaging with laser-induced postionization. Science 2015, 348, 211–215. [Google Scholar] [CrossRef]
- Yagnik, G.B.; Hansen, R.L.; Korte, A.R.; Reichert, M.D.; Vela, J.; Lee, Y.J. Large Scale Nanoparticle Screening for Small Molecule Analysis in Laser Desorption Ionization Mass Spectrometry. Anal. Chem. 2016, 88, 8926–8930. [Google Scholar] [CrossRef]
- Hansen, R.L.; Dueñas, M.E.; Lee, Y.J. Sputter-Coated Metal Screening for Small Molecule Analysis and High-Spatial Resolution Imaging in Laser Desorption Ionization Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2019, 30, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Römpp, A.; Schäfer, K.C.; Guenther, S.; Wang, Z.; Köstler, M.; Leisner, A.; Paschke, C.; Schramm, T.; Spengler, B. High-resolution atmospheric pressure infrared laser desorption/ionization mass spectrometry imaging of biological tissue. Anal. Bioanal. Chem. 2013, 405, 6959–6968. [Google Scholar] [CrossRef] [PubMed]
- Touboul, D.; Brunelle, A. What more can TOF-SIMS bring than other MS imaging methods? Bioanalysis 2016, 8, 367–369. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Passarelli, M.K.; Pirkl, A.; Moellers, R.; Grinfeld, D.; Kollmer, F.; Havelund, R.; Newman, C.F.; Marshall, P.S.; Arlinghaus, H.; Alexander, M.R.; et al. The 3D OrbiSIMS—Label-free metabolic imaging with subcellular lateral resolution and high mass-resolving power. Nat. Methods 2017, 14, 1175–1183. [Google Scholar] [CrossRef] [PubMed]
- Newell, C.L.; Vorng, J.-L.; MacRae, J.I.; Gilmore, I.S.; Gould, A.P. Cryogenic OrbiSIMS Localizes Semi-Volatile Molecules in Biological Tissues. Angew. Chem. Int. Ed. 2020, 59, 18194–18200. [Google Scholar] [CrossRef]
- Wiseman, J.M.; Ifa, D.R.; Song, Q.; Cooks, R.G. Tissue Imaging at Atmospheric Pressure Using Desorption Electrospray Ionization (DESI) Mass Spectrometry. Angew. Chem. Int. Ed. 2006, 45, 7188–7192. [Google Scholar] [CrossRef]
- Kulkarni, P.; Wilschut, R.A.; Verhoeven, K.J.F.; van der Putten, W.H.; Garbeva, P. LAESI mass spectrometry imaging as a tool to differentiate the root metabolome of native and range-expanding plant species. Planta 2018, 248, 1515–1523. [Google Scholar] [CrossRef]
- Samarah, L.Z.; Khattar, R.; Tran, T.H.; Stopka, S.A.; Brantner, C.A.; Parlanti, P.; Veličković, D.; Shaw, J.B.; Agtuca, B.J.; Stacey, G.; et al. Single-Cell Metabolic Profiling: Metabolite Formulas from Isotopic Fine Structures in Heterogeneous Plant Cell Populations. Anal. Chem. 2020, 92, 7289–7298. [Google Scholar] [CrossRef]
- Etalo, D.W.; De Vos, R.C.H.; Joosten, M.H.A.J.; Hall, R.D. Spatially Resolved Plant Metabolomics: Some Potentials and Limitations of Laser-Ablation Electrospray Ionization Mass Spectrometry Metabolite Imaging. Plant Physiol. 2015, 169, 1424–1435. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, T. Spatial Metabolomics and Imaging Mass Spectrometry in the Age of Artificial Intelligence. Annu. Rev. Biomed. Data Sci. 2020, 3, 61–87. [Google Scholar] [CrossRef]
- Schramm, T.; Hester, Z.; Klinkert, I.; Both, J.-P.; Heeren, R.M.A.; Brunelle, A.; Laprévote, O.; Desbenoit, N.; Robbe, M.-F.; Stoeckli, M.; et al. imzML—A common data format for the flexible exchange and processing of mass spectrometry imaging data. J. Proteom. 2012, 75, 5106–5110. [Google Scholar] [CrossRef] [PubMed]
- Shroff, R.; Schramm, K.; Jeschke, V.; Nemes, P.; Vertes, A.; Gershenzon, J.; Svatoš, A. Quantification of plant surface metabolites by matrix-assisted laser desorption–ionization mass spectrometry imaging: Glucosinolates on Arabidopsis thaliana leaves. Plant J. 2015, 81, 961–972. [Google Scholar] [CrossRef]
- Yamamoto, K.; Takahashi, K.; Caputi, L.; Mizuno, H.; Rodriguez-Lopez, C.E.; Iwasaki, T.; Ishizaki, K.; Fukaki, H.; Ohnishi, M.; Yamazaki, M.; et al. The complexity of intercellular localisation of alkaloids revealed by single-cell metabolomics. New Phytol. 2019, 224, 848–859. [Google Scholar] [CrossRef]
- Yamamoto, K.; Takahashi, K.; Mizuno, H.; Anegawa, A.; Ishizaki, K.; Fukaki, H.; Ohnishi, M.; Yamazaki, M.; Masujima, T.; Mimura, T. Cell-specific localization of alkaloids in Catharanthus roseus stem tissue measured with Imaging MS and Single-cell MS. Proc. Natl. Acad. Sci. USA 2016, 113, 3891–3896. [Google Scholar] [CrossRef]
- Livingston, S.J.; Quilichini, T.D.; Booth, J.K.; Wong, D.C.J.; Rensing, K.H.; Laflamme-Yonkman, J.; Castellarin, S.D.; Bohlmann, J.; Page, J.E.; Samuels, A.L. Cannabis glandular trichomes alter morphology and metabolite content during flower maturation. Plant J. 2020, 101, 37–56. [Google Scholar] [CrossRef]
- Sugahara, K.; Kitao, K.; Watanabe, T.; Yamagaki, T. Imaging Mass Spectrometry Analysis of Flavonoids in Blue Viola Petals and Their Enclosure Effects on Violanin during Color Expression. Anal. Chem. 2019, 91, 896–902. [Google Scholar] [CrossRef]
- Goto, T.; Kondo, T. Structure and Molecular Stacking of Anthocyanins—Flower Color Variation. Angew. Chem. Int. Ed. 1991, 30, 17–33. [Google Scholar] [CrossRef]
- Bertea, C.M.; Freije, J.R.; van der Woude, H.; Verstappen, F.W.A.; Perk, L.; Marquez, V.; De Kraker, J.W.; Posthumus, M.A.; Jansen, B.J.M.; de Groot, A.; et al. Identification of Intermediates and Enzymes Involved in the Early Steps of Artemisinin Biosynthesis in Artemisia annua. Planta Med. 2005, 71, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Frerigmann, H.; Böttcher, C.; Baatout, D.; Gigolashvili, T. Glucosinolates are produced in trichomes of Arabidopsis thaliana. Front. Plant Sci. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Schilmiller, A.; Shi, F.; Kim, J.; Charbonneau, A.L.; Holmes, D.; Daniel Jones, A.; Last, R.L. Mass spectrometry screening reveals widespread diversity in trichome specialized metabolites of tomato chromosomal substitution lines. Plant J. 2010, 62, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Mohana Kumara, P.; Uma Shaanker, R.; Pradeep, T. UPLC and ESI-MS analysis of metabolites of Rauvolfia tetraphylla L. and their spatial localization using desorption electrospray ionization (DESI) mass spectrometric imaging. Phytochemistry 2019, 159, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.; Flanigan, P.M.; Archer, J.J.; Levis, R.J. Ambient Molecular Analysis of Biological Tissue Using Low-Energy, Femtosecond Laser Vaporization and Nanospray Postionization Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2016, 27, 542–551. [Google Scholar] [CrossRef]
- Thunig, J.; Hansen, S.H.; Janfelt, C. Analysis of Secondary Plant Metabolites by Indirect Desorption Electrospray Ionization Imaging Mass Spectrometry. Anal. Chem. 2011, 83, 3256–3259. [Google Scholar] [CrossRef]
- Hölscher, D.; Shroff, R.; Knop, K.; Gottschaldt, M.; Crecelius, A.; Schneider, B.; Heckel, D.G.; Schubert, U.S.; Svatoš, A. Matrix-free UV-laser desorption/ionization (LDI) mass spectrometric imaging at the single-cell level: Distribution of secondary metabolites of Arabidopsis thaliana and Hypericum species. Plant J. 2009, 60, 907–918. [Google Scholar] [CrossRef]
- Bhatia, S.; Naved, T.; Sardana, S. Introduction to Pharmaceutical Biotechnology, Volume 3. In Animal Tissue Culture and Biopharmaceuticals; IOP Publishing: Bristol, UK, 2019. [Google Scholar]
- Rappez, L.; Stadler, M.; Triana, S.; Phapale, P.; Heikenwalder, M.; Alexandrov, T. Spatial single-cell profiling of intracellular metabolomes in situ. bioRxiv 2019, 510222. [Google Scholar] [CrossRef]
- Fehér, A. Callus, Dedifferentiation, Totipotency, Somatic Embryogenesis: What These Terms Mean in the Era of Molecular Plant Biology? Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- O’Neill, K.C.; Lee, Y.J. Visualizing Genotypic and Developmental Differences of Free Amino Acids in Maize Roots With Mass Spectrometry Imaging. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Feenstra, A.D.; Dueñas, M.E.; Lee, Y.J. Five Micron High Resolution MALDI Mass Spectrometry Imaging with Simple, Interchangeable, Multi-Resolution Optical System. J. Am. Soc. Mass Spectrom. 2017, 28, 434–442. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Bhandari, D.R.; Janfelt, C.; Römpp, A.; Spengler, B. Natural products in Glycyrrhiza glabra (licorice) rhizome imaged at the cellular level by atmospheric pressure matrix-assisted laser desorption/ionization tandem mass spectrometry imaging. Plant J. 2014, 80, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Doppler, M.; Kluger, B.; Bueschl, C.; Steiner, B.; Buerstmayr, H.; Lemmens, M.; Krska, R.; Adam, G.; Schuhmacher, R. Stable Isotope-Assisted Plant Metabolomics: Investigation of Phenylalanine-Related Metabolic Response in Wheat Upon Treatment With the Fusarium Virulence Factor Deoxynivalenol. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Horn, P.J.; Silva, J.E.; Anderson, D.; Fuchs, J.; Borisjuk, L.; Nazarenus, T.J.; Shulaev, V.; Cahoon, E.B.; Chapman, K.D. Imaging heterogeneity of membrane and storage lipids in transgenic Camelina sativa seeds with altered fatty acid profiles. Plant J. 2013, 76, 138–150. [Google Scholar] [CrossRef] [PubMed]
- Marmon, S.; Sturtevant, D.; Herrfurth, C.; Chapman, K.; Stymne, S.; Feussner, I. Two Acyltransferases Contribute Differently to Linolenic Acid Levels in Seed Oil. Plant Physiol. 2017, 173, 2081–2095. [Google Scholar] [CrossRef]
- Usher, S.; Han, L.; Haslam, R.P.; Michaelson, L.V.; Sturtevant, D.; Aziz, M.; Chapman, K.D.; Sayanova, O.; Napier, J.A. Tailoring seed oil composition in the real world: Optimising omega-3 long chain polyunsaturated fatty acid accumulation in transgenic Camelina sativa. Sci. Rep. 2017, 7, 6570. [Google Scholar] [CrossRef]
- Woodfield, H.K.; Sturtevant, D.; Borisjuk, L.; Munz, E.; Guschina, I.A.; Chapman, K.; Harwood, J.L. Spatial and Temporal Mapping of Key Lipid Species in Brassica napus Seeds. Plant Physiol. 2017, 173, 1998–2009. [Google Scholar] [CrossRef]
- Sturtevant, D.; Dueñas, M.E.; Lee, Y.-J.; Chapman, K.D. Three-dimensional visualization of membrane phospholipid distributions in Arabidopsis thaliana seeds: A spatial perspective of molecular heterogeneity. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2017, 1862, 268–281. [Google Scholar] [CrossRef]
- Gorzolka, K.; Kölling, J.; Nattkemper, T.W.; Niehaus, K. Spatio-Temporal Metabolite Profiling of the Barley Germination Process by MALDI MS Imaging. PLoS ONE 2016, 11, e0150208. [Google Scholar] [CrossRef]
- Rolletschek, H.; Melkus, G.; Grafahrend-Belau, E.; Fuchs, J.; Heinzel, N.; Schreiber, F.; Jakob, P.M.; Borisjuk, L. Combined Noninvasive Imaging and Modeling Approaches Reveal Metabolic Compartmentation in the Barley Endosperm. Plant Cell 2011, 23, 3041–3054. [Google Scholar] [CrossRef]
- Shewry, P.R.; Wan, Y.; Hawkesford, M.J.; Tosi, P. Spatial distribution of functional components in the starchy endosperm of wheat grains. J. Cereal Sci. 2020, 91, 102869. [Google Scholar] [CrossRef] [PubMed]
- Feenstra, A.D.; Alexander, L.E.; Song, Z.; Korte, A.R.; Yandeau-Nelson, M.D.; Nikolau, B.J.; Lee, Y.J. Spatial Mapping and Profiling of Metabolite Distributions during Germination. Plant Physiol. 2017, 174, 2532–2548. [Google Scholar] [CrossRef] [PubMed]
- Li, F.-S.; Phyo, P.; Jacobowitz, J.; Hong, M.; Weng, J.-K. The molecular structure of plant sporopollenin. Nat. Plants 2019, 5, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Mikhael, A.; Jurcic, K.; Schneider, C.; Karr, D.; Fisher, G.L.; Fridgen, T.D.; Diego-Taboada, A.; Georghiou, P.E.; Mackenzie, G.; Banoub, J. Demystifying and unravelling the molecular structure of the biopolymer sporopollenin. Front. Plant Sci. 2020, 34, e8740. [Google Scholar] [CrossRef] [PubMed]
- Diehn, S.; Zimmermann, B.; Tafintseva, V.; Seifert, S.; Bağcıoğlu, M.; Ohlson, M.; Weidner, S.; Fjellheim, S.; Kohler, A.; Kneipp, J. Combining Chemical Information From Grass Pollen in Multimodal Characterization. Plant Cell 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Grunewald, S.; Marillonnet, S.; Hause, G.; Haferkamp, I.; Neuhaus, H.E.; Veß, A.; Hollemann, T.; Vogt, T. The Tapetal Major Facilitator NPF2.8 Is Required for Accumulation of Flavonol Glycosides on the Pollen Surface in Arabidopsis thaliana. Plant Cell 2020, 32, 1727–1748. [Google Scholar] [CrossRef] [PubMed]
- Kenđel, A.; Zimmermann, B. Chemical Analysis of Pollen by FT-Raman and FTIR Spectroscopies. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Wan, X.; Wu, S.; Li, Z.; An, X.; Tian, Y. Lipid Metabolism: Critical Roles in Male Fertility and Other Aspects of Reproductive Development in Plants. Mol. Plant 2020, 13, 955–983. [Google Scholar] [CrossRef]
- Selinski, J.; Scheibe, R. Pollen tube growth: Where does the energy come from? Plant Signal. Behav. 2014, 9, e977200. [Google Scholar] [CrossRef]
- Borghi, M.; Fernie, A.R. Outstanding questions in flower metabolism. Plant J. 2020, 103, 1275–1288. [Google Scholar] [CrossRef]
- Baumeister, T.U.H.; Vallet, M.; Kaftan, F.; Svatoš, A.; Pohnert, G. Live Single-Cell Metabolomics With Matrix-Free Laser/Desorption Ionization Mass Spectrometry to Address Microalgal Physiology. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Jaschinski, T.; Helfrich, E.J.N.; Bock, C.; Wolfram, S.; Svatoš, A.; Hertweck, C.; Pohnert, G. Matrix-free single-cell LDI-MS investigations of the diatoms Coscinodiscus granii and Thalassiosira pseudonana. J. Mass Spectrom. 2014, 49, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Yang, Z.; Wawrik, B. Metabolomic Fingerprints of Individual Algal Cells Using the Single-Probe Mass Spectrometry Technique. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Baumeister, T.U.H.; Vallet, M.; Kaftan, F.; Guillou, L.; Svatoš, A.; Pohnert, G. Identification to species level of live single microalgal cells from plankton samples with matrix-free laser/desorption ionization mass spectrometry. Metabolomics 2020, 16, 28. [Google Scholar] [CrossRef] [PubMed]
- Bölling, C.; Fiehn, O. Metabolite Profiling of Chlamydomonas reinhardtii under Nutrient Deprivation. Plant Physiol. 2005, 139, 1995–2005. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.Y.; Park, J.-J.; Barupal, D.K.; Fiehn, O. System response of metabolic networks in Chlamydomonas reinhardtii to total available ammonium. Mol. Cell Proteom. 2012, 11, 973–988. [Google Scholar] [CrossRef]
- Schreiber, F.; Ackermann, M. Environmental drivers of metabolic heterogeneity in clonal microbial populations. Curr. Opin. Biotechnol. 2020, 62, 202–211. [Google Scholar] [CrossRef]
- Krismer, J.; Tamminen, M.; Fontana, S.; Zenobi, R.; Narwani, A. Single-cell mass spectrometry reveals the importance of genetic diversity and plasticity for phenotypic variation in nitrogen-limited Chlamydomonas. ISME J. 2017, 11, 988–998. [Google Scholar] [CrossRef]
- Cha, S.; Song, Z.; Nikolau, B.J.; Yeung, E.S. Direct Profiling and Imaging of Epicuticular Waxes on Arabidopsis thaliana by Laser Desorption/Ionization Mass Spectrometry Using Silver Colloid as a Matrix. Anal. Chem. 2009, 81, 2991–3000. [Google Scholar] [CrossRef]
- Günl, M.; Neumetzler, L.; Kraemer, F.; de Souza, A.; Schultink, A.; Pena, M.; York, W.S.; Pauly, M. AXY8 Encodes an α-Fucosidase, Underscoring the Importance of Apoplastic Metabolism on the Fine Structure of Arabidopsis Cell Wall Polysaccharides. Plant Cell 2011, 23, 4025–4040. [Google Scholar] [CrossRef]
- Korte, A.R.; Song, Z.; Nikolau, B.J.; Lee, Y.J. Mass spectrometric imaging as a high-spatial resolution tool for functional genomics: Tissue-specific gene expression of TT7 inferred from heterogeneous distribution of metabolites in Arabidopsis flowers. Anal. Methods 2012, 4, 474–481. [Google Scholar] [CrossRef]
- Hansen, R.L.; Guo, H.; Yin, Y.; Lee, Y.J. FERONIA mutation induces high levels of chloroplast-localized Arabidopsides which are involved in root growth. Plant J. 2019, 97, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.T.; Yagnik, G.B.; Hohenstein, J.D.; Ji, Z.; Zi, J.; Reichert, M.D.; MacIntosh, G.C.; Yang, B.; Peters, R.J.; Vela, J.; et al. Investigation of the Chemical Interface in the Soybean–Aphid and Rice–Bacteria Interactions Using MALDI-Mass Spectrometry Imaging. Anal. Chem. 2015, 87, 5294–5301. [Google Scholar] [CrossRef] [PubMed]
- Hohenstein, J.D.; Studham, M.E.; Klein, A.; Kovinich, N.; Barry, K.; Lee, Y.-J.; MacIntosh, G.C. Transcriptional and Chemical Changes in Soybean Leaves in Response to Long-Term Aphid Colonization. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Gemperline, E.; Jayaraman, D.; Maeda, J.; Ané, J.-M.; Li, L. Multifaceted investigation of metabolites during nitrogen fixation in Medicago via high resolution MALDI-MS imaging and ESI-MS. J. Am. Soc. Mass Spectrom. 2015, 26, 149–158. [Google Scholar] [CrossRef]
- Ye, H.; Gemperline, E.; Venkateshwaran, M.; Chen, R.; Delaux, P.-M.; Howes-Podoll, M.; Ané, J.-M.; Li, L. MALDI mass spectrometry-assisted molecular imaging of metabolites during nitrogen fixation in the Medicago truncatula–Sinorhizobium meliloti symbiosis. Plant J. 2013, 75, 130–145. [Google Scholar] [CrossRef]
- Veličković, D.; Agtuca, B.J.; Stopka, S.A.; Vertes, A.; Koppenaal, D.W.; Paša-Tolić, L.; Stacey, G.; Anderton, C.R. Observed metabolic asymmetry within soybean root nodules reflects unexpected complexity in rhizobacteria-legume metabolite exchange. ISME J. 2018, 12, 2335–2338. [Google Scholar] [CrossRef]
- Agtuca, B.J.; Stopka, S.A.; Evans, S.; Samarah, L.; Liu, Y.; Xu, D.; Stacey, M.G.; Koppenaal, D.W.; Paša-Tolić, L.; Anderton, C.R.; et al. Metabolomic profiling of wild-type and mutant soybean root nodules using laser-ablation electrospray ionization mass spectrometry reveals altered metabolism. Plant J. 2020, 103, 1937–1958. [Google Scholar] [CrossRef]
- Boughton, B.A.; Thinagaran, D.; Sarabia, D.; Bacic, A.; Roessner, U. Mass spectrometry imaging for plant biology: A review. Phytochem. Rev. 2016, 15, 445–488. [Google Scholar] [CrossRef]
- Tarkowská, D.; Novák, O.; Oklestkova, J.; Strnad, M. The determination of 22 natural brassinosteroids in a minute sample of plant tissue by UHPLC–ESI–MS/MS. Anal. Bioanal. Chem. 2016, 408, 6799–6812. [Google Scholar] [CrossRef]
- Shimizu, T.; Miyakawa, S.; Esaki, T.; Mizuno, H.; Masujima, T.; Koshiba, T.; Seo, M. Live Single-Cell Plant Hormone Analysis by Video-Mass Spectrometry. Plant Cell Physiol. 2015, 56, 1287–1296. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.; Bhatt, K.; Baumann, K. Shaping in plant cells. Curr. Opin. Plant Biol. 2001, 4, 540–549. [Google Scholar] [CrossRef]
- Gago, J.; Daloso, D.d.M.; Figueroa, C.M.; Flexas, J.; Fernie, A.R.; Nikoloski, Z. Relationships of Leaf Net Photosynthesis, Stomatal Conductance, and Mesophyll Conductance to Primary Metabolism: A Multispecies Meta-Analysis Approach. Plant Physiol. 2016, 171, 265–279. [Google Scholar] [CrossRef] [PubMed]
- Wen, W.; Li, D.; Li, X.; Gao, Y.; Li, W.; Li, H.; Liu, J.; Liu, H.; Chen, W.; Luo, J.; et al. Metabolome-based genome-wide association study of maize kernel leads to novel biochemical insights. Nat. Commun. 2014, 5, 3438. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Shi, J.; Quan, S.; Cui, B.; Kleessen, S.; Nikoloski, Z.; Tohge, T.; Alexander, D.; Guo, L.; Lin, H.; et al. Metabolic variation between japonica and indica rice cultivars as revealed by non-targeted metabolomics. Sci. Rep. 2014, 4, 5067. [Google Scholar] [CrossRef] [PubMed]
- Robaina-Estévez, S.; Daloso, D.M.; Zhang, Y.; Fernie, A.R.; Nikoloski, Z. Resolving the central metabolism of Arabidopsis guard cells. Sci. Rep. 2017, 7, 8307. [Google Scholar] [CrossRef] [PubMed]
- Töpfer, N.; Braam, T.; Shameer, S.; Ratcliffe, R.G.; Sweetlove, L.J. Alternative CAM Modes Provide Environment-Specific Water-Saving Benefits in a Leaf Metabolic Model. Plant Cell 2020. [Google Scholar] [CrossRef]
- Kim-Hellmuth, S.; Aguet, F.; Oliva, M.; Muñoz-Aguirre, M.; Kasela, S.; Wucher, V.; Castel, S.E.; Hamel, A.R.; Viñuela, A.; Roberts, A.L.; et al. Cell type–specific genetic regulation of gene expression across human tissues. Science 2020, 369, eaaz8528. [Google Scholar] [CrossRef]
- Shrestha, B. Ten Major Future Challenges in Single-Cell Metabolomics. In Single Cell Metabolism: Methods and Protocols; Shrestha, B., Ed.; Springer: New York, NY, USA, 2020; pp. 219–223. [Google Scholar]
- Macaulay, I.C.; Ponting, C.P.; Voet, T. Single-Cell Multiomics: Multiple Measurements from Single Cells. Trends Genet. 2017, 33, 155–168. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Souza, L.P.; Borghi, M.; Fernie, A. Plant Single-Cell Metabolomics—Challenges and Perspectives. Int. J. Mol. Sci. 2020, 21, 8987. https://doi.org/10.3390/ijms21238987
de Souza LP, Borghi M, Fernie A. Plant Single-Cell Metabolomics—Challenges and Perspectives. International Journal of Molecular Sciences. 2020; 21(23):8987. https://doi.org/10.3390/ijms21238987
Chicago/Turabian Stylede Souza, Leonardo Perez, Monica Borghi, and Alisdair Fernie. 2020. "Plant Single-Cell Metabolomics—Challenges and Perspectives" International Journal of Molecular Sciences 21, no. 23: 8987. https://doi.org/10.3390/ijms21238987
APA Stylede Souza, L. P., Borghi, M., & Fernie, A. (2020). Plant Single-Cell Metabolomics—Challenges and Perspectives. International Journal of Molecular Sciences, 21(23), 8987. https://doi.org/10.3390/ijms21238987