Plant Volatile Organic Compounds Evolution: Transcriptional Regulation, Epigenetics and Polyploidy
Abstract
1. Plant Volatile Organic Compounds in the Epigenetic Era
2. Biosynthesis and Epigenetic Regulation of VOCs
2.1. Genetic Roadmap to Terpenoid Biosynthesis and Regulation
2.2. Regulation of Transcription Factors Affects Phenylpropanoid/Benzenoid Biosynthesis
2.3. Crosstalk between Fatty Acid Derivatives, Biosynthesis and Epigenetics
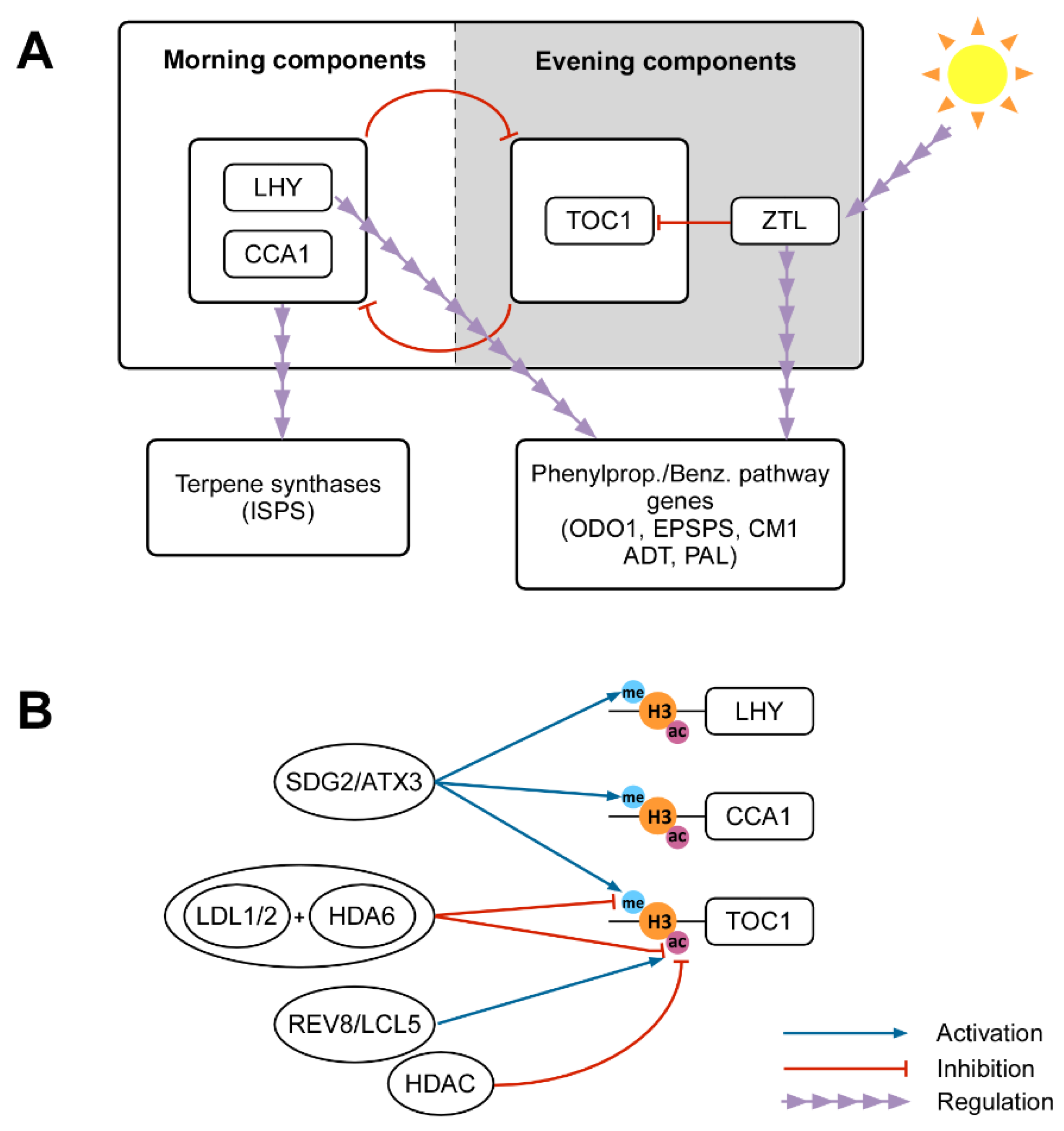
3. The Temporal Pattern of Emissions and Its Epigenetic Regulation
4. Evolution of VOCs: The Role of Polyploidy and Transgenerational Memory
5. Final Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pichersky, E.; Gang, D.R. Genetics and biochemistry of secondary metabolites in plants: An evolutionary perspective. Trends Plant Sci. 2000, 5, 439–445. [Google Scholar] [CrossRef]
- Pichersky, E.; Lewinsohn, E. Convergent Evolution in Plant Specialized Metabolism. Annu. Rev. Plant Biol. 2011, 62, 549–566. [Google Scholar] [CrossRef] [PubMed]
- Pichersky, E.; Noel, J.P.; Dudareva, N. Biosynthesis of plant volatiles: Nature’s diversity and ingenuity. Science 2006, 311, 808–811. [Google Scholar] [CrossRef] [PubMed]
- Dudareva, N.; Klempien, A.; Muhlemann, J.K.; Kaplan, I. Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 2013, 198, 16–32. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, J.T.; Eriksson, R.; Gershenzon, J.; Ståhl, B. Diversity and distribution of floral scent. Bot. Rev. 2006, 72, 1–120. [Google Scholar] [CrossRef]
- Farré-Armengol, G.; Filella, I.; Llusia, J.; Peñuelas, J. Bidirectional Interaction between Phyllospheric Microbiotas and Plant Volatile Emissions. Trends Plant Sci. 2016, 21, 854–860. [Google Scholar] [CrossRef]
- Kessler, A.; Baldwin, I.T. Defensive function of herbivore-induced plant volatile emissions in nature. Science 2001, 291, 2141–2144. [Google Scholar] [CrossRef]
- Heil, M.; Bueno, J.C.S. Within-plant signaling by volatiles leads to induction and priming of an indirect plant defense in nature. Proc. Natl. Acad. Sci. USA 2007, 104, 5467–5472. [Google Scholar] [CrossRef]
- Farré-Armengol, G.; Filella, I.; Llusia, J.; Peñuelas, J. Floral volatile organic compounds: Between attraction and deterrence of visitors under global change. Perspect. Plant Ecol. Evol. Syst. 2013, 15, 56–67. [Google Scholar] [CrossRef]
- Raguso, R.A. Wake Up and Smell the Roses: The Ecology and Evolution of Floral Scent. Annu. Rev. Ecol. Evol. Syst. 2008, 39, 549–569. [Google Scholar] [CrossRef]
- Austin, A.T.; Ballaré, C.L. Plant interactions with other organisms: Molecules, ecology and evolution. New Phytol. 2014, 204, 257–260. [Google Scholar] [CrossRef] [PubMed]
- Kessler, D.; Kallenbach, M.; Diezel, C.; Rothe, E.; Murdock, M.; Baldwin, I.T. How scent and nectar influence floral antagonists and mutualists. Elife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Grüter, C.; Ratnieks, F.L.W. Flower constancy in insect pollinators: Adaptive foraging behavior or cognitive limitation? Commun. Integr. Biol. 2011, 4, 633–636. [Google Scholar] [CrossRef] [PubMed]
- Koornneef, A.; Pieterse, C.M.J. Cross talk in defense signaling. Plant Physiol. 2008, 146, 839–844. [Google Scholar] [CrossRef] [PubMed]
- Junker, R.R.; Blüthgen, N. Floral scents repel potentially nectar-thieving ants. Evol. Ecol. Res. 2008, 10, 295–308. [Google Scholar]
- Balao, F.; Herrera, J.; Talavera, S.; Dötterl, S. Spatial and temporal patterns of floral scent emission in Dianthus inoxianus and electroantennographic responses of its hawkmoth pollinator. Phytochemistry 2011, 72, 601–609. [Google Scholar] [CrossRef]
- Huang, M.; Sanchez-Moreiras, A.M.; Abel, C.; Sohrabi, R.; Lee, S.; Gershenzon, J.; Tholl, D. The major volatile organic compound emitted from Arabidopsis thaliana flowers, the sesquiterpene (E)-β-caryophyllene, is a defense against a bacterial pathogen. New Phytol. 2012, 193, 997–1008. [Google Scholar] [CrossRef]
- Baldwin, I.T.; Halitschke, R.; Paschold, A.; Von Dahl, C.C.; Preston, C.A. Volatile signaling in plant-plant interactions: “Talking trees” in the genomics era. Science 2006, 311, 812–815. [Google Scholar] [CrossRef]
- Kegge, W.; Pierik, R. Biogenic volatile organic compounds and plant competition. Trends Plant Sci. 2010, 15, 126–132. [Google Scholar] [CrossRef]
- Rodríguez, A.; Alquézar, B.; Peña, L. Fruit aromas in mature fleshy fruits as signals of readiness for predation and seed dispersal. New Phytol. 2013, 197, 36–48. [Google Scholar] [CrossRef]
- Hansen, U.; Seufert, G. Temperature and light dependence of β-caryophyllene emission rates. J. Geophys. Res. D Atmos. 2003, 108. [Google Scholar] [CrossRef]
- Truong, D.H.; Delory, B.M.; Vanderplanck, M.; Brostaux, Y.; Vandereycken, A.; Heuskin, S.; Delaplace, P.; Francis, F.; Lognay, G. Temperature regimes and aphid density interactions differentially influence VOC emissions in Arabidopsis. Arthropod. Plant. Interact. 2014, 8, 317–327. [Google Scholar] [CrossRef][Green Version]
- Holopainen, J.K.; Gershenzon, J. Multiple stress factors and the emission of plant VOCs. Trends Plant Sci. 2010, 15, 176–184. [Google Scholar] [CrossRef]
- Sharkey, T.D.; Chen, X.; Yeh, S. Isoprene increases thermotolerance of fosmidomycin-fed leaves. Plant Physiol. 2001, 125, 2001–2006. [Google Scholar] [CrossRef]
- Vickers, C.E.; Possell, M.; Cojocariu, C.I.; Velikova, V.B.; Laothawornkitkul, J.; Ryan, A.; Mullineaux, P.M.; Nicholas Hewitt, C. Isoprene synthesis protects transgenic tobacco plants from oxidative stress. Plant Cell Environ. 2009, 32, 520–531. [Google Scholar] [CrossRef]
- Gibney, E.R.; Nolan, C.M. Epigenetics and gene expression. Heredity (Edinb.) 2010, 105, 4–13. [Google Scholar] [CrossRef]
- Ramya, M.; Jang, S.; An, H.-R.; Lee, S.-Y.; Park, P.-M.; Park, P.H. Volatile Organic Compounds from Orchids: From Synthesis and Function to Gene Regulation. Int. J. Mol. Sci. 2020, 21, 1160. [Google Scholar] [CrossRef]
- Dupont, C.; Armant, D.R.; Brenner, C.A. Epigenetics: Definition, mechanisms and clinical perspective. Semin. Reprod. Med. 2009, 27, 351–357. [Google Scholar] [CrossRef]
- Newell-Price, J.; Clark, A.J.L.; King, P. DNA methylation and silencing of gene expression. Trends Endocrinol. Metab. 2000, 11, 142–148. [Google Scholar] [CrossRef]
- Wade, P.A. Methyl CpG binding proteins: Coupling chromatin architecture to gene regulation. Oncogene 2001, 20, 3166–3173. [Google Scholar] [CrossRef]
- Boyko, A.; Kovalchuk, I. Genome instability and epigenetic modification-heritable responses to environmental stress? Curr. Opin. Plant Biol. 2011, 14, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Blomen, V.A.; Boonstra, J. Stable transmission of reversible modifications: Maintenance of epigenetic information through the cell cycle. Cell. Mol. Life Sci. 2011, 68, 27–44. [Google Scholar] [CrossRef] [PubMed]
- Moazed, D. Small RNAs in transcriptional gene silencing and genome defence. Nature 2009, 457, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Cech, T.R.; Steitz, J.A. The noncoding RNA revolution-Trashing old rules to forge new ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef]
- Holoch, D.; Moazed, D. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 2015, 16, 71–84. [Google Scholar] [CrossRef]
- Lämke, J.; Bäurle, I. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 2017, 18, 1–11. [Google Scholar] [CrossRef]
- Alonso, C.; Ramos-Cruz, D.; Becker, C. The role of plant epigenetics in biotic interactions. New Phytol. 2019, 221, 731–737. [Google Scholar] [CrossRef]
- Madlung, A.; Wendel, J.F. Genetic and epigenetic aspects of polyploid evolution in plants. Cytogenet. Genome Res. 2013, 140, 270–285. [Google Scholar] [CrossRef]
- Kellenberger, R.T.; Schlüter, P.M.; Schiestl, F.P. Herbivore-Induced DNA Demethylation Changes Floral Signalling and Attractiveness to Pollinators in Brassica rapa. PLoS ONE 2016, 11, e0166646. [Google Scholar] [CrossRef]
- Asakawa, Y.; Ludwiczuk, A.; Nagashima, F. Phytochemical and biological studies of bryophytes. Phytochemistry 2013, 91, 52–80. [Google Scholar] [CrossRef]
- Chen, F.; Ludwiczuk, A.; Wei, G.; Chen, X.; Crandall-Stotler, B.; Bowman, J.L. Terpenoid Secondary Metabolites in Bryophytes: Chemical Diversity, Biosynthesis and Biological Functions. CRC. Crit. Rev. Plant Sci. 2018, 37, 210–231. [Google Scholar] [CrossRef]
- Muhlemann, J.K.; Klempien, A.; Dudareva, N. Floral volatiles: From biosynthesis to function. Plant Cell Environ. 2014, 37, 1936–1949. [Google Scholar] [CrossRef] [PubMed]
- McGarvey, D.J.; Croteau, R. Terpenoid metabolism. Plant Cell 1995, 7, 1015–1026. [Google Scholar] [PubMed]
- Hsieh, M.H.; Chang, C.Y.; Hsu, S.J.; Chen, J.J. Chloroplast localization of methylerythritol 4-phosphate pathway enzymes and regulation of mitochondrial genes in ispD and ispE albino mutants in Arabidopsis. Plant Mol. Biol. 2008, 66, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Simkin, A.J.; Guirimand, G.; Papon, N.; Courdavault, V.; Thabet, I.; Ginis, O.; Bouzid, S.; Giglioli-Guivarc’h, N.; Clastre, M. Peroxisomal localisation of the final steps of the mevalonic acid pathway in planta. Planta 2011, 234, 903–914. [Google Scholar] [CrossRef]
- Pulido, P.; Perello, C.; Rodriguez-Concepcion, M. New Insights into Plant Isoprenoid Metabolism. Mol. Plant 2012, 5, 964–967. [Google Scholar] [CrossRef]
- Laule, O.; Fürholz, A.; Chang, H.S.; Zhu, T.; Wang, X.; Heifetz, P.B.; Gruissem, W.; Lange, B.M. Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2003, 100, 6866–6871. [Google Scholar] [CrossRef]
- Ward, J.L.; Baker, J.M.; Llewellyn, A.M.; Hawkins, N.D.; Beale, M.H. Metabolomic analysis of Arabidopsis reveals hemiterpenoid glycosides as products of a nitrate ion-regulated, carbon flux overflow. Proc. Natl. Acad. Sci. USA 2011, 108, 10762–10767. [Google Scholar] [CrossRef]
- Gutensohn, M.; Nagegowda, D.A.; Dudareva, N. Involvement of compartmentalization in monoterpene and sesquiterpene biosynthesis in plants. In Isoprenoid Synthesis in Plants and Microorganisms: New Concepts and Experimental Approaches; Springer: New York, NY, USA, 2013; pp. 159–169. ISBN 9781461440635. [Google Scholar]
- Zhao, L.; Chang, W.; Xiao, Y.; Liu, H.; Liu, P. Methylerythritol Phosphate Pathway of Isoprenoid Biosynthesis. Annu. Rev. Biochem. 2013, 82, 497–530. [Google Scholar] [CrossRef]
- Degenhardt, J.; Köllner, T.G.; Gershenzon, J. Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 2009, 70, 1621–1637. [Google Scholar] [CrossRef]
- Bleeker, P.M.; Spyropoulou, E.A.; Diergaarde, P.J.; Volpin, H.; De Both, M.T.J.; Zerbe, P.; Bohlmann, J.; Falara, V.; Matsuba, Y.; Pichersky, E.; et al. RNA-seq discovery, functional characterization, and comparison of sesquiterpene synthases from Solanum lycopersicum and Solanum habrochaites trichomes. Plant Mol. Biol. 2011, 77, 323–336. [Google Scholar] [CrossRef] [PubMed]
- Tholl, D. Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr. Opin. Plant Biol. 2006, 9, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Nagegowda, D.A.; Gutensohn, M.; Wilkerson, C.G.; Dudareva, N. Two nearly identical terpene synthases catalyze the formation of nerolidol and linalool in snapdragon flowers. Plant J. 2008, 55, 224–239. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Tholl, D.; Bohlmann, J.; Pichersky, E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011, 66, 212–229. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Liu, B.; Li, M.; Gao, X.; Fang, Q.; Liu, C.; Ding, H.; Wang, L.; Gao, X. Identification and characterization of terpene synthase genes accounting for volatile terpene emissions in flowers of Freesia x hybrida. J. Exp. Bot. 2018, 69, 4249–4265. [Google Scholar] [CrossRef]
- Byers, K.J.R.P.; Vela, J.P.; Peng, F.; Riffell, J.A.; Bradshaw, H.D. Floral volatile alleles can contribute to pollinator-mediated reproductive isolation in monkeyflowers (Mimulus). Plant J. 2014, 80, 1031–1042. [Google Scholar] [CrossRef]
- Hong, G.J.; Xue, X.Y.; Mao, Y.B.; Wang, L.J.; Chen, X.Y. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 2012, 24, 2635–2648. [Google Scholar] [CrossRef]
- Samad, A.F.A.; Sajad, M.; Nazaruddin, N.; Fauzi, I.A.; Murad, A.M.A.; Zainal, Z.; Ismail, I. MicroRNA and transcription factor: Key players in plant regulatory network. Front. Plant Sci. 2017, 8, 565. [Google Scholar] [CrossRef]
- Tan, H.; Xiao, L.; Gao, S.; Li, Q.; Chen, J.; Xiao, Y.; Ji, Q.; Chen, R.; Chen, W.; Zhang, L. TRICHOME and ARTEMISININ REGULATOR 1 is required for trichome development and artemisinin biosynthesis in Artemisia annua. Mol. Plant 2015, 8, 1396–1411. [Google Scholar] [CrossRef]
- Reeves, P.H.; Ellis, C.M.; Ploense, S.E.; Wu, M.F.; Yadav, V.; Tholl, D.; Chételat, A.; Haupt, I.; Kennerley, B.J.; Hodgens, C.; et al. A regulatory network for coordinated flower maturation. PLoS Genet. 2012, 8, e1002506. [Google Scholar] [CrossRef]
- Bedon, F.; Bomal, C.; Caron, S.; Levasseur, C.; Boyle, B.; Mansfield, S.D.; Schmidt, A.; Gershenzon, J.; Grima-Pettenati, J.; Séguin, A.; et al. Subgroup 4 R2R3-MYBs in conifer trees: Gene family expansion and contribution to the isoprenoid-and flavonoid-oriented responses. J. Exp. Bot. 2010, 61, 3847–3864. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Li, Y.; Gao, F.; Jin, W.; Li, S.; Kimani, S.; Yang, S.; Bao, T.; Gao, X.; Wang, L. MYB21 interacts with MYC2 to control the expression of terpene synthase genes in flowers of Freesia hybrida and Arabidopsis thaliana. J. Exp. Bot. 2020, 71, 4140–4158. [Google Scholar] [CrossRef]
- Qi, T.; Huang, H.; Song, S.; Xie, D. Regulation of jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in arabidopsis. Plant Cell 2015, 27, 1620–1633. [Google Scholar] [CrossRef]
- Skibbe, M.; Qu, N.; Galis, I.; Baldwin, I.T. Induced plant defenses in the natural environment: Nicotiana attenuata WRKY3 and WRKY6 coordinate responses to herbivory. Plant Cell 2008, 20, 1984–2000. [Google Scholar] [CrossRef]
- Loivamäki, M.; Louis, S.; Cinege, G.; Zimmer, I.; Fischbach, R.J.; Schnitzler, J.P. Circadian rhythms of isoprene biosynthesis in grey poplar leaves. Plant Physiol. 2007, 143, 540–551. [Google Scholar] [CrossRef]
- Narnoliya, L.K.; Kaushal, G.; Singh, S.P. Long noncoding RNAs and miRNAs regulating terpene and tartaric acid biosynthesis in rose-scented geranium. FEBS Lett. 2019, 593, 2235–2249. [Google Scholar] [CrossRef]
- Ye, J.; Zhang, X.; Tan, J.; Xu, F.; Cheng, S.; Chen, Z.; Zhang, W.; Liao, Y. Global identification of Ginkgo biloba microRNAs and insight into their role in metabolism regulatory network of terpene trilactones by high-throughput sequencing and degradome analysis. Ind. Crops Prod. 2020, 148, 112289. [Google Scholar] [CrossRef]
- Maeda, H.; Dudareva, N. The Shikimate Pathway and Aromatic Amino Acid Biosynthesis in Plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef]
- Schnepp, J.; Dudareva, N. Floral Scent: Biosynthesis, Regulation and Genetic Modifications. In Flowering and its Manipulation; Blackwell Publishing Ltd.: Oxford, UK, 2007; pp. 240–257. ISBN 9780470988602. [Google Scholar]
- Orlova, I.; Marshall-Colón, A.; Schnepp, J.; Wood, B.; Varbanova, M.; Fridman, E.; Blakeslee, J.J.; Peer, W.A.; Murphy, A.S.; Rhodes, D.; et al. Reduction of benzenoid synthesis in petunia flowers reveals multiple pathways to benzoic acid and enhancement in auxin transport. Plant Cell 2006, 18, 3458–3475. [Google Scholar] [CrossRef]
- Boatright, J.; Negre, F.; Chen, X.; Kish, C.M.; Wood, B.; Peel, G.; Orlova, I.; Gang, D.; Rhodes, D.; Dudareva, N. Understanding in vivo benzenoid metabolism in petunia petal tissue. Plant Physiol. 2004, 135, 1993–2011. [Google Scholar] [CrossRef]
- D’Auria, J.C. Acyltransferases in plants: A good time to be BAHD. Curr. Opin. Plant Biol. 2006, 9, 331–340. [Google Scholar] [CrossRef] [PubMed]
- D’Auria, J.C.; Chen, F.; Pichersky, E. The SABATH family of MTS in Arabidopsis Thaliana and other plant species. Recent Adv. Phytochem. 2003, 37, 253–283. [Google Scholar] [CrossRef]
- Dexter, R.; Qualley, A.; Kish, C.M.; Ma, C.J.; Koeduka, T.; Nagegowda, D.A.; Dudareva, N.; Pichersky, E.; Clark, D. Characterization of a petunia acetyltransferase involved in the biosynthesis of the floral volatile isoeugenol. Plant J. 2007, 49, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Koeduka, T.; Fridman, E.; Gang, D.R.; Vassão, D.G.; Jackson, B.L.; Kish, C.M.; Orlova, I.; Spassova, S.M.; Lewis, N.G.; Noel, J.P.; et al. Eugenol and isoeugenol, characteristic aromatic constituents of spices, are biosynthesized via reduction of a coniferyl alcohol ester. Proc. Natl. Acad. Sci. USA 2006, 103, 10128–10133. [Google Scholar] [CrossRef] [PubMed]
- Koeduka, T.; Louie, G.V.; Orlova, I.; Kish, C.M.; Ibdah, M.; Wilkerson, C.G.; Bowman, M.E.; Baiga, T.J.; Noel, J.P.; Dudareva, N.; et al. The multiple phenylpropene synthases in both Clarkia breweri and Petunia hybrida represent two distinct protein lineages. Plant J. 2008, 54, 362–374. [Google Scholar] [CrossRef]
- Kaminaga, Y.; Schnepp, J.; Peel, G.; Kish, C.M.; Ben-Nissan, G.; Weiss, D.; Orlova, I.; Lavie, O.; Rhodes, D.; Wood, K.; et al. Plant phenylacetaldehyde synthase is a bifunctional homotetrameric enzyme that catalyzes phenylalanine decarboxylation and oxidation. J. Biol. Chem. 2006, 281, 23357–23366. [Google Scholar] [CrossRef]
- Tieman, D.M.; Loucas, H.M.; Kim, J.Y.; Clark, D.G.; Klee, H.J. Tomato phenylacetaldehyde reductases catalyze the last step in the synthesis of the aroma volatile 2-phenylethanol. Phytochemistry 2007, 68, 2660–2669. [Google Scholar] [CrossRef]
- Farhi, M.; Lavie, O.; Masci, T.; Hendel-Rahmanim, K.; Weiss, D.; Abeliovich, H.; Vainstein, A. Identification of rose phenylacetaldehyde synthase by functional complementation in yeast. Plant Mol. Biol. 2010, 72, 235–245. [Google Scholar] [CrossRef]
- Hirata, H.; Ohnishi, T.; Ishida, H.; Tomida, K.; Sakai, M.; Hara, M.; Watanabe, N. Functional characterization of aromatic amino acid aminotransferase involved in 2-phenylethanol biosynthesis in isolated rose petal protoplasts. J. Plant Physiol. 2012, 169, 444–451. [Google Scholar] [CrossRef]
- Gonda, I.; Bar, E.; Portnoy, V.; Lev, S.; Burger, J.; Schaffer, A.A.; Tadmor, Y.; Gepstein, S.; Giovannoni, J.J.; Katzir, N.; et al. Branched-chain and aromatic amino acid catabolism into aroma volatiles in Cucumis melo L. fruit. J. Exp. Bot. 2010, 61, 1111–1123. [Google Scholar] [CrossRef]
- Sakai, M.; Hirata, H.; Sayama, H.; Sekiguchi, K.; Itano, H.; Asai, T.; Dohra, H.; Hara, M.; Watanabe, N. Production of 2-phenylethanol in roses as the dominant floral scent compound from L-phenylalanine by two key enzymes, a PLP-dependent decarboxylase and a phenylacetaldehyde reductase. Biosci. Biotechnol. Biochem. 2007, 71, 2408–2419. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.M.; Kobayashi, H.; Sakai, M.; Hirata, H.; Asai, T.; Ohnishi, T.; Baldermann, S.; Watanabe, N. Functional characterization of rose phenylacetaldehyde reductase (PAR), an enzyme involved in the biosynthesis of the scent compound 2-phenylethanol. J. Plant Physiol. 2011, 168, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Osbourn, A.; Ma, P. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 2015, 8, 689–708. [Google Scholar] [CrossRef] [PubMed]
- Spitzer-Rimon, B.; Farhi, M.; Albo, B.; Cna’ani, A.; Ben Zvi, M.M.; Masci, T.; Edelbaum, O.; Yu, Y.; Shklarman, E.; Ovadis, M.; et al. The R2R3-MYB-like regulatory factor EOBI, acting downstream of EOBII, regulates scent production by activating ODO1 and structural scent-related genes in Petunia. Plant Cell 2012, 24, 5089–5105. [Google Scholar] [CrossRef] [PubMed]
- Verdonk, J.C.; Haring, M.A.; Van Tunen, A.J.; Schuurink, R.C. Odorant1 regulates fragrance biosynthesis in Petunia flowers. Plant Cell 2005, 17, 1612–1624. [Google Scholar] [CrossRef]
- Van Moerkercke, A.; Haring, M.A.; Schuurink, R.C. The transcription factor EMISSION OF BENZENOIDS II activates the MYB ODORANT1 promoter at a MYB binding site specific for fragrant petunias. Plant J. 2011, 67, 917–928. [Google Scholar] [CrossRef]
- Colquhoun, T.A.; Clark, D.G. Unraveling the regulation of floral fragrance biosynthesis. Plant Signal. Behav. 2011, 6, 378–381. [Google Scholar] [CrossRef]
- Shan, X.; Li, Y.; Yang, S.; Yang, Z.; Qiu, M.; Gao, R.; Han, T.; Meng, X.; Xu, Z.; Wang, L.; et al. The spatio-temporal biosynthesis of floral flavonols is controlled by differential phylogenetic MYB regulators in Freesia hybrida. New Phytol. 2020. [Google Scholar] [CrossRef]
- Allan, A.C.; Espley, R.V. MYBs Drive Novel Consumer Traits in Fruits and Vegetables. Trends Plant Sci. 2018, 23, 693–705. [Google Scholar] [CrossRef]
- Sollars, E.S.A.; Buggs, R.J.A. Genome-wide epigenetic variation among ash trees differing in susceptibility to a fungal disease. BMC Genom. 2018, 19, 502. [Google Scholar] [CrossRef]
- D’Amelia, V.; Villano, C.; Batelli, G.; Çobanoğlu, Ö.; Carucci, F.; Melito, S.; Chessa, M.; Chiaiese, P.; Aversano, R.; Carputo, D. Genetic and epigenetic dynamics affecting anthocyanin biosynthesis in potato cell culture. Plant Sci. 2020, 298, 110597. [Google Scholar] [CrossRef] [PubMed]
- Robbins, M.L.; Roy, A.; Wang, P.H.; Gaffoor, I.; Sekhon, R.S.; Buanafina, M.M.d.O.; Rohila, J.S.; Chopra, S. Comparative proteomics analysis by DIGE and iTRAQ provides insight into the regulation of phenylpropanoids in maize. J. Proteom. 2013, 93, 254–275. [Google Scholar] [CrossRef] [PubMed]
- Harmer, S.L. The circadian system in higher plants. Annu. Rev. Plant Biol. 2009, 60, 357–377. [Google Scholar] [CrossRef] [PubMed]
- Song, M.S.; Dong, G.K.; Lee, S.H. Isolation and characterization of a jasmonic acid carboxyl methyltransferase gene from hot pepper (Capsicum annuum L.). J. Plant Biol. 2005, 48, 292–297. [Google Scholar] [CrossRef]
- Gigot, C.; Ongena, M.; Fauconnier, M.L.; Wathelet, J.P.; du Jardin, P.; Thonart, P. The lipoxygenase metabolic pathway in plants: Potential for industrial production of natural green leaf volatiles. Biotechnol. Agron. Soc. Environ. 2010, 14, 451–460. [Google Scholar]
- Schade, F.; Legge, R.L.; Thompson, J.E. Fragrance volatiles of developing and senescing carnation flowers. Phytochemistry 2001, 56, 703–710. [Google Scholar] [CrossRef]
- Suchet, C.; Dormont, L.; Schatz, B.; Giurfa, M.; Simon, V.; Raynaud, C.; Chave, J. Floral scent variation in two Antirrhinum majus subspecies influences the choice of naïve bumblebees. Behav. Ecol. Sociobiol. 2011, 65, 1015–1027. [Google Scholar] [CrossRef]
- Schäffler, I.; Balao, F.; Dötterl, S. Floral and vegetative cues in oil-secreting and non-oil-secreting Lysimachia species. Ann. Bot. 2012, 110, 125–138. [Google Scholar] [CrossRef]
- Browse, J. The power of mutants for investigating jasmonate biosynthesis and signaling. Phytochemistry 2009, 70, 1539–1546. [Google Scholar] [CrossRef]
- Pirrello, J.; Leclercq, J.; Dessailly, F.; Rio, M.; Piyatrakul, P.; Kuswanhadi, K.; Tang, C.; Montoro, P. Transcriptional and post-transcriptional regulation of the jasmonate signalling pathway in response to abiotic and harvesting stress in Hevea brasiliensis. BMC Plant Biol. 2014, 14, 1–17. [Google Scholar] [CrossRef]
- De Geyter, N.; Gholami, A.; Goormachtig, S.; Goossens, A. Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci. 2012, 17, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Issakidis-Bourguet, E.; Zhou, D.X. Perspectives on the interactions between metabolism, redox, and epigenetics in plants. J. Exp. Bot. 2016, 67, 5291–5300. [Google Scholar] [CrossRef] [PubMed]
- Lindermayr, C.; Rudolf, E.E.; Durner, J.; Groth, M. Interactions between metabolism and chromatin in plant models. Mol. Metab. 2020, 38, 100951. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, C.; Liu, X.; Cheng, J.; Li, S.; Zhu, J.K.; Gong, Z. Peroxisomal β-oxidation regulates histone acetylation and DNA methylation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 10576–10585. [Google Scholar] [CrossRef]
- He, M.; Qin, C.X.; Wang, X.; Ding, N.Z. Plant Unsaturated Fatty Acids: Biosynthesis and Regulation. Front. Plant Sci. 2020, 11, 390. [Google Scholar] [CrossRef]
- Wang, T.; Xing, J.; Liu, X.; Liu, Z.; Yao, Y.; Hu, Z.; Peng, H.; Xin, M.; Zhou, D.X.; Zhang, Y.; et al. Histone acetyltransferase general control non-repressed protein 5 (GCN5) affects the fatty acid composition of Arabidopsis thaliana seeds by acetylating fatty acid desaturase3 (FAD3). Plant J. 2016, 88, 794–808. [Google Scholar] [CrossRef]
- Kong, L.; Zhi, P.; Liu, J.; Li, H.; Zhang, X.; Xu, J.; Zhou, J.; Wang, X.; Chang, C. Epigenetic activation of enoyl-coa reductase by an acetyltransferase complex triggers wheat wax biosynthesis. Plant Physiol. 2020, 183, 1250–1267. [Google Scholar] [CrossRef]
- Fenske, M.P.; Imaizumi, T. Circadian rhythms in floral scent emission. Front. Plant Sci. 2016, 7, 1–6. [Google Scholar] [CrossRef]
- Zeng, L.; Wang, X.; Kang, M.; Dong, F.; Yang, Z. Regulation of the rhythmic emission of plant volatiles by the circadian clock. Int. J. Mol. Sci. 2017, 18, 2408. [Google Scholar] [CrossRef]
- Dudareva, N.; Pichersky, E. Biochemical and molecular genetic aspects of floral scents. Plant Physiol. 2000, 122, 627–633. [Google Scholar] [CrossRef]
- Terry, M.I.; Pérez-Sanz, F.; Díaz-Galián, M.V.; Pérez de los Cobos, F.; Navarro, P.J.; Egea-Cortines, M.; Weiss, J. The Petunia CHANEL Gene is a ZEITLUPE Ortholog Coordinating Growth and Scent Profiles. Cells 2019, 8, 343. [Google Scholar] [CrossRef] [PubMed]
- Fenske, M.P.; Hewett Hazelton, K.D.; Hempton, A.K.; Shim, J.S.; Yamamoto, B.M.; Riffell, J.A.; Imaizumi, T. Circadian clock gene LATE ELONGATED HYPOCOTYL directly regulates the timing of floral scent emission in Petunia. Proc. Natl. Acad. Sci. USA 2015, 112, 9775–9780. [Google Scholar] [CrossRef] [PubMed]
- Kolosova, N.; Gorenstein, N.; Kish, C.M.; Dudareva, N. Regulation of Circadian Methyl Benzoate Emission in Diurnally and Nocturnally Emitting Plants. Plant Cell 2001, 13, 2333–2347. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.A.; Nemchenko, A.; Borrego, E.; Murray, I.; Sobhy, I.S.; Bosak, L.; Deblasio, S.; Erb, M.; Robert, C.A.M.; Vaughn, K.A.; et al. The maize lipoxygenase, ZmLOX10, mediates green leaf volatile, jasmonate and herbivore-induced plant volatile production for defense against insect attack. Plant J. 2013, 74, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Nagel, D.H.; Kay, S.A. Complexity in the wiring and regulation of plant circadian networks. Curr. Biol. 2012, 22, R648–R657. [Google Scholar] [CrossRef]
- McClung, C.R. Beyond Arabidopsis: The circadian clock in non-model plant species. Semin. Cell Dev. Biol. 2013, 24, 430–436. [Google Scholar] [CrossRef]
- Alabadí, D.; Oyama, T.; Yanovsky, M.J.; Harmon, F.G.; Más, P.; Kay, S.A. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 2001, 293, 880–883. [Google Scholar] [CrossRef]
- Yon, F.; Kessler, D.; Joo, Y.; Cortés Llorca, L.; Kim, S.G.; Baldwin, I.T. Fitness consequences of altering floral circadian oscillations for Nicotiana attenuata. J. Integr. Plant Biol. 2017, 59, 180–189. [Google Scholar] [CrossRef]
- Barneche, F.; Malapeira, J.; Mas, P. The impact of chromatin dynamics on plant light responses and circadian clock function. J. Exp. Bot. 2014, 65, 2895–2913. [Google Scholar] [CrossRef]
- Yon, F.; Joo, Y.; Cortés Llorca, L.; Rothe, E.; Baldwin, I.T.; Kim, S.G. Silencing Nicotiana attenuata LHY and ZTL alters circadian rhythms in flowers. New Phytol. 2015, 209, 1058–1066. [Google Scholar] [CrossRef]
- Malapeira, J.; Khaitova, L.C.; Mas, P. Ordered changes in histone modifications at the core of the Arabidopsis circadian clock. Proc. Natl. Acad. Sci. USA 2012, 109, 21540–21545. [Google Scholar] [CrossRef] [PubMed]
- Henriques, R.; Mas, P. Chromatin remodeling and alternative splicing: Pre- and post-transcriptional regulation of the Arabidopsis circadian clock. Semin. Cell Dev. Biol. 2013, 24, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Du, S.; Chen, L.; Ge, L.; Huang, W. A novel loop: Mutual regulation between epigenetic modification and the circadian clock. Front. Plant Sci. 2019, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Farinas, B.; Mas, P. Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J. 2011, 66, 318–329. [Google Scholar] [CrossRef]
- Song, H.R.; Noh, Y.S. Rhythmic oscillation of histone acetylation and methylation at the arabidopsis central clock loci. Mol. Cells 2012, 34, 279–287. [Google Scholar] [CrossRef]
- Hung, F.Y.; Chen, F.F.; Li, C.; Chen, C.; Lai, Y.C.; Chen, J.H.; Cui, Y.; Wu, K. The Arabidopsis LDL1/2-HDA6 histone modification complex is functionally associated with CCA1/LHY in regulation of circadian clock genes. Nucleic Acids Res. 2018, 46, 10669–10681. [Google Scholar] [CrossRef]
- Baulcombe, D.C.; Dean, C. Epigenetic regulation in plant responses to the environment. Cold Spring Harb. Perspect. Biol. 2014, 6, a019471. [Google Scholar] [CrossRef]
- Weiss, J.; Mühlemann, J.K.; Ruiz-Hernández, V.; Dudareva, N.; Egea-Cortines, M. Phenotypic Space and Variation of Floral Scent Profiles during Late Flower Development in Antirrhinum. Front. Plant Sci. 2016, 7, 1903. [Google Scholar] [CrossRef]
- Prieto-Benítez, S.; Millanes, A.M.; Dötterl, S.; Giménez-Benavides, L. Comparative analyses of flower scent in Sileneae reveal a contrasting phylogenetic signal between night and day emissions. Ecol. Evol. 2016, 6, 7869–7881. [Google Scholar] [CrossRef]
- Amrad, A.; Moser, M.; Mandel, T.; de Vries, M.; Schuurink, R.C.; Freitas, L.; Kuhlemeier, C. Gain and loss of floral scent croduction through changes in structural genes during pollinator-mediated Speciation. Curr. Biol. 2016, 26, 3303–3312. [Google Scholar] [CrossRef]
- Jantzen, F.; Lynch, J.H.; Kappel, C.; Höfflin, J.; Skaliter, O.; Wozniak, N.; Sicard, A.; Sas, C.; Adebesin, F.; Ravid, J.; et al. Retracing the molecular basis and evolutionary history of the loss of benzaldehyde emission in the genus Capsella. New Phytol. 2019, 224, 1349–1360. [Google Scholar] [CrossRef] [PubMed]
- Cseke, L.; Dudareva, N.; Pichersky, E. Structure and Evolution of Linalool Synthase. Mol. Biol. Evol. 1998, 15, 1491–1498. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Quadrana, L.; Colot, V. Plant Transgenerational Epigenetics. Annu. Rev. Genet. 2016, 50, 467–491. [Google Scholar] [CrossRef] [PubMed]
- Kellenberger, R.T.; Desurmont, G.A.; Schlüter, P.M.; Schiestl, F.P. Trans-generational inheritance of herbivory-induced phenotypic changes in Brassica rapa. Sci. Rep. 2018, 8, 3536. [Google Scholar] [CrossRef] [PubMed]
- Farré-Armengol, G.; Fernández-Martínez, M.; Filella, I.; Junker, R.R.; Peñuelas, J. Deciphering the Biotic and Climatic Factors That Influence Floral Scents: A Systematic Review of Floral Volatile Emissions. Front. Plant Sci. 2020, 11, 1154. [Google Scholar] [CrossRef] [PubMed]
- Ramos, S.E.; Schiestl, F.P. Evolution of Floral Fragrance Is Compromised by Herbivory. Front. Ecol. Evol. 2020, 8, 1–8. [Google Scholar] [CrossRef]
- Baniaga, A.E.; Marx, H.E.; Arrigo, N.; Barker, M.S. Polyploid plants have faster rates of multivariate niche differentiation than their diploid relatives. Ecol. Lett. 2020, 23, 68–78. [Google Scholar] [CrossRef]
- Landis, J.B.; Soltis, D.E.; Li, Z.; Marx, H.E.; Barker, M.S.; Tank, D.C.; Soltis, P.S. Impact of whole-genome duplication events on diversification rates in angiosperms. Am. J. Bot. 2018, 105, 348–363. [Google Scholar] [CrossRef]
- Tank, D.C.; Eastman, J.M.; Pennell, M.W.; Soltis, P.S.; Soltis, D.E.; Hinchliff, C.E.; Brown, J.W.; Sessa, E.B.; Harmon, L.J. Nested radiations and the pulse of angiosperm diversification: Increased diversification rates often follow whole genome duplications. New Phytol. 2015, 207, 454–467. [Google Scholar] [CrossRef]
- Doyle, J.J.; Flagel, L.E.; Paterson, A.H.; Rapp, R.A.; Soltis, D.E.; Soltis, P.S.; Wendel, J.F. Evolutionary Genetics of Genome Merger and Doubling in Plants. Annu. Rev. Genet. 2008, 42, 443–461. [Google Scholar] [CrossRef]
- Feliner, G.N.; Casacuberta, J.; Wendel, J.F. Genomics of Evolutionary Novelty in Hybrids and Polyploids. Front. Genet. 2020, 11, 792. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, Y.; Xia, E.H.; Yao, Q.Y.; Liu, X.D.; Gao, L.Z. Autotetraploid rice methylome analysis reveals methylation variation of transposable elements and their effects on gene expression. Proc. Natl. Acad. Sci. USA 2015, 112, E7022–E7029. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Bombarely, A.; Xu, B.; Wu, B.; Frazier, T.P.; Zhang, X.; Chen, J.; Chen, P.; Sun, M.; Feng, G.; et al. Autopolyploidization in switchgrass alters phenotype and flowering time via epigenetic and transcription regulation. J. Exp. Bot. 2019, 70, 5673–5686. [Google Scholar] [CrossRef] [PubMed]
- Cavé-Radet, A.; Giraud, D.; Lima, O.; El Amrani, A.; Aïnouche, M.; Salmon, A. Evolution of small RNA expression following hybridization and allopolyploidization: Insights from Spartina species (Poaceae, Chloridoideae). Plant Mol. Biol. 2020, 102, 55–72. [Google Scholar] [CrossRef]
- Ng, D.W.K.; Lu, J.; Chen, Z.J. Big roles for small RNAs in polyploidy, hybrid vigor, and hybrid incompatibility. Curr. Opin. Plant Biol. 2012, 15, 154–161. [Google Scholar] [CrossRef]
- Zhang, H.; Zheng, R.; Wang, Y.; Zhang, Y.; Hong, P.; Fang, Y.; Li, G.; Fang, Y. The effects of Arabidopsis genome duplication on the chromatin organization and transcriptional regulation. Nucleic Acids Res. 2019, 47, 7857–7869. [Google Scholar] [CrossRef]
- Doyle, J.J.; Coate, J.E. Autopolyploidy: An epigenetic macromutation. Am. J. Bot. 2020, 107, 1097–1100. [Google Scholar] [CrossRef]
- Vicient, C.M.; Casacuberta, J.M. Impact of transposable elements on polyploid plant genomes. Ann. Bot. 2017, 120, 195–207. [Google Scholar] [CrossRef]
- Arango, J.; Beltrán, J.; Nuñez, J.; Chavarriaga, P. Evidence of epigenetic mechanisms affecting carotenoids. Subcell. Biochem. 2016, 79, 295–307. [Google Scholar] [CrossRef]
- Gross, K.; Schiestl, F.P. Are tetraploids more successful? Floral signals, reproductive success and floral isolation in mixed-ploidy populations of a terrestrial orchid. Ann. Bot. 2015, 115, 263–273. [Google Scholar] [CrossRef]
- Jersáková, J.; Castro, S.; Sonk, N.; Milchreit, K.; Schödelbauerová, I.; Tolasch, T.; Dötterl, S.; Jersáková, J.; Schödelbauerová, I.; Schödelbauerová, Á.I.; et al. Absence of pollinator-mediated premating barriers in mixed-ploidy populations of Gymnadenia conopsea s.l. (Orchidaceae). Evol. Ecol. 2010, 24, 1199–1218. [Google Scholar] [CrossRef]
- Klahre, U.; Gurba, A.; Hermann, K.; Saxenhofer, M.; Bossolini, E.; Guerin, P.M.; Kuhlemeier, C. Pollinator choice in petunia depends on two major genetic loci for floral scent production. Curr. Biol. 2011, 21, 730–739. [Google Scholar] [CrossRef] [PubMed]
- Barkman, T.J. Evidence for positive selection on the floral scent gene isoeugenol-O-methyltransferase. Mol. Biol. Evol. 2003, 20, 168–172. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bao, F.; Ding, A.; Zhang, T.; Luo, L.; Wang, J.; Cheng, T.; Zhang, Q. Expansion of PmBEAT genes in the Prunus mume genome induces characteristic floral scent production. Hortic. Res. 2019, 6, 1–18. [Google Scholar] [CrossRef]
- Balao, F.; Valente, L.M.; Vargas, P.; Herrera, J.; Talavera, S. Radiative evolution of polyploid races of the Iberian carnation Dianthus broteri (Caryophyllaceae). New Phytol. 2010, 187, 542–551. [Google Scholar] [CrossRef]
- Balao, F.; Casimiro-Soriguer, R.; Talavera, M.; Herrera, J.; Talavera, S. Distribution and diversity of cytotypes in Dianthus broteri as evidenced by genome size variations. Ann. Bot. 2009, 104, 965–973. [Google Scholar] [CrossRef]
- López-Jurado, J.; Balao, F.; Mateos-Naranjo, E. Polyploidy-mediated divergent light-harvesting and photoprotection strategies under temperature stress in a Mediterranean carnation complex. Environ. Exp. Bot. 2020, 171, 103956. [Google Scholar] [CrossRef]
- López-Jurado, J.; Mateos-Naranjo, E.; Balao, F. Niche divergence and limits to expansion in the high polyploid Dianthus broteri complex. New Phytol. 2019, 222, 1076–1087. [Google Scholar] [CrossRef]
- Alonso, C.; Balao, F.; Bazaga, P.; Pérez, R. Epigenetic contribution to successful polyploidizations: Variation in global cytosine methylation along an extensive ploidy series in Dianthus broteri (Caryophyllaceae). New Phytol. 2016, 212, 571–576. [Google Scholar] [CrossRef]
- Balao, F.; Paun, O.; Alonso, C. Uncovering the contribution of epigenetics to plant phenotypic variation in Mediterranean ecosystems. Plant Biol. 2018, 20, 38–49. [Google Scholar] [CrossRef]
- Mirouze, M.; Paszkowski, J. Epigenetic contribution to stress adaptation in plants. Curr. Opin. Plant Biol. 2011, 14, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Herrera, J.; Balao, F. A test of phenotypic selection on petal form in the wild carnation, Dianthus inoxianus. Plant Biol. 2015, 17, 1227–1232. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.Y.H.; Trumble, J.T.; Kumamoto, J. Activity of volatile compounds in glandular trichomes of Lycopersicon species against two insect herbivores. J. Chem. Ecol. 1987, 13, 837–850. [Google Scholar] [CrossRef] [PubMed]
- Balao, F. (University of Seville, Seville, Spain). Personal communication, 2019. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Picazo-Aragonés, J.; Terrab, A.; Balao, F. Plant Volatile Organic Compounds Evolution: Transcriptional Regulation, Epigenetics and Polyploidy. Int. J. Mol. Sci. 2020, 21, 8956. https://doi.org/10.3390/ijms21238956
Picazo-Aragonés J, Terrab A, Balao F. Plant Volatile Organic Compounds Evolution: Transcriptional Regulation, Epigenetics and Polyploidy. International Journal of Molecular Sciences. 2020; 21(23):8956. https://doi.org/10.3390/ijms21238956
Chicago/Turabian StylePicazo-Aragonés, Jesús, Anass Terrab, and Francisco Balao. 2020. "Plant Volatile Organic Compounds Evolution: Transcriptional Regulation, Epigenetics and Polyploidy" International Journal of Molecular Sciences 21, no. 23: 8956. https://doi.org/10.3390/ijms21238956
APA StylePicazo-Aragonés, J., Terrab, A., & Balao, F. (2020). Plant Volatile Organic Compounds Evolution: Transcriptional Regulation, Epigenetics and Polyploidy. International Journal of Molecular Sciences, 21(23), 8956. https://doi.org/10.3390/ijms21238956




