An Odorant Binding Protein (SaveOBP9) Involved in Chemoreception of the Wheat Aphid Sitobion avenae
Abstract
1. Introduction
2. Results
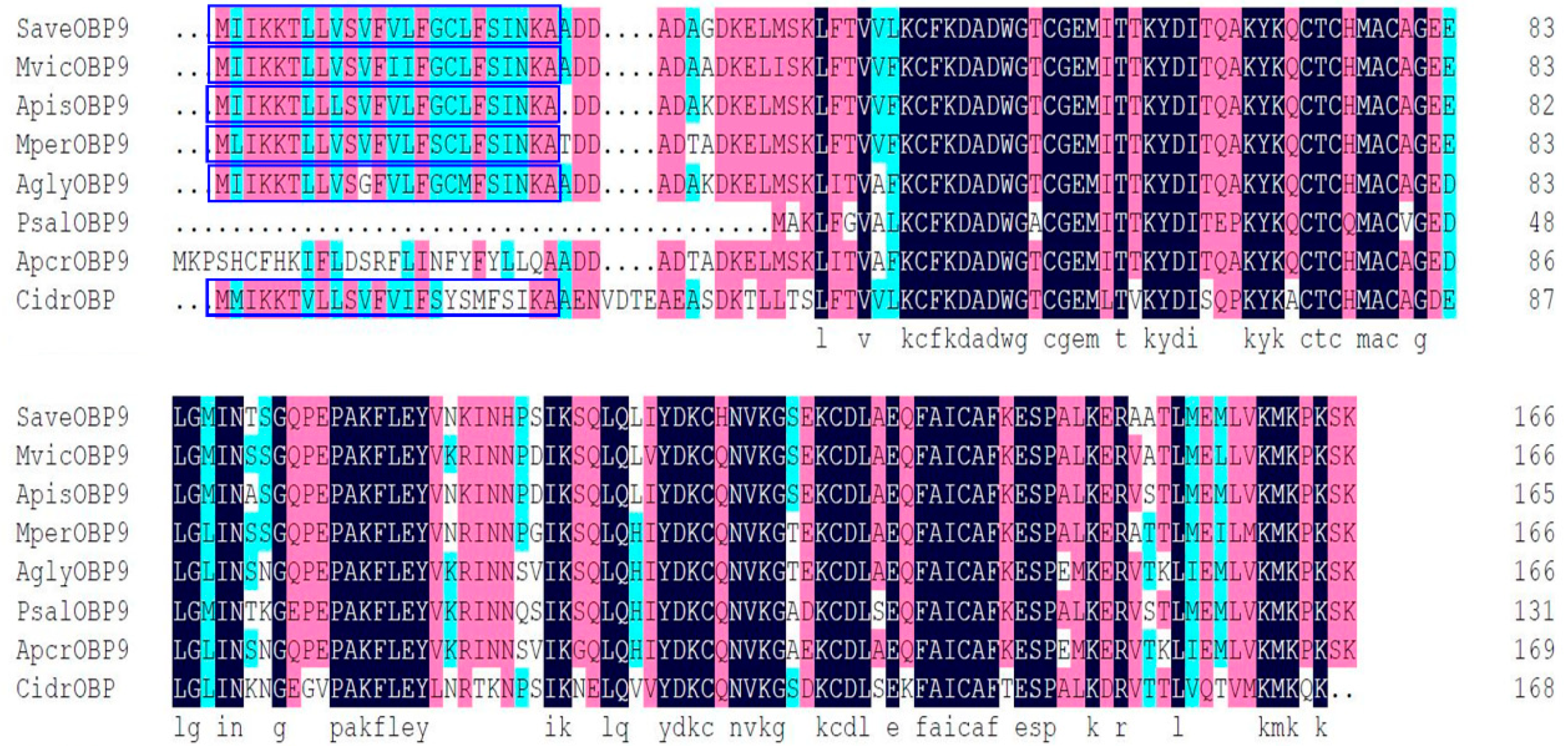
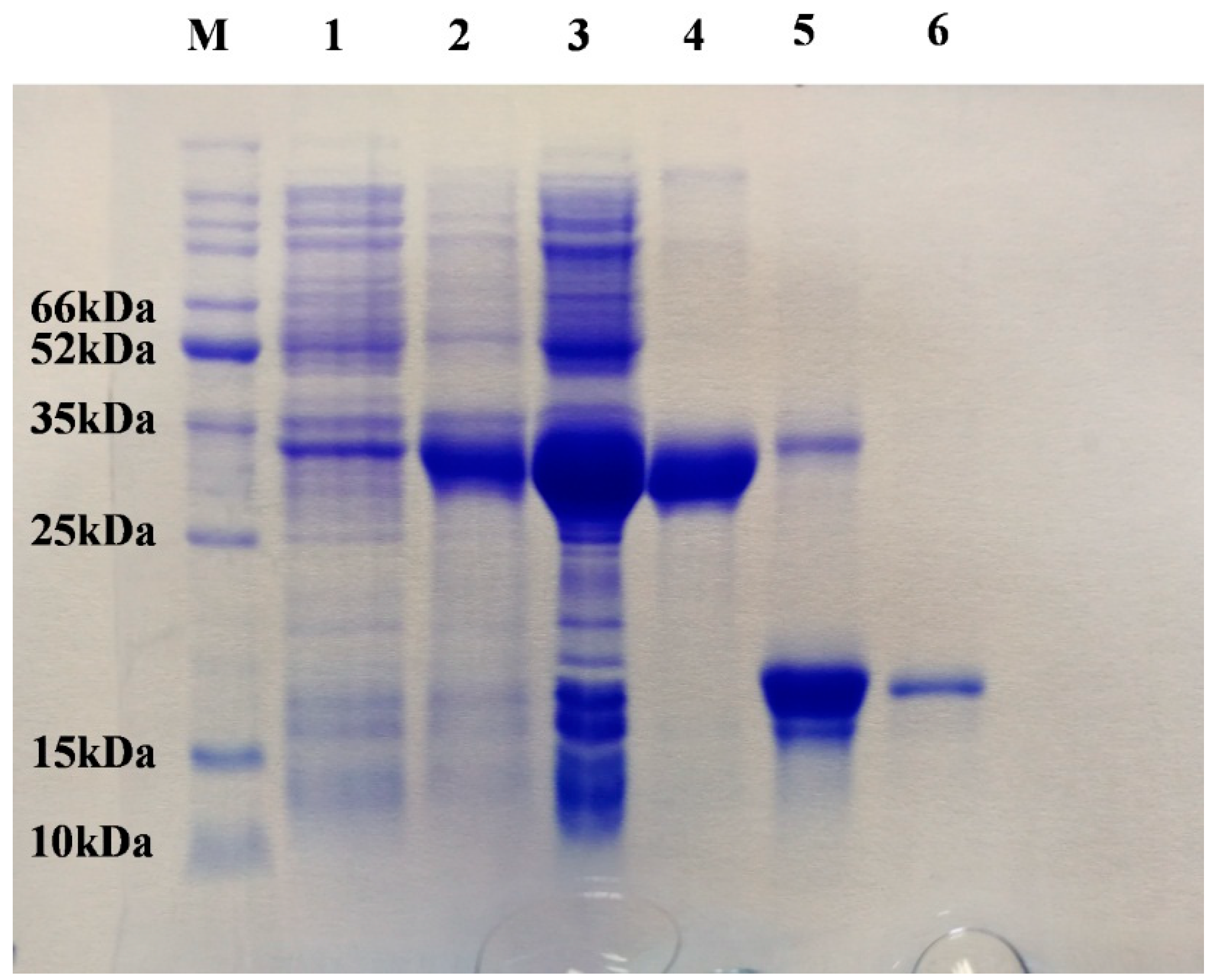
2.1. Identification and Characterization of the SaveOBP9
2.2. Fluorescence Binding Assay
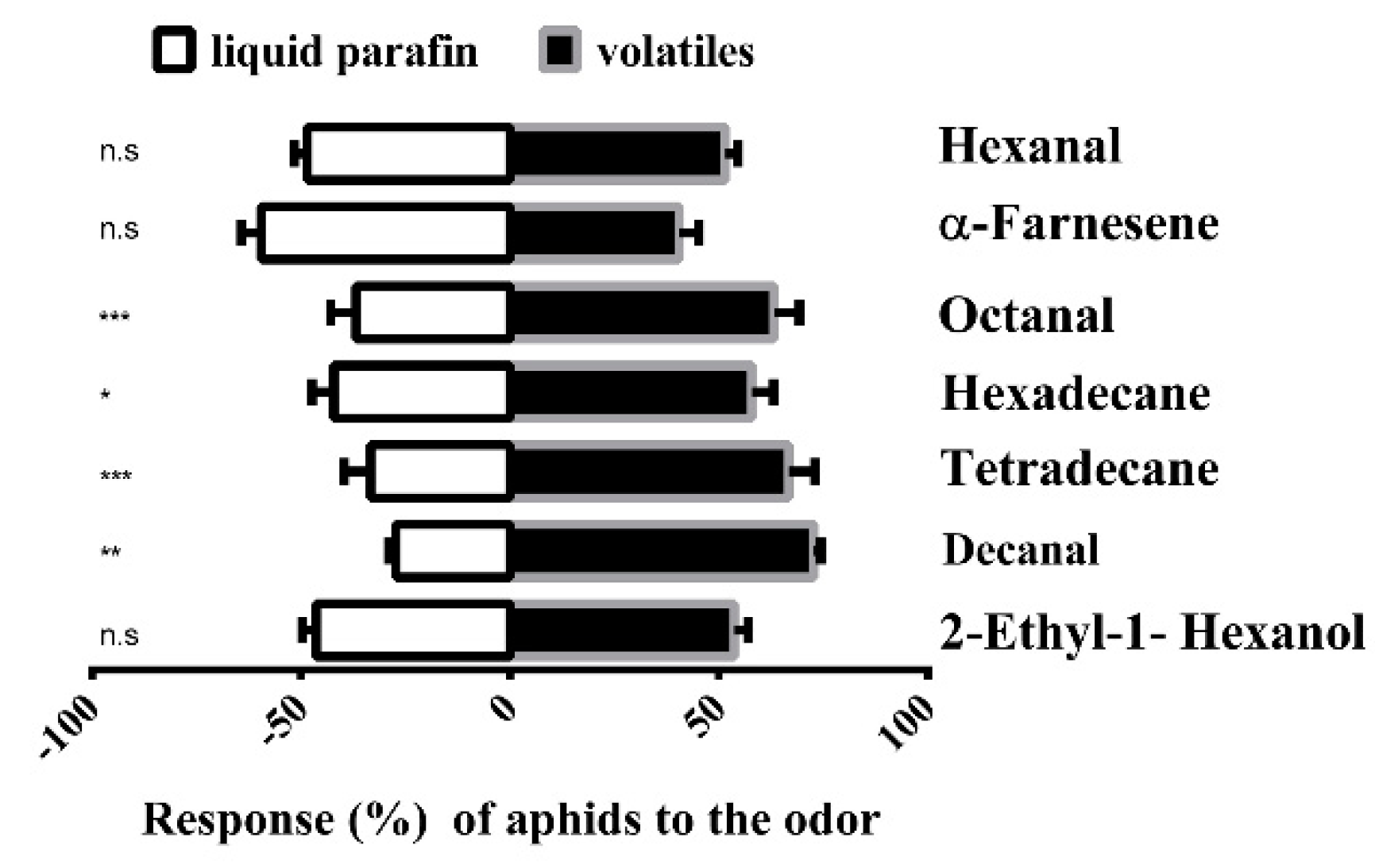
2.3. Behavioral Trials
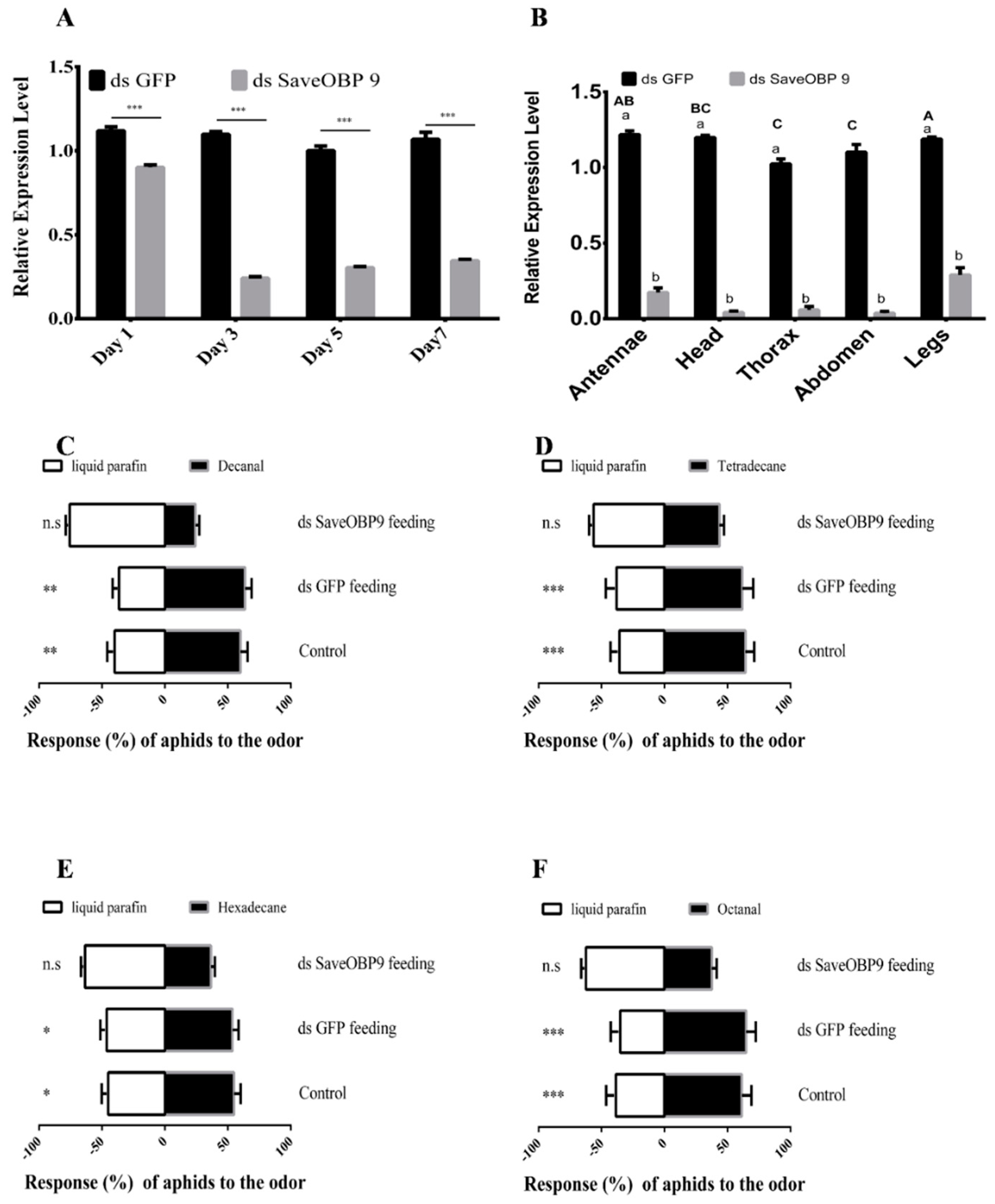
2.4. RNAi-Based Silencing and Post-RNAi Behavior of Sitobion avenae
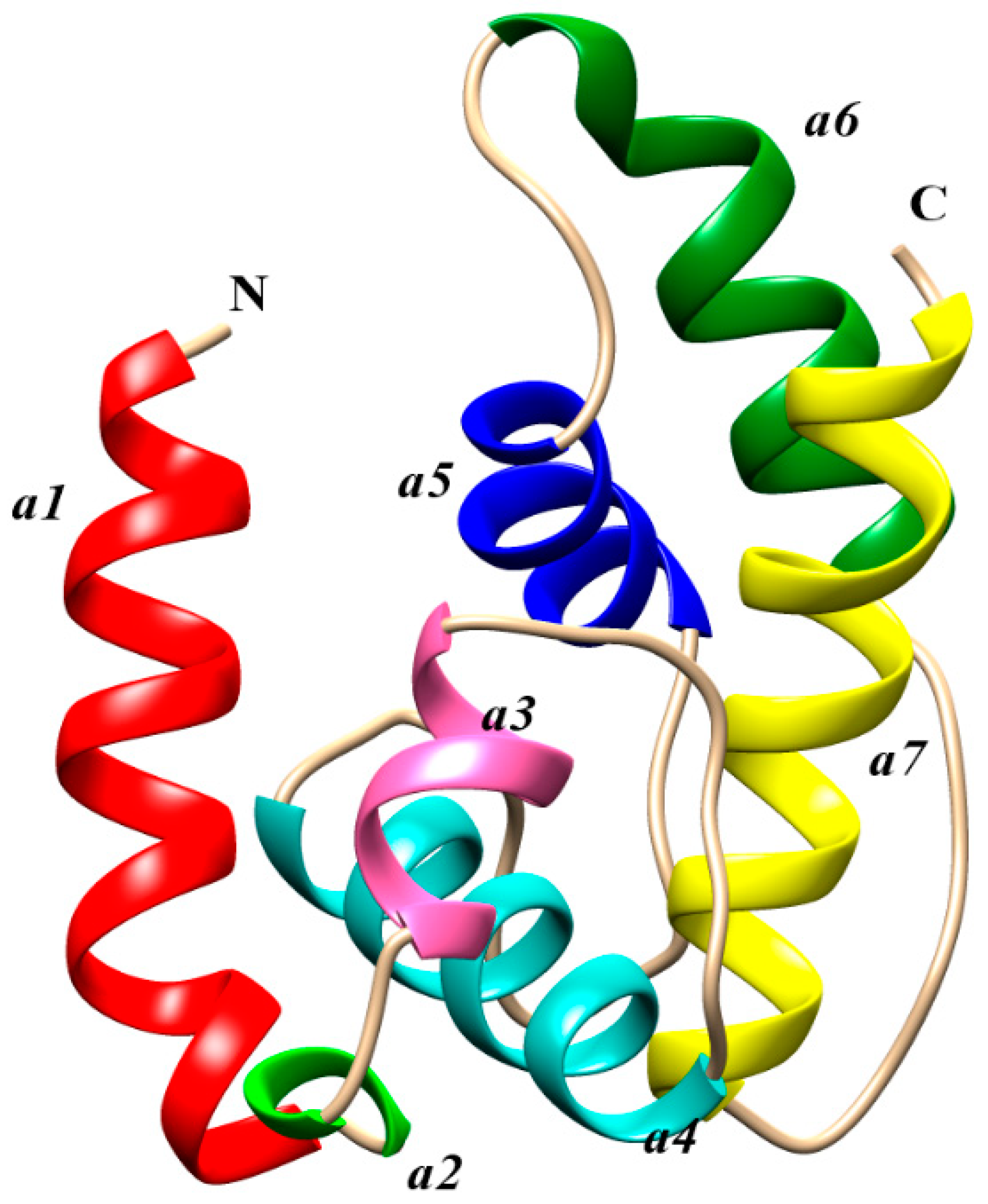
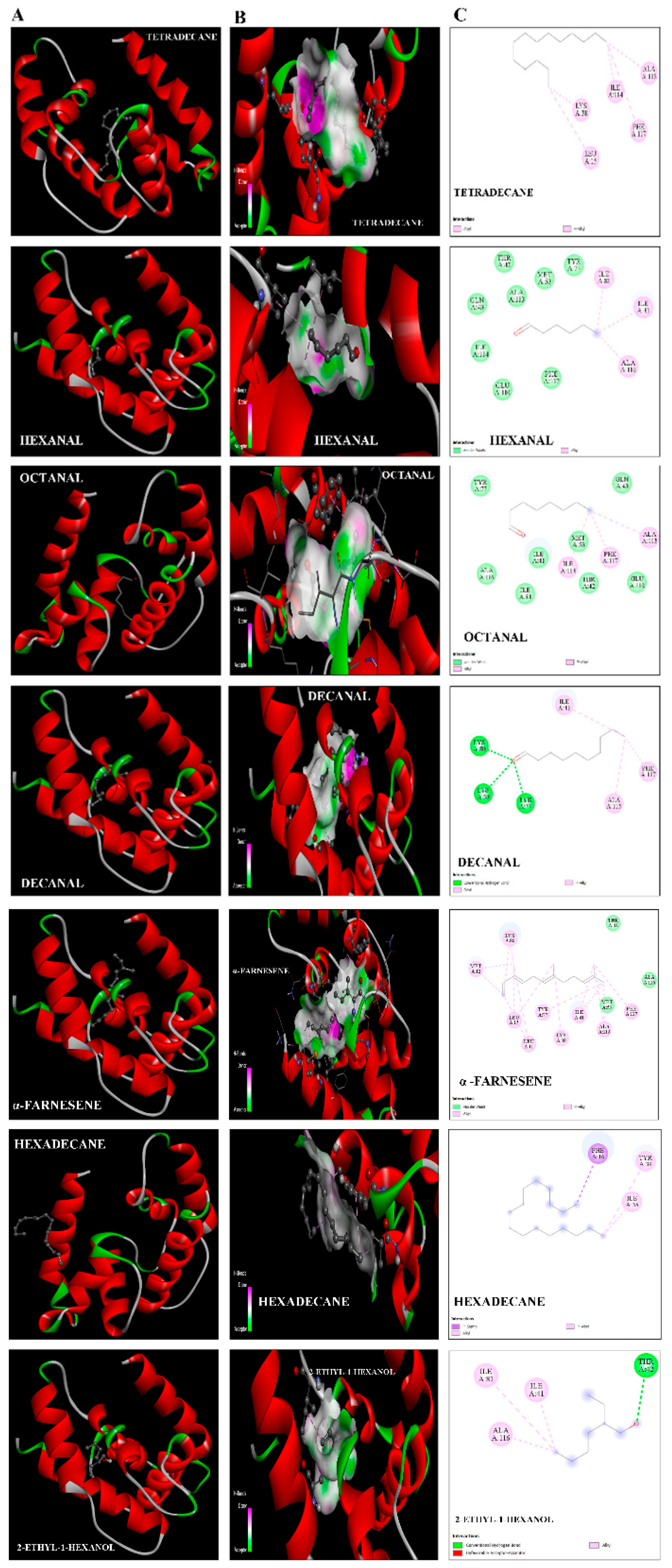
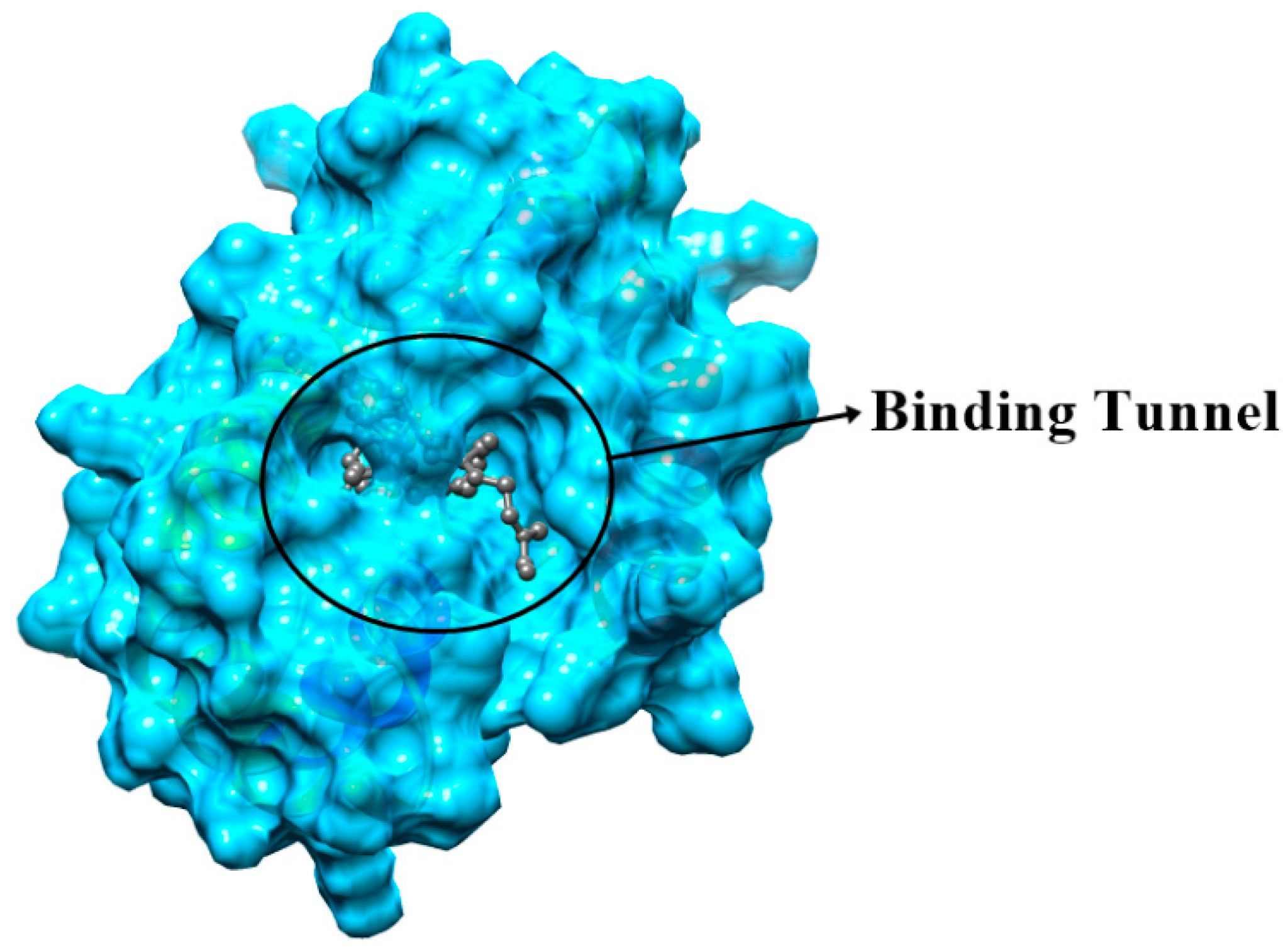
2.5. Protein Structure and Interaction Analysis by 3-Dimensional Docking
3. Discussion
4. Materials and Methods
4.1. Sitobion avenae Rearing
4.2. Cloning, Expression, and Purification of SaveOBP9
4.3. Fluorescence Ligand Binding Assays
4.4. Double-Stranded RNA Synthesis
4.5. Dietary RNAi and Gene Expression Analysis
4.6. Olfactometer Bioassay
4.7. Modeling of Three-Dimensional (3D) Structure and Molecular Docking of Ligands
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhou, J.J. Odorant-binding proteins in insects. Vitam. Horm. 2010, 83, 241–272. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.; Tuccori, E.; He, X.; Gazzano, A.; Field, L.; Zhou, J.J.; Pelosi, P. Discrimination of alarm pheromone (E)-β-farnesene by aphid odorant-binding proteins. Insect Biochem. Mol. Biol. 2009, 39, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Lassance, J.M.; Löfstedt, C. Chemical communication: A jewel sheds light on signal evolution. Curr. Biol. 2013, 23, R346–R348. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, Q.; Shang, Y.; Hilton, D.S.; Inthavong, K.; Zhang, D.; Elgar, M.A. Antennal scales improve signal detection efficiency in moths. Proc. R. Soc. B Biol. Sci. 2018, 285. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, P.; Iovinella, I.; Zhu, J.; Wang, G.; Dani, F.R. Beyond chemoreception: Diverse tasks of soluble olfactory proteins in insects. Biol. Rev. 2018, 93, 184–200. [Google Scholar] [CrossRef]
- Fan, J.; Francis, F.; Liu, Y.; Chen, J.L.; Cheng, D.F. An overview of odorant-binding protein functions in insect peripheral olfactory reception. Genet. Mol. Res. 2011, 10, 3056–3069. [Google Scholar] [CrossRef]
- Vandermoten, S.; Francis, F.; Haubruge, E.; Leal, W.S. Conserved odorant-binding proteins from aphids and eavesdropping predators. PLoS ONE 2012, 7, e23608. [Google Scholar] [CrossRef]
- Zhong, T.; Yin, J.; Deng, S.; Li, K.; Cao, Y. Fluorescence competition assay for the assessment of green leaf volatiles and trans-β-farnesene bound to three odorant-binding proteins in the wheat aphid Sitobion avenae (Fabricius). J. Insect Physiol. 2012, 58, 771–781. [Google Scholar] [CrossRef]
- YJ, Z.; Zeng, J. Occurring trends of major crop pests in national significances in 2009. China Plant Prot. 2009, 29, 33–36. [Google Scholar]
- Webster, B. The role of olfaction in aphid host location. Physiol. Entomol. 2012, 37, 10–18. [Google Scholar] [CrossRef]
- Conchou, L.; Lucas, P.; Meslin, C.; Proffit, M.; Staudt, M.; Renou, M. Insect odorscapes: From plant volatiles to natural olfactory scenes. Front. Physiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Riffell, J.A.; Lei, H.; Christensen, T.A. Report Characterization and Coding of Behaviorally Significant Odor Mixtures. Curr. Biol. 2009, 19, 335–340. [Google Scholar] [CrossRef]
- Bruce, T.J.A. Interplay between insects and plants: Dynamic and complex interactions that have coevolved over millions of years but act in milliseconds. J. Exp. Bot. 2015, 66, 455–465. [Google Scholar] [CrossRef]
- Pickett, J.A.; Khan, Z.R. Plant volatile-mediated signalling and its application in agriculture: Successes and challenges. New Phytol. 2016, 212, 856–870. [Google Scholar] [CrossRef]
- Sobhy, I.S.; Caulfield, J.C.; Pickett, J.A.; Birkett, M.A. Sensing the Danger Signals: Cis-Jasmone Reduces Aphid Performance on Potato and Modulates the Magnitude of Released Volatiles. Front. Ecol. Evol. 2020, 7, 1–14. [Google Scholar] [CrossRef]
- Xue, W.; Fan, J.; Zhang, Y.; Xu, Q.; Han, Z.; Sun, J.; Chen, J. Identification and expression analysis of candidate odorant-binding protein and chemosensory protein genes by antennal transcriptome of sitobion avenae. PLoS ONE 2016, 11, e161839. [Google Scholar] [CrossRef]
- Younas, A.; Waris, M.I.; Chang, X.Q.; Shaaban, M.; Abdelnabby, H.; Ul Qamar, M.T.; Wang, M.Q. A chemosensory protein MsepCSP5 involved in chemoreception of oriental armyworm Mythimna separata. Int. J. Biol. Sci. 2018, 14, 1935–1949. [Google Scholar] [CrossRef]
- Li, Z.Q.; Zhang, S.; Luo, J.Y.; Cui, J.J.; Ma, Y.; Dong, S.L. Two Minus-C odorant binding proteins from Helicoverpa armigera display higher ligand binding affinity at acidic pH than neutral pH. J. Insect Physiol. 2013, 59, 263–272. [Google Scholar] [CrossRef]
- Damberger, F.; Horst, R.; Wüthrich, K.; Peng, G.; Nikonova, L.; Leal, W.S. NMR characterization of a pH-dependent equilibrium between two folded solution conformations of the pheromone-binding protein from Bombyx mori. Protein Sci. 2000, 9, 1038–1041. [Google Scholar] [CrossRef]
- Horst, R.; Damberger, F.; Luginbühl, P.; Güntert, P.; Peng, G.; Nikonova, L.; Leal, W.S.; Wüthrich, K. NMR structure reveals intramolecular regulation mechanism for pheromone binding and release. Proc. Natl. Acad. Sci. USA 2001, 98, 14374–14379. [Google Scholar] [CrossRef] [PubMed]
- Hekmat-Scafe, D.S.; Scafe, C.R.; McKinney, A.J.; Tanouye, M.A. Genome-Wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Res. 2002, 12, 1357–1369. [Google Scholar] [CrossRef] [PubMed]
- Robertson, H.M.; Baits, R.L.; Walden, K.K.O.; Wada-Katsumata, A.; Schal, C. Enormous expansion of the chemosensory gene repertoire in the omnivorous German cockroach Blattella germanica. J. Exp. Zool. Part B Mol. Dev. Evol. 2018, 330, 265–278. [Google Scholar] [CrossRef]
- Gong, D.P.; Zhang, H.J.; Zhao, P.; Xia, Q.Y.; Xiang, Z.H. The odorant binding protein gene family from the genome of silkworm, bombyx mori. BMC Genom. 2009, 10, 332. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Winberg, G.; Ren, H.; Zhang, S. Expression, purification and functional analysis of an odorant binding protein AaegOBP22 from Aedes aegypti. Protein Expr. Purif. 2011, 75, 165–171. [Google Scholar] [CrossRef]
- Bruce, T.J.A.; Wadhams, L.J.; Woodcock, C.M. Insect host location: A volatile situation. Trends Plant Sci. 2005, 10, 269–274. [Google Scholar] [CrossRef]
- Visser, J.H.; Yan, F.-S. Electroantennogram responses of the grain aphids Sitobion avenae (F.) and Metopolophium dirhodum (Walk.) (Hom., Aphididae) to plant odour components. J. Appl. Entomol. 1995, 119, 539–542. [Google Scholar] [CrossRef]
- Lautenschlager, C.; Leal, W.S.; Clardy, J. Pheromone Ligands: Implications for Pheromone Recognition. Structure 2007, 15, 1148–1154. [Google Scholar]
- Leal, W.S.; Chen, A.M.; Ishida, Y.; Chiang, V.P.; Erickson, M.L.; Morgan, T.I.; Tsuruda, J.M. Kinetics and molecular properties of pheromone binding and release. Proc. Natl. Acad. Sci. USA 2005, 102, 5386–5391. [Google Scholar] [CrossRef]
- Zhou, J.J.; Robertson, G.; He, X.; Dufour, S.; Hooper, A.M.; Pickett, J.A.; Keep, N.H.; Field, L.M. Characterisation of Bombyx mori Odorant-binding Proteins Reveals that a General Odorant-binding Protein Discriminates between Sex Pheromone Components. J. Mol. Biol. 2009, 389, 529–545. [Google Scholar] [CrossRef]
- Sun, X.; Zeng, F.F.; Yan, M.J.; Zhang, A.; Lu, Z.X.; Wang, M.Q. Interactions of two odorant-binding proteins influence insect chemoreception. Insect Mol. Biol. 2016, 25, 712–723. [Google Scholar] [CrossRef]
- Li, Z.Q.; Zhang, S.; Cai, X.M.; Luo, J.Y.; Dong, S.L.; Cui, J.J.; Chen, Z.M. Three odorant binding proteins may regulate the behavioural response of Chrysopa pallens to plant volatiles and the aphid alarm pheromone (E)-β-farnesene. Insect Mol. Biol. 2017, 26, 255–265. [Google Scholar] [CrossRef]
- Clyne, P.J.; Warr, C.G.; Freeman, M.R.; Lessing, D.; Kim, J.; Carlson, J.R.; Haven, N. A Novel Family of Divergent Seven-Transmembrane Proteins: Candidate Odorant Receptors in Drosophila. Neuron 1999, 22, 327–338. [Google Scholar] [CrossRef]
- Vosshall, L.B.; Wong, A.M.; Axel, R. An Olfactory Sensory Map in the Fly Brain. Cell 2000, 102, 147–159. [Google Scholar] [CrossRef]
- Visser, J.H.; Piron, P.G.M.; Hardie, J. The aphids’ peripheral perception of plant volatiles. Exp. Appl. 1996, 80, 35–38. [Google Scholar] [CrossRef]
- Webster, B.; Bruce, T.; Dufour, S.; Birkemeyer, C.; Birkett, M.; Hardie, J.; Pickett, J. Identification of volatile compounds used in host location by the black bean aphid, Aphis fabae. J. Chem. Ecol. 2008, 34, 1153–1161. [Google Scholar] [CrossRef]
- Gu, S.; Wang, W.; Wang, G.-R.; Zhang, X.-Y.; Guo, Y.-Y.; Zhang, Z.; Zhou, J.-J.; Zhang, Y.-J. Functional characterization and immunolocalization of odorant binding protein 1 in the lucerne plant bug, Adelphocoris lineolatus (Goeze). Arch. Insect Biochem. Physiol. 2011, 77, 81–98. [Google Scholar] [CrossRef] [PubMed]
- Harada, E.; Nakagawa, J.; Asano, T.; Taoka, M.; Sorimachi, H.; Ito, Y. Functional Evolution of Duplicated Odorant-Binding Protein Genes, Obp57d and Obp57e, in Drosophila. PLoS ONE 2012, 7, e29710. [Google Scholar] [CrossRef]
- Pesenti, M.E.; Spinelli, S.; Bezirard, V.; Briand, L.; Pernollet, J.; Campanacci, V.; Tegoni, M.; Cambillau, C. Queen Bee Pheromone Binding Protein pH-Induced Domain Swapping Favors Pheromone Release. J. Mol. Biol. 2009, 390, 981–990. [Google Scholar] [CrossRef]
- Pesenti, M.E.; Spinelli, S.; Bezirard, V.; Briand, L.; Pernollet, J.; Tegoni, M.; Cambillau, C. Structural Basis of the Honey Bee PBP Pheromone and pH-induced Conformational Change. J. Mol. Biol. 2008, 380, 158–169. [Google Scholar] [CrossRef]
- Li, N.; Sun, X.; Wang, M. Expression pattern and ligand-binding properties of odorant- binding protein 13 from Monochamus alternatus hope. J. Appl. Entomol. 2017, 141, 751–757. [Google Scholar] [CrossRef]
- Sandler, B.H.; Nikonova, L.; Leal, W.S.; Clardy, J. Sexual attraction in the silkworm moth: Structure of the pheromone-binding-protein—bombykol complex. Chem. Biol. 2000, 7, 143–151. [Google Scholar] [CrossRef]
- Leite, N.R.; Krogh, R.; Xu, W.; Ishida, Y.; Iulek, J.; Leal, W.S.; Oliva, G. Structure of an Odorant-Binding Protein from the Mosquito Aedes aegypti Suggests a Binding Pocket Covered by a pH-Sensitive “Lid”. PLoS ONE 2009, 4, e8006. [Google Scholar] [CrossRef] [PubMed]
- Michel, E.; Damberger, F.F.; Ishida, Y.; Fiorito, F.; Lee, D.; Leal, W.S.; Wüthrich, K. Dynamic Conformational Equilibria in the Physiological Function of the Bombyx mori Pheromone-Binding Protein. J. Mol. Biol. 2011, 408, 922–931. [Google Scholar] [CrossRef]
- Farag, M.A.; Paré, P.W. C6-green leaf volatiles trigger local and systemic VOC emissions in tomato. Phytochemistry 2002, 61, 545–554. [Google Scholar] [CrossRef]
- Tholl, D.; Boland, W.; Hansel, A.; Loreto, F.; Röse, U.S.R.; Schnitzler, J.P. Practical approaches to plant volatile analysis. Plant J. 2006, 45, 540–560. [Google Scholar] [CrossRef]
- Francis, F.; Vandermoten, S.; Verheggen, F.; Lognay, G.; Haubruge, E. Is the (E)-β-farnesene only volatile terpenoid in aphids? J. Appl. Entomol. 2005, 129, 6–11. [Google Scholar] [CrossRef]
- Das, A.; Lee, S.H.; Hyun, T.K.; Kim, S.W.; Kim, J.Y. Plant volatiles as method of communication. Plant Biotechnol. Rep. 2013, 7, 9–26. [Google Scholar] [CrossRef]
- Waris, M.I.; Younas, A.; ul Qamar, M.T.; Hao, L.; Ameen, A.; Ali, S.; Abdelnabby, H.E.; Zeng, F.F.; Wang, M.Q. Silencing of chemosensory protein gene NlugCSP8 by RNAi induces declining behavioral responses of Nilaparvata lugens. Front. Physiol. 2018, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Wang, B.; Grossi, G.; Falabella, P.; Liu, Y.; Yan, S.; Lu, J.; Xi, J.; Wang, G. Molecular Basis of Alarm Pheromone Detection in Aphids. Curr. Biol. 2017, 27, 55–61. [Google Scholar] [CrossRef]
- Zhang, Y.; Shen, C.; Xia, D.; Wang, J.; Tang, Q. Characterization of the Expression and Functions of Two Odorant-Binding Proteins of Sitophilus zeamais Motschulsky (Coleoptera: Curculionoidea). Insects 2019, 10, 409. [Google Scholar] [CrossRef]
- Li, Q.L.; Yi, S.C.; Li, D.Z.; Nie, X.P.; Li, S.Q.; Wang, M.; Zhou, A.M. Optimization of reverse chemical ecology method: False positive binding of Aenasius bambawalei odorant binding protein 1 caused by uncertain binding mechanism. Insect Mol. Biol. 2018, 27, 305–318. [Google Scholar] [CrossRef]
- Yang, R.; Li, D.; Yu, G.; Yi, S.; Zhang, Y. Structural Transformation Detection Contributes to Screening of Behaviorally Active Compounds: Dynamic Binding Process Analysis of DhelOBP21 from Dastarcus helophoroides. J. Chem. Ecol. 2017, 43, 1033–1045. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Bao, W.; Wuriyanghan, H. Silencing of Target Chitinase Genes via Oral Delivery of dsRNA Caused Lethal Phenotypic Effects in Mythimna separata (Lepidoptera: Noctuidae). Appl. Biochem. Biotechnol. 2017, 181, 860–866. [Google Scholar] [CrossRef]
- Deng, F.; Zhao, Z. Influence of catalase gene silencing on the survivability of Sitobion avenae. Arch. Insect Biochem. Physiol. 2014, 86, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Dallakyan, S.; Olson, A.J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015, 1263, 243–250. [Google Scholar] [CrossRef] [PubMed]

| Ligands | CAS No. | Purity % | pH 5.0 | pH 7.4 | ||
|---|---|---|---|---|---|---|
| IC50 (μM) | Ki (μM) | IC50 (μM) | Ki (μM) | |||
| Compounds from the Wheat Plants Identified by GC-MS | ||||||
| 2-Ethyl-1-hexanol | 104-76-7 | 99 | 43.85 | 39.61 | 11.18 | 9.93 |
| Nonanal | 124-19-6 | 95 | 12.32 | 11.13 | 39.85 | 35.41 |
| Naphthalene | 91-20-3 | 98 | 30.34 | 27.41 | 12.97 | 11.53 |
| Decanal | 112-31-2 | 97 | 19.77 | 17.86 | 9.95 | 8.84 |
| Tetradecane | 629-59-4 | 98 | 11.87 | 10.73 | 7.96 | 7.07 |
| Pentadecane | 629-62-9 | 97 | 15.12 | 13.66 | 12.82 | 11.39 |
| Butylated Hydroxytoluene | 128-37-0 | 100 | 8.80 | 7.95 | 45.58 | 40.49 |
| Hexadecane | 544-76-3 | 98 | 14.64 | 13.22 | 10.90 | 9.68 |
| General Odorants and Phenylpropanoids | ||||||
| Acetophenone | 98-86-2 | 99 | 30.75 | 27.78 | 12.98 | 11.53 |
| Octanal | 124-13-0 | 99 | 20.28 | 18.32 | 8.79 | 7.80 |
| 3-Butenyl isothiocyanate | 3386-97-8 | 99 | 26.32 | 23.78 | 52.70 | 46.82 |
| Methyl salicylate | 119-36-8 | 98 | 24.20 | 21.86 | 13.42 | 11.92 |
| Benzaldehyde | 100-52-7 | 99 | 40.99 | 37.03 | 108.63 | 96.51 |
| Terpenoids | ||||||
| α-Pinene | 7785-70-8 | 99 | 27.19 | 24.56 | 37.97 | 33.73 |
| β -Pinene | 18172-67-3 | 99 | 27.73 | 25.05 | 37.42 | 33.25 |
| β-Myrcene, | 123-35-3 | 97 | 10.70 | 9.67 | 12.19 | 10.83 |
| (+)-Limonene oxide | 203719-54-4 | 97 | 37.47 | 33.85 | 24.31 | 21.60 |
| α-Farnesene | 502-61-4 | 98 | 9.51 | 8.59 | 10.26 | 9.11 |
| S-(−)-Limonene | 5989-54-8 | 99 | 11.52 | 10.41 | 32.10 | 28.52 |
| Linalool | 78-70-6 | 97 | 11.78 | 10.65 | 24.10 | 21.41 |
| β-Caryophyllene | 87-44-5 | 99 | 622.0954 | 561.97 | 52.43 | 46.58 |
| (+)-3-Carene | 13466-78-9 | 99 | 39.94 | 36.08 | 14.67 | 13.04 |
| (±)-Camphor | 76-22-2 | 95 | 17.37 | 15.70 | 48.17 | 42.79 |
| Green Leaf Volatiles and Alcohols | ||||||
| trans-2-Hexen-1-al | 6728-26-3 | 97 | 21.56 | 19.48 | 41.87 | 37.20 |
| cis-3-Hexenyl acetate | 3634-71-8 | 97 | - | - | 29.73 | 26.41 |
| cis-3-Hexen-1-ol | 928-96-1 | 90 | 36.88 | 33.32 | 13.22 | 11.75 |
| Hexanal | 66-25-1 | 98 | 16.81 | 15.18 | 8.23 | 7.31 |
| α-Terpineol | 10482-56-1 | 99 | 37.38 | 33.77 | 26.19 | 23.26 |
| trans-2-Hexen-1-ol | 928-95-0 | 96 | 21.69 | 19.60 | 14.48 | 12.86 |
| 1-Octen-3-ol | 3391-86-4 | 98 | 34.23 | 30.92 | 24.24 | 21.53 |
| PubChem IDs | Ligands | S-Score | Residues Interacting with H-Bonding | Covalent Bonds (Pi Alkyls and Sigma Alkayls) | Van der Waals Interactions |
|---|---|---|---|---|---|
| 629-59-4 | Tetradecane | −4.7 | Lys38, Leu15, Phe117, Ile114, Ala113 | ||
| 66-25-1 | Hexanal | −5.6 | Ile81, Ile41, Ala116 | Gln43, Ile114,Glu110, Phe117,Ala113,Met53,Tyr77 | |
| 124-13-0 | Octanal | −4.8 | Ile114, Phe117,Ala113, | Tyr77,Ala116,Ile81,Ile41, Met53, Gln43, Glu110 | |
| 112-31-2 | Decanal | −5.4 | Lys38, Tyr77, Lys 80 | Ile41, Phe117,Ala113 | |
| 502-61-4 | α-farnesene | −5.7 | Met12,Lys80,Leu15,Leu11,Tyr77, Lys38,Ile41,Ala113, Phe117 | Mer53, Ala116, Thr42 | |
| 544-76-3 | Hexadecane | −3.9 | Phe16, Tyr39, Ile35 | ||
| 104-76-7 | 2-ethyl-1-hexanal | −4.6 | Thr42 | Ile81,Ile41,Ala116 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ullah, R.M.K.; Quershi, S.R.; Adeel, M.M.; Abdelnabby, H.; Waris, M.I.; Duan, S.-G.; Wang, M.-Q. An Odorant Binding Protein (SaveOBP9) Involved in Chemoreception of the Wheat Aphid Sitobion avenae. Int. J. Mol. Sci. 2020, 21, 8331. https://doi.org/10.3390/ijms21218331
Ullah RMK, Quershi SR, Adeel MM, Abdelnabby H, Waris MI, Duan S-G, Wang M-Q. An Odorant Binding Protein (SaveOBP9) Involved in Chemoreception of the Wheat Aphid Sitobion avenae. International Journal of Molecular Sciences. 2020; 21(21):8331. https://doi.org/10.3390/ijms21218331
Chicago/Turabian StyleUllah, Rana Muhammad Kaleem, Sundas Rana Quershi, Muhammad Muzammal Adeel, Hazem Abdelnabby, Muhammad Irfan Waris, Shuang-Gang Duan, and Man-Qun Wang. 2020. "An Odorant Binding Protein (SaveOBP9) Involved in Chemoreception of the Wheat Aphid Sitobion avenae" International Journal of Molecular Sciences 21, no. 21: 8331. https://doi.org/10.3390/ijms21218331
APA StyleUllah, R. M. K., Quershi, S. R., Adeel, M. M., Abdelnabby, H., Waris, M. I., Duan, S.-G., & Wang, M.-Q. (2020). An Odorant Binding Protein (SaveOBP9) Involved in Chemoreception of the Wheat Aphid Sitobion avenae. International Journal of Molecular Sciences, 21(21), 8331. https://doi.org/10.3390/ijms21218331







