Substitution of the Native Zn(II) with Cd(II), Co(II) and Ni(II) Changes the Downhill Unfolding Mechanism of Ros87 to a Completely Different Scenario
Abstract
:1. Introduction
2. Results
2.1. Co(II)—Ros87 Thermal Unfolding
2.2. Ni(II)-Ros87 Thermal Unfolding
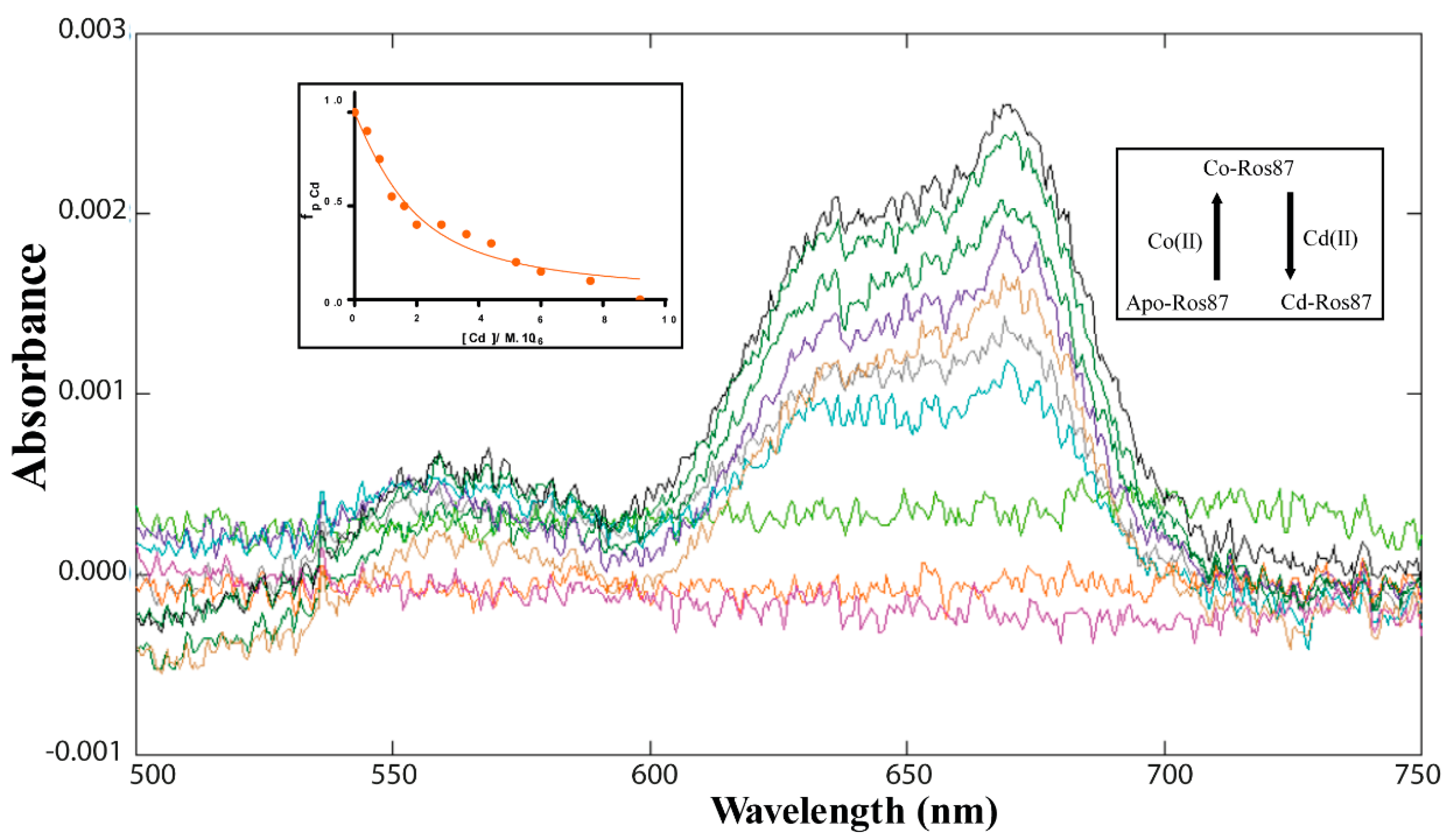
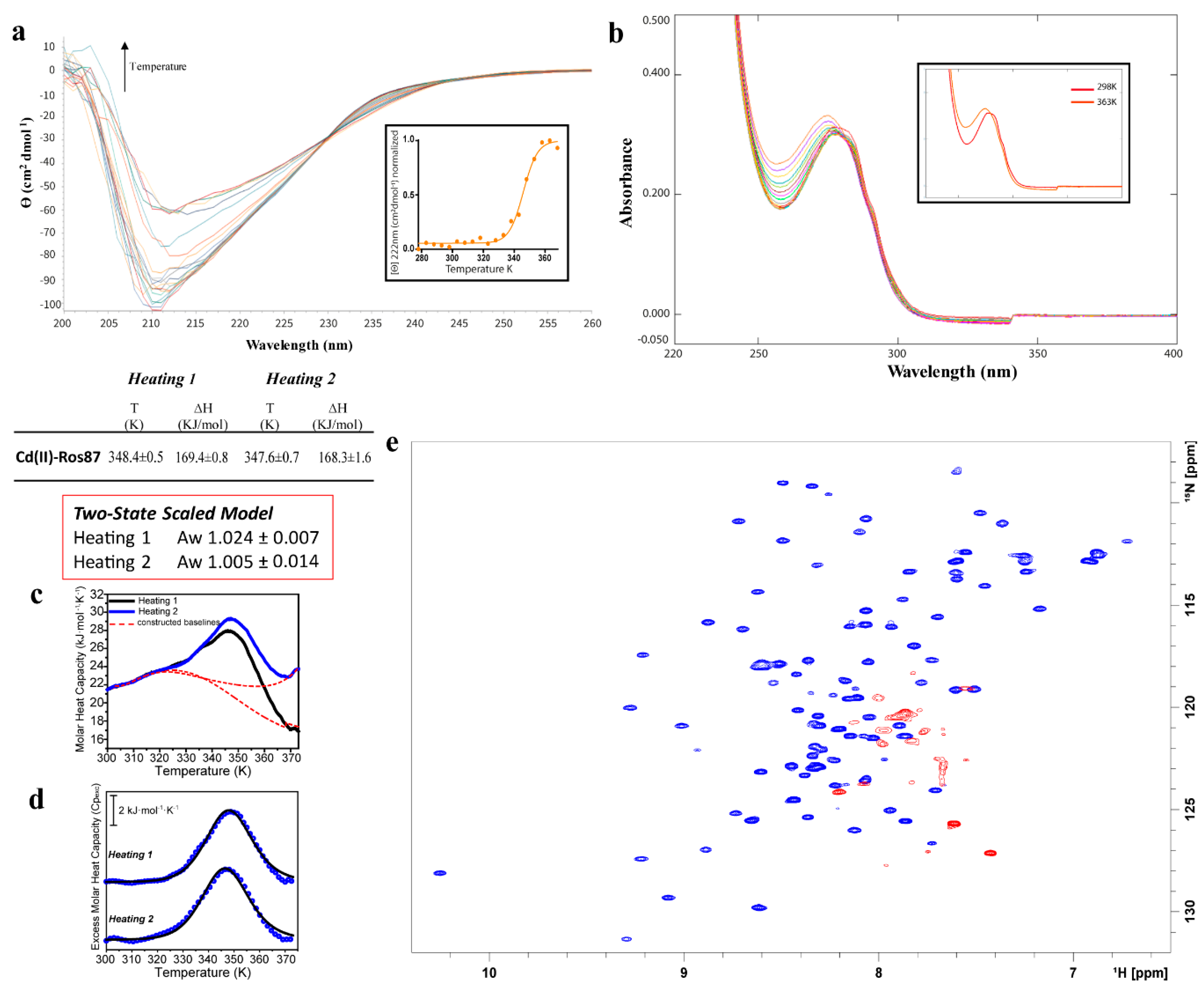
2.3. Cd(II)—Ros87 Thermal Unfolding
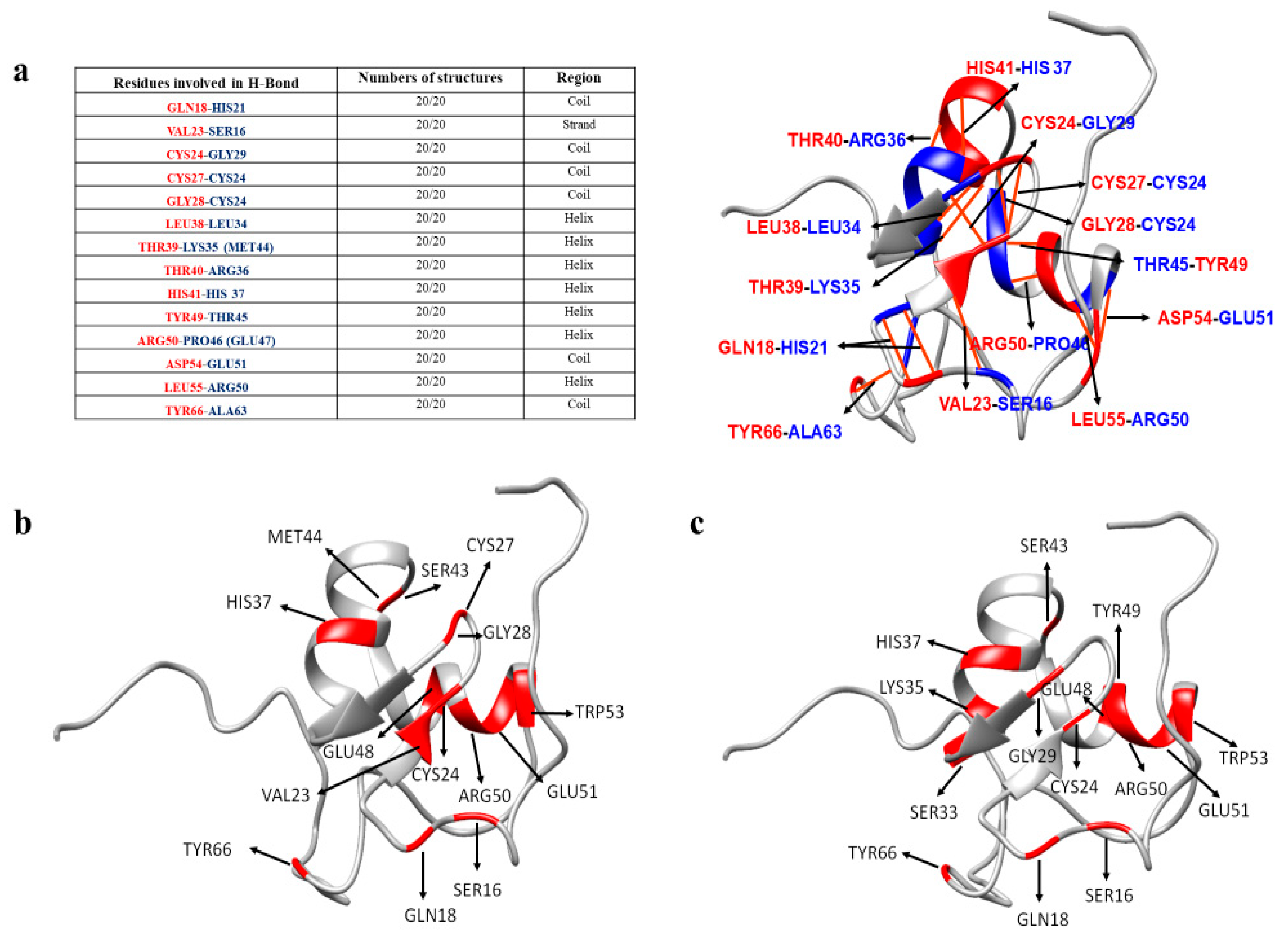
3. Discussion
4. Experimental Section
4.1. Protein Expression and Purification
4.2. UV-Vis Spectroscopy
4.3. CD Spectroscopy
4.4. NMR Spectroscopy
4.5. Differential Scanning Calorimetry (DSC)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wilson, C.J.; Apiyo, D.; Wittung-Stafshede, P. Role of cofactors in metalloprotein folding. Q. Rev. Biophys. 2004, 37, 285–314. [Google Scholar] [CrossRef]
- Kluska, K.; Adamczyk, J.; Krężel, A. Metal binding properties, stability and reactivity of zinc fingers. Coord. Chem. Rev. 2018, 367, 18–64. [Google Scholar] [CrossRef]
- Green, L.M.; Berg, J.M. Retroviral nucleocapsid protein-metal ion interactions: Folding and sequence variants. Proc. Natl. Acad. Sci. USA 1990, 87, 6403–6407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coyne, H.J.; Ciofi-Baffoni, S.; Banci, L.; Bertini, I.; Zhang, L.; George, G.N.; Winge, D.R. The characterization and role of zinc binding in yeast Cox4. J. Biol. Chem. 2007, 282, 8926–8934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malgieri, G.; Grasso, G. The clearance of misfolded proteins in neurodegenerative diseases by zinc metalloproteases: An inorganic perspective. Coord. Chem. Rev. 2014, 260, 139–155. [Google Scholar] [CrossRef]
- Wittung-Stafshede, P. Role of cofactors in protein folding. Acc. Chem. Res. 2002, 35, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, J.; Wang, J.; Wang, W. Metal-coupled folding of Cys2His2 zinc-finger. J. Am. Chem. Soc. 2008, 130, 892–900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeguchi, M.; Kuwajima, K.; Sugai, S. Ca2+-induced alteration in the unfolding behavior of alpha-lactalbumin. J. Biochem. 1986, 99, 1191–1201. [Google Scholar] [CrossRef]
- Sadqi, M.; Fushman, D.; Muñoz, V. Atom-by-atom analysis of global downhill protein folding. Nature 2006, 442, 317–321. [Google Scholar] [CrossRef]
- Palmieri, M.; Malgieri, G.; Russo, L.; Baglivo, I.; Esposito, S.; Netti, F.; Del Gatto, A.; de Paola, I.; Zaccaro, L.; Pedone, P.V.; et al. Structural Zn(II) implies a switch from fully cooperative to partly downhill folding in highly homologous proteins. J. Am. Chem. Soc. 2013, 135, 5220–5228. [Google Scholar] [CrossRef]
- Garcia-Mira, M.M.; Sadqi, M.; Fischer, N.; Sanchez-Ruiz, J.M.; Muñoz, V. Experimental identification of downhill protein folding. Science 2002, 298, 2191–2195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz, V. Thermodynamics and kinetics of downhill protein folding investigated with a simple statistical mechanical model. Int. J. Quantum Chem. 2002, 90, 1522–1528. [Google Scholar] [CrossRef]
- Sanchez-Ruiz, J.M. Probing free-energy surfaces with differential scanning calorimetry. Annu. Rev. Phys. Chem. 2011, 62, 231–255. [Google Scholar] [CrossRef]
- Farber, P.; Darmawan, H.; Sprules, T.; Mittermaier, A. Analyzing protein folding cooperativity by differential scanning calorimetry and NMR spectroscopy. J. Am. Chem. Soc. 2010, 132, 6214–6222. [Google Scholar] [CrossRef]
- Chou, A.Y.; Archdeacon, J.; Kado, C.I. Agrobacterium transcriptional regulator Ros is a prokaryotic zinc finger protein that regulates the plant oncogene ipt. Proc. Natl. Acad. Sci. USA 1998, 95, 5293–5298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esposito, S.; Baglivo, I.; Malgieri, G.; Russo, L.; Zaccaro, L.; D’Andrea, L.D.; Mammucari, M.; Di Blasio, B.; Isernia, C.; Fattorusso, R.; et al. A novel type of zinc finger DNA binding domain in the Agrobacterium tumefaciens transcriptional regulator Ros. Biochemistry 2006, 45, 10394–10405. [Google Scholar] [CrossRef]
- Malgieri, G.; Palmieri, M.; Russo, L.; Fattorusso, R.; Pedone, P.V.; Isernia, C. The prokaryotic zinc-finger: Structure, function and comparison with the eukaryotic counterpart. FEBS J. 2015, 282, 4480–4496. [Google Scholar] [CrossRef] [Green Version]
- Pirone, L.; Pitzer, J.E.; D’Abrosca, G.; Fattorusso, R.; Malgieri, G.; Pedone, E.M.; Pedone, P.V.; Roop, R.M.; Baglivo, I. Identifying the region responsible for Brucella abortus MucR higher-order oligomer formation and examining its role in gene regulation. Sci. Rep. 2018, 8, 17238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baglivo, I.; Russo, L.; Esposito, S.; Malgieri, G.; Renda, M.; Salluzzo, A.; Di Blasio, B.; Isernia, C.; Fattorusso, R.; Pedone, P.V. The structural role of the zinc ion can be dispensable in prokaryotic zinc-finger domains. Proc. Natl. Acad. Sci. USA 2009, 106, 6933–6938. [Google Scholar] [CrossRef] [Green Version]
- Isernia, C.; Malgieri, G.; Russo, L.; D’Abrosca, G.; Baglivo, I.; Pedone, P.V.; Fattorusso, R. Zinc Fingers. Met. Ions. Life Sci. 2020, 20. [Google Scholar] [CrossRef]
- Malgieri, G.; Russo, L.; Esposito, S.; Baglivo, I.; Zaccaro, L.; Pedone, E.M.; Di Blasio, B.; Isernia, C.; Pedone, P.V.; Fattorusso, R. The prokaryotic Cys2His2 zinc-finger adopts a novel fold as revealed by the NMR structure of Agrobacterium tumefaciens Ros DNA-binding domain. Proc. Natl. Acad. Sci. USA 2007, 104, 17341–17346. [Google Scholar] [CrossRef] [Green Version]
- D’Abrosca, G.; Russo, L.; Palmieri, M.; Baglivo, I.; Netti, F.; de Paola, I.; Zaccaro, L.; Farina, B.; Iacovino, R.; Pedone, P.V.; et al. The (unusual) aspartic acid in the metal coordination sphere of the prokaryotic zinc finger domain. J. Inorg. Biochem. 2016, 161, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, M.; Russo, L.; Malgieri, G.; Esposito, S.; Baglivo, I.; Rivellino, A.; Farina, B.; de Paola, I.; Zaccaro, L.; Milardi, D.; et al. Deciphering the zinc coordination properties of the prokaryotic zinc finger domain: The solution structure characterization of Ros87 H42A functional mutant. J. Inorg. Biochem. 2014, 131, 30–36. [Google Scholar] [CrossRef] [PubMed]
- D’Abrosca, G.; Paladino, A.; Baglivo, I.; Russo, L.; Sassano, M.; Grazioso, R.; Iacovino, R.; Pirone, L.; Pedone, E.M.; Pedone, P.V.; et al. Structural Insight of the Full-Length Ros Protein: A Prototype of the Prokaryotic Zinc-Finger Family. Sci. Rep. 2020, 10, 9283. [Google Scholar] [CrossRef]
- Waldron, K.J.; Robinson, N.J. How do bacterial cells ensure that metalloproteins get the correct metal? Nat. Rev. Microbiol. 2009, 7, 25–35. [Google Scholar] [CrossRef]
- Hartwig, A. Zinc finger proteins as potential targets for toxic metal ions: Differential effects on structure and function. Antioxid Redox Signal 2001, 3, 625–634. [Google Scholar] [CrossRef]
- Malgieri, G.; Zaccaro, L.; Leone, M.; Bucci, E.; Esposito, S.; Baglivo, I.; Del Gatto, A.; Russo, L.; Scandurra, R.; Pedone, P.V.; et al. Zinc to cadmium replacement in the A. thaliana SUPERMAN Cys₂ His₂ zinc finger induces structural rearrangements of typical DNA base determinant positions. Biopolymers 2011, 95, 801–810. [Google Scholar] [CrossRef]
- Jan, A.T.; Azam, M.; Siddiqui, K.; Ali, A.; Choi, I.; Haq, Q.M. Heavy Metals and Human Health: Mechanistic Insight into Toxicity and Counter Defense System of Antioxidants. Int. J. Mol. Sci. 2015, 16, 29592–29630. [Google Scholar] [CrossRef] [Green Version]
- Malgieri, G.; Palmieri, M.; Esposito, S.; Maione, V.; Russo, L.; Baglivo, I.; de Paola, I.; Milardi, D.; Diana, D.; Zaccaro, L.; et al. Zinc to cadmium replacement in the prokaryotic zinc-finger domain. Metallomics 2014, 6, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Sivo, V.; D’Abrosca, G.; Russo, L.; Iacovino, R.; Pedone, P.V.; Fattorusso, R.; Isernia, C.; Malgieri, G. Co(II) Coordination in Prokaryotic Zinc Finger Domains as Revealed by UV-Vis Spectroscopy. Bioinorg. Chem. Appl. 2017, 2017, 1527247. [Google Scholar] [CrossRef] [Green Version]
- Sivo, V.; D’Abrosca, G.; Baglivo, I.; Iacovino, R.; Pedone, P.V.; Fattorusso, R.; Russo, L.; Malgieri, G.; Isernia, C. Ni(II), Hg(II), and Pb(II) Coordination in the Prokaryotic Zinc-Finger Ros87. Inorg. Chem. 2019, 58, 1067–1080. [Google Scholar] [CrossRef] [PubMed]
- Shishido, H.; Yoon, J.S.; Yang, Z.; Skach, W.R. CFTR trafficking mutations disrupt cotranslational protein folding by targeting biosynthetic intermediates. Nat. Commun. 2020, 11, 4258. [Google Scholar] [CrossRef]
- Shi, Y.; Beger, R.D.; Berg, J.M. Metal binding properties of single amino acid deletion mutants of zinc finger peptides: Studies using cobalt(II) as a spectroscopic probe. Biophys. J. 1993, 64, 749–753. [Google Scholar] [CrossRef] [Green Version]
- La Rosa, C.; Milardi, D.; Grasso, D.; Guzzi, R.; Sportelli, L. Thermodynamics of the thermal unfolding of azurin. J. Phys. Chem. 1995, 99, 14864–14870. [Google Scholar] [CrossRef]
- Kochańczyk, T.; Nowakowski, M.; Wojewska, D.; Kocyła, A.; Ejchart, A.; Koźmiński, W.; Krężel, A. Metal-coupled folding as the driving force for the extreme stability of Rad50 zinc hook dimer assembly. Sci. Rep. 2016, 6, 36346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kornhaber, G.J.; Snyder, D.; Moseley, H.N.; Montelione, G.T. Identification of zinc-ligated cysteine residues based on 13Calpha and 13Cbeta chemical shift data. J. Biomol. Nmr. 2006, 34, 259–269. [Google Scholar] [CrossRef]
- Palm-Espling, M.E.; Niemiec, M.S.; Wittung-Stafshede, P. Role of metal in folding and stability of copper proteins in vitro. Biochim. Biophys. Acta 2012, 1823, 1594–1603. [Google Scholar] [CrossRef] [Green Version]
- Arai, M. Unified understanding of folding and binding mechanisms of globular and intrinsically disordered proteins. Biophys. Rev. 2018, 10, 163–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Netti, F.; Malgieri, G.; Esposito, S.; Palmieri, M.; Baglivo, I.; Isernia, C.; Omichinski, J.G.; Pedone, P.V.; Lartillot, N.; Fattorusso, R. An experimentally tested scenario for the structural evolution of eukaryotic Cys2His2 zinc fingers from eubacterial ros homologs. Mol. Biol. Evol. 2013, 30, 1504–1513. [Google Scholar] [CrossRef] [Green Version]
- Malgieri, G.; D’Abrosca, G.; Pirone, L.; Toto, A.; Palmieri, M.; Russo, L.; Sciacca, M.F.M.; Tatè, R.; Sivo, V.; Baglivo, I.; et al. Folding mechanisms steer the amyloid fibril formation propensity of highly homologous proteins. Chem. Sci. 2018, 9, 3290–3298. [Google Scholar] [CrossRef] [Green Version]
- Baglivo, I.; Palmieri, M.; Rivellino, A.; Netti, F.; Russo, L.; Esposito, S.; Iacovino, R.; Farina, B.; Isernia, C.; Fattorusso, R.; et al. Molecular strategies to replace the structural metal site in the prokaryotic zinc finger domain. Biochim. Biophys. Acta 2014, 1844, 497–504. [Google Scholar] [CrossRef]
- Vassall, K.A.; Stubbs, H.R.; Primmer, H.A.; Tong, M.S.; Sullivan, S.M.; Sobering, R.; Srinivasan, S.; Briere, L.A.; Dunn, S.D.; Colón, W.; et al. Decreased stability and increased formation of soluble aggregates by immature superoxide dismutase do not account for disease severity in ALS. Proc. Natl. Acad. Sci. USA 2011, 108, 2210–2215. [Google Scholar] [CrossRef] [Green Version]
- Isernia, C.; Bucci, E.; Leone, M.; Zaccaro, L.; Di Lello, P.; Digilio, G.; Esposito, S.; Saviano, M.; Di Blasio, B.; Pedone, C.; et al. NMR structure of the single QALGGH zinc finger domain from the Arabidopsis thaliana SUPERMAN protein. Chembiochem 2003, 4, 171–180. [Google Scholar] [CrossRef]
- Muhandiram, D.R.; Kay, L.E. Gradient-Enhanced Triple-Resonance Three-Dimensional NMR Experiments with Improved Sensitivity. J. Magn. Reson. Ser. B 1994, 103, 203–216. [Google Scholar] [CrossRef]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. Nmr. 1995, 6, 277–293. [Google Scholar] [CrossRef]
- Keller, R. The Computer Aided Resonance Assignment; Cantina: Goldau, Switzerland, 2004. [Google Scholar]
- Lee, W.; Tonelli, M.; Markley, J.L. NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 2015, 31, 1325–1327. [Google Scholar] [CrossRef] [Green Version]
- Grasso, D.; La Rosa, C.; Milardi, D.; Fasone, S. The effects of scan rate and protein concentration on DSC thermograms of bovine superoxide dismutase. Thermochim. Acta 1995, 265, 163–175. [Google Scholar] [CrossRef]
- Sharma, S.K.; Goloubinoff, P.; Christen, P. Heavy metal ions are potent inhibitors of protein folding. Biochem. Biophys. Res. Commun. 2008, 372, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, D.; Rancy, P.C.; Nagarkar, R.P.; Schneider, J.P.; Thorpe, C. Arsenic(III) species inhibit oxidative protein folding in vitro. Biochemistry 2009, 48, 424–432. [Google Scholar] [CrossRef] [Green Version]
- Jacobson, T.; Navarrete, C.; Sharma, S.K.; Sideri, T.C.; Ibstedt, S.; Priya, S.; Grant, C.M.; Christen, P.; Goloubinoff, P.; Tamás, M.J. Arsenite interferes with protein folding and triggers formation of protein aggregates in yeast. J. Cell Sci. 2012, 125, 5073–5083. [Google Scholar] [CrossRef] [Green Version]
- Tamás, M.J.; Sharma, S.K.; Ibstedt, S.; Jacobson, T.; Christen, P. Heavy metals and metalloids as a cause for protein misfolding and aggregation. Biomolecules 2014, 4, 252–267. [Google Scholar] [CrossRef] [Green Version]
- Diana, D.; Ziaco, B.; Scarabelli, G.; Pedone, C.; Colombo, G.; D’Andrea, L.D.; Fattorusso, R. Structural analysis of a helical peptide unfolding pathway. Chemistry 2010, 16, 5400–5407. [Google Scholar] [CrossRef] [PubMed]
- Gaeta, A.; Hider, R.C. The crucial role of metal ions in neurodegeneration: The basis for a promising therapeutic strategy. Br. J. Pharm. 2005, 146, 1041–1059. [Google Scholar] [CrossRef] [Green Version]
- Travaglia, A.; Arena, G.; Fattorusso, R.; Isernia, C.; La Mendola, D.; Malgieri, G.; Nicoletti, V.G.; Rizzarelli, E. The inorganic perspective of nerve growth factor: Interactions of Cu2+ and Zn2+ with the N-terminus fragment of nerve growth factor encompassing the recognition domain of the TrkA receptor. Chemistry 2011, 17, 3726–3738. [Google Scholar] [CrossRef]
- Travaglia, A.; La Mendola, D.; Magrì, A.; Pietropaolo, A.; Nicoletti, V.G.; Grasso, G.; Malgieri, G.; Fattorusso, R.; Isernia, C.; Rizzarelli, E. Zinc(II) interactions with brain-derived neurotrophic factor N-terminal peptide fragments: Inorganic features and biological perspectives. Inorg. Chem. 2013, 52, 11075–11083. [Google Scholar] [CrossRef] [PubMed]
- Quintanar, L.; Lim, M.H. Metal ions and degenerative diseases. J. Biol. Inorg. Chem. 2019, 24, 1137–1139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witkiewicz-Kucharczyk, A.; Bal, W. Damage of zinc fingers in DNA repair proteins, a novel molecular mechanism in carcinogenesis. Toxicol. Lett. 2006, 162, 29–42. [Google Scholar] [CrossRef]
- Posewitz, M.C.; Wilcox, D.E. Properties of the Sp1 zinc finger 3 peptide: Coordination chemistry, redox reactions, and metal binding competition with metallothionein. Chem. Res. Toxicol. 1995, 8, 1020–1028. [Google Scholar] [CrossRef]
- Miłoch, A.; Krężel, A. Metal binding properties of the zinc finger metallome--insights into variations in stability. Metallomics 2014, 6, 2015–2024. [Google Scholar] [CrossRef]
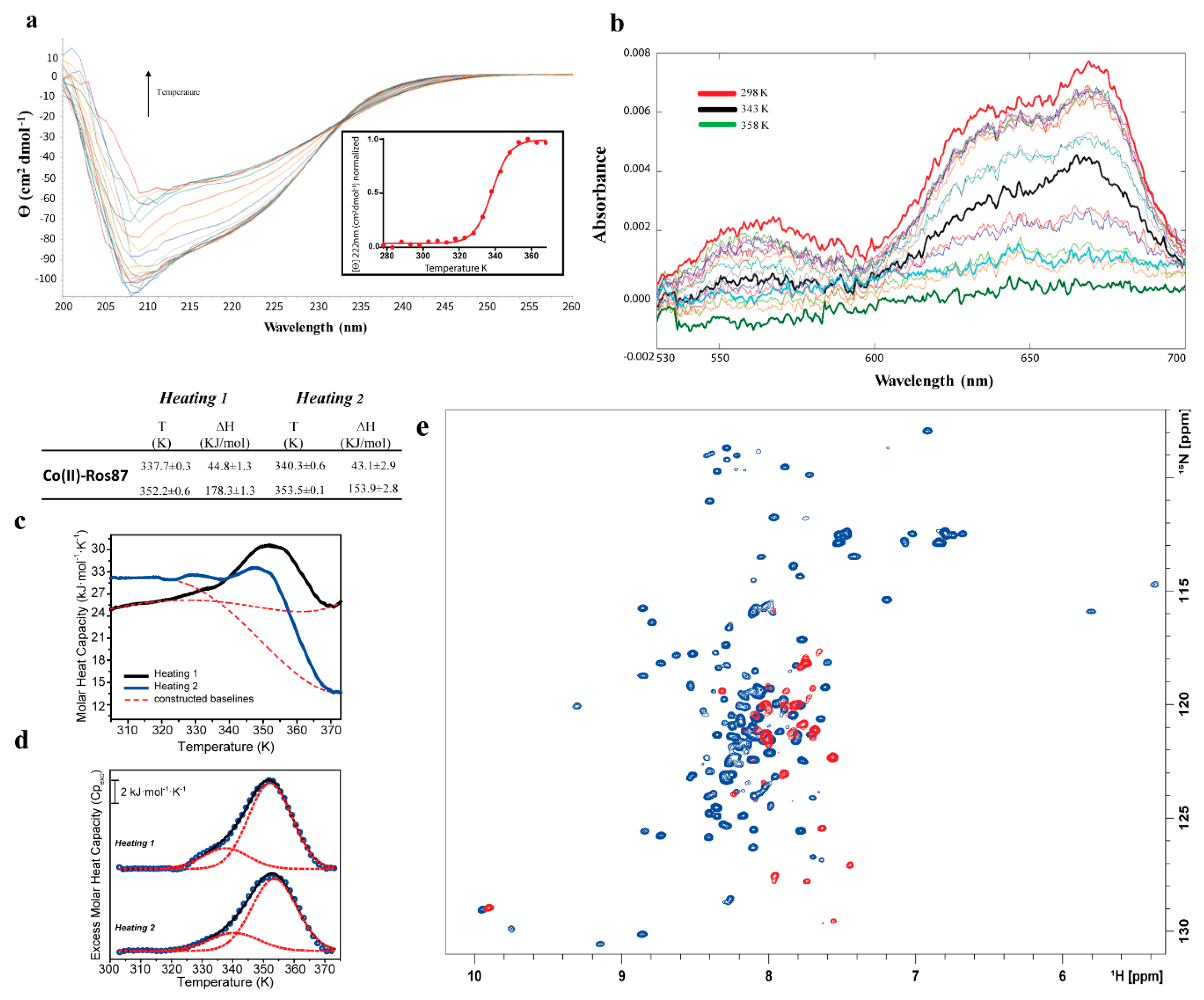
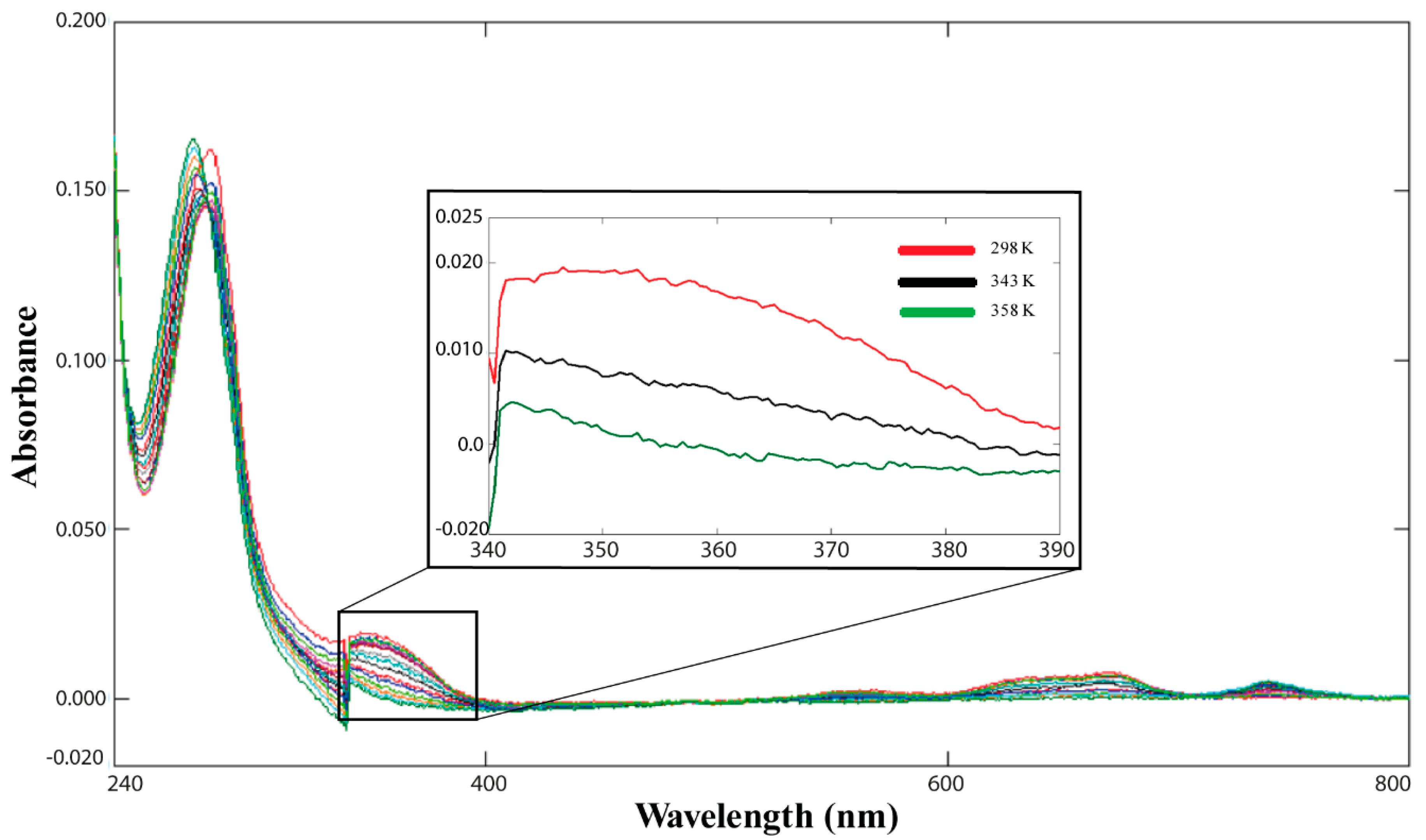
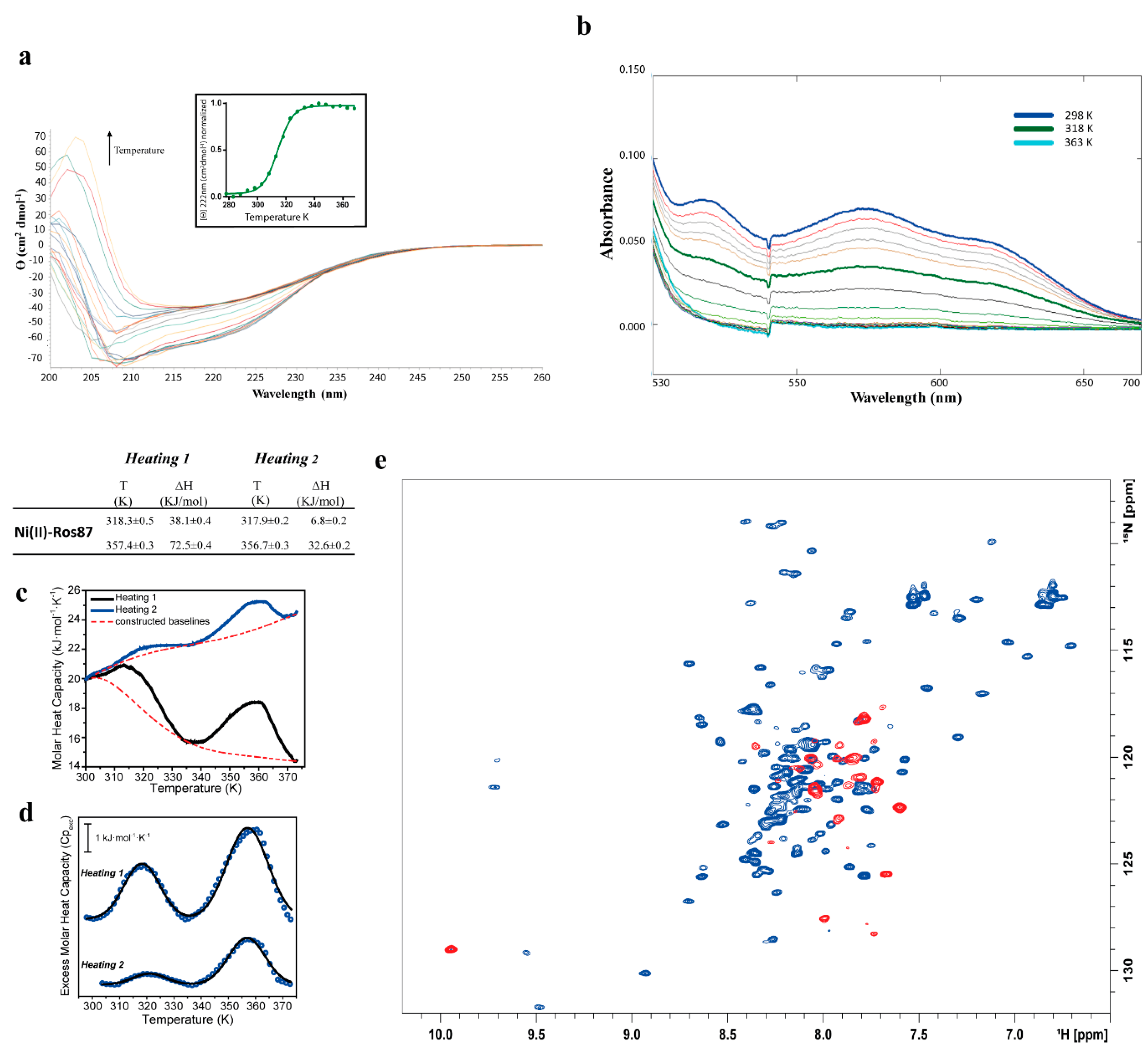
| Chemical Shift at 298 K (ppm) | Chemical Shift at 308 K (ppm) | Chemical Shift at 328 K (ppm) | Chemical Shift at 343 K (ppm) | |
|---|---|---|---|---|
| Cβ Cys 24 | 32.63 (±0.4) | 33.05 (±0.4) | 33.24 (±0.4) | 33.12 (±0.4) |
| Cβ Cys 27 | 33.68 (±0.4) | 33.08 (±0.4) | 33.79 (±0.4) | 33.64 (±0.4) |
| Tm (K) | ΔH (kJ/mol) | |
|---|---|---|
| Co(II)-Ros87 | CD: 338 ± 1 | |
| UV-Vis: 341 ± 2 | ||
| DSC 1st transition 1st Cycle: 337.7 ± 0.3 | 1st Cycle: 44.8 ± 1.3 | |
| DSC 1st transition 2nd Cycle: 340.3 ± 0.6 | 2nd Cycle: 43.1 ± 2.9 | |
| DSC 2nd transition 1st Cycle: 352.2 ± 0.6 | 1st Cycle: 178.3 ± 1.3 | |
| DSC 2nd transition 2nd Cycle: 353.5± 0.1 | 2nd Cycle: 153.9 ± 2.8 | |
| NMR: 338 | ||
| Ni(II)-Ros87 | CD: 314 ± 0.7 | |
| UV-Vis: 317 ± 1.3 | ||
| DSC 1st transition 1st Cycle: 318.3 ± 0.5 | 1st Cycle: 38.1 ± 0.4 | |
| DSC 1st transition 2nd Cycle: 317.9 ± 0.2 | 2nd Cycle: 6.7 ± 0.2 | |
| DSC 2nd transition 1st Cycle: 357.4 ± 0.3 | 1st Cycle: 72.5 ± 0.4 | |
| DSC 2nd transition 2nd Cycle: 356.7 ± 0.3 | 2nd Cycle: 32.6 ± 0.2 | |
| NMR: 318 | ||
| Cd(II)-Ros87 | CD: 345 ± 1.4 | |
| UV-Vis: None | ||
| DSC 1st transition 1st Cycle: 348.4 ± 0.5 | 1st Cycle: 169.4 ± 0.8 | |
| DSC 1st transition 2nd Cycle: 347.6 ± 0.7 | 2nd Cycle: 168.3 ± 1.6 | |
| NMR: 343 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grazioso, R.; García-Viñuales, S.; Russo, L.; D’Abrosca, G.; Esposito, S.; Zaccaro, L.; Iacovino, R.; Milardi, D.; Fattorusso, R.; Malgieri, G.; et al. Substitution of the Native Zn(II) with Cd(II), Co(II) and Ni(II) Changes the Downhill Unfolding Mechanism of Ros87 to a Completely Different Scenario. Int. J. Mol. Sci. 2020, 21, 8285. https://doi.org/10.3390/ijms21218285
Grazioso R, García-Viñuales S, Russo L, D’Abrosca G, Esposito S, Zaccaro L, Iacovino R, Milardi D, Fattorusso R, Malgieri G, et al. Substitution of the Native Zn(II) with Cd(II), Co(II) and Ni(II) Changes the Downhill Unfolding Mechanism of Ros87 to a Completely Different Scenario. International Journal of Molecular Sciences. 2020; 21(21):8285. https://doi.org/10.3390/ijms21218285
Chicago/Turabian StyleGrazioso, Rinaldo, Sara García-Viñuales, Luigi Russo, Gianluca D’Abrosca, Sabrina Esposito, Laura Zaccaro, Rosa Iacovino, Danilo Milardi, Roberto Fattorusso, Gaetano Malgieri, and et al. 2020. "Substitution of the Native Zn(II) with Cd(II), Co(II) and Ni(II) Changes the Downhill Unfolding Mechanism of Ros87 to a Completely Different Scenario" International Journal of Molecular Sciences 21, no. 21: 8285. https://doi.org/10.3390/ijms21218285
APA StyleGrazioso, R., García-Viñuales, S., Russo, L., D’Abrosca, G., Esposito, S., Zaccaro, L., Iacovino, R., Milardi, D., Fattorusso, R., Malgieri, G., & Isernia, C. (2020). Substitution of the Native Zn(II) with Cd(II), Co(II) and Ni(II) Changes the Downhill Unfolding Mechanism of Ros87 to a Completely Different Scenario. International Journal of Molecular Sciences, 21(21), 8285. https://doi.org/10.3390/ijms21218285









