In Vivo Imaging with Genetically Encoded Redox Biosensors
Abstract
1. Introduction
- This technology is not invasive and does not introduce artifacts caused by sample preparation. A classic example in this area is measurements of glutathione redox potential (EGSH) using redox active fluorescent proteins (FPs). In contrast to traditional approaches, these sensors revealed that the cellular glutathione pool is highly reducing, and reaches a reduced/oxidized glutathione (GSH/GSSG) ratio of between 50,000:1 and 500,000:1 [11].
- GEFIs can be targeted to different subcellular compartments via the host sorting machinery. There are studies where sensors were expressed in the matrix [12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30] and in the intermembrane space [31,32,33] of mitochondria (IMS), in the endoplasmic reticulum (ER) [34,35,36,37,38,39,40,41,42,43], peroxisomes [12,19,24,44], apicoplasts [16,45] and the nucleus [46]. Since many redox processes proceed in specific parts of the cell, whole cell approaches often lack sufficient sensitivity to record pronounced but localized events. Moreover, even the traditional analytical methods that take subcellular heterogeneity into account can suffer from unequal behavior of different organelles during sample preparation.
- The genes coding for GEFIs can be placed under the control of specific promoters. Therefore, it is possible to express these instruments in only the desired subsets of cells. Some examples include keratinocytes [53,54], pancreatic beta cells [55], muscle cells [22,23,56], neutrophils [57], heart tissue [58] and neurons [59,60,61,62].
- Modern genetic engineering approaches allow transgenic organisms to be created, which provides both signal stability and reproducibility of results. This approach has already been implemented in many traditional model organisms: Mus musculus [20,21,53,54,59,60,62], Drosophila melanogaster [61], Danio rerio [57,58,63,64,65,66,67,68,69], Caenorhabditis elegans [22,23,50,56,70], Xenopus laevis [71,72], Pasmodium falciparum [17,73], and Corynebacterium glutamicum [74] among others.
- Finally, GEFIs are compatible with low molecular weight chemical dyes and many other experimental approaches, therefore, facilitating study design.
2. A Short Overview of Redox GEFIs Applied In Vivo
3. Redox Biosensors in Animals
3.1. Embryogenesis, Development and Aging
3.2. Inflammation
3.3. Regeneration
3.4. Neuroscience
3.5. Cancer
3.6. Some Other Interesting Examples in Mammals
4. Redox Biosensors in Plants
4.1. Redox Metabolism of Chloroplasts
4.2. Redox Metabolism of Peroxisomes
4.3. Stress Conditions
Glutathione Metabolism during Stress
4.4. Growth and Development
4.4.1. Root Growth
4.4.2. Pollen Germination
4.5. Redox Processes Regulating Stomata Function
4.6. Interaction of Plants and Phytopathogens
4.7. Symbiosis
4.8. Analysis of the Topology of Transmembrane Proteins
5. Redox Biosensors in Microorganisms
5.1. Oxidative Stress Caused by External Factors and Genetic Landscape
5.1.1. Oxidative Stress in Bacteria
5.1.2. Oxidative Stress in S. cerevisiae
5.2. Glutathione and Thioredoxin Systems
5.3. The UPR in Yeast
5.4. Inheritance of Mitochondria in Yeast
5.5. Redox Regulation of Transport Proteins in Microorganisms
5.6. Redox Processes during Pathogenic Bacteria and Host Interaction
5.7. Redox Processes during Interactions of Pathogenic Microorganisms and Drugs
5.7.1. P. falciparum
5.7.2. Trypanosomatidae
5.7.3. Mycobacteria
5.8. Biotechnology
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 2CP | 2-Cys peroxiredoxin |
| AA | arachidonic acid |
| ATCUN | Amino Terminal Copper and Nickel binding motif |
| AXP | total pool of ATP and ADP |
| benzylIMD | 3-[substituted-benzyl]-menadiones |
| BSH | reduced bacillithiol |
| BSSB | oxidized bacillithiol |
| CFU | colony-forming unit |
| CNS | central nervous system |
| cp | circularly permuted |
| CQ | chloroquine |
| CuOOH | cumene hydroperoxide |
| DBMIB | 2,5-dibromo-6-isopropyl-3-methyl-1,4-benzoquinone |
| DCMU | 3-(3,4-dichlorophenyl)-1,1-dimethylurea |
| DPI | diphenyleneiodonium |
| dppf | 1,1′-bis(diphenylphosphino) ferrocene |
| DPS | 4,4′-dipyridyl disulfide |
| DR | diet-restricted |
| EB | elementary bodies |
| EBSH | bacillithiol redox potential |
| EGSH | glutathione redox potential |
| EMB | ethambutol |
| EMSH | mycothiol redox potential |
| ET(SH)2 | trypanothione redox potential |
| ER | endoplasmic reticulum |
| ETC | electron transport chain |
| FD | ferredoxin |
| FF | fully fed |
| FLIM | fluorescence lifetime imaging |
| FNR | ferredoxin-NADP+ reductase |
| FP | fluorescent protein |
| FRET | Förster resonance energy transfer |
| FTR | (Fd)-dependent Trx reductase |
| G6PD | glucose-6-phosphate dehydrogenase |
| GEFIs | genetically encoded fluorescent indicators |
| GFP | green fluorescent protein |
| GR | glutathione reductase |
| Grx1 | glutaredoxin 1 |
| GSH | reduced glutathione |
| GSSG | oxidized glutathione |
| hpi | hours post infection |
| IFN | interferon |
| IMS | intermembrane space of mitochondria |
| INH | isoniazid |
| iNOS | inducible NO synthase |
| L | lineage |
| LPS | lipopolysaccharides |
| MetT | 4-Methoxy-2,2,6,6-tetramethylpiperidine 1-oxyl |
| mlt | meltdown |
| MPO | myeloperoxidase |
| MSH | reduced mycothiol |
| Msm | Mycobacterium smegmatis |
| MSSM | oxidized mycothiol |
| Mtb | Mycobacterium tuberculosis |
| MTH1 | mutT homologue |
| Myo | myosin |
| NCs | normal/long-sized cells |
| NOX | NADPH-oxidase |
| NTRC | NADPH-dependent thioredoxin reductase |
| OxyR-RD | regulatory domain of OxyR transcription factor |
| PI3K | phosphoinositide 3-kinase |
| PKC | protein kinase C |
| PLB | plumbagin |
| PNS | peripheral nervous system |
| PMA | phorbol 12-myristate 13-acetate |
| RACF | retrograde actin cable flow |
| RB | reticulate bodies |
| RBCs | red blood cells |
| RIF | rifampicin |
| RNS | reactive nitrogen species |
| roGFP | redox-sensitive green fluorescent protein |
| ROS | reactive oxygen species |
| rxRFP | redox sensitive red fluorescent protein |
| rxYFP | redox-sensitive yellow fluorescent protein |
| SCs | short-sized cells |
| SCV | Salmonella-containing vacuole |
| SEM | standard error of the mean |
| SHAM | salicylhydroxamic acid |
| sigH | SigmaH factor |
| SPI2 | Salmonella pathogenicity island |
| SOD | superoxide dismutase |
| T3SS | type 3 secretion system |
| T-REX | Rex protein from Thermus aquaticus |
| T(SH)2 | reduced trypanothione |
| Tpx | tryparedoxin |
| Trx | thioredoxin |
| TS2 | oxidized trypanothione |
| UPR | unfolded protein response |
| WT | wild type |
| YFP | yellow fluorescent protein |
References
- Trebst, A. Energy Conservation in Photosynthetic Electron Transport of Chloroplasts. Annu. Rev. Plant. Physiol. 1974, 25, 423–458. [Google Scholar] [CrossRef]
- Hatefi, Y. The mitochondrial electron transport and oxidative phosphorylation system. Annu. Rev. Biochem. 1985, 54, 1015–1069. [Google Scholar] [CrossRef] [PubMed]
- Cantó, C.; Menzies, K.J.; Auwerx, J. NAD(+) Metabolism and the Control of Energy Homeostasis: A Balancing Act between Mitochondria and the Nucleus. Cell Metab. 2015, 22, 31–53. [Google Scholar] [CrossRef] [PubMed]
- Agledal, L.; Niere, M.; Ziegler, M. The phosphate makes a difference: Cellular functions of NADP. Redox Rep. 2010, 15, 2–10. [Google Scholar] [CrossRef]
- Forman, H.J.; Zhang, H.; Rinna, A. Glutathione: Overview of its protective roles, measurement, and biosynthesis. Mol. Asp. Med. 2009, 30, 1–12. [Google Scholar] [CrossRef]
- Collet, J.-F.; Messens, J. Structure, Function, and Mechanism of Thioredoxin Proteins. Antioxid. Redox Signal. 2010, 13, 1205–1216. [Google Scholar] [CrossRef]
- Collin, F. Chemical Basis of Reactive Oxygen Species Reactivity and Involvement in Neurodegenerative Diseases. Int. J. Mol. Sci. 2019, 20, 2407. [Google Scholar] [CrossRef]
- Yang, Y.; Bazhin, A.V.; Werner, J.; Karakhanova, S. Reactive Oxygen Species in the Immune System. Int. Rev. Immunol. 2013, 32, 249–270. [Google Scholar] [CrossRef]
- Finkel, T. Signal transduction by reactive oxygen species. J. Cell Biol. 2011, 194, 7–15. [Google Scholar] [CrossRef]
- Alfadda, A.A.; Sallam, R.M. Reactive oxygen species in health and disease. J. Biomed. Biotechnol. 2012, 2012, 936486. [Google Scholar] [CrossRef]
- Schwarzländer, M.; Dick, T.P.; Meyer, A.J.; Morgan, B. Dissecting Redox Biology Using Fluorescent Protein Sensors. Antioxid. Redox Signal. 2015, 24, 680–712. [Google Scholar] [CrossRef] [PubMed]
- Ayer, A.; Sanwald, J.; Pillay, B.A.; Meyer, A.J.; Perrone, G.G.; Dawes, I.W. Distinct Redox Regulation in Sub-Cellular Compartments in Response to Various Stress Conditions in Saccharomyces cerevisiae. PLoS ONE 2013, 8, e65240. [Google Scholar] [CrossRef]
- Christ, S.; Leichert, L.I.; Willms, A.; Lill, R.; Mühlenhoff, U. Defects in Mitochondrial Iron–Sulfur Cluster Assembly Induce Cysteine S-Polythiolation on Iron–Sulfur Apoproteins. Antioxid. Redox Signal. 2016, 25, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Pastor-Flores, D.; Becker, K.; Dick, T.P. Monitoring yeast mitochondria with peroxiredoxin-based redox probes: The influence of oxygen and glucose availability. Interface Focus 2017, 7, 20160143. [Google Scholar] [CrossRef]
- Calabrese, G.; Peker, E.; Amponsah, P.S.; Hoehne, M.N.; Riemer, T.; Mai, M.; Bienert, G.P.; Deponte, M.; Morgan, B.; Riemer, J. Hyperoxidation of mitochondrial peroxiredoxin limits H2O2-induced cell death in yeast. EMBO J. 2019, 38, e101552. [Google Scholar] [CrossRef]
- Mohring, F.; Rahbari, M.; Zechmann, B.; Rahlfs, S.; Przyborski, J.M.; Meyer, A.J.; Becker, K. Determination of glutathione redox potential and pH value in subcellular compartments of malaria parasites. Free Radic. Biol. Med. 2017, 104, 104–117. [Google Scholar] [CrossRef]
- Rahbari, M.; Rahlfs, S.; Przyborski, J.M.; Schuh, A.K.; Hunt, N.H.; Fidock, D.A.; Grau, G.E.; Becker, K. Hydrogen peroxide dynamics in subcellular compartments of malaria parasites using genetically encoded redox probes. Sci. Rep. 2017, 7, 10449. [Google Scholar] [CrossRef]
- Albrecht, S.C.; Barata, A.G.; Großhans, J.; Teleman, A.A.; Dick, T.P. In Vivo Mapping of Hydrogen Peroxide and Oxidized Glutathione Reveals Chemical and Regional Specificity of Redox Homeostasis. Cell Metab. 2011, 14, 819–829. [Google Scholar] [CrossRef]
- Ayer, A.; Fellermeier, S.; Fife, C.; Li, S.S.; Smits, G.; Meyer, A.J.; Dawes, I.W.; Perrone, G.G. A Genome-Wide Screen in Yeast Identifies Specific Oxidative Stress Genes Required for the Maintenance of Sub-Cellular Redox Homeostasis. PLoS ONE 2012, 7, e44278. [Google Scholar] [CrossRef]
- Galvan, D.L.; Badal, S.S.; Long, J.; Chang, B.H.; Schumacker, P.T.; Overbeek, P.A.; Danesh, F.R. Real-time in vivo mitochondrial redox assessment confirms enhanced mitochondrial reactive oxygen species in diabetic nephropathy. Kidney Int. 2017, 92, 1282–1287. [Google Scholar] [CrossRef]
- Galvan, D.L.; Long, J.; Green, N.; Chang, B.H.; Lin, J.S.; Schumacker, P.; Truong, L.D.; Overbeek, P.; Danesh, F.R. Drp1S600 phosphorylation regulates mitochondrial fission and progression of nephropathy in diabetic mice. J. Clin. Investig. 2019, 129, 2807–2823. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.; Allman, E.; Nehrke, K. Regulation of acid-base transporters by reactive oxygen species following mitochondrial fragmentation. Am. J. Physiol. Cell Physiol. 2012, 302, C1045. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.; Nehrke, K. Mitochondrial Fragmentation Leads to Intracellular Acidification in Caenorhabditis elegans and Mammalian Cells. Mol. Biol. Cell 2010, 21, 2191–2201. [Google Scholar] [CrossRef]
- Elbaz-Alon, Y.; Morgan, B.; Clancy, A.; Amoako, T.N.E.; Zalckvar, E.; Dick, T.P.; Schwappach, B.; Schuldiner, M. The yeast oligopeptide transporter Opt2 is localized to peroxisomes and affects glutathione redox homeostasis. FEMS Yeast Res. 2014, 14, 1055–1067. [Google Scholar] [CrossRef] [PubMed]
- McFaline-Figueroa, J.R.; Vevea, J.; Swayne, T.C.; Zhou, C.; Liu, C.; Leung, G.; Boldogh, I.R.; Pon, L.A. Mitochondrial quality control during inheritance is associated with lifespan and mother–daughter age asymmetry in budding yeast. Aging Cell 2011, 10, 885–895. [Google Scholar] [CrossRef]
- Higuchi, R.; Vevea, J.D.; Swayne, T.C.; Chojnowski, R.; Hill, V.; Boldogh, I.R.; Pon, L.A. Actin Dynamics Affect Mitochondrial Quality Control and Aging in Budding Yeast. Curr. Biol. 2013, 23, 2417–2422. [Google Scholar] [CrossRef]
- Pernice, W.M.; Vevea, J.D.; Pon, L.A. A role for Mfb1p in region-specific anchorage of high-functioning mitochondria and lifespan in Saccharomyces cerevisiae. Nat. Commun. 2016, 7, 10595. [Google Scholar] [CrossRef]
- McInnes, J.; Rehders, M.; McFaline-Figueroa, J.R.; Brix, K.; Pon, L.A.; Nevoigt, E. Defects in mitochondrial distribution during the prolonged lag phase of Saccharomyces cerevisiae preceding growth in glycerol as the sole source of carbon. FEMS Yeast Res. 2013, 13, 706–710. [Google Scholar] [CrossRef][Green Version]
- Garcia, E.J.; de Jonge, J.J.; Liao, P.-C.; Stivison, E.; Sing, C.N.; Higuchi-Sanabria, R.; Boldogh, I.R.; Pon, L.A. Reciprocal interactions between mtDNA and lifespan control in budding yeast. Mol. Biol. Cell 2019, 30, 2943–2952. [Google Scholar] [CrossRef]
- Manzano-López, J.; Matellán, L.; Álvarez-Llamas, A.; Blanco-Mira, J.C.; Monje-Casas, F. Asymmetric inheritance of spindle microtubule-organizing centres preserves replicative lifespan. Nat. Cell Biol. 2019, 21, 952–965. [Google Scholar] [CrossRef]
- Hu, J.; Dong, L.; Outten, C.E. The redox environment in the mitochondrial intermembrane space is maintained separately from the cytosol and matrix. J. Biol. Chem. 2008, 283, 29126–29134. [Google Scholar] [CrossRef]
- Kojer, K.; Bien, M.; Gangel, H.; Morgan, B.; Dick, T.P.; Riemer, J. Glutathione redox potential in the mitochondrial intermembrane space is linked to the cytosol and impacts the Mia40 redox state. EMBO J. 2012, 31, 3169–3182. [Google Scholar] [CrossRef] [PubMed]
- Kojer, K.; Peleh, V.; Calabrese, G.; Herrmann, J.M.; Riemer, J. Kinetic control by limiting glutaredoxin amounts enables thiol oxidation in the reducing mitochondrial intermembrane space. Mol. Biol. Cell 2014, 26, 195–204. [Google Scholar] [CrossRef]
- Kirstein, J.; Morito, D.; Kakihana, T.; Sugihara, M.; Minnen, A.; Hipp, M.S.; Nussbaum-Krammer, C.; Kasturi, P.; Hartl, F.U.; Nagata, K.; et al. Proteotoxic stress and ageing triggers the loss of redox homeostasis across cellular compartments. EMBO J. 2015, 34, 2334–2349. [Google Scholar] [CrossRef] [PubMed]
- Merksamer, P.I.; Trusina, A.; Papa, F.R. Real-Time Redox Measurements during Endoplasmic Reticulum Stress Reveal Interlinked Protein Folding Functions. Cell 2008, 135, 933–947. [Google Scholar] [CrossRef]
- Rubio, C.; Pincus, D.; Korennykh, A.; Schuck, S.; El-Samad, H.; Walter, P. Homeostatic adaptation to endoplasmic reticulum stress depends on Ire1 kinase activity. J. Cell Biol. 2011, 193, 171–184. [Google Scholar] [CrossRef]
- Delic, M.; Rebnegger, C.; Wanka, F.; Puxbaum, V.; Haberhauer-Troyer, C.; Hann, S.; Köllensperger, G.; Mattanovich, D.; Gasser, B. Oxidative protein folding and unfolded protein response elicit differing redox regulation in endoplasmic reticulum and cytosol of yeast. Free Radic. Biol. Med. 2012, 52, 2000–2012. [Google Scholar] [CrossRef] [PubMed]
- Igbaria, A.; Merksamer, P.I.; Trusina, A.; Tilahun, F.; Johnson, J.R.; Brandman, O.; Krogan, N.J.; Weissman, J.S.; Papa, F.R. Chaperone-mediated reflux of secretory proteins to the cytosol during endoplasmic reticulum stress. Proc. Natl. Acad. Sci. USA 2019, 116, 11291. [Google Scholar] [CrossRef]
- Le, Q.G.; Ishiwata-Kimata, Y.; Kohno, K.; Kimata, Y. Cadmium impairs protein folding in the endoplasmic reticulum and induces the unfolded protein response. FEMS Yeast Res. 2016, 16. [Google Scholar] [CrossRef]
- Mai, C.T.; Le, Q.G.; Ishiwata-Kimata, Y.; Takagi, H.; Kohno, K.; Kimata, Y. 4-Phenylbutyrate suppresses the unfolded protein response without restoring protein folding in Saccharomyces cerevisiae. FEMS Yeast Res. 2018, 18. [Google Scholar] [CrossRef]
- Navarro-Tapia, E.; Pérez-Torrado, R.; Querol, A. Ethanol Effects Involve Non-canonical Unfolded Protein Response Activation in Yeast Cells. Front. Microbiol. 2017, 8, 383. [Google Scholar] [CrossRef]
- Tran, D.M.; Ishiwata-Kimata, Y.; Mai, T.C.; Kubo, M.; Kimata, Y. The unfolded protein response alongside the diauxic shift of yeast cells and its involvement in mitochondria enlargement. Sci. Rep. 2019, 9, 12780. [Google Scholar] [CrossRef] [PubMed]
- Ponsero, A.J.; Igbaria, A.; Darch, M.A.; Miled, S.; Outten, C.E.; Winther, J.R.; Palais, G.; D’Autréaux, B.; Delaunay-Moisan, A.; Toledano, M.B. Endoplasmic Reticulum Transport of Glutathione by Sec61 Is Regulated by Ero1 and Bip. Mol. Cell 2017, 67, 962–973.e5. [Google Scholar] [CrossRef]
- Yano, T.; Oku, M.; Akeyama, N.; Itoyama, A.; Yurimoto, H.; Kuge, S.; Fujiki, Y.; Sakai, Y. A Novel Fluorescent Sensor Protein for Visualization of Redox States in the Cytoplasm and in Peroxisomes. Mol. Cell. Biol. 2010, 30, 3758. [Google Scholar] [CrossRef]
- Biddau, M.; Bouchut, A.; Major, J.; Saveria, T.; Tottey, J.; Oka, O.; van-Lith, M.; Jennings, K.E.; Ovciarikova, J.; DeRocher, A.; et al. Two essential Thioredoxins mediate apicoplast biogenesis, protein import, and gene expression in Toxoplasma gondii. PLoS Pathog. 2018, 14, e1006836. [Google Scholar] [CrossRef] [PubMed]
- Dardalhon, M.; Kumar, C.; Iraqui, I.; Vernis, L.; Kienda, G.; Banach-Latapy, A.; He, T.; Chanet, R.; Faye, G.; Outten, C.E.; et al. Redox-sensitive YFP sensors monitor dynamic nuclear and cytosolic glutathione redox changes. Free Radic. Biol. Med. 2012, 52, 2254–2265. [Google Scholar] [CrossRef][Green Version]
- Radzinski, M.; Fassler, R.; Yogev, O.; Breuer, W.; Shai, N.; Gutin, J.; Ilyas, S.; Geffen, Y.; Tsytkin-Kirschenzweig, S.; Nahmias, Y.; et al. Temporal profiling of redox-dependent heterogeneity in single cells. eLife 2018, 7, e37623. [Google Scholar] [CrossRef] [PubMed]
- Radzinski, M.; Yogev, O.; Yesharim, Y.; Brielle, E.S.; Israeli, R.; Fassler, R.; Melamed-Book, N.; Shai, N.; Arkin, I.T.; Pick, E.; et al. A molecular switch for Cdc48 activity and localization during oxidative stress and aging. BioRxiv 2019, 733709. [Google Scholar] [CrossRef]
- Mishra, R.; Kohli, S.; Malhotra, N.; Bandyopadhyay, P.; Mehta, M.; Munshi, M.; Adiga, V.; Ahuja, V.K.; Shandil, R.K.; Rajmani, R.S.; et al. Targeting redox heterogeneity to counteract drug tolerance in replicating Mycobacterium tuberculosis. Sci. Transl. Med. 2019, 11, eaaw6635. [Google Scholar] [CrossRef]
- Romero-Aristizabal, C.; Marks, D.S.; Fontana, W.; Apfeld, J. Regulated spatial organization and sensitivity of cytosolic protein oxidation in Caenorhabditis elegans. Nat. Commun. 2014, 5, 5020. [Google Scholar] [CrossRef]
- Xie, K.; Varatnitskaya, M.; Maghnouj, A.; Bader, V.; Winklhofer, K.F.; Hahn, S.; Leichert, L.I. Activation leads to a significant shift in the intracellular redox homeostasis of neutrophil-like cells. Redox Biol. 2020, 28, 101344. [Google Scholar] [CrossRef]
- Bhaskar, A.; Chawla, M.; Mehta, M.; Parikh, P.; Chandra, P.; Bhave, D.; Kumar, D.; Carroll, K.S.; Singh, A. Reengineering Redox Sensitive GFP to Measure Mycothiol Redox Potential of Mycobacterium tuberculosis during Infection. PLoS Pathog. 2014, 10, e1003902. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, Y.; Ohta, S.; Wolf, A.M. Blue light-induced oxidative stress in live skin. Free Radic. Biol. Med. 2017, 108, 300–310. [Google Scholar] [CrossRef]
- Wolf, A.M.; Nishimaki, K.; Kamimura, N.; Ohta, S. Real-Time Monitoring of Oxidative Stress in Live Mouse Skin. J. Investig. Dermatol. 2014, 134, 1701–1709. [Google Scholar] [CrossRef]
- Reissaus, C.A.; Piñeros, A.R.; Twigg, A.N.; Orr, K.S.; Conteh, A.M.; Martinez, M.M.; Kamocka, M.M.; Day, R.N.; Tersey, S.A.; Mirmira, R.G.; et al. A Versatile, Portable Intravital Microscopy Platform for Studying Beta-cell Biology In Vivo. Sci. Rep. 2019, 9, 8449. [Google Scholar] [CrossRef]
- Kang, D.; Kirienko, D.R.; Webster, P.; Fisher, A.L.; Kirienko, N.V. Pyoverdine, a siderophore from Pseudomonas aeruginosa, translocates into C. elegans, removes iron, and activates a distinct host response. Virulence 2018, 9, 804–817. [Google Scholar] [CrossRef]
- Pase, L.; Layton, J.E.; Wittmann, C.; Ellett, F.; Nowell, C.J.; Reyes-Aldasoro, C.C.; Varma, S.; Rogers, K.L.; Hall, C.J.; Keightley, M.C.; et al. Neutrophil-Delivered Myeloperoxidase Dampens the Hydrogen Peroxide Burst after Tissue Wounding in Zebrafish. Curr. Biol. 2012, 22, 1818–1824. [Google Scholar] [CrossRef]
- Han, P.; Zhou, X.-H.; Chang, N.; Xiao, C.-L.; Yan, S.; Ren, H.; Yang, X.-Z.; Zhang, M.-L.; Wu, Q.; Tang, B.; et al. Hydrogen peroxide primes heart regeneration with a derepression mechanism. Cell Res. 2014, 24, 1091–1107. [Google Scholar] [CrossRef] [PubMed]
- Wagener, K.C.; Kolbrink, B.; Dietrich, K.; Kizina, K.M.; Terwitte, L.S.; Kempkes, B.; Bao, G.; Müller, M. Redox Indicator Mice Stably Expressing Genetically Encoded Neuronal roGFP: Versatile Tools to Decipher Subcellular Redox Dynamics in Neuropathophysiology. Antioxid. Redox Signal. 2016, 25, 41–58. [Google Scholar] [CrossRef]
- Guzman, J.N.; Sanchez-Padilla, J.; Wokosin, D.; Kondapalli, J.; Ilijic, E.; Schumacker, P.T.; Surmeier, D.J. Oxidant stress evoked by pacemaking in dopaminergic neurons is attenuated by DJ-1. Nature 2010, 468, 696–700. [Google Scholar] [CrossRef]
- Liu, Z.; Celotto, A.M.; Romero, G.; Wipf, P.; Palladino, M.J. Genetically encoded redox sensor identifies the role of ROS in degenerative and mitochondrial disease pathogenesis. Neurobiol. Dis 2012, 45, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Breckwoldt, M.O.; Pfister, F.M.J.; Bradley, P.M.; Marinković, P.; Williams, P.R.; Brill, M.S.; Plomer, B.; Schmalz, A.; St Clair, D.K.; Naumann, R.; et al. Multiparametric optical analysis of mitochondrial redox signals during neuronal physiology and pathology in vivo. Nat. Med. 2014, 20, 555–560. [Google Scholar] [CrossRef] [PubMed]
- Meda, F.; Gauron, C.; Rampon, C.; Teillon, J.; Volovitch, M.; Vriz, S. Nerves Control Redox Levels in Mature Tissues Through Schwann Cells and Hedgehog Signaling. Antioxid. Redox Signal. 2016, 24, 299–311. [Google Scholar] [CrossRef]
- Wong, H.-T.C.; Zhang, Q.; Beirl, A.J.; Petralia, R.S.; Wang, Y.-X.; Kindt, K. Synaptic mitochondria regulate hair-cell synapse size and function. Elife 2019, 8, e48914. [Google Scholar] [CrossRef]
- Jelcic, M.; Enyedi, B.; Xavier, J.B.; Niethammer, P. Image-Based Measurement of H2O2 Reaction-Diffusion in Wounded Zebrafish Larvae. Biophys. J. 2017, 112, 2011–2018. [Google Scholar] [CrossRef]
- Bilan, D.S.; Pase, L.; Joosen, L.; Gorokhovatsky, A.Y.; Ermakova, Y.G.; Gadella, T.W.J.; Grabher, C.; Schultz, C.; Lukyanov, S.; Belousov, V.V. HyPer-3: A Genetically Encoded H2O2 Probe with Improved Performance for Ratiometric and Fluorescence Lifetime Imaging. ACS Chem. Biol. 2013, 8, 535–542. [Google Scholar] [CrossRef]
- Gault, W.J.; Enyedi, B.; Niethammer, P. Osmotic surveillance mediates rapid wound closure through nucleotide release. J. Cell Biol. 2014, 207, 767–782. [Google Scholar] [CrossRef]
- Enyedi, B.; Kala, S.; Nikolich-Zugich, T.; Niethammer, P. Tissue damage detection by osmotic surveillance. Nat. Cell Biol. 2013, 15, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Esterberg, R.; Linbo, T.; Pickett, S.B.; Wu, P.; Ou, H.C.; Rubel, E.W.; Raible, D.W. Mitochondrial calcium uptake underlies ROS generation during aminoglycoside-induced hair cell death. J. Clin. Investig. 2016, 126, 3556–3566. [Google Scholar] [CrossRef]
- Toledano, M.; Toledano-Osorio, M.; Navarro-Hortal, M.D.; Varela-López, A.; Osorio, R.; Quiles, J.L. Novel Polymeric Nanocarriers Reduced Zinc and Doxycycline Toxicity in the Nematode Caenorhabditis elegans. Antioxidants 2019, 8, 550. [Google Scholar] [CrossRef]
- Love, N.R.; Chen, Y.; Ishibashi, S.; Kritsiligkou, P.; Lea, R.; Koh, Y.; Gallop, J.L.; Dorey, K.; Amaya, E. Amputation-induced reactive oxygen species are required for successful Xenopus tadpole tail regeneration. Nat. Cell Biol. 2013, 15, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, F.; Raghunathan, V.; Luxardi, G.; Zhu, K.; Zhao, M. Early redox activities modulate Xenopus tail regeneration. Nat. Commun. 2018, 9, 4296. [Google Scholar] [CrossRef] [PubMed]
- Schuh, A.K.; Rahbari, M.; Heimsch, K.C.; Mohring, F.; Gabryszewski, S.J.; Weder, S.; Buchholz, K.; Rahlfs, S.; Fidock, D.A.; Becker, K. Stable Integration and Comparison of hGrx1-roGFP2 and sfroGFP2 Redox Probes in the Malaria Parasite Plasmodium falciparum. ACS Infect. Dis. 2018, 4, 1601–1612. [Google Scholar] [CrossRef]
- Tung, Q.N.; Loi, V.V.; Busche, T.; Nerlich, A.; Mieth, M.; Milse, J.; Kalinowski, J.; Hocke, A.C.; Antelmann, H. Stable integration of the Mrx1-roGFP2 biosensor to monitor dynamic changes of the mycothiol redox potential in Corynebacterium glutamicum. Redox Biol. 2019, 20, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Tao, R.; Zhao, Y.; Chu, H.; Wang, A.; Zhu, J.; Chen, X.; Zou, Y.; Shi, M.; Liu, R.; Su, N.; et al. Genetically encoded fluorescent sensors reveal dynamic regulation of NADPH metabolism. Nat. Methods 2017, 14, 720–728. [Google Scholar] [CrossRef] [PubMed]
- Díaz-García, C.M.; Mongeon, R.; Lahmann, C.; Koveal, D.; Zucker, H.; Yellen, G. Neuronal Stimulation Triggers Neuronal Glycolysis and Not Lactate Uptake. Cell Metab. 2017, 26, 361–374.e4. [Google Scholar] [CrossRef]
- Kostyuk, A.I.; Panova, A.S.; Bilan, D.S.; Belousov, V.V. Redox biosensors in a context of multiparameter imaging. Free Radic. Biol. Med. 2018, 128, 23–39. [Google Scholar] [CrossRef]
- Meyer, A.J.; Dick, T.P. Fluorescent Protein-Based Redox Probes. Antioxid. Redox Signal. 2010, 13, 621–650. [Google Scholar] [CrossRef]
- Kostyuk, A.I.; Demidovich, A.D.; Kotova, D.A.; Belousov, V.V.; Bilan, D.S. Circularly Permuted Fluorescent Protein-Based Indicators: History, Principles, and Classification. Int. J. Mol. Sci. 2019, 20, 4200. [Google Scholar] [CrossRef]
- Bilan, D.S.; Belousov, V.V. New tools for redox biology: From imaging to manipulation. Free Radic. Biol. Med. 2017, 109, 167–188. [Google Scholar] [CrossRef]
- Gökerküçük, E.B.; Tramier, M.; Bertolin, G. Imaging Mitochondrial Functions: From Fluorescent Dyes to Genetically-Encoded Sensors. Genes 2020, 11, 125. [Google Scholar] [CrossRef]
- O’Banion, C.P.; Yasuda, R. Fluorescent sensors for neuronal signaling. Curr. Opin. Neurobiol. 2020, 63, 31–41. [Google Scholar] [CrossRef]
- Bilan, D.S.; Belousov, V.V. In Vivo Imaging of Hydrogen Peroxide with HyPer Probes. Antioxid. Redox Signal. 2018, 29, 569–584. [Google Scholar] [CrossRef]
- Tung, Q.N.; Linzner, N.; Loi, V.V.; Antelmann, H. Application of genetically encoded redox biosensors to measure dynamic changes in the glutathione, bacillithiol and mycothiol redox potentials in pathogenic bacteria. Free Radic. Biol. Med. 2018, 128, 84–96. [Google Scholar] [CrossRef]
- Hanson, G.T.; Aggeler, R.; Oglesbee, D.; Cannon, M.; Capaldi, R.A.; Tsien, R.Y.; Remington, S.J. Investigating Mitochondrial Redox Potential with Redox-sensitive Green Fluorescent Protein Indicators. J. Biol. Chem. 2004, 279, 13044–13053. [Google Scholar] [CrossRef] [PubMed]
- Cannon, M.B.; Remington, S.J. Re-engineering redox-sensitive green fluorescent protein for improved response rate. Protein Sci. 2006, 15, 45–57. [Google Scholar] [CrossRef]
- Lohman, J.R.; Remington, S.J. Development of a family of redox-sensitive green fluorescent protein indicators for use in relatively oxidizing subcellular environments. Biochemistry 2008, 47, 8678–8688. [Google Scholar] [CrossRef]
- Gutscher, M.; Pauleau, A.-L.; Marty, L.; Brach, T.; Wabnitz, G.H.; Samstag, Y.; Meyer, A.J.; Dick, T.P. Real-time imaging of the intracellular glutathione redox potential. Nat. Methods 2008, 5, 553–559. [Google Scholar] [CrossRef]
- Loi, V.V.; Harms, M.; Müller, M.; Huyen, N.T.T.; Hamilton, C.J.; Hochgräfe, F.; Pané-Farré, J.; Antelmann, H. Real-Time Imaging of the Bacillithiol Redox Potential in the Human Pathogen Staphylococcus aureus Using a Genetically Encoded Bacilliredoxin-Fused Redox Biosensor. Antioxid. Redox Signal. 2017, 26, 835–848. [Google Scholar] [CrossRef]
- Ebersoll, S.; Bogacz, M.; Günter, L.M.; Dick, T.P.; Krauth-Siegel, R.L. A tryparedoxin-coupled biosensor reveals a mitochondrial trypanothione metabolism in trypanosomes. Elife 2020, 9, e53227. [Google Scholar] [CrossRef]
- Gutscher, M.; Sobotta, M.C.; Wabnitz, G.H.; Ballikaya, S.; Meyer, A.J.; Samstag, Y.; Dick, T.P. Proximity-based protein thiol oxidation by H2O2-scavenging peroxidases. J. Biol. Chem. 2009, 284, 31532–31540. [Google Scholar] [CrossRef] [PubMed]
- Morgan, B.; Van Laer, K.; Owusu, T.N.E.; Ezeriņa, D.; Pastor-Flores, D.; Amponsah, P.S.; Tursch, A.; Dick, T.P. Real-time monitoring of basal H2O2 levels with peroxiredoxin-based probes. Nat. Chem. Biol. 2016, 12, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Østergaard, H.; Henriksen, A.; Hansen, F.G.; Winther, J.R. Shedding light on disulfide bond formation: Engineering a redox switch in green fluorescent protein. EMBO J. 2001, 20, 5853–5862. [Google Scholar] [CrossRef]
- Müller, A.; Schneider, J.F.; Degrossoli, A.; Lupilova, N.; Dick, T.P.; Leichert, L.I. Systematic in vitro assessment of responses of roGFP2-based probes to physiologically relevant oxidant species. Free Radic. Biol. Med. 2017, 106, 329–338. [Google Scholar] [CrossRef]
- Sugiura, K.; Nagai, T.; Nakano, M.; Ichinose, H.; Nakabayashi, T.; Ohta, N.; Hisabori, T. Redox sensor proteins for highly sensitive direct imaging of intracellular redox state. Biochem. Biophys. Res. Commun. 2015, 457, 242–248. [Google Scholar] [CrossRef]
- Fan, Y.; Chen, Z.; Ai, H. Monitoring redox dynamics in living cells with a redox-sensitive red fluorescent protein. Anal. Chem. 2015, 87, 2802–2810. [Google Scholar] [CrossRef]
- Shokhina, A.G.; Kostyuk, A.I.; Ermakova, Y.G.; Panova, A.S.; Staroverov, D.B.; Egorov, E.S.; Baranov, M.S.; van Belle, G.J.; Katschinski, D.M.; Belousov, V.V.; et al. Red fluorescent redox-sensitive biosensor Grx1-roCherry. Redox Biol. 2019, 21, 101071. [Google Scholar] [CrossRef]
- Kumagai, A.; Ando, R.; Miyatake, H.; Greimel, P.; Kobayashi, T.; Hirabayashi, Y.; Shimogori, T.; Miyawaki, A. A bilirubin-inducible fluorescent protein from eel muscle. Cell 2013, 153, 1602–1611. [Google Scholar] [CrossRef]
- Hu, H.; Wang, A.; Huang, L.; Zou, Y.; Gu, Y.; Chen, X.; Zhao, Y.; Yang, Y. Monitoring cellular redox state under hypoxia using a fluorescent sensor based on eel fluorescent protein. Free Radic. Biol. Med. 2018, 120, 255–265. [Google Scholar] [CrossRef]
- Belousov, V.V.; Fradkov, A.F.; Lukyanov, K.A.; Staroverov, D.B.; Shakhbazov, K.S.; Terskikh, A.V.; Lukyanov, S. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat. Methods 2006, 3, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Markvicheva, K.N.; Bilan, D.S.; Mishina, N.M.; Gorokhovatsky, A.Y.; Vinokurov, L.M.; Lukyanov, S.; Belousov, V.V. A genetically encoded sensor for H2O2 with expanded dynamic range. Bioorg. Med. Chem. 2011, 19, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Pak, V.V.; Ezeriņa, D.; Lyublinskaya, O.G.; Pedre, B.; Tyurin-Kuzmin, P.A.; Mishina, N.M.; Thauvin, M.; Young, D.; Wahni, K.; Gache, S.A.M.; et al. Ultrasensitive Genetically Encoded Indicator for Hydrogen Peroxide Identifies Roles for the Oxidant in Cell Migration and Mitochondrial Function. Cell Metab. 2020, 31, 642–653.e6. [Google Scholar] [CrossRef] [PubMed]
- Ermakova, Y.G.; Bilan, D.S.; Matlashov, M.E.; Mishina, N.M.; Markvicheva, K.N.; Subach, O.M.; Subach, F.V.; Bogeski, I.; Hoth, M.; Enikolopov, G.; et al. Red fluorescent genetically encoded indicator for intracellular hydrogen peroxide. Nat. Commun. 2014, 5, 5222. [Google Scholar] [CrossRef]
- Melo, E.P.; Lopes, C.; Gollwitzer, P.; Lortz, S.; Lenzen, S.; Mehmeti, I.; Kaminski, C.F.; Ron, D.; Avezov, E. TriPer, an optical probe tuned to the endoplasmic reticulum tracks changes in luminal H2O2. BMC Biol. 2017, 15, 24. [Google Scholar] [CrossRef]
- Subach, O.M.; Kunitsyna, T.A.; Mineyeva, O.A.; Lazutkin, A.A.; Bezryadnov, D.V.; Barykina, N.V.; Piatkevich, K.D.; Ermakova, Y.G.; Bilan, D.S.; Belousov, V.V.; et al. Slowly Reducible Genetically Encoded Green Fluorescent Indicator for In Vivo and Ex Vivo Visualization of Hydrogen Peroxide. Int. J. Mol. Sci. 2019, 20, 3138. [Google Scholar] [CrossRef]
- McLaughlin, K.J.; Strain-Damerell, C.M.; Xie, K.; Brekasis, D.; Soares, A.S.; Paget, M.S.B.; Kielkopf, C.L. Structural basis for NADH/NAD+ redox sensing by a Rex family repressor. Mol. Cell 2010, 38, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Hung, Y.P.; Albeck, J.G.; Tantama, M.; Yellen, G. Imaging cytosolic NADH-NAD(+) redox state with a genetically encoded fluorescent biosensor. Cell Metab. 2011, 14, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Tejwani, V.; Schmitt, F.-J.; Wilkening, S.; Zebger, I.; Horch, M.; Lenz, O.; Friedrich, T. Investigation of the NADH/NAD+ ratio in Ralstonia eutropha using the fluorescence reporter protein Peredox. Biochim. Biophys. Acta Bioenerg. 2017, 1858, 86–94. [Google Scholar] [CrossRef]
- Zhao, Y.; Jin, J.; Hu, Q.; Zhou, H.-M.; Yi, J.; Yu, Z.; Xu, L.; Wang, X.; Yang, Y.; Loscalzo, J. Genetically encoded fluorescent sensors for intracellular NADH detection. Cell Metab. 2011, 14, 555–566. [Google Scholar] [CrossRef]
- Bilan, D.S.; Matlashov, M.E.; Gorokhovatsky, A.Y.; Schultz, C.; Enikolopov, G.; Belousov, V.V. Genetically encoded fluorescent indicator for imaging NAD(+)/NADH ratio changes in different cellular compartments. Biochim. Biophys. Acta 2014, 1840, 951–957. [Google Scholar] [CrossRef]
- Zhao, Y.; Hu, Q.; Cheng, F.; Su, N.; Wang, A.; Zou, Y.; Hu, H.; Chen, X.; Zhou, H.-M.; Huang, X.; et al. SoNar, a Highly Responsive NAD+/NADH Sensor, Allows High-Throughput Metabolic Screening of Anti-tumor Agents. Cell Metab. 2015, 21, 777–789. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Wang, A.; Huang, L.; Zhu, X.; Hu, Q.; Zhang, Y.; Chen, X.; Li, F.; Wang, Q.; Wang, H.; et al. Illuminating NAD+ Metabolism in Live Cells and In Vivo Using a Genetically Encoded Fluorescent Sensor. Dev. Cell 2020, 53, 240–252.e7. [Google Scholar] [CrossRef]
- Tarrago, L.; Péterfi, Z.; Lee, B.C.; Michel, T.; Gladyshev, V.N. Monitoring methionine sulfoxide with stereospecific mechanism-based fluorescent sensors. Nat. Chem. Biol. 2015, 11, 332–338. [Google Scholar] [CrossRef]
- Simen Zhao, B.; Liang, Y.; Song, Y.; Zheng, C.; Hao, Z.; Chen, P.R. A Highly Selective Fluorescent Probe for Visualization of Organic Hydroperoxides in Living Cells. J. Am. Chem. Soc. 2010, 132, 17065–17067. [Google Scholar] [CrossRef]
- Zhao, F.-L.; Zhang, C.; Zhang, C.; Tang, Y.; Ye, B.-C. A genetically encoded biosensor for in vitro and in vivo detection of NADP+. Biosens. Bioelectron. 2016, 77, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, K.; Yokochi, Y.; Fu, N.; Fukaya, Y.; Yoshida, K.; Mihara, S.; Hisabori, T. The thioredoxin (Trx) redox state sensor protein can visualize Trx activities in the light/dark response in chloroplasts. J. Biol. Chem. 2019, 294, 12091–12098. [Google Scholar] [CrossRef]
- Dooley, C.T.; Dore, T.M.; Hanson, G.T.; Jackson, W.C.; Remington, S.J.; Tsien, R.Y. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J. Biol. Chem. 2004, 279, 22284–22293. [Google Scholar] [CrossRef]
- Morsci, N.S.; Hall, D.H.; Driscoll, M.; Sheng, Z.-H. Age-Related Phasic Patterns of Mitochondrial Maintenance in Adult Caenorhabditis elegans Neurons. J. Neurosci. 2016, 36, 1373–1385. [Google Scholar] [CrossRef]
- Jiang, H.-C.; Hsu, J.-M.; Yen, C.-P.; Chao, C.-C.; Chen, R.-H.; Pan, C.-L. Neural activity and CaMKII protect mitochondria from fragmentation in aging Caenorhabditis elegans neurons. Proc. Natl. Acad. Sci. USA 2015, 112, 8768–8773. [Google Scholar] [CrossRef] [PubMed]
- Back, P.; De Vos, W.H.; Depuydt, G.G.; Matthijssens, F.; Vanfleteren, J.R.; Braeckman, B.P. Exploring real-time in vivo redox biology of developing and aging Caenorhabditis elegans. Free Radic. Biol. Med. 2012, 52, 850–859. [Google Scholar] [CrossRef]
- Henderson, D.; Huebner, C.; Markowitz, M.; Taube, N.; Harvanek, Z.M.; Jakob, U.; Knoefler, D. Do developmental temperatures affect redox level and lifespan in C. elegans through upregulation of peroxiredoxin? Redox Biol. 2017, 14, 386–390. [Google Scholar] [CrossRef]
- Bazopoulou, D.; Knoefler, D.; Zheng, Y.; Ulrich, K.; Oleson, B.J.; Xie, L.; Kim, M.; Kaufmann, A.; Lee, Y.-T.; Dou, Y.; et al. Developmental ROS individualizes organismal stress resistance and lifespan. Nature 2019, 576, 301–305. [Google Scholar] [CrossRef]
- Knoefler, D.; Thamsen, M.; Koniczek, M.; Niemuth, N.J.; Diederich, A.-K.; Jakob, U. Quantitative in vivo redox sensors uncover oxidative stress as an early event in life. Mol. Cell 2012, 47, 767–776. [Google Scholar] [CrossRef] [PubMed]
- Cabreiro, F.; Ackerman, D.; Doonan, R.; Araiz, C.; Back, P.; Papp, D.; Braeckman, B.P.; Gems, D. Increased life span from overexpression of superoxide dismutase in Caenorhabditis elegans is not caused by decreased oxidative damage. Free Radic. Biol. Med. 2011, 51, 1575–1582. [Google Scholar] [CrossRef] [PubMed]
- Rieckher, M.; Markaki, M.; Princz, A.; Schumacher, B.; Tavernarakis, N. Maintenance of Proteostasis by P Body-Mediated Regulation of eIF4E Availability during Aging in Caenorhabditis elegans. Cell Rep. 2018, 25, 199–211.e6. [Google Scholar] [CrossRef]
- Ewald, C.Y.; Hourihan, J.M.; Bland, M.S.; Obieglo, C.; Katic, I.; Moronetti Mazzeo, L.E.; Alcedo, J.; Blackwell, T.K.; Hynes, N.E. NADPH oxidase-mediated redox signaling promotes oxidative stress resistance and longevity through memo-1 in C. elegans. eLife 2017, 6, e19493. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Chisholm, A.D. C. elegans epidermal wounding induces a mitochondrial ROS burst that promotes wound repair. Dev. Cell 2014, 31, 48–60. [Google Scholar] [CrossRef]
- De Henau, S.; Tilleman, L.; Vangheel, M.; Luyckx, E.; Trashin, S.; Pauwels, M.; Germani, F.; Vlaeminck, C.; Vanfleteren, J.R.; Bert, W.; et al. A redox signalling globin is essential for reproduction in Caenorhabditis elegans. Nat. Commun. 2015, 6, 8782. [Google Scholar] [CrossRef]
- Henau, S.D.; Pagès-Gallego, M.; Pannekoek, W.-J.; Dansen, T.B. Mitochondria-Derived H2O2 Promotes Symmetry Breaking of the C. elegans Zygote. Dev. Cell 2020, 53, 263–271.e6. [Google Scholar] [CrossRef]
- Castelein, N.; Muschol, M.; Dhondt, I.; Cai, H.; De Vos, W.H.; Dencher, N.A.; Braeckman, B.P. Mitochondrial efficiency is increased in axenically cultured Caenorhabditis elegans. Exp. Gerontol. 2014, 56, 26–36. [Google Scholar] [CrossRef]
- Bohnert, K.A.; Kenyon, C. A lysosomal switch triggers proteostasis renewal in the immortal C. elegans germ lineage. Nature 2017, 551, 629–633. [Google Scholar] [CrossRef]
- Terhzaz, S.; Cabrero, P.; Brinzer, R.A.; Halberg, K.A.; Dow, J.A.T.; Davies, S.-A. A novel role of Drosophila cytochrome P450-4e3 in permethrin insecticide tolerance. Insect Biochem. Mol. Biol. 2015, 67, 38–46. [Google Scholar] [CrossRef]
- O’Donnell, K.C.; Vargas, M.E.; Sagasti, A. WldS and PGC-1 Regulate Mitochondrial Transport and Oxidation State after Axonal Injury. J. Neurosci. 2013, 33, 14778–14790. [Google Scholar] [CrossRef]
- Yadav, S.; Chawla, B.; Khursheed, M.A.; Ramachandran, R.; Bachhawat, A.K. The glutathione degrading enzyme, Chac1, is required for calcium signaling in developing zebrafish: Redox as an upstream activator of calcium. Biochem. J. 2019, 476, 1857–1873. [Google Scholar] [CrossRef]
- Seiler, C.; Davuluri, G.; Abrams, J.; Byfield, F.J.; Janmey, P.A.; Pack, M. Smooth Muscle Tension Induces Invasive Remodeling of the Zebrafish Intestine. PLoS Biol. 2012, 10, e1001386. [Google Scholar] [CrossRef]
- Bräutigam, L.; Pudelko, L.; Jemth, A.-S.; Gad, H.; Narwal, M.; Gustafsson, R.; Karsten, S.; Carreras Puigvert, J.; Homan, E.; Berndt, C.; et al. Hypoxic Signaling and the Cellular Redox Tumor Environment Determine Sensitivity to MTH1 Inhibition. Cancer Res. 2016, 76, 2366–2375. [Google Scholar] [CrossRef] [PubMed]
- Panieri, E.; Millia, C.; Santoro, M.M. Real-time quantification of subcellular H2O2 and glutathione redox potential in living cardiovascular tissues. Free Radic. Biol. Med. 2017, 109, 189–200. [Google Scholar] [CrossRef]
- Panieri, E.; Santoro, M.M. Data on metabolic-dependent antioxidant response in the cardiovascular tissues of living zebrafish under stress conditions. Data Brief. 2017, 12, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Wang, A.; Shi, M.; Chen, X.; Liu, R.; Li, T.; Zhang, C.; Zhang, Z.; Zhu, L.; Ju, Z.; et al. Analysis of redox landscapes and dynamics in living cells and in vivo using genetically encoded fluorescent sensors. Nat. Protoc. 2018, 13, 2362–2386. [Google Scholar] [CrossRef]
- Gauron, C.; Meda, F.; Dupont, E.; Albadri, S.; Quenech’Du, N.; Ipendey, E.; Volovitch, M.; Del Bene, F.; Joliot, A.; Rampon, C.; et al. Hydrogen peroxide (H2O2) controls axon pathfinding during zebrafish development. Dev. Biol. 2016, 414, 133–141. [Google Scholar] [CrossRef]
- Niethammer, P.; Grabher, C.; Look, A.T.; Mitchison, T.J. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature 2009, 459, 996–999. [Google Scholar] [CrossRef]
- Deng, Q.; Harvie, E.A.; Huttenlocher, A. Distinct signalling mechanisms mediate neutrophil attraction to bacterial infection and tissue injury: H2O2-independent neutrophil attraction to infection. Cell. Microbiol. 2012, 14, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Guan, J.; Borrelli, L.A.; Xu, J.; Serrano-Pozo, A.; Bacskai, B.J. Mitochondrial Alterations near Amyloid Plaques in an Alzheimer’s Disease Mouse Model. J. Neurosci. 2013, 33, 17042–17051. [Google Scholar] [CrossRef] [PubMed]
- Can, K.; Menzfeld, C.; Rinne, L.; Rehling, P.; Kügler, S.; Golubiani, G.; Dudek, J.; Müller, M. Neuronal Redox-Imbalance in Rett Syndrome Affects Mitochondria as Well as Cytosol, and Is Accompanied by Intensified Mitochondrial O2 Consumption and ROS Release. Front. Physiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Galvan, D.L.; Green, N.H.; Danesh, F.R. The hallmarks of mitochondrial dysfunction in chronic kidney disease. Kidney Int. 2017, 92, 1051–1057. [Google Scholar] [CrossRef] [PubMed]
- Haga, S.; Morita, N.; Irani, K.; Fujiyoshi, M.; Ogino, T.; Ozawa, T.; Ozaki, M. p66Shc has a pivotal function in impaired liver regeneration in aged mice by a redox-dependent mechanism. Lab. Investig. 2010, 90, 1718–1726. [Google Scholar] [CrossRef]
- Haga, S.; Ozawa, T.; Yamada, Y.; Morita, N.; Nagashima, I.; Inoue, H.; Inaba, Y.; Noda, N.; Abe, R.; Umezawa, K.; et al. p62/SQSTM1 Plays a Protective Role in Oxidative Injury of Steatotic Liver in a Mouse Hepatectomy Model. Antioxid. Redox Signal. 2014, 21, 2515–2530. [Google Scholar] [CrossRef]
- Hao, X.; Gu, H.; Chen, C.; Huang, D.; Zhao, Y.; Xie, L.; Zou, Y.; Shu, H.S.; Zhang, Y.; He, X.; et al. Metabolic Imaging Reveals a Unique Preference of Symmetric Cell Division and Homing of Leukemia-Initiating Cells in an Endosteal Niche. Cell Metab. 2019, 29, 950–965.e6. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, A.; Zou, Y.; Su, N.; Loscalzo, J.; Yang, Y. In vivo monitoring of cellular energy metabolism using SoNar, a highly responsive sensor for NAD+/NADH redox state. Nat. Protoc. 2016, 11, 1345–1359. [Google Scholar] [CrossRef]
- Van Hameren, G.; Campbell, G.; Deck, M.; Berthelot, J.; Gautier, B.; Quintana, P.; Chrast, R.; Tricaud, N. In vivo real-time dynamics of ATP and ROS production in axonal mitochondria show decoupling in mouse models of peripheral neuropathies. Acta Neuropathol. Commun. 2019, 7, 86. [Google Scholar] [CrossRef]
- Han, Y.; Ishibashi, S.; Iglesias-Gonzalez, J.; Chen, Y.; Love, N.R.; Amaya, E. Ca2+-Induced Mitochondrial ROS Regulate the Early Embryonic Cell Cycle. Cell Rep. 2018, 22, 218–231. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Moe-Lange, J.; Hennet, L.; Feldman, L.J. Salt Stress Affects the Redox Status of Arabidopsis Root Meristems. Front. Plant. Sci. 2016, 7, 81. [Google Scholar] [CrossRef] [PubMed]
- Schwarzländer, M.; Fricker, M.D.; Müller, C.; Marty, L.; Brach, T.; Novak, J.; Sweetlove, L.J.; Hell, R.; Meyer, A.J. Confocal imaging of glutathione redox potential in living plant cells. J. Microsc. 2008, 231, 299–316. [Google Scholar] [CrossRef]
- Jiang, K.; Schwarzer, C.; Lally, E.; Zhang, S.; Ruzin, S.; Machen, T.; Remington, S.J.; Feldman, L. Expression and Characterization of a Redox-Sensing Green Fluorescent Protein (Reduction-Oxidation-Sensitive Green Fluorescent Protein) in Arabidopsis. Plant. Physiol. 2006, 141, 397–403. [Google Scholar] [CrossRef]
- Jubany-Mari, T.; Alegre-Batlle, L.; Jiang, K.; Feldman, L.J. Use of a redox-sensing GFP (c-roGFP1) for real-time monitoring of cytosol redox status in Arabidopsis thaliana water-stressed plants. FEBS Lett. 2010, 584, 889–897. [Google Scholar] [CrossRef]
- Au, K.K.C.; Pérez-Gómez, J.; Neto, H.; Müller, C.; Meyer, A.J.; Fricker, M.D.; Moore, I. A perturbation in glutathione biosynthesis disrupts endoplasmic reticulum morphology and secretory membrane traffic in Arabidopsis thaliana. Plant J. 2012, 71, 881–894. [Google Scholar] [CrossRef]
- Brossa, R.; Pintó-Marijuan, M.; Jiang, K.; Alegre, L.; Feldman, L.J. Assessing the regulation of leaf redox status under water stress conditions in Arabidopsis thaliana. Plant. Signal. Behav. 2013, 8, e24781. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Csiszár, J.; Brunner, S.; Horváth, E.; Bela, K.; Ködmön, P.; Riyazuddin, R.; Gallé, Á.; Hurton, Á.; Papdi, C.; Szabados, L.; et al. Exogenously applied salicylic acid maintains redox homeostasis in salt-stressed Arabidopsis gr1 mutants expressing cytosolic roGFP1. Plant. Growth Regul. 2018, 86, 181–194. [Google Scholar] [CrossRef]
- Meyer, A.J.; Brach, T.; Marty, L.; Kreye, S.; Rouhier, N.; Jacquot, J.-P.; Hell, R. Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J. 2007, 52, 973–986. [Google Scholar] [CrossRef]
- Schwarzländer, M.; Fricker, M.D.; Sweetlove, L.J. Monitoring the in vivo redox state of plant mitochondria: Effect of respiratory inhibitors, abiotic stress and assessment of recovery from oxidative challenge. Biochim. Biophys. Acta Bioenerg. 2009, 1787, 468–475. [Google Scholar] [CrossRef]
- Maughan, S.C.; Pasternak, M.; Cairns, N.; Kiddle, G.; Brach, T.; Jarvis, R.; Haas, F.; Nieuwland, J.; Lim, B.; Müller, C.; et al. Plant homologs of the Plasmodium falciparum chloroquine-resistance transporter, PfCRT, are required for glutathione homeostasis and stress responses. Proc. Natl. Acad. Sci. USA 2010, 107, 2331–2336. [Google Scholar] [CrossRef] [PubMed]
- Shibata, M.; Oikawa, K.; Yoshimoto, K.; Kondo, M.; Mano, S.; Yamada, K.; Hayashi, M.; Sakamoto, W.; Ohsumi, Y.; Nishimura, M. Highly Oxidized Peroxisomes Are Selectively Degraded via Autophagy in Arabidopsis. Plant. Cell 2013, 25, 4967–4983. [Google Scholar] [CrossRef] [PubMed]
- Brunkard, J.O.; Runkel, A.M.; Zambryski, P.C. Chloroplasts extend stromules independently and in response to internal redox signals. Proc. Natl. Acad. Sci. USA 2015, 112, 10044–10049. [Google Scholar] [CrossRef] [PubMed]
- Bratt, A.; Rosenwasser, S.; Meyer, A.; Fluhr, R. Organelle redox autonomy during environmental stress. Plant Cell Environ. 2016, 39, 1909–1919. [Google Scholar] [CrossRef]
- Lai, Y.-S.; Renna, L.; Yarema, J.; Ruberti, C.; He, S.Y.; Brandizzi, F. Salicylic acid-independent role of NPR1 is required for protection from proteotoxic stress in the plant endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2018, 115, E5203–E5212. [Google Scholar] [CrossRef]
- García-Quirós, E.; de Alché, J.D.; Karpinska, B.; Foyerde Alché, C.H. Glutathione redox state plays a key role in flower development and pollen vigour. J. Exp. Bot. 2020, 71, 730–741. [Google Scholar] [CrossRef]
- Haber, Z.; Rosenwasser, S. Resolving the dynamics of photosynthetically produced ROS by high-resolution monitoring of chloroplastic EGSH in Arabidopsis. BioRxiv 2020. [Google Scholar] [CrossRef]
- Marty, L.; Siala, W.; Schwarzländer, M.; Fricker, M.D.; Wirtz, M.; Sweetlove, L.J.; Meyer, Y.; Meyer, A.J.; Reichheld, J.-P.; Hell, R. The NADPH-dependent thioredoxin system constitutes a functional backup for cytosolic glutathione reductase in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 9109–9114. [Google Scholar] [CrossRef]
- Dubreuil-Maurizi, C.; Vitecek, J.; Marty, L.; Branciard, L.; Frettinger, P.; Wendehenne, D.; Meyer, A.J.; Mauch, F.; Poinssot, B. Glutathione Deficiency of the Arabidopsis Mutant pad2-1 Affects Oxidative Stress-Related Events, Defense Gene Expression, and the Hypersensitive Response. Plant Physiol. 2011, 157, 2000–2012. [Google Scholar] [CrossRef]
- Yu, X.; Pasternak, T.; Eiblmeier, M.; Ditengou, F.; Kochersperger, P.; Sun, J.; Wang, H.; Rennenberg, H.; Teale, W.; Paponov, I.; et al. Plastid-Localized Glutathione Reductase2–Regulated Glutathione Redox Status Is Essential for Arabidopsis Root Apical Meristem Maintenance. Plant Cell 2013, 25, 4451–4468. [Google Scholar] [CrossRef]
- Cheng, M.-C.; Ko, K.; Chang, W.-L.; Kuo, W.-C.; Chen, G.-H.; Lin, T.-P. Increased glutathione contributes to stress tolerance and global translational changes in Arabidopsis. Plant J. 2015, 83, 926–939. [Google Scholar] [CrossRef]
- Lytvyn, D.I.; Raynaud, C.; Yemets, A.I.; Bergounioux, C.; Blume, Y.B. Involvement of Inositol Biosynthesis and Nitric Oxide in the Mediation of UV-B Induced Oxidative Stress. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Vogelsang, L.; Seidel, T.; Schmidt, R.; Weber, M.; Reichelt, M.; Meyer, A.; Clemens, S.; Sharma, S.S.; Dietz, K.-J. Interference between arsenic-induced toxicity and hypoxia. Plant Cell Environ. 2019, 42, 574–590. [Google Scholar] [CrossRef] [PubMed]
- Scuffi, D.; Nietzel, T.; Fino, L.M.D.; Meyer, A.J.; Lamattina, L.; Schwarzländer, M.; Laxalt, A.M.; García-Mata, C. Hydrogen Sulfide Increases Production of NADPH Oxidase-Dependent Hydrogen Peroxide and Phospholipase D-Derived Phosphatidic Acid in Guard Cell Signaling. Plant Physiol. 2018, 176, 2532–2542. [Google Scholar] [CrossRef] [PubMed]
- Horváth, E.; Bela, K.; Holinka, B.; Riyazuddin, R.; Gallé, Á.; Hajnal, Á.; Hurton, Á.; Fehér, A.; Csiszár, J. The Arabidopsis glutathione transferases, AtGSTF8 and AtGSTU19 are involved in the maintenance of root redox homeostasis affecting meristem size and salt stress sensitivity. Plant Sci. 2019, 283, 366–374. [Google Scholar] [CrossRef]
- Anoman, A.D.; Flores-Tornero, M.; Benstein, R.M.; Blau, S.; Rosa-Téllez, S.; Bräutigam, A.; Fernie, A.R.; Muñoz-Bertomeu, J.; Schilasky, S.; Meyer, A.J.; et al. Deficiency in the Phosphorylated Pathway of Serine Biosynthesis Perturbs Sulfur Assimilation. Plant Physiol. 2019, 180, 153–170. [Google Scholar] [CrossRef]
- Waadt, R.; Köster, P.; Andrés, Z.; Waadt, C.; Bradamante, G.; Lampou, K.; Kudla, J.; Schumacher, K. Dual-sensing genetically encoded fluorescent indicators resolve the spatiotemporal coordination of cytosolic abscisic acid and second messenger dynamics in Arabidopsis. BioRxiv 2019, 844118. [Google Scholar] [CrossRef]
- Aller, I.; Rouhier, N.; Meyer, A.J. Development of roGFP2-derived redox probes for measurement of the glutathione redox potential in the cytosol of severely glutathione-deficient rml1 seedlings. Front. Plant Sci. 2013, 4. [Google Scholar] [CrossRef]
- Nietzel, T.; Elsässer, M.; Ruberti, C.; Steinbeck, J.; Ugalde, J.M.; Fuchs, P.; Wagner, S.; Ostermann, L.; Moseler, A.; Lemke, P.; et al. The fluorescent protein sensor roGFP2-Orp1 monitors in vivo H2O2 and thiol redox integration and elucidates intracellular H2O2 dynamics during elicitor-induced oxidative burst in Arabidopsis. New Phytol. 2019, 221, 1649–1664. [Google Scholar] [CrossRef]
- Costa, A.; Drago, I.; Behera, S.; Zottini, M.; Pizzo, P.; Schroeder, J.I.; Pozzan, T.; Schiavo, F.L. H2O2 in plant peroxisomes: An in vivo analysis uncovers a Ca2+-dependent scavenging system. Plant J. 2010, 62, 760–772. [Google Scholar] [CrossRef]
- Boisson-Dernier, A.; Lituiev, D.; Nestorova, A.; Franck, C.; Thirugnanarajah, S.; Grossniklaus, U. ANXUR Receptor-Like Kinases Coordinate Cell Wall Integrity with Growth at the Pollen Tube Tip Via NADPH Oxidases. PLoS Biol. 2013, 11, e1001719. [Google Scholar] [CrossRef] [PubMed]
- Exposito-Rodriguez, M.; Laissue, P.P.; Littlejohn, G.R.; Smirnoff, N.; Mullineaux, P.M. Chapter Ten—The Use of HyPer to Examine Spatial and Temporal Changes in H2O2 in High Light-Exposed Plants. In Methods in Enzymology; Cadenas, E., Packer, L., Eds.; Hydrogen Peroxide and cell signaling, Part B.; Academic Press: Cambridge, MA, USA, 2013; Volume 527, pp. 185–201. [Google Scholar]
- Hernández-Barrera, A.; Velarde-Buendía, A.; Zepeda, I.; Sanchez, F.; Quinto, C.; Sánchez-Lopez, R.; Cheung, A.Y.; Wu, H.-M.; Cardenas, L. Hyper, a Hydrogen Peroxide Sensor, Indicates the Sensitivity of the Arabidopsis Root Elongation Zone to Aluminum Treatment. Sensors 2015, 15, 855–867. [Google Scholar] [CrossRef]
- Jaipargas, E.-A.; Mathur, N.; Bou Daher, F.; Wasteneys, G.O.; Mathur, J. High Light Intensity Leads to Increased Peroxule-Mitochondria Interactions in Plants. Front. Cell Dev. Biol. 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, O.; Reshetnyak, G.; Grondin, A.; Saijo, Y.; Leonhardt, N.; Maurel, C.; Verdoucq, L. Aquaporins facilitate hydrogen peroxide entry into guard cells to mediate ABA- and pathogen-triggered stomatal closure. Proc. Natl. Acad. Sci. USA 2017, 114, 9200–9205. [Google Scholar] [CrossRef] [PubMed]
- Andrio, E.; Marino, D.; Marmeys, A.; de Segonzac, M.D.; Damiani, I.; Genre, A.; Huguet, S.; Frendo, P.; Puppo, A.; Pauly, N. Hydrogen peroxide-regulated genes in the Medicago truncatula–Sinorhizobium meliloti symbiosis. New Phytol. 2013, 198, 179–189. [Google Scholar] [CrossRef]
- Caplan, J.L.; Kumar, A.S.; Park, E.; Padmanabhan, M.S.; Hoban, K.; Modla, S.; Czymmek, K.; Dinesh-Kumar, S.P. Chloroplast stromules function during innate immunity. Dev. Cell 2015, 34, 45–57. [Google Scholar] [CrossRef]
- Exposito-Rodriguez, M.; Laissue, P.P.; Yvon-Durocher, G.; Smirnoff, N.; Mullineaux, P.M. Photosynthesis-dependent H 2 O 2 transfer from chloroplasts to nuclei provides a high-light signalling mechanism. Nat. Commun. 2017, 8, 49. [Google Scholar] [CrossRef]
- Brach, T.; Soyk, S.; Müller, C.; Hinz, G.; Hell, R.; Brandizzi, F.; Meyer, A.J. Non-invasive topology analysis of membrane proteins in the secretory pathway. Plant J. 2009, 57, 534–541. [Google Scholar] [CrossRef]
- Huang, W.-J.; Liu, H.-K.; McCormick, S.; Tang, W.-H. Tomato Pistil Factor STIG1 Promotes in Vivo Pollen Tube Growth by Binding to Phosphatidylinositol 3-Phosphate and the Extracellular Domain of the Pollen Receptor Kinase LePRK2. Plant Cell 2014, 26, 2505–2523. [Google Scholar] [CrossRef]
- Heller, J.; Meyer, A.J.; Tudzynski, P. Redox-sensitive GFP2: Use of the genetically encoded biosensor of the redox status in the filamentous fungus Botrytis cinerea. Mol. Plant Pathol. 2012, 13, 935–947. [Google Scholar] [CrossRef]
- Marschall, R.; Schumacher, J.; Siegmund, U.; Tudzynski, P. Chasing stress signals—Exposure to extracellular stimuli differentially affects the redox state of cell compartments in the wild type and signaling mutants of Botrytis cinerea. Fungal Genet. Biol. 2016, 90, 12–22. [Google Scholar] [CrossRef]
- Ronen, M.; Shalaby, S.; Horwitz, B.A. Role of the transcription factor ChAP1 in cytoplasmic redox homeostasis: Imaging with a genetically encoded sensor in the maize pathogen Cochliobolus heterostrophus. Mol. Plant. Pathol. 2013, 14, 786–790. [Google Scholar] [CrossRef] [PubMed]
- Mentges, M.; Bormann, J. Real-time imaging of hydrogen peroxide dynamics in vegetative and pathogenic hyphae of Fusarium graminearum. Sci. Rep. 2015, 5, 14980. [Google Scholar] [CrossRef] [PubMed]
- Samalova, M.; Meyer, A.J.; Gurr, S.J.; Fricker, M.D. Robust anti-oxidant defences in the rice blast fungus Magnaporthe oryzae confer tolerance to the host oxidative burst. New Phytol. 2014, 201, 556–573. [Google Scholar] [CrossRef]
- Huang, K.; Caplan, J.; Sweigard, J.A.; Czymmek, K.J.; Donofrio, N.M. Optimization of the HyPer sensor for robust real-time detection of hydrogen peroxide in the rice blast fungus. Mol. Plant. Pathol. 2017, 18, 298–307. [Google Scholar] [CrossRef]
- Delic, M.; Graf, A.B.; Koellensperger, G.; Haberhauer-Troyer, C.; Hann, S.; Mattanovich, D.; Gasser, B. Overexpression of the transcription factor Yap1 modifies intracellular redox conditions and enhances recombinant protein secretion. Microb. Cell 2014, 1, 376–386. [Google Scholar] [CrossRef]
- Delic, M.; Mattanovich, D.; Gasser, B. Monitoring intracellular redox conditions in the endoplasmic reticulum of living yeasts. FEMS Microbiol. Lett. 2010, 306, 61–66. [Google Scholar] [CrossRef]
- Østergaard, H.; Tachibana, C.; Winther, J.R. Monitoring disulfide bond formation in the eukaryotic cytosol. J. Cell Biol. 2004, 166, 337–345. [Google Scholar] [CrossRef] [PubMed]
- López-Mirabal, H.R.; Thorsen, M.; Kielland-Brandt, M.C.; Toledano, M.B.; Winther, J.R. Cytoplasmic glutathione redox status determines survival upon exposure to the thiol-oxidant 4,4′-dipyridyl disulfide. FEMS Yeast Res. 2007, 7, 391–403. [Google Scholar] [CrossRef]
- López-Mirabal, H.R.; Winther, J.R. The thiol oxidant dipyridyl disulfide can supply the PDI-Ero1p pathway with additional oxidative equivalents. Antonie Van Leeuwenhoek 2007, 92, 463–472. [Google Scholar] [CrossRef]
- Higuchi-Sanabria, R.; Vevea, J.; Charalel, J.; Sapar, M.; Pon, L. The transcriptional repressor Sum1p counteracts Sir2p in regulation of the actin cytoskeleton, mitochondrial quality control and replicative lifespan in Saccharomyces cerevisiae. Microb. Cell 2016, 3, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Qin, W.; Zhuang, G.; Zhang, X.; Chen, G.; Liu, W. Monitoring Oxidative Stress and DNA Damage Induced by Heavy Metals in Yeast Expressing a Redox-Sensitive Green Fluorescent Protein. Curr. Microbiol. 2009, 58, 504–510. [Google Scholar] [CrossRef]
- Gruhlke, M.C.H.; Portz, D.; Stitz, M.; Anwar, A.; Schneider, T.; Jacob, C.; Schlaich, N.L.; Slusarenko, A.J. Allicin disrupts the cell’s electrochemical potential and induces apoptosis in yeast. Free Radic. Biol. Med. 2010, 49, 1916–1924. [Google Scholar] [CrossRef]
- Ayer, A.; Tan, S.-X.; Grant, C.M.; Meyer, A.J.; Dawes, I.W.; Perrone, G.G. The critical role of glutathione in maintenance of the mitochondrial genome. Free Radic. Biol. Med. 2010, 49, 1956–1968. [Google Scholar] [CrossRef]
- Kritsiligkou, P.; Rand, J.D.; Weids, A.J.; Wang, X.; Kershaw, C.J.; Grant, C.M. Endoplasmic reticulum (ER) stress–induced reactive oxygen species (ROS) are detrimental for the fitness of a thioredoxin reductase mutant. J. Biol. Chem. 2018, 293, 11984–11995. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Zhang, Y.; Zou, K.; Brandman, O.; Luo, C.; Ouyang, Q.; Li, H. Molecular phenotyping of aging in single yeast cells using a novel microfluidic device. Aging Cell 2012, 11, 599–606. [Google Scholar] [CrossRef]
- Puigpinós, J.; Casas, C.; Herrero, E. Altered intracellular calcium homeostasis and endoplasmic reticulum redox state in Saccharomyces cerevisiae cells lacking Grx6 glutaredoxin. Mol. Biol. Cell 2015, 26, 104–116. [Google Scholar] [CrossRef]
- Chandel, A.; Das, K.K.; Bachhawat, A.K. Glutathione depletion activates the yeast vacuolar transient receptor potential channel, Yvc1p, by reversible glutathionylation of specific cysteines. Mol. Biol. Cell 2016, 27, 3913–3925. [Google Scholar] [CrossRef]
- Braun, N.A.; Morgan, B.; Dick, T.P.; Schwappach, B. The yeast CLC protein counteracts vesicular acidification during iron starvation. J. Cell Sci. 2010, 123, 2342–2350. [Google Scholar] [CrossRef]
- Chandel, A.; Bachhawat, A.K. Redox regulation of the yeast voltage-gated Ca 2+ channel homolog Cch1p by glutathionylation of specific cysteine residues. J. Cell Sci. 2017, 130, 2317–2328. [Google Scholar] [CrossRef]
- Morgan, B.; Ezeriņa, D.; Amoako, T.N.E.; Riemer, J.; Seedorf, M.; Dick, T.P. Multiple glutathione disulfide removal pathways mediate cytosolic redox homeostasis. Nat. Chem. Biol. 2013, 9, 119–125. [Google Scholar] [CrossRef]
- Zhao, H.; Zhang, Y.; Pan, M.; Song, Y.; Bai, L.; Miao, Y.; Huang, Y.; Zhu, X.; Song, C.-P. Dynamic imaging of cellular pH and redox homeostasis with a genetically encoded dual-functional biosensor, pHaROS, in yeast. J. Biol. Chem. 2019, 294, 15768–15780. [Google Scholar] [CrossRef]
- Knieß, R.A.; Mayer, M.P. The oxidation state of the cytoplasmic glutathione redox system does not correlate with replicative lifespan in yeast. NPJ Aging Mech. Dis. 2016, 2, 1–11. [Google Scholar] [CrossRef]
- Oku, M.; Hoseki, J.; Ichiki, Y.; Sakai, Y. A fluorescence resonance energy transfer (FRET)-based redox sensor reveals physiological role of thioredoxin in the yeast Saccharomyces cerevisiae. FEBS Lett. 2013, 587, 793–798. [Google Scholar] [CrossRef] [PubMed]
- Bodvard, K.; Peeters, K.; Roger, F.; Romanov, N.; Igbaria, A.; Welkenhuysen, N.; Palais, G.; Reiter, W.; Toledano, M.B.; Käll, M.; et al. Light-sensing via hydrogen peroxide and a peroxiredoxin. Nat. Commun. 2017, 8, 14791. [Google Scholar] [CrossRef]
- Braymer, J.J.; Stümpfig, M.; Thelen, S.; Mühlenhoff, U.; Lill, R. Depletion of thiol reducing capacity impairs cytosolic but not mitochondrial iron-sulfur protein assembly machineries. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 240–251. [Google Scholar] [CrossRef]
- Carmona, M.; de Cubas, L.; Bautista, E.; Moral-Blanch, M.; Medraño-Fernández, I.; Sitia, R.; Boronat, S.; Ayté, J.; Hidalgo, E. Monitoring cytosolic H 2 O 2 fluctuations arising from altered plasma membrane gradients or from mitochondrial activity. Nat. Commun. 2019, 10, 4526. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, S.K.; Stockdreher, Y.; Wandrey, G.; Hosseinpour Tehrani, H.; Zambanini, T.; Meyer, A.J.; Büchs, J.; Blank, L.M.; Schwarzländer, M.; Wierckx, N. Online in vivo monitoring of cytosolic NAD redox dynamics in Ustilago maydis. Biochim. Biophys. Bioenerg. 2018, 1859, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, K.; Nishimaki, Y.; Owa, M.; Hisabori, T.; Wakabayashi, K.-I. Assessment of the flagellar redox potential in Chlamydomonas reinhardtii using a redox-sensitive fluorescent protein, Oba-Qc. Biochem. Biophys. Res. Commun. 2018, 503, 2083–2088. [Google Scholar] [CrossRef]
- van Creveld, S.G.; Rosenwasser, S.; Schatz, D.; Koren, I.; Vardi, A. Early perturbation in mitochondria redox homeostasis in response to environmental stress predicts cell fate in diatoms. ISME J. 2015, 9, 385–395. [Google Scholar] [CrossRef]
- Rosenwasser, S.; van Creveld, S.G.; Schatz, D.; Malitsky, S.; Tzfadia, O.; Aharoni, A.; Levin, Y.; Gabashvili, A.; Feldmesser, E.; Vardi, A. Mapping the diatom redox-sensitive proteome provides insight into response to nitrogen stress in the marine environment. Proc. Natl. Acad. Sci. USA 2014, 111, 2740–2745. [Google Scholar] [CrossRef]
- Kasozi, D.; Mohring, F.; Rahlfs, S.; Meyer, A.J.; Becker, K. Real-time imaging of the intracellular glutathione redox potential in the malaria parasite Plasmodium falciparum. PLoS Pathog 2013, 9, e1003782. [Google Scholar] [CrossRef]
- Bielitza, M.; Belorgey, D.; Ehrhardt, K.; Johann, L.; Lanfranchi, D.A.; Gallo, V.; Schwarzer, E.; Mohring, F.; Jortzik, E.; Williams, D.L.; et al. Antimalarial NADPH-Consuming Redox-Cyclers As Superior Glucose-6-Phosphate Dehydrogenase Deficiency Copycats. Antioxid. Redox Signal. 2015, 22, 1337–1351. [Google Scholar] [CrossRef]
- Mohring, F.; Jortzik, E.; Becker, K. Comparison of methods probing the intracellular redox milieu in Plasmodium falciparum. Mol. Biochem. Parasitol. 2016, 206, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Rahbari, M.; Rahlfs, S.; Jortzik, E.; Bogeski, I.; Becker, K. H2O2 dynamics in the malaria parasite Plasmodium falciparum. PLoS ONE 2017, 12, e0174837. [Google Scholar] [CrossRef]
- Franco, J.; Medeiros, A.; Benítez, D.; Perelmuter, K.; Serra, G.; Comini, M.A.; Scarone, L. In vitro activity and mode of action of distamycin analogues against African trypanosomes. Eur. J. Med. Chem. 2017, 126, 776–788. [Google Scholar] [CrossRef]
- Franco, J.; Sardi, F.; Szilágyi, L.; Kövér, K.E.; Fehér, K.; Comini, M.A. Diglycosyl diselenides alter redox homeostasis and glucose consumption of infective African trypanosomes. Int. J. Parasitol. Drugs Drug Resist. 2017, 7, 303–313. [Google Scholar] [CrossRef]
- Rivas, F.; Medeiros, A.; Rodríguez Arce, E.; Comini, M.; Ribeiro, C.M.; Pavan, F.R.; Gambino, D. New heterobimetallic ferrocenyl derivatives: Evaluation of their potential as prospective agents against trypanosomatid parasites and Mycobacterium tuberculosis. J. Inorg. Biochem. 2018, 187, 73–84. [Google Scholar] [CrossRef]
- Rodríguez Arce, E.; Putzu, E.; Lapier, M.; Maya, J.D.; Olea Azar, C.; Echeverría, G.A.; Piro, O.E.; Medeiros, A.; Sardi, F.; Comini, M.; et al. New heterobimetallic ferrocenyl derivatives are promising antitrypanosomal agents. Dalton Trans. 2019, 48, 7644–7658. [Google Scholar] [CrossRef]
- Franco, J.; Scarone, L.; Comini, M.A. Novel distamycin analogues that block the cell cycle of African trypanosomes with high selectivity and potency. Eur. J. Med. Chem. 2020, 189, 112043. [Google Scholar] [CrossRef]
- Bogacz, M.; Dirdjaja, N.; Wimmer, B.; Habich, C.; Krauth-Siegel, R.L. The mitochondrial peroxiredoxin displays distinct roles in different developmental stages of African trypanosomes. Redox Biol. 2020, 34, 101547. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, S.; Janakiraman, B.; Kumar, L.; Radhakrishnan, S.K. A cell cycle-controlled redox switch regulates the topoisomerase IV activity. Genes Dev. 2015, 29, 1175–1187. [Google Scholar] [CrossRef]
- Wang, X.; Schwarzer, C.; Hybiske, K.; Machen, T.E.; Stephens, R.S. Developmental stage oxidoreductive states of Chlamydia and infected host cells. MBio 2014, 5, e01924. [Google Scholar] [CrossRef]
- Van der Heijden, J.; Vogt, S.L.; Reynolds, L.A.; Peña-Díaz, J.; Tupin, A.; Aussel, L.; Finlay, B.B. Exploring the redox balance inside gram-negative bacteria with redox-sensitive GFP. Free Radic. Biol. Med. 2016, 91, 34–44. [Google Scholar] [CrossRef]
- Zhang, W.; Zheng, W.; Mao, M.; Yang, Y. Highly efficient folding of multi-disulfide proteins in superoxidizing Escherichia coli cytoplasm. Biotechnol. Bioeng. 2014, 111, 2520–2527. [Google Scholar] [CrossRef]
- Arias-Barreiro, C.R.; Okazaki, K.; Koutsaftis, A.; Inayat-Hussain, S.H.; Tani, A.; Katsuhara, M.; Kimbara, K.; Mori, I.C. A bacterial biosensor for oxidative stress using the constitutively expressed redox-sensitive protein roGFP2. Sensors 2010, 10, 6290–6306. [Google Scholar] [CrossRef] [PubMed]
- Ooi, L.; Heng, L.Y.; Mori, I.C. A high-throughput oxidative stress biosensor based on Escherichia coli roGFP2 cells immobilized in a k-carrageenan matrix. Sensors 2015, 15, 2354–2368. [Google Scholar] [CrossRef]
- Degrossoli, A.; Müller, A.; Xie, K.; Schneider, J.F.; Bader, V.; Winklhofer, K.F.; Meyer, A.J.; Leichert, L.I. Neutrophil-generated HOCl leads to non-specific thiol oxidation in phagocytized bacteria. eLife 2018, 7, e32288. [Google Scholar] [CrossRef]
- Reuter, W.H.; Masuch, T.; Ke, N.; Lenon, M.; Radzinski, M.; Van Loi, V.; Ren, G.; Riggs, P.; Antelmann, H.; Reichmann, D.; et al. Utilizing redox-sensitive GFP fusions to detect in vivo redox changes in a genetically engineered prokaryote. Redox Biol. 2019, 26, 101280. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Zhang, N.; Yang, Y.; Wang, Y.; Chu, J.; Zhuang, Y.; Zhang, S. The effect of redox environment on l-lactic acid production by Lactobacillus paracasei—A proof by genetically encoded in vivo NADH biosensor. Process. Biochem. 2015, 50, 2029–2034. [Google Scholar] [CrossRef]
- Chen, J.; Shen, J.; Solem, C.; Jensen, P.R. Oxidative Stress at High Temperatures in Lactococcus lactis Due to an Insufficient Supply of Riboflavin. Appl. Env. Microbiol. 2013, 79, 6140. [Google Scholar] [CrossRef]
- Ishikawa, M.; Tanaka, Y.; Suzuki, R.; Kimura, K.; Tanaka, K.; Kamiya, K.; Ito, H.; Kato, S.; Kamachi, T.; Hori, K.; et al. Real-time monitoring of intracellular redox changes in Methylococcus capsulatus (Bath) for efficient bioconversion of methane to methanol. Bioresour. Technol. 2017, 241, 1157–1161. [Google Scholar] [CrossRef]
- Black, H.D.; Xu, W.; Hortle, E.; Robertson, S.I.; Britton, W.J.; Kaur, A.; New, E.J.; Witting, P.K.; Chami, B.; Oehlers, S.H. The cyclic nitroxide antioxidant 4-methoxy-TEMPO decreases mycobacterial burden in vivo through host and bacterial targets. Free Radic. Biol. Med. 2019, 135, 157–166. [Google Scholar] [CrossRef]
- Nambi, S.; Long, J.E.; Mishra, B.B.; Baker, R.; Murphy, K.C.; Olive, A.J.; Nguyen, H.P.; Shaffer, S.A.; Sassetti, C.M. The Oxidative Stress Network of Mycobacterium tuberculosis Reveals Coordination between Radical Detoxification Systems. Cell Host Microbe 2015, 17, 829–837. [Google Scholar] [CrossRef]
- Libardo, M.D.J.; de la Fuente-Nuñez, C.; Anand, K.; Krishnamoorthy, G.; Kaiser, P.; Pringle, S.C.; Dietz, C.; Pierce, S.; Smith, M.B.; Barczak, A.; et al. Phagosomal Copper-Promoted Oxidative Attack on Intracellular Mycobacterium tuberculosis. ACS Infect. Dis. 2018, 4, 1623–1634. [Google Scholar] [CrossRef] [PubMed]
- Nair, R.R.; Sharan, D.; Sebastian, J.; Swaminath, S.; Ajitkumar, P. Heterogeneity of ROS levels in antibiotic-exposed mycobacterial subpopulations confers differential susceptibility. Microbiology 2019, 165, 668–682. [Google Scholar] [CrossRef]
- Nair, R.R.; Sharan, D.; Ajitkumar, P. A Minor Subpopulation of Mycobacteria Inherently Produces High Levels of Reactive Oxygen Species That Generate Antibiotic Resisters at High Frequency From Itself and Enhance Resister Generation From Its Major Kin Subpopulation. Front. Microbiol. 2019, 10, 1842. [Google Scholar] [CrossRef]
- Swaminath, S.; Paul, A.; Pradhan, A.; Sebastian, J.; Nair, R.R.; Ajitkumar, P. Mycobacterium smegmatis moxifloxacin persister cells produce high levels of hydroxyl radical, generating genetic resisters selectable not only with moxifloxacin, but also with ethambutol and isoniazid. Microbiology 2020, 166, 180–198. [Google Scholar] [CrossRef] [PubMed]
- Bhat, S.A.; Iqbal, I.K.; Kumar, A. Imaging the NADH:NAD+ Homeostasis for Understanding the Metabolic Response of Mycobacterium to Physiologically Relevant Stresses. Front. Cell. Infect. Microbiol. 2016, 6, 145. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, P.; Shankaran, D.; Bothra, A.; Gandotra, S.; Rao, V. The MmpS6-MmpL6 Operon Is an Oxidative Stress Response System Providing Selective Advantage to Mycobacterium tuberculosis in Stress. J. Infect. Dis. 2019, 219, 459–469. [Google Scholar] [CrossRef]
- Nandy, A.; Mondal, A.K.; Pandey, R.; Arumugam, P.; Dawa, S.; Jaisinghani, N.; Rao, V.; Dash, D.; Gandotra, S. Adipocyte Model of Mycobacterium tuberculosis Infection Reveals Differential Availability of Iron to Bacilli in the Lipid-Rich Caseous Environment. Infect. Immun. 2018, 86, e00041-18. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, J.; Swaminath, S.; Nair, R.R.; Jakkala, K.; Pradhan, A.; Ajitkumar, P. De Novo Emergence of Genetically Resistant Mutants of Mycobacterium tuberculosis from the Persistence Phase Cells Formed against Antituberculosis Drugs In Vitro. Antimicrob. Agents Chemother. 2017, 61, e01343-16. [Google Scholar] [CrossRef] [PubMed]
- Baker, J.J.; Johnson, B.K.; Abramovitch, R.B. Slow growth of Mycobacterium tuberculosis at acidic pH is regulated by phoPR and host-associated carbon sources. Mol. Microbiol. 2014, 94, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Coulson, G.B.; Johnson, B.K.; Zheng, H.; Colvin, C.J.; Fillinger, R.J.; Haiderer, E.R.; Hammer, N.D.; Abramovitch, R.B. Targeting Mycobacterium tuberculosis Sensitivity to Thiol Stress at Acidic pH Kills the Bacterium and Potentiates Antibiotics. Cell Chem. Biol. 2017, 24, 993–1004.e4. [Google Scholar] [CrossRef]
- Liu, T.-H.; Yaghmour, M.A.; Lee, M.-H.; Gradziel, T.M.; Leveau, J.H.J.; Bostock, R.M. An roGFP2-Based Bacterial Bioreporter for Redox Sensing of Plant Surfaces. Phytopathology 2019, 110, 297–308. [Google Scholar] [CrossRef]
- Wilkening, S.; Schmitt, F.-J.; Lenz, O.; Zebger, I.; Horch, M.; Friedrich, T. Discriminating changes in intracellular NADH/NAD+ levels due to anoxicity and H2 supply in R. eutropha cells using the Frex fluorescence sensor. Biochim. Biophys. Acta Bioenerg. 2019, 1860, 148062. [Google Scholar] [CrossRef]
- Van der Heijden, J.; Reynolds, L.A.; Deng, W.; Mills, A.; Scholz, R.; Imami, K.; Foster, L.J.; Duong, F.; Finlay, B.B. Salmonella Rapidly Regulates Membrane Permeability To Survive Oxidative Stress. mBio 2016, 7, e01238-16. [Google Scholar] [CrossRef]
- Van der Heijden, J.; Bosman, E.S.; Reynolds, L.A.; Finlay, B.B. Direct measurement of oxidative and nitrosative stress dynamics in Salmonella inside macrophages. Proc. Natl. Acad. Sci. USA 2014, 112, 560–565. [Google Scholar] [CrossRef]
- Linzner, N.; Loi, V.V.; Fritsch, V.N.; Tung, Q.N.; Stenzel, S.; Wirtz, M.; Hell, R.; Hamilton, C.J.; Tedin, K.; Fulde, M.; et al. Staphylococcus aureus Uses the Bacilliredoxin (BrxAB)/Bacillithiol Disulfide Reductase (YpdA) Redox Pathway to Defend Against Oxidative Stress Under Infections. Front. Microbiol. 2019, 10, 1355. [Google Scholar] [CrossRef]
- Loi, V.V.; Busche, T.; Preuß, T.; Kalinowski, J.; Bernhardt, J.; Antelmann, H. The AGXX® Antimicrobial Coating Causes a Thiol-Specific Oxidative Stress Response and Protein S-bacillithiolation in Staphylococcus aureus. Front. Microbiol. 2018, 9, 3037. [Google Scholar] [CrossRef]
- Fritsch, V.N.; Loi, V.V.; Busche, T.; Sommer, A.; Tedin, K.; Nürnberg, D.J.; Kalinowski, J.; Bernhardt, J.; Fulde, M.; Antelmann, H. The MarR-Type Repressor MhqR Confers Quinone and Antimicrobial Resistance in Staphylococcus aureus. Antioxid. Redox Signal. 2019, 31, 1235–1252. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.; Wang, X.; Dong, Y.; Hu, Q.; Zhao, Z.; Zhu, Y.; Dong, L.; Bai, F.; Dong, X. A Streptococcus aquaporin acts as peroxiporin for efflux of cellular hydrogen peroxide and alleviation of oxidative stress. J. Biol. Chem. 2019, 294, 4583–4595. [Google Scholar] [CrossRef]
- Chen, A.H.; Robinson-Mosher, A.; Savage, D.F.; Silver, P.A.; Polka, J.K. The Bacterial Carbon-Fixing Organelle Is Formed by Shell Envelopment of Preassembled Cargo. PLoS ONE 2013, 8, e76127. [Google Scholar] [CrossRef]
- Morlino, G.; Barreiro, O.; Baixauli, F.; Robles-Valero, J.; González-Granado, J.M.; Villa-Bellosta, R.; Cuenca, J.; Sánchez-Sorzano, C.O.; Veiga, E.; Martín-Cófreces, N.B.; et al. Miro-1 Links Mitochondria and Microtubule Dynein Motors To Control Lymphocyte Migration and Polarity. Mol. Cell. Biol. 2014, 34, 1412–1426. [Google Scholar] [CrossRef] [PubMed]
- Coffman, J.A.; Coluccio, A.; Planchart, A.; Robertson, A.J. Oral-aboral axis specification in the sea urchin embryo III. Role of mitochondrial redox signaling via H2O2. Dev. Biol. 2009, 330, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.C. Antimicrobial Actions of Reactive Oxygen Species. mBio 2011, 2, e00141-11. [Google Scholar] [CrossRef]
- Forrester, S.J.; Kikuchi, D.S.; Hernandes, M.S.; Xu, Q.; Griendling, K.K. Reactive Oxygen Species in Metabolic and Inflammatory Signaling. Circ. Res. 2018, 122, 877–902. [Google Scholar] [CrossRef]
- Mittal, M.; Siddiqui, M.R.; Tran, K.; Reddy, S.P.; Malik, A.B. Reactive oxygen species in inflammation and tissue injury. Antioxid. Redox Signal. 2014, 20, 1126–1167. [Google Scholar] [CrossRef]
- Panday, A.; Sahoo, M.K.; Osorio, D.; Batra, S. NADPH oxidases: An overview from structure to innate immunity-associated pathologies. Cell Mol. Immunol. 2015, 12, 5–23. [Google Scholar] [CrossRef]
- Yoo, S.K.; Starnes, T.W.; Deng, Q.; Huttenlocher, A. Lyn is a redox sensor that mediates leukocyte wound attraction in vivo. Nature 2011, 480, 109–112. [Google Scholar] [CrossRef]
- Hoffmann, E.K.; Lambert, I.H.; Pedersen, S.F. Physiology of Cell Volume Regulation in Vertebrates. Physiol. Rev. 2009, 89, 193–277. [Google Scholar] [CrossRef] [PubMed]
- Quatresous, E.; Legrand, C.; Pouvreau, S. Mitochondria-targeted cpYFP: pH or superoxide sensor? J. Gen. Physiol. 2012, 140, 567–570. [Google Scholar] [CrossRef]
- Michalopoulos, G.K. Liver Regeneration. Science 1997, 276, 60–66. [Google Scholar] [CrossRef]
- Rieger, S.; Sagasti, A. Hydrogen Peroxide Promotes Injury-Induced Peripheral Sensory Axon Regeneration in the Zebrafish Skin. PLoS Biol. 2011, 9, e1000621. [Google Scholar] [CrossRef]
- Raichle, M.E.; Gusnard, D.A. Appraising the brain’s energy budget. Proc. Natl. Acad. Sci. USA 2002, 99, 10237–10239. [Google Scholar] [CrossRef] [PubMed]
- Shulman, R.G.; Rothman, D.L.; Behar, K.L.; Hyder, F. Energetic basis of brain activity: Implications for neuroimaging. Trends Neurosci. 2004, 27, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Hyder, F.; Rothman, D.L.; Bennett, M.R. Cortical energy demands of signaling and nonsignaling components in brain are conserved across mammalian species and activity levels. Proc. Natl. Acad. Sci. USA 2013, 110, 3549–3554. [Google Scholar] [CrossRef]
- Quijano, C.; Trujillo, M.; Castro, L.; Trostchansky, A. Interplay between oxidant species and energy metabolism. Redox Biol. 2016, 8, 28–42. [Google Scholar] [CrossRef]
- Watts, M.E.; Pocock, R.; Claudianos, C. Brain Energy and Oxygen Metabolism: Emerging Role in Normal Function and Disease. Front. Mol. Neurosci. 2018, 11, 216. [Google Scholar] [CrossRef]
- Van Battum, E.Y.; Gunput, R.-A.F.; Lemstra, S.; Groen, E.J.N.; Yu, K.L.; Adolfs, Y.; Zhou, Y.; Hoogenraad, C.C.; Yoshida, Y.; Schachner, M.; et al. The intracellular redox protein MICAL-1 regulates the development of hippocampal mossy fibre connections. Nat. Commun. 2014, 5, 4317. [Google Scholar] [CrossRef]
- Valek, L.; Häussler, A.; Dröse, S.; Eaton, P.; Schröder, K.; Tegeder, I. Redox-guided axonal regrowth requires cyclic GMP dependent protein kinase 1: Implication for neuropathic pain. Redox Biol. 2017, 11, 176–191. [Google Scholar] [CrossRef]
- Kumar, A.; Yegla, B.; Foster, T.C. Redox Signaling in Neurotransmission and Cognition During Aging. Antioxid. Redox Signal. 2018, 28, 1724–1745. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, G.; Colín-González, A.L.; Rangel-López, E.; Chavarría, A.; Santamaría, A. Redox Signaling, Neuroinflammation, and Neurodegeneration. Antioxid. Redox Signal. 2018, 28, 1626–1651. [Google Scholar] [CrossRef]
- Olguín-Albuerne, M.; Morán, J. Redox Signaling Mechanisms in Nervous System Development. Antioxid. Redox Signal. 2018, 28, 1603–1625. [Google Scholar] [CrossRef]
- Maciel-Barón, L.Á.; Moreno-Blas, D.; Morales-Rosales, S.L.; González-Puertos, V.Y.; López-Díazguerrero, N.E.; Torres, C.; Castro-Obregón, S.; Königsberg, M. Cellular Senescence, Neurological Function, and Redox State. Antioxid. Redox Signal. 2018, 28, 1704–1723. [Google Scholar] [CrossRef] [PubMed]
- Bórquez, D.A.; Urrutia, P.J.; Wilson, C.; van Zundert, B.; Núñez, M.T.; González-Billault, C. Dissecting the role of redox signaling in neuronal development. J. Neurochem. 2016, 137, 506–517. [Google Scholar] [CrossRef] [PubMed]
- Noble, M.; Smith, J.; Power, J.; Mayer-Pröschel, M. Redox State as a Central Modulator of Precursor Cell Function. Ann. N. Y. Acad. Sci. USA 2006, 991, 251–271. [Google Scholar] [CrossRef]
- Noble, M.; Mayer-Pröschel, M.; Pröschel, C. Redox Regulation of Precursor Cell Function: Insights and Paradoxes. Antioxid. Redox Signal. 2005, 7, 1456–1467. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Huang, S.; Gao, H.; Liu, Y.; Qi, J.; Chen, P.; Wang, C.; Scragg, J.L.; Vakurov, A.; Peers, C.; et al. Redox-Dependent Modulation of T-Type Ca2+ Channels in Sensory Neurons Contributes to Acute Anti-Nociceptive Effect of Substance P. Antioxid. Redox Signal. 2016, 25, 233–251. [Google Scholar] [CrossRef]
- Yang, Y.-J.; Wu, P.-F.; Long, L.-H.; Yu, D.-F.; Wu, W.-N.; Hu, Z.-L.; Fu, H.; Xie, N.; Jin, Y.; Ni, L.; et al. Reversal of aging-associated hippocampal synaptic plasticity deficits by reductants via regulation of thiol redox and NMDA receptor function: Reductants reverse aging-associated impairment of LTP. Aging Cell 2010, 9, 709–721. [Google Scholar] [CrossRef]
- Bodhinathan, K.; Kumar, A.; Foster, T.C. Intracellular Redox State Alters NMDA Receptor Response during Aging through Ca2+/Calmodulin-Dependent Protein Kinase II. J. Neurosci. 2010, 30, 1914–1924. [Google Scholar] [CrossRef]
- Bernard, C.; Hirsch, J.C.; Khazipov, R.; Ben-Ari, Y.; Gozlan, H. Redox modulation of synaptic responses and plasticity in rat CA1 hippocampal neurons. Exp. Brain Res. 1997, 113, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Thinschmidt, J.S.; Foster, T.C. Subunit contribution to NMDA receptor hypofunction and redox sensitivity of hippocampal synaptic transmission during aging. Aging 2019, 11, 5140–5157. [Google Scholar] [CrossRef]
- Munnamalai, V.; Suter, D.M. Reactive oxygen species regulate F-actin dynamics in neuronal growth cones and neurite outgrowth. J. Neurochem. 2009, 108, 644–661. [Google Scholar] [CrossRef]
- Soerensen, J.; Jakupoglu, C.; Beck, H.; Förster, H.; Schmidt, J.; Schmahl, W.; Schweizer, U.; Conrad, M.; Brielmeier, M. The Role of Thioredoxin Reductases in Brain Development. PLoS ONE 2008, 3, e1813. [Google Scholar] [CrossRef]
- Celotto, A.M.; Liu, Z.; VanDemark, A.P.; Palladino, M.J. A novel Drosophila SOD2 mutant demonstrates a role for mitochondrial ROS in neurodevelopment and disease. Brain Behav. 2012, 2, 424–434. [Google Scholar] [CrossRef] [PubMed]
- Borchert, A.; Wang, C.C.; Ufer, C.; Schiebel, H.; Savaskan, N.E.; Kuhn, H. The Role of Phospholipid Hydroperoxide Glutathione Peroxidase Isoforms in Murine Embryogenesis. J. Biol. Chem. 2006, 281, 19655–19664. [Google Scholar] [CrossRef]
- Dickinson, B.C.; Peltier, J.; Stone, D.; Schaffer, D.V.; Chang, C.J. Nox2 redox signaling maintains essential cell populations in the brain. Nat. Chem. Biol. 2011, 7, 106–112. [Google Scholar] [CrossRef]
- Cobley, J.N.; Fiorello, M.L.; Bailey, D.M. 13 reasons why the brain is susceptible to oxidative stress. Redox Biol. 2018, 15, 490–503. [Google Scholar] [CrossRef] [PubMed]
- Serrano, F.; Klann, E. Reactive oxygen species and synaptic plasticity in the aging hippocampus. Ageing Res. Rev. 2004, 3, 431–443. [Google Scholar] [CrossRef]
- Liu, R.; Liu, I.Y.; Bi, X.; Thompson, R.F.; Doctrow, S.R.; Malfroy, B.; Baudry, M. Reversal of age-related learning deficits and brain oxidative stress in mice with superoxide dismutase/catalase mimetics. Proc. Natl. Acad. Sci. USA 2003, 100, 8526–8531. [Google Scholar] [CrossRef]
- Tabrizi, S.J.; Workman, J.; Hart, P.E.; Mangiarini, L.; Mahal, A.; Bates, G.; Cooper, J.M.; Schapira, A.H. Mitochondrial dysfunction and free radical damage in the Huntington R6/2 transgenic mouse. Ann. Neurol. 2000, 47, 80–86. [Google Scholar] [CrossRef]
- Zhou, C.; Huang, Y.; Przedborski, S. Oxidative Stress in Parkinson’s Disease: A Mechanism of Pathogenic and Therapeutic Significance. Ann. N. Y. Acad. Sci. 2008, 1147, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Tönnies, E.; Trushina, E. Oxidative Stress, Synaptic Dysfunction, and Alzheimer’s Disease. J. Alzheimer’s Dis. 2017, 57, 1105–1121. [Google Scholar] [CrossRef]
- Butterfield, D.A.; Halliwell, B. Oxidative stress, dysfunctional glucose metabolism and Alzheimer disease. Nat. Rev. Neurosci. 2019, 20, 148–160. [Google Scholar] [CrossRef]
- Pollari, E.; Goldsteins, G.; Bart, G.; Koistinaho, J.; Giniatullin, R. The role of oxidative stress in degeneration of the neuromuscular junction in amyotrophic lateral sclerosis. Front. Cell. Neurosci. 2014, 8. [Google Scholar] [CrossRef]
- Carrì, M.T.; Valle, C.; Bozzo, F.; Cozzolino, M. Oxidative stress and mitochondrial damage: Importance in non-SOD1 ALS. Front. Cell. Neurosci. 2015, 9. [Google Scholar] [CrossRef] [PubMed]
- Allen, C.L.; Bayraktutan, U. Oxidative Stress and Its Role in the Pathogenesis of Ischaemic Stroke. Int. J. Stroke 2009, 4, 461–470. [Google Scholar] [CrossRef]
- Abdul-Muneer, P.M.; Chandra, N.; Haorah, J. Interactions of Oxidative Stress and Neurovascular Inflammation in the Pathogenesis of Traumatic Brain Injury. Mol. Neurobiol. 2015, 51, 966–979. [Google Scholar] [CrossRef]
- De Felice, C.; Ciccoli, L.; Leoncini, S.; Signorini, C.; Rossi, M.; Vannuccini, L.; Guazzi, G.; Latini, G.; Comporti, M.; Valacchi, G.; et al. Systemic oxidative stress in classic Rett syndrome. Free Radic. Biol. Med. 2009, 47, 440–448. [Google Scholar] [CrossRef]
- Bjørklund, G.; Meguid, N.A.; El-Bana, M.A.; Tinkov, A.A.; Saad, K.; Dadar, M.; Hemimi, M.; Skalny, A.V.; Hosnedlová, B.; Kizek, R.; et al. Oxidative Stress in Autism Spectrum Disorder. Mol. Neurobiol. 2020, 57, 2314–2332. [Google Scholar] [CrossRef] [PubMed]
- Kulak, A.; Steullet, P.; Cabungcal, J.-H.; Werge, T.; Ingason, A.; Cuenod, M.; Do, K.Q. Redox Dysregulation in the Pathophysiology of Schizophrenia and Bipolar Disorder: Insights from Animal Models. Antioxid. Redox Signal. 2013, 18, 1428–1443. [Google Scholar] [CrossRef]
- Cabungcal, J.-H.; Steullet, P.; Kraftsik, R.; Cuenod, M.; Do, K.Q. A developmental redox dysregulation leads to spatio-temporal deficit of parvalbumin neuron circuitry in a schizophrenia mouse model. Schizophr. Res. 2019, 213, 96–106. [Google Scholar] [CrossRef]
- Kandlur, A.; Satyamoorthy, K.; Gangadharan, G. Oxidative Stress in Cognitive and Epigenetic Aging: A Retrospective Glance. Front. Mol. Neurosci. 2020, 13, 41. [Google Scholar] [CrossRef]
- Salim, S. Oxidative stress and psychological disorders. Curr. Neuropharmacol. 2014, 12, 140–147. [Google Scholar] [CrossRef]
- Su, K.G.; Banker, G.; Bourdette, D.; Forte, M. Axonal degeneration in multiple sclerosis: The mitochondrial hypothesis. Curr. Neurol. Neurosci. Rep. 2009, 9, 411–417. [Google Scholar] [CrossRef]
- Lei, X.G.; Zhu, J.-H.; Cheng, W.-H.; Bao, Y.; Ho, Y.-S.; Reddi, A.R.; Holmgren, A.; Arnér, E.S.J. Paradoxical Roles of Antioxidant Enzymes: Basic Mechanisms and Health Implications. Physiol. Rev. 2016, 96, 307–364. [Google Scholar] [CrossRef]
- Hatori, Y.; Yan, Y.; Schmidt, K.; Furukawa, E.; Hasan, N.M.; Yang, N.; Liu, C.-N.; Sockanathan, S.; Lutsenko, S. Neuronal differentiation is associated with a redox-regulated increase of copper flow to the secretory pathway. Nat. Commun. 2016, 7, 10640. [Google Scholar] [CrossRef]
- Yin, B.; Barrionuevo, G.; Weber, S.G. Mitochondrial GSH Systems in CA1 Pyramidal Cells and Astrocytes React Differently during Oxygen-Glucose Deprivation and Reperfusion. ACS Chem. Neurosci. 2018, 9, 738–748. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Hou, S.; Jiang, J.; Sekutowicz, M.; Kelly, J.; Bacskai, B.J. Rapid cell death is preceded by amyloid plaque-mediated oxidative stress. Proc. Natl. Acad. Sci. USA 2013, 110, 7904–7909. [Google Scholar] [CrossRef]
- Graves, S.M.; Xie, Z.; Stout, K.A.; Zampese, E.; Burbulla, L.F.; Shih, J.C.; Kondapalli, J.; Patriarchi, T.; Tian, L.; Brichta, L.; et al. Dopamine metabolism by a monoamine oxidase mitochondrial shuttle activates the electron transport chain. Nat. Neurosci. 2020, 23, 15–20. [Google Scholar] [CrossRef]
- Yin, B.; Barrionuevo, G.; Weber, S.G. Optimized Real-Time Monitoring of Glutathione Redox Status in Single Pyramidal Neurons in Organotypic Hippocampal Slices during Oxygen–Glucose Deprivation and Reperfusion. ACS Chem. Neurosci. 2015, 6, 1838–1848. [Google Scholar] [CrossRef]
- Großer, E.; Hirt, U.; Janc, O.A.; Menzfeld, C.; Fischer, M.; Kempkes, B.; Vogelgesang, S.; Manzke, T.U.; Opitz, L.; Salinas-Riester, G.; et al. Oxidative burden and mitochondrial dysfunction in a mouse model of Rett syndrome. Neurobiol. Dis. 2012, 48, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Miller, N.; Shi, H.; Zelikovich, A.S.; Ma, Y.-C. Motor neuron mitochondrial dysfunction in spinal muscular atrophy. Hum. Mol. Genet. 2016, 25, 3395–3406. [Google Scholar] [CrossRef]
- Bebensee, D.F.; Can, K.; Müller, M. Increased Mitochondrial Mass and Cytosolic Redox Imbalance in Hippocampal Astrocytes of a Mouse Model of Rett Syndrome: Subcellular Changes Revealed by Ratiometric Imaging of JC-1 and roGFP1 Fluorescence. Oxidative Med. Cell. Longev. 2017, 2017, 1–15. [Google Scholar] [CrossRef]
- Adam, Y.; Kim, J.J.; Lou, S.; Zhao, Y.; Xie, M.E.; Brinks, D.; Wu, H.; Mostajo-Radji, M.A.; Kheifets, S.; Parot, V.; et al. Voltage imaging and optogenetics reveal behaviour-dependent changes in hippocampal dynamics. Nature 2019, 569, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Borden, P.M.; Zhang, P.; Shivange, A.V.; Marvin, J.S.; Cichon, J.; Dan, C.; Podgorski, K.; Figueiredo, A.; Novak, O.; Tanimoto, M.; et al. A fast genetically encoded fluorescent sensor for faithful in vivo acetylcholine detection in mice, fish, worms and flies. BioRxiv 2020. [Google Scholar] [CrossRef]
- Feng, J.; Zhang, C.; Lischinsky, J.E.; Jing, M.; Zhou, J.; Wang, H.; Zhang, Y.; Dong, A.; Wu, Z.; Wu, H.; et al. A Genetically Encoded Fluorescent Sensor for Rapid and Specific In Vivo Detection of Norepinephrine. Neuron 2019, 102, 745–761.e8. [Google Scholar] [CrossRef] [PubMed]
- Marvin, J.S.; Shimoda, Y.; Magloire, V.; Leite, M.; Kawashima, T.; Jensen, T.P.; Kolb, I.; Knott, E.L.; Novak, O.; Podgorski, K.; et al. A genetically encoded fluorescent sensor for in vivo imaging of GABA. Nat. Methods 2019, 16, 763–770. [Google Scholar] [CrossRef]
- Marvin, J.S.; Borghuis, B.G.; Tian, L.; Cichon, J.; Harnett, M.T.; Akerboom, J.; Gordus, A.; Renninger, S.L.; Chen, T.-W.; Bargmann, C.I.; et al. An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat. Methods 2013, 10, 162–170. [Google Scholar] [CrossRef]
- Sun, F.; Zeng, J.; Jing, M.; Zhou, J.; Feng, J.; Owen, S.F.; Luo, Y.; Li, F.; Wang, H.; Yamaguchi, T.; et al. A Genetically Encoded Fluorescent Sensor Enables Rapid and Specific Detection of Dopamine in Flies, Fish, and Mice. Cell 2018, 174, 481–496.e19. [Google Scholar] [CrossRef]
- Díaz-García, C.M.; Lahmann, C.; Martínez-François, J.R.; Li, B.; Koveal, D.; Nathwani, N.; Rahman, M.; Keller, J.P.; Marvin, J.S.; Looger, L.L.; et al. Quantitative in vivo imaging of neuronal glucose concentrations with a genetically encoded fluorescence lifetime sensor. J. Neurosci. Res. 2019, 97, 946–960. [Google Scholar] [CrossRef]
- Harada, K.; Ito, M.; Wang, X.; Tanaka, M.; Wongso, D.; Konno, A.; Hirai, H.; Hirase, H.; Tsuboi, T.; Kitaguchi, T. Red fluorescent protein-based cAMP indicator applicable to optogenetics and in vivo imaging. Sci. Rep. 2017, 7, 7351. [Google Scholar] [CrossRef]
- Subach, O.M.; Sotskov, V.P.; Plusnin, V.V.; Gruzdeva, A.M.; Barykina, N.V.; Ivashkina, O.I.; Anokhin, K.V.; Nikolaeva, A.Y.; Korzhenevskiy, D.A.; Vlaskina, A.V.; et al. Novel Genetically Encoded Bright Positive Calcium Indicator NCaMP7 Based on the mNeonGreen Fluorescent Protein. Int. J. Mol. Sci. 2020, 21, 1644. [Google Scholar] [CrossRef]
- Ziv, Y.; Burns, L.D.; Cocker, E.D.; Hamel, E.O.; Ghosh, K.K.; Kitch, L.J.; Gamal, A.E.; Schnitzer, M.J. Long-term dynamics of CA1 hippocampal place codes. Nat. Neurosci. 2013, 16, 264–266. [Google Scholar] [CrossRef]
- Szabo, V.; Ventalon, C.; De Sars, V.; Bradley, J.; Emiliani, V. Spatially Selective Holographic Photoactivation and Functional Fluorescence Imaging in Freely Behaving Mice with a Fiberscope. Neuron 2014, 84, 1157–1169. [Google Scholar] [CrossRef] [PubMed]
- Akerboom, J.; Carreras Calderón, N.; Tian, L.; Wabnig, S.; Prigge, M.; Tolö, J.; Gordus, A.; Orger, M.B.; Severi, K.E.; Macklin, J.J.; et al. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front. Mol. Neurosci. 2013, 6, 2. [Google Scholar] [CrossRef]
- Chen, T.-W.; Wardill, T.J.; Sun, Y.; Pulver, S.R.; Renninger, S.L.; Baohan, A.; Schreiter, E.R.; Kerr, R.A.; Orger, M.B.; Jayaraman, V.; et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 2013, 499, 295–300. [Google Scholar] [CrossRef]
- Zhao, Y.; Araki, S.; Wu, J.; Teramoto, T.; Chang, Y.-F.; Nakano, M.; Abdelfattah, A.S.; Fujiwara, M.; Ishihara, T.; Nagai, T.; et al. An expanded palette of genetically encoded Ca2+ indicators. Science 2011, 333, 1888–1891. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Hires, S.A.; Mao, T.; Huber, D.; Chiappe, M.E.; Chalasani, S.H.; Petreanu, L.; Akerboom, J.; McKinney, S.A.; Schreiter, E.R.; et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat. Methods 2009, 6, 875–881. [Google Scholar] [CrossRef]
- Poburko, D.; Santo-Domingo, J.; Demaurex, N. Dynamic Regulation of the Mitochondrial Proton Gradient during Cytosolic Calcium Elevations. J. Biol. Chem. 2011, 286, 11672–11684. [Google Scholar] [CrossRef] [PubMed]
- Imamura, H.; Nhat, K.P.H.; Togawa, H.; Saito, K.; Iino, R.; Kato-Yamada, Y.; Nagai, T.; Noji, H. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc. Natl. Acad. Sci. USA 2009, 106, 15651–15656. [Google Scholar] [CrossRef]
- Slabbaert, J.R.; Kuenen, S.; Swerts, J.; Maes, I.; Uytterhoeven, V.; Kasprowicz, J.; Fernandes, A.C.; Blust, R.; Verstreken, P. Shawn, the Drosophila Homolog of SLC25A39/40, Is a Mitochondrial Carrier That Promotes Neuronal Survival. J. Neurosci. 2016, 36, 1914–1929. [Google Scholar] [CrossRef] [PubMed]
- Yin, B.; Barrionuevo, G.; Batinic-Haberle, I.; Sandberg, M.; Weber, S.G. Differences in Reperfusion-Induced Mitochondrial Oxidative Stress and Cell Death Between Hippocampal CA1 and CA3 Subfields Are Due to the Mitochondrial Thioredoxin System. Antioxid. Redox Signal. 2017, 27, 534–549. [Google Scholar] [CrossRef] [PubMed]
- Ricke, K.M.; Paß, T.; Kimoloi, S.; Fährmann, K.; Jüngst, C.; Schauss, A.; Baris, O.R.; Aradjanski, M.; Trifunovic, A.; Eriksson Faelker, T.M.; et al. Mitochondrial Dysfunction Combined with High Calcium Load Leads to Impaired Antioxidant Defense Underlying the Selective Loss of Nigral Dopaminergic Neurons. J. Neurosci. 2020, 40, 1975–1986. [Google Scholar] [CrossRef]
- Wang, B.; Liu, Q.; Shan, H.; Xia, C.; Liu, Z. Nrf2 inducer and cncC overexpression attenuates neurodegeneration due to α-synuclein in Drosophila. Biochem. Cell Biol. 2015, 93, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Stapper, Z.A.; Jahn, T.R. Changes in Glutathione Redox Potential Are Linked to Aβ42-Induced Neurotoxicity. Cell Rep. 2018, 24, 1696–1703. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Chandel, N.S. We need to talk about the Warburg effect. Nat. Metab. 2020, 2, 127–129. [Google Scholar] [CrossRef]
- Hsu, P.P.; Sabatini, D.M. Cancer Cell Metabolism: Warburg and Beyond. Cell 2008, 134, 703–707. [Google Scholar] [CrossRef]
- Rodic, S.; Vincent, M.D. Reactive oxygen species (ROS) are a key determinant of cancer’s metabolic phenotype: ROS are a key determinant of cancer’s metabolic phenotype. Int. J. Cancer 2018, 142, 440–448. [Google Scholar] [CrossRef]
- Wang, K.; Jiang, J.; Lei, Y.; Zhou, S.; Wei, Y.; Huang, C. Targeting Metabolic–Redox Circuits for Cancer Therapy. Trends Biochem. Sci. 2019, 44, 401–414. [Google Scholar] [CrossRef]
- Sabharwal, S.S.; Schumacker, P.T. Mitochondrial ROS in cancer: Initiators, amplifiers or an Achilles’ heel? Nat. Rev. Cancer 2014, 14, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Chio, I.I.C.; Tuveson, D.A. ROS in Cancer: The Burning Question. Trends Mol. Med. 2017, 23, 411–429. [Google Scholar] [CrossRef]
- Sullivan, L.B.; Chandel, N.S. Mitochondrial reactive oxygen species and cancer. Cancer Metab. 2014, 2, 17. [Google Scholar] [CrossRef] [PubMed]
- Davalli, P.; Marverti, G.; Lauriola, A.; D’Arca, D. Targeting Oxidatively Induced DNA Damage Response in Cancer: Opportunities for Novel Cancer Therapies. Oxidative Med. Cell. Longev. 2018, 2018, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Tubbs, A.; Nussenzweig, A. Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell 2017, 168, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Joseph, P. Cadmium-Induced Cell Transformation and Tumorigenesis Are Associated with Transcriptional Activation of c-fos, c-jun, and c-myc Proto-Oncogenes: Role of Cellular Calcium and Reactive Oxygen Species. Toxicol. Sci. 2001, 61, 295–303. [Google Scholar] [CrossRef]
- Wei, H. Activation of oncogenes and/or inactivation of anti-oncogenes by reactive oxygen species. Med. Hypotheses 1992, 39, 267–270. [Google Scholar] [CrossRef]
- Vafa, O.; Wade, M.; Kern, S.; Beeche, M.; Pandita, T.K.; Hampton, G.M.; Wahl, G.M. c-Myc Can Induce DNA Damage, Increase Reactive Oxygen Species, and Mitigate p53 Function. Mol. Cell 2002, 9, 1031–1044. [Google Scholar] [CrossRef]
- Maya-Mendoza, A.; Ostrakova, J.; Kosar, M.; Hall, A.; Duskova, P.; Mistrik, M.; Merchut-Maya, J.M.; Hodny, Z.; Bartkova, J.; Christensen, C.; et al. Myc and Ras oncogenes engage different energy metabolism programs and evoke distinct patterns of oxidative and DNA replication stress. Mol. Oncol. 2015, 9, 601–616. [Google Scholar] [CrossRef]
- Carnero, A. MAP17, a ROS-dependent oncogene. Front. Oncol. 2012, 2, 112. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.; Chua, D.; Tan, N.S. Reactive oxygen species: A volatile driver of field cancerization and metastasis. Mol. Cancer 2019, 18, 65. [Google Scholar] [CrossRef]
- Ishikawa, K.; Takenaga, K.; Akimoto, M.; Koshikawa, N.; Yamaguchi, A.; Imanishi, H.; Nakada, K.; Honma, Y.; Hayashi, J.-I. ROS-Generating Mitochondrial DNA Mutations Can Regulate Tumor Cell Metastasis. Science 2008, 320, 661–664. [Google Scholar] [CrossRef]
- Lam, C.R.I.; Tan, C.; Teo, Z.; Tay, C.Y.; Phua, T.; Wu, Y.L.; Cai, P.Q.; Tan, L.P.; Chen, X.; Zhu, P.; et al. Loss of TAK1 increases cell traction force in a ROS-dependent manner to drive epithelial–mesenchymal transition of cancer cells. Cell Death Dis. 2013, 4, e848. [Google Scholar] [CrossRef] [PubMed]
- DeNicola, G.M.; Karreth, F.A.; Humpton, T.J.; Gopinathan, A.; Wei, C.; Frese, K.; Mangal, D.; Yu, K.H.; Yeo, C.J.; Calhoun, E.S.; et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 2011, 475, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Sayin, V.I.; Ibrahim, M.X.; Larsson, E.; Nilsson, J.A.; Lindahl, P.; Bergo, M.O. Antioxidants accelerate lung cancer progression in mice. Sci. Transl. Med. 2014, 6, 221ra15. [Google Scholar] [CrossRef]
- Van Zandwijk, N. EUROSCAN, a Randomized Trial of Vitamin A and N-Acetylcysteine in Patients With Head and Neck Cancer or Lung Cancer. J. Natl. Cancer Inst. 2000, 92, 977–986. [Google Scholar] [CrossRef]
- Liou, G.-Y.; Storz, P. Reactive oxygen species in cancer. Free Radic. Res. 2010, 44, 479–496. [Google Scholar] [CrossRef]
- Yang, H.; Villani, R.M.; Wang, H.; Simpson, M.J.; Roberts, M.S.; Tang, M.; Liang, X. The role of cellular reactive oxygen species in cancer chemotherapy. J. Exp. Clin. Cancer Res. 2018, 37, 266. [Google Scholar] [CrossRef]
- Kim, W.; Lee, S.; Seo, D.; Kim, D.; Kim, K.; Kim, E.; Kang, J.; Seong, K.M.; Youn, H.; Youn, B. Cellular Stress Responses in Radiotherapy. Cells 2019, 8, 1105. [Google Scholar] [CrossRef]
- Luo, J.; Solimini, N.L.; Elledge, S.J. Principles of Cancer Therapy: Oncogene and Non-oncogene Addiction. Cell 2009, 136, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Imamura, T.; Saitou, T.; Kawakami, R. In vivo optical imaging of cancer cell function and tumor microenvironment. Cancer Sci. 2018, 109, 912–918. [Google Scholar] [CrossRef]
- Kolenc, O.I.; Quinn, K.P. Evaluating Cell Metabolism Through Autofluorescence Imaging of NAD(P)H and FAD. Antioxid. Redox Signal. 2019, 30, 875–889. [Google Scholar] [CrossRef]
- Shen, J.P.; Zhao, D.; Sasik, R.; Luebeck, J.; Birmingham, A.; Bojorquez-Gomez, A.; Licon, K.; Klepper, K.; Pekin, D.; Beckett, A.N.; et al. Combinatorial CRISPR–Cas9 screens for de novo mapping of genetic interactions. Nat. Methods 2017, 14, 573–576. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Badur, M.G.; Luebeck, J.; Magaña, J.H.; Birmingham, A.; Sasik, R.; Ahn, C.S.; Ideker, T.; Metallo, C.M.; Mali, P. Combinatorial CRISPR-Cas9 Metabolic Screens Reveal Critical Redox Control Points Dependent on the KEAP1-NRF2 Regulatory Axis. Mol. Cell 2018, 69, 699–708.e7. [Google Scholar] [CrossRef]
- Fletcher, N.M.; Belotte, J.; Saed, M.G.; Memaj, I.; Diamond, M.P.; Morris, R.T.; Saed, G.M. Specific point mutations in key redox enzymes are associated with chemoresistance in epithelial ovarian cancer. Free Radic. Biol. Med. 2017, 102, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Zhan, T.; Rindtorff, N.; Betge, J.; Ebert, M.P.; Boutros, M. CRISPR/Cas9 for cancer research and therapy. Semin. Cancer Biol. 2019, 55, 106–119. [Google Scholar] [CrossRef]
- Kraus, F.; Ryan, M.T. The constriction and scission machineries involved in mitochondrial fission. J. Cell Sci. 2017, 130, 2953–2960. [Google Scholar] [CrossRef]
- Wang, W.; Wang, Y.; Long, J.; Wang, J.; Haudek, S.B.; Overbeek, P.; Chang, B.H.J.; Schumacker, P.T.; Danesh, F.R. Mitochondrial Fission Triggered by Hyperglycemia Is Mediated by ROCK1 Activation in Podocytes and Endothelial Cells. Cell Metab. 2012, 15, 186–200. [Google Scholar] [CrossRef]
- Narayanan, D.L.; Saladi, R.N.; Fox, J.L. Review: Ultraviolet radiation and skin cancer: UVR and skin cancer. Int. J. Dermatol. 2010, 49, 978–986. [Google Scholar] [CrossRef]
- Li, R.; Jia, Z.; Trush, M.A. Defining ROS in Biology and Medicine. React. Oxyg. Species 2016, 1, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Foreman, J.; Demidchik, V.; Bothwell, J.H.F.; Mylona, P.; Miedema, H.; Torres, M.A.; Linstead, P.; Costa, S.; Brownlee, C.; Jones, J.D.G.; et al. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 2003, 422, 442–446. [Google Scholar] [CrossRef]
- Huang, H.; Ullah, F.; Zhou, D.-X.; Yi, M.; Zhao, Y. Mechanisms of ROS Regulation of Plant Development and Stress Responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef]
- Dröge, W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002, 82, 47–95. [Google Scholar] [CrossRef] [PubMed]
- Janků, M.; Luhová, L.; Petřivalský, M. On the Origin and Fate of Reactive Oxygen Species in Plant Cell Compartments. Antioxidants 2019, 8, 105. [Google Scholar] [CrossRef] [PubMed]
- Kamada, T.; Nito, K.; Hayashi, H.; Mano, S.; Hayashi, M.; Nishimura, M. Functional Differentiation of Peroxisomes Revealed by Expression Profiles of Peroxisomal Genes in Arabidopsis thaliana. Plant Cell Physiol. 2003, 44, 1275–1289. [Google Scholar] [CrossRef]
- Kirsch, T.; Löffler, H.G.; Kindl, H. Plant acyl-CoA oxidase. Purification, characterization, and monomeric apoprotein. J. Biol. Chem. 1986, 261, 8570–8575. [Google Scholar]
- Nishimura, M.; Akhmedov, Y.D.; Strzalka, K.; Akazawa, T. Purification and characterization of glycolate oxidase from pumpkin cotyledons. Arch. Biochem. Biophys. 1983, 222, 397–402. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Breusegem, F.V. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Zhang, H.; Sonnewald, U. Differences and commonalities of plant responses to single and combined stresses. Plant J. 2017, 90, 839–855. [Google Scholar] [CrossRef]
- May, M.J.; Vernoux, T.; Sánchez-Fernández, R.; Montagu, M.V.; Inzé, D. Evidence for posttranscriptional activation of γ-glutamylcysteine synthetase during plant stress responses. Proc. Natl. Acad. Sci. USA 1998, 95, 12049–12054. [Google Scholar] [CrossRef] [PubMed]
- Galant, A.; Preuss, M.L.; Cameron, J.C.; Jez, J.M. Plant Glutathione Biosynthesis: Diversity in Biochemical Regulation and Reaction Products. Front. Plant Sci. 2011, 2. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Anjum, N.A.; Hasanuzzaman, M.; Gill, R.; Trivedi, D.K.; Ahmad, I.; Pereira, E.; Tuteja, N. Glutathione and glutathione reductase: A boon in disguise for plant abiotic stress defense operations. Plant Physiol. Biochem. 2013, 70, 204–212. [Google Scholar] [CrossRef]
- Xu, L.; Carrie, C.; Law, S.R.; Murcha, M.W.; Whelan, J. Acquisition, Conservation, and Loss of Dual-Targeted Proteins in Land Plants. Plant Physiol. 2013, 161, 644–662. [Google Scholar] [CrossRef] [PubMed]
- Kochian, L.V.; Hoekenga, O.A.; Pineros, M.A. How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu. Rev. Plant Biol. 2004, 55, 459–493. [Google Scholar] [CrossRef]
- Considine, M.J.; Foyer, C.H. Redox Regulation of Plant Development. Antioxid. Redox Signal. 2013, 21, 1305–1326. [Google Scholar] [CrossRef]
- Zechmann, B.; Müller, M. Subcellular compartmentation of glutathione in dicotyledonous plants. Protoplasma 2010, 246, 15–24. [Google Scholar] [CrossRef]
- Buckley, T.N. How do stomata respond to water status? New Phytol. 2019, 224, 21–36. [Google Scholar] [CrossRef]
- Kwak, J.M.; Mori, I.C.; Pei, Z.-M.; Leonhardt, N.; Torres, M.A.; Dangl, J.L.; Bloom, R.E.; Bodde, S.; Jones, J.D.G.; Schroeder, J.I. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 2003, 22, 2623–2633. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, H.; Zhang, Q.; Li, M.; Yan, M.; Wang, R.; Wang, L.; Welti, R.; Zhang, W.; Wang, X. Phospholipase Dα1 and Phosphatidic Acid Regulate NADPH Oxidase Activity and Production of Reactive Oxygen Species in ABA-Mediated Stomatal Closure in Arabidopsis. Plant Cell 2009, 21, 2357–2377. [Google Scholar] [CrossRef]
- Garcia-Mata, C.; Lamattina, L. Abscisic acid (ABA) inhibits light-induced stomatal opening through calcium- and nitric oxide-mediated signaling pathways. Nitric Oxide 2007, 17, 143–151. [Google Scholar] [CrossRef]
- Scuffi, D.; Lamattina, L.; García-Mata, C. Gasotransmitters and Stomatal Closure: Is There Redundancy, Concerted Action, or Both? Front. Plant Sci. 2016, 7, 277. [Google Scholar] [CrossRef] [PubMed]
- Melotto, M.; Underwood, W.; Koczan, J.; Nomura, K.; He, S.Y. Plant stomata function in innate immunity against bacterial invasion. Cell 2006, 126, 969–980. [Google Scholar] [CrossRef]
- Wang, W.; Chen, D.; Zhang, X.; Liu, D.; Cheng, Y.; Shen, F. Role of plant respiratory burst oxidase homologs in stress responses. Free Radic. Res. 2018, 52, 826–839. [Google Scholar] [CrossRef]
- Mengiste, T. Plant Immunity to Necrotrophs. Annu. Rev. Phytopathol. 2011, 50, 267–294. [Google Scholar] [CrossRef]
- Chi, M.-H.; Park, S.-Y.; Kim, S.; Lee, Y.-H. A novel pathogenicity gene is required in the rice blast fungus to suppress the basal defenses of the host. PLoS Pathog. 2009, 5, e1000401. [Google Scholar] [CrossRef]
- Guo, M.; Chen, Y.; Du, Y.; Dong, Y.; Guo, W.; Zhai, S.; Zhang, H.; Dong, S.; Zhang, Z.; Wang, Y.; et al. The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 2011, 7, e1001302. [Google Scholar] [CrossRef] [PubMed]
- Scott, B.; Eaton, C.J. Role of reactive oxygen species in fungal cellular differentiations. Curr. Opin. Microbiol. 2008, 11, 488–493. [Google Scholar] [CrossRef]
- Heller, J.; Tudzynski, P. Reactive Oxygen Species in Phytopathogenic Fungi: Signaling, Development, and Disease. Annu. Rev. Phytopathol. 2011, 49, 369–390. [Google Scholar] [CrossRef]
- Leach, J.E.; Triplett, L.R.; Argueso, C.T.; Trivedi, P. Communication in the Phytobiome. Cell 2017, 169, 587–596. [Google Scholar] [CrossRef]
- Marino, D.; Andrio, E.; Danchin, E.G.J.; Oger, E.; Gucciardo, S.; Lambert, A.; Puppo, A.; Pauly, N. A Medicago truncatula NADPH oxidase is involved in symbiotic nodule functioning. New Phytol. 2011, 189, 580–592. [Google Scholar] [CrossRef]
- MacNeil, A.; Glaziou, P.; Sismanidis, C.; Maloney, S.; Floyd, K. Global Epidemiology of Tuberculosis and Progress Toward Achieving Global Targets—2017. MMWR Morb. Mortal. Wkly. Rep. 2019, 68, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Comas, I.; Coscolla, M.; Luo, T.; Borrell, S.; Holt, K.E.; Kato-Maeda, M.; Parkhill, J.; Malla, B.; Berg, S.; Thwaites, G.; et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat. Genet. 2013, 45, 1176–1182. [Google Scholar] [CrossRef]
- Melly, G.; Purdy, G.E. MmpL Proteins in Physiology and Pathogenesis of M. tuberculosis. Microorganisms 2019, 7, 70. [Google Scholar] [CrossRef] [PubMed]
- Bailo, R.; Bhatt, A.; Aínsa, J.A. Lipid transport in Mycobacterium tuberculosis and its implications in virulence and drug development. Biochem. Pharm. 2015, 96, 159–167. [Google Scholar] [CrossRef]
- Chandrangsu, P.; Loi, V.V.; Antelmann, H.; Helmann, J.D. The Role of Bacillithiol in Gram-Positive Firmicutes. Antioxid. Redox Signal. 2018, 28, 445–462. [Google Scholar] [CrossRef] [PubMed]
- Gaballa, A.; Chi, B.K.; Roberts, A.A.; Becher, D.; Hamilton, C.J.; Antelmann, H.; Helmann, J.D. Redox regulation in Bacillus subtilis: The bacilliredoxins BrxA(YphP) and BrxB(YqiW) function in de-bacillithiolation of S-bacillithiolated OhrR and MetE. Antioxid. Redox Signal. 2014, 21, 357–367. [Google Scholar] [CrossRef]
- Gaballa, A.; Newton, G.L.; Antelmann, H.; Parsonage, D.; Upton, H.; Rawat, M.; Claiborne, A.; Fahey, R.C.; Helmann, J.D. Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proc. Natl. Acad. Sci. USA 2010, 107, 6482. [Google Scholar] [CrossRef]
- Mikheyeva, I.V.; Thomas, J.M.; Kolar, S.L.; Corvaglia, A.-R.; Gaϊa, N.; Leo, S.; Francois, P.; Liu, G.Y.; Rawat, M.; Cheung, A.L. YpdA, a putative bacillithiol disulfide reductase, contributes to cellular redox homeostasis and virulence in Staphylococcus aureus. Mol. Microbiol. 2019, 111, 1039–1056. [Google Scholar] [CrossRef]
- Padilla-Benavides, T.; Long, J.E.; Raimunda, D.; Sassetti, C.M.; Argüello, J.M. A novel P(1B)-type Mn2+-transporting ATPase is required for secreted protein metallation in mycobacteria. J. Biol. Chem. 2013, 288, 11334–11347. [Google Scholar] [CrossRef]
- Lobstein, J.; Emrich, C.A.; Jeans, C.; Faulkner, M.; Riggs, P.; Berkmen, M. SHuffle, a novel Escherichia coli protein expression strain capable of correctly folding disulfide bonded proteins in its cytoplasm. Microb. Cell Fact. 2012, 11, 56. [Google Scholar] [CrossRef]
- Hwang, C.; Sinskey, A.J.; Lodish, H.F. Oxidized redox state of glutathione in the endoplasmic reticulum. Science 1992, 257, 1496–1502. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.; Gießelmann, G.; Hoffmann, S.L.; Wittmann, C. Corynebacterium glutamicum for Sustainable Bioproduction: From Metabolic Physiology to Systems Metabolic Engineering. Adv. Biochem. Eng. Biotechnol. 2018, 162, 217–263. [Google Scholar] [CrossRef] [PubMed]
- Frunzke, J.; Bramkamp, M.; Schweitzer, J.-E.; Bott, M. Population Heterogeneity in Corynebacterium glutamicum ATCC 13032 Caused by Prophage CGP3. J. Bacteriol. 2008, 190, 5111. [Google Scholar] [CrossRef]
- Newton, G.L.; Ta, P.; Fahey, R.C. A mycothiol synthase mutant of Mycobacterium smegmatis produces novel thiols and has an altered thiol redox status. J. Bacteriol. 2005, 187, 7309–7316. [Google Scholar] [CrossRef][Green Version]
- Cavanagh, D.; Fitzgerald, G.F.; McAuliffe, O. From field to fermentation: The origins of Lactococcus lactis and its domestication to the dairy environment. Food Microbiol. 2015, 47, 45–61. [Google Scholar] [CrossRef] [PubMed]
- Abramovitch, R.B.; Rohde, K.H.; Hsu, F.-F.; Russell, D.G. aprABC: A Mycobacterium tuberculosis complex-specific locus that modulates pH-driven adaptation to the macrophage phagosome. Mol. Microbiol. 2011, 80, 678–694. [Google Scholar] [CrossRef]
- Vandal, O.H.; Pierini, L.M.; Schnappinger, D.; Nathan, C.F.; Ehrt, S. A membrane protein preserves intrabacterial pH in intraphagosomal Mycobacterium tuberculosis. Nat. Med. 2008, 14, 849–854. [Google Scholar] [CrossRef]
- Asensio, J.G.; Maia, C.; Ferrer, N.L.; Barilone, N.; Laval, F.; Soto, C.Y.; Winter, N.; Daffé, M.; Gicquel, B.; Martín, C.; et al. The Virulence-associated Two-component PhoP-PhoR System Controls the Biosynthesis of Polyketide-derived Lipids in Mycobacterium tuberculosis. J. Biol. Chem. 2006, 281, 1313–1316. [Google Scholar] [CrossRef]
- Sirakova, T.D.; Thirumala, A.K.; Dubey, V.S.; Sprecher, H.; Kolattukudy, P.E. The Mycobacterium tuberculosis pks2 Gene Encodes the Synthase for the Hepta- and Octamethyl-branched Fatty Acids Required for Sulfolipid Synthesis. J. Biol. Chem. 2001, 276, 16833–16839. [Google Scholar] [CrossRef]
- Van der Veen, B.S.; de Winther, M.P.J.; Heeringa, P. Myeloperoxidase: Molecular mechanisms of action and their relevance to human health and disease. Antioxid. Redox Signal. 2009, 11, 2899–2937. [Google Scholar] [CrossRef] [PubMed]
- Kohanski, M.A.; Dwyer, D.J.; Collins, J.J. How antibiotics kill bacteria: From targets to networks. Nat. Rev. Microbiol. 2010, 8, 423–435. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Hayete, B.; Lawrence, C.A.; Collins, J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 2007, 130, 797–810. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, D.J.; Belenky, P.A.; Yang, J.H.; MacDonald, I.C.; Martell, J.D.; Takahashi, N.; Chan, C.T.Y.; Lobritz, M.A.; Braff, D.; Schwarz, E.G.; et al. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. USA 2014, 111, E2100. [Google Scholar] [CrossRef] [PubMed]
- Guridi, A.; Diederich, A.-K.; Aguila-Arcos, S.; Garcia-Moreno, M.; Blasi, R.; Broszat, M.; Schmieder, W.; Clauss-Lendzian, E.; Sakinc-Gueler, T.; Andrade, R.; et al. New antimicrobial contact catalyst killing antibiotic resistant clinical and waterborne pathogens. Mater. Sci. Eng. C 2015, 50, 1–11. [Google Scholar] [CrossRef]
- López-Mirabal, H.R.; Winther, J.R. Redox characteristics of the eukaryotic cytosol. Biochim. Biophys. Acta Mol. Cell Res. 2008, 1783, 629–640. [Google Scholar] [CrossRef]
- Orij, R.; Postmus, J.; Ter Beek, A.; Brul, S.; Smits, G.J. In vivo measurement of cytosolic and mitochondrial pH using a pH-sensitive GFP derivative in Saccharomyces cerevisiae reveals a relation between intracellular pH and growth. Microbiology 2009, 155, 268–278. [Google Scholar] [CrossRef]
- Davidson, J.F.; Whyte, B.; Bissinger, P.H.; Schiestl, R.H. Oxidative stress is involved in heat-induced cell death in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1996, 93, 5116–5121. [Google Scholar] [CrossRef]
- Sugiyama, K.; Izawa, S.; Inoue, Y. The Yap1p-dependent Induction of Glutathione Synthesis in Heat Shock Response of Saccharomyces cerevisiae. J. Biol. Chem. 2000, 275, 15535–15540. [Google Scholar] [CrossRef]
- Hohmann, S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 2002, 66, 300–372. [Google Scholar] [CrossRef]
- Ng, C.-H.; Tan, S.-X.; Perrone, G.G.; Thorpe, G.W.; Higgins, V.J.; Dawes, I.W. Adaptation to hydrogen peroxide in Saccharomyces cerevisiae: The role of NADPH-generating systems and the SKN7 transcription factor. Free Radic. Biol. Med. 2008, 44, 1131–1145. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.J.; Hawkins, C.L.; Pattison, D.I.; Rees, M.D. Mammalian heme peroxidases: From molecular mechanisms to health implications. Antioxid. Redox Signal. 2008, 10, 1199–1234. [Google Scholar] [CrossRef] [PubMed]
- Pattison, D.I.; Davies, M.J. Reactions of myeloperoxidase-derived oxidants with biological substrates: Gaining chemical insight into human inflammatory diseases. Curr. Med. Chem. 2006, 13, 3271–3290. [Google Scholar] [CrossRef]
- Davies, M.J. Protein oxidation and peroxidation. Biochem. J. 2016, 473, 805–825. [Google Scholar] [CrossRef]
- Wood, Z.A.; Poole, L.B.; Karplus, P.A. Peroxiredoxin Evolution and the Regulation of Hydrogen Peroxide Signaling. Science 2003, 300, 650. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.; Karplus, P.A.; Poole, L.B. Typical 2-Cys peroxiredoxins--structures, mechanisms and functions. FEBS J. 2009, 276, 2469–2477. [Google Scholar] [CrossRef]
- Brocklehurst, K. Specific covalent modification of thiols: Applications in the study of enzymes and other biomolecules. Int. J. Biochem. 1979, 10, 259–274. [Google Scholar] [CrossRef]
- PEDERSEN, A.O.; JACOBSEN, J. Reactivity of the Thiol Group in Human and Bovine Albumin at pH 3–9, as Measured by Exchange with 2,2′-Dithiodipyridine. Eur. J. Biochem. 1980, 106, 291–295. [Google Scholar] [CrossRef]
- Wu, X.-B.; Brüne, B.; von Appen, F.; Ullrich, V. Reversible activation of soluble guanylate cyclase by oxidizing agents. Arch. Biochem. Biophys. 1992, 294, 75–82. [Google Scholar] [CrossRef]
- Hampton, R.Y. ER-associated degradation in protein quality control and cellular regulation. Curr. Opin. Cell Biol. 2002, 14, 476–482. [Google Scholar] [CrossRef]
- Heo, J.-M.; Livnat-Levanon, N.; Taylor, E.B.; Jones, K.T.; Dephoure, N.; Ring, J.; Xie, J.; Brodsky, J.L.; Madeo, F.; Gygi, S.P.; et al. A stress-responsive system for mitochondrial protein degradation. Mol. Cell 2010, 40, 465–480. [Google Scholar] [CrossRef]
- Bodnar, N.O.; Rapoport, T.A. Molecular Mechanism of Substrate Processing by the Cdc48 ATPase Complex. Cell 2017, 169, 722–735.e9. [Google Scholar] [CrossRef] [PubMed]
- Brandes, N.; Tienson, H.; Lindemann, A.; Vitvitsky, V.; Reichmann, D.; Banerjee, R.; Jakob, U. Time line of redox events in aging postmitotic cells. elife 2013, 2, e00306. [Google Scholar] [CrossRef] [PubMed]
- Baek, G.H.; Cheng, H.; Choe, V.; Bao, X.; Shao, J.; Luo, S.; Rao, H. Cdc48: A swiss army knife of cell biology. J. Amino Acids 2013, 2013, 183421. [Google Scholar] [CrossRef]
- Imlay, J.A. Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 2006, 59, 1073–1082. [Google Scholar] [CrossRef] [PubMed]
- Rouault, T.A. Biogenesis of iron-sulfur clusters in mammalian cells: New insights and relevance to human disease. Dis. Models Mech. 2012, 5, 155. [Google Scholar] [CrossRef] [PubMed]
- Idnurm, A.; Verma, S.; Corrochano, L.M. A glimpse into the basis of vision in the kingdom Mycota. Fungal Genet. Biol. 2010, 47, 881–892. [Google Scholar] [CrossRef] [PubMed]
- Jacquet, M.; Renault, G.; Lallet, S.; De Mey, J.; Goldbeter, A. Oscillatory nucleocytoplasmic shuttling of the general stress response transcriptional activators Msn2 and Msn4 in Saccharomyces cerevisiae. J. Cell Biol. 2003, 161, 497–505. [Google Scholar] [CrossRef]
- Hockberger, P.E.; Skimina, T.A.; Centonze, V.E.; Lavin, C.; Chu, S.; Dadras, S.; Reddy, J.K.; White, J.G. Activation of flavin-containing oxidases underlies light-induced production of H2O2 in mammalian cells. Proc. Natl. Acad. Sci. USA 1999, 96, 6255. [Google Scholar] [CrossRef]
- Bass, J.; Takahashi, J.S. Circadian rhythms: Redox redux. Nature 2011, 469, 476–478. [Google Scholar] [CrossRef]
- O’Neill, J.S.; Reddy, A.B. Circadian clocks in human red blood cells. Nature 2011, 469, 498–503. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J.S.; van Ooijen, G.; Dixon, L.E.; Troein, C.; Corellou, F.; Bouget, F.-Y.; Reddy, A.B.; Millar, A.J. Circadian rhythms persist without transcription in a eukaryote. Nature 2011, 469, 554–558. [Google Scholar] [CrossRef]
- Edgar, R.S.; Green, E.W.; Zhao, Y.; van Ooijen, G.; Olmedo, M.; Qin, X.; Xu, Y.; Pan, M.; Valekunja, U.K.; Feeney, K.A.; et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature 2012, 485, 459–464. [Google Scholar] [CrossRef]
- Liu, C.Y. The unfolded protein response. J. Cell Sci. 2003, 116, 1861–1862. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Ng, B.S.H.; Thibault, G. Endoplasmic reticulum stress response in yeast and humans. Biosci. Rep. 2014, 34, e00118. [Google Scholar] [CrossRef]
- Promlek, T.; Ishiwata-Kimata, Y.; Shido, M.; Sakuramoto, M.; Kohno, K.; Kimata, Y. Membrane aberrancy and unfolded proteins activate the endoplasmic reticulum stress sensor Ire1 in different ways. Mol. Biol. Cell 2011, 22, 3520–3532. [Google Scholar] [CrossRef] [PubMed]
- Tu, B.P.; Weissman, J.S. Oxidative protein folding in eukaryotes. J. Cell Biol. 2004, 164, 341–346. [Google Scholar] [CrossRef]
- Klecker, T.; Westermann, B. Asymmetric inheritance of mitochondria in yeast. Biol. Chem. 2020, 401, 779–791. [Google Scholar] [CrossRef] [PubMed]
- Higuchi-Sanabria, R.; Pernice, W.M.A.; Vevea, J.D.; Alessi Wolken, D.M.; Boldogh, I.R.; Pon, L.A. Role of asymmetric cell division in lifespan control in Saccharomyces cerevisiae. FEMS Yeast Res. 2014, 14, 1133–1146. [Google Scholar] [CrossRef]
- Lang, S.; Pfeffer, S.; Lee, P.-H.; Cavalié, A.; Helms, V.; Förster, F.; Zimmermann, R. An Update on Sec61 Channel Functions, Mechanisms, and Related Diseases. Front. Physiol. 2017, 8, 887. [Google Scholar] [CrossRef]
- Oestreicher, J.; Morgan, B. Glutathione: Subcellular distribution and membrane transport. Biochem. Cell Biol. 2019, 97, 270–289. [Google Scholar] [CrossRef] [PubMed]
- Zito, E. ERO1: A protein disulfide oxidase and H2O2 producer. Free Radic. Biol. Med. 2015, 83, 299–304. [Google Scholar] [CrossRef]
- Bastidas, R.J.; Elwell, C.A.; Engel, J.N.; Valdivia, R.H. Chlamydial intracellular survival strategies. Cold Spring Harb. Perspect. Med. 2013, 3, a010256. [Google Scholar] [CrossRef]
- Newhall, W.J. Biosynthesis and disulfide cross-linking of outer membrane components during the growth cycle of Chlamydia trachomatis. Infect Immun. 1987, 55, 162–168. [Google Scholar] [CrossRef]
- Hatch, T.P.; Miceli, M.; Sublett, J.E. Synthesis of disulfide-bonded outer membrane proteins during the developmental cycle of Chlamydia psittaci and Chlamydia trachomatis. J. Bacteriol. 1986, 165, 379–385. [Google Scholar] [CrossRef]
- Caldwell, H.D.; Kromhout, J.; Schachter, J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun. 1981, 31, 1161–1176. [Google Scholar] [CrossRef]
- Russell, D.G.; Cardona, P.-J.; Kim, M.-J.; Allain, S.; Altare, F. Foamy macrophages and the progression of the human tuberculosis granuloma. Nat. Immunol. 2009, 10, 943–948. [Google Scholar] [CrossRef] [PubMed]
- Voskuil, M.I.; Bartek, I.L.; Visconti, K.; Schoolnik, G.K. The response of mycobacterium tuberculosis to reactive oxygen and nitrogen species. Front Microbiol. 2011, 2, 105. [Google Scholar] [CrossRef] [PubMed]
- Figueira, R.; Holden, D.W. Functions of the Salmonella pathogenicity island 2 (SPI-2) type III secretion system effectors. Microbiology 2012, 158, 1147–1161. [Google Scholar] [CrossRef]
- Hensel, M. Salmonella Pathogenicity Island 2. Mol. Microbiol. 2000, 36, 1015–1023. [Google Scholar] [CrossRef]
- Chakravortty, D.; Hansen-Wester, I.; Hensel, M. Salmonella pathogenicity island 2 mediates protection of intracellular Salmonella from reactive nitrogen intermediates. J. Exp. Med. 2002, 195, 1155–1166. [Google Scholar] [CrossRef]
- Vazquez-Torres, A.; Xu, Y.; Jones-Carson, J.; Holden, D.W.; Lucia, S.M.; Dinauer, M.C.; Mastroeni, P.; Fang, F.C. Salmonella Pathogenicity Island 2-Dependent Evasion of the Phagocyte NADPH Oxidase. Science 2000, 287, 1655. [Google Scholar] [CrossRef] [PubMed]
- Aussel, L.; Zhao, W.; Hébrard, M.; Guilhon, A.-A.; Viala, J.P.M.; Henri, S.; Chasson, L.; Gorvel, J.-P.; Barras, F.; Méresse, S. Salmonella detoxifying enzymes are sufficient to cope with the host oxidative burst. Mol. Microbiol. 2011, 80, 628–640. [Google Scholar] [CrossRef] [PubMed]
- Vilchèze, C.; Weisbrod, T.R.; Chen, B.; Kremer, L.; Hazbón, M.H.; Wang, F.; Alland, D.; Sacchettini, J.C.; Jacobs, W.R., Jr. Altered NADH/NAD+ ratio mediates coresistance to isoniazid and ethionamide in mycobacteria. Antimicrob. Agents Chemother. 2005, 49, 708–720. [Google Scholar] [CrossRef] [PubMed]
- Schaible, U.E.; Sturgill-Koszycki, S.; Schlesinger, P.H.; Russell, D.G. Cytokine Activation Leads to Acidification and Increases Maturation of Mycobacterium avium-Containing Phagosomes in Murine Macrophages. J. Immunol. 1998, 160, 1290. [Google Scholar]
- Midwinter, R.G.; Cheah, F.-C.; Moskovitz, J.; Vissers, M.C.; Winterbourn, C.C. IkappaB is a sensitive target for oxidation by cell-permeable chloramines: Inhibition of NF-kappaB activity by glycine chloramine through methionine oxidation. Biochem. J. 2006, 396, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.E.; Boyle, K.B.; Davidson, K.; Chessa, T.A.M.; Kulkarni, S.; Jarvis, G.E.; Sindrilaru, A.; Scharffetter-Kochanek, K.; Rausch, O.; Stephens, L.R.; et al. CD18-dependent activation of the neutrophil NADPH oxidase during phagocytosis of Escherichia coli or Staphylococcus aureus is regulated by class III but not class I or II PI3Ks. Blood 2008, 112, 5202–5211. [Google Scholar] [CrossRef]
- Rastogi, R.; Geng, X.; Li, F.; Ding, Y. NOX Activation by Subunit Interaction and Underlying Mechanisms in Disease. Front. Cell. Neurosci. 2017, 10, 301. [Google Scholar] [CrossRef]
- Belambri, S.A.; Rolas, L.; Raad, H.; Hurtado-Nedelec, M.; Dang, P.M.-C.; El-Benna, J. NADPH oxidase activation in neutrophils: Role of the phosphorylation of its subunits. Eur. J. Clin. Investig. 2018, 48, e12951. [Google Scholar] [CrossRef]
- Ashley, E.A.; Pyae Phyo, A.; Woodrow, C.J. Malaria. Lancet 2018, 391, 1608–1621. [Google Scholar] [CrossRef]
- World Health Organization. World Malaria Report. 2017. Available online: https://www.who.int/malaria/publications/world-malaria-report-2017/report/en/ (accessed on 25 August 2020).
- World Health Organization. World Malaria Report. 2018. Available online: https://www.who.int/malaria/publications/world-malaria-report-2018/report/en/ (accessed on 25 August 2020).
- Cowman, A.F.; Healer, J.; Marapana, D.; Marsh, K. Malaria: Biology and Disease. Cell 2016, 167, 610–624. [Google Scholar] [CrossRef] [PubMed]
- Conrad, M.D.; Rosenthal, P.J. Antimalarial drug resistance in Africa: The calm before the storm? Lancet Infect. Dis. 2019, 19, e338–e351. [Google Scholar] [CrossRef]
- Müller, S. Role and Regulation of Glutathione Metabolism in Plasmodium falciparum. Molecules 2015, 20, 10511–10534. [Google Scholar] [CrossRef] [PubMed]
- Atamna, H.; Ginsburg, H. Origin of reactive oxygen species in erythrocytes infected with Plasmodium falciparum. Mol. Biochem. Parasitol. 1993, 61, 231–241. [Google Scholar] [CrossRef]
- Fong, K.Y.; Wright, D.W. Hemozoin and antimalarial drug discovery. Future Med. Chem. 2013, 5, 1437–1450. [Google Scholar] [CrossRef]
- Atamna, H.; Krugliak, M.; Shalmiev, G.; Deharo, E.; Pescarmona, G.; Ginsburg, H. Mode of antimalarial effect of methylene blue and some of its analogues on Plasmodium falciparum in culture and their inhibition of P. vinckei petteri and P. yoelii nigeriensis in vivo. Biochem. Pharmacol. 1996, 51, 693–700. [Google Scholar] [CrossRef]
- Famin, O.; Krugliak, M.; Ginsburg, H. Kinetics of inhibition of glutathione-mediated degradation of ferriprotoporphyrin IX by antimalarial drugs. Biochem. Pharmacol. 1999, 58, 59–68. [Google Scholar] [CrossRef]
- Krishna, S.; Bustamante, L.; Haynes, R.K.; Staines, H.M. Artemisinins: Their growing importance in medicine. Trends Pharm. Sci. 2008, 29, 520–527. [Google Scholar] [CrossRef]
- Schirmer, R.H.; Adler, H.; Pickhardt, M.; Mandelkow, E. Lest we forget you—Methylene blue …. Neurobiol. Aging 2011, 32, 2325.e7–2325.e16. [Google Scholar] [CrossRef]
- Färber, P.M.; Arscott, L.D.; Williams, C.H.; Becker, K.; Schirmer, R.H. Recombinant Plasmodium falciparum glutathione reductase is inhibited by the antimalarial dye methylene blue. FEBS Lett. 1998, 422, 311–314. [Google Scholar] [CrossRef]
- Rahbari, M.; Diederich, K.; Becker, K.; Krauth-Siegel, R.L.; Jortzik, E. Detection of thiol-based redox switch processes in parasites—Facts and future. Biol. Chem. 2015, 396, 445–463. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Rahlfs, S.; Nickel, C.; Schirmer, R.H. Glutathione--functions and metabolism in the malarial parasite Plasmodium falciparum. Biol. Chem. 2003, 384, 551–566. [Google Scholar] [CrossRef]
- Meierjohann, S.; Walter, R.D.; Müller, S. Regulation of intracellular glutathione levels in erythrocytes infected with chloroquine-sensitive and chloroquine-resistant Plasmodium falciparum. Biochem. J. 2002, 368, 761–768. [Google Scholar] [CrossRef]
- Wang, J.; Huang, L.; Li, J.; Fan, Q.; Long, Y.; Li, Y.; Zhou, B. Artemisinin directly targets malarial mitochondria through its specific mitochondrial activation. PLoS ONE 2010, 5, e9582. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, I.K.; Rottenberg, H.; Vaidya, A.B. Atovaquone, a Broad Spectrum Antiparasitic Drug, Collapses Mitochondrial Membrane Potential in a Malarial Parasite. J. Biol. Chem. 1997, 272, 3961–3966. [Google Scholar] [CrossRef] [PubMed]
- Ke, H.; Lewis, I.A.; Morrisey, J.M.; McLean, K.J.; Ganesan, S.M.; Painter, H.J.; Mather, M.W.; Jacobs-Lorena, M.; Llinás, M.; Vaidya, A.B. Genetic investigation of tricarboxylic acid metabolism during the Plasmodium falciparum life cycle. Cell Rep. 2015, 11, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Arese, P.; Gallo, V.; Pantaleo, A.; Turrini, F. Life and Death of Glucose-6-Phosphate Dehydrogenase (G6PD) Deficient Erythrocytes—Role of Redox Stress and Band 3 Modifications. Transfus. Med. Hemother. 2012, 39, 328–334. [Google Scholar] [CrossRef]
- Pantaleo, A.; Ferru, E.; Giribaldi, G.; Mannu, F.; Carta, F.; Matte, A.; de Franceschi, L.; Turrini, F. Oxidized and poorly glycosylated band 3 is selectively phosphorylated by Syk kinase to form large membrane clusters in normal and G6PD-deficient red blood cells. Biochem. J. 2009, 418, 359–367. [Google Scholar] [CrossRef]
- Cappellini, M.D.; Tavazzi, D.; Duca, L.; Graziadei, G.; Mannu, F.; Turrini, F.; Arese, P.; Fiorelli, G. Metabolic indicators of oxidative stress correlate with haemichrome attachment to membrane, band 3 aggregation and erythrophagocytosis in β-thalassaemia intermedia. Br. J. Haematol. 1999, 104, 504–512. [Google Scholar] [CrossRef]
- Cappadoro, M.; Giribaldi, G.; O’Brien, E.; Turrini, F.; Mannu, F.; Ulliers, D.; Simula, G.; Luzzatto, L.; Arese, P. Early Phagocytosis of Glucose-6-Phosphate Dehydrogenase (G6PD)-Deficient Erythrocytes Parasitized by Plasmodium falciparum May Explain Malaria Protection in G6PD Deficiency. Blood 1998, 92, 2527–2534. [Google Scholar] [CrossRef]
- Ehrhardt, K.; Davioud-Charvet, E.; Ke, H.; Vaidya, A.B.; Lanzer, M.; Deponte, M. The antimalarial activities of methylene blue and the 1,4-naphthoquinone 3-[4-(trifluoromethyl)benzyl]-menadione are not due to inhibition of the mitochondrial electron transport chain. Antimicrob. Agents Chemother. 2013, 57, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Müller, T.; Johann, L.; Jannack, B.; Brückner, M.; Lanfranchi, D.A.; Bauer, H.; Sanchez, C.; Yardley, V.; Deregnaucourt, C.; Schrével, J.; et al. Glutathione Reductase-Catalyzed Cascade of Redox Reactions To Bioactivate Potent Antimalarial 1,4-Naphthoquinones—A New Strategy to Combat Malarial Parasites. J. Am. Chem. Soc. 2011, 133, 11557–11571. [Google Scholar] [CrossRef] [PubMed]
- Lukeš, J.; Butenko, A.; Hashimi, H.; Maslov, D.A.; Votýpka, J.; Yurchenko, V. Trypanosomatids Are Much More than Just Trypanosomes: Clues from the Expanded Family Tree. Trends Parasitol. 2018, 34, 466–480. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.C.F.; Godinho, J.L.P.; de Souza, W. Biology of Human Pathogenic Trypanosomatids: Epidemiology, Lifecycle and Ultrastructure. In Proteins and Proteomics of Leishmania and Trypanosoma; Santos, A.L.S., Branquinha, M.H., d’Avila-Levy, C.M., Kneipp, L.F., Sodré, C.L., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 1–42. ISBN 978-94-007-7305-9. [Google Scholar]
- Matthews, K.R. The developmental cell biology of Trypanosoma brucei. J. Cell Sci. 2005, 118, 283–290. [Google Scholar] [CrossRef]
- Burza, S.; Croft, S.L.; Boelaert, M. Leishmaniasis. Lancet 2018, 392, 951–970. [Google Scholar] [CrossRef]
- Tyler, K.M.; Olson, C.L.; Engman, D.M. The Life Cycle Of Trypanosoma Cruzi. In American Trypanosomiasis; Tyler, K.M., Miles, M.A., Eds.; Springer: Boston, MA, USA, 2003; pp. 1–11. ISBN 978-1-4419-9206-2. [Google Scholar]
- World Health Organization & TDR Disease Reference Group on Chagas Disease, Human African Trypanosomiasis and Leishmaniasis. Research Priorities for Chagas Disease, Human African Trypanosomiasis and Leishmaniasis. Available online: https://apps.who.int/iris/handle/10665/77472 (accessed on 25 August 2020).
- Babokhov, P.; Sanyaolu, A.O.; Oyibo, W.A.; Fagbenro-Beyioku, A.F.; Iriemenam, N.C. A current analysis of chemotherapy strategies for the treatment of human African trypanosomiasis. Pathog. Glob. Health 2013, 107, 242–252. [Google Scholar] [CrossRef]
- Franco, J.; Scarone, L.; Comini, M.A. Chapter Three—Drugs and Drug Resistance in African and American Trypanosomiasis. In Annual Reports in Medicinal Chemistry; Botta, M., Ed.; Academic Press: Cambridge, MA, USA, 2018; Volume 51, pp. 97–133. ISBN 0065-7743. [Google Scholar]
- Lang, S.; Khalaf, A.I.; Breen, D.; Huggan, J.K.; Clements, C.J.; MacKay, S.P.; Suckling, C.J. Oligoamides of 2-amino-5-alkylthiazole 4-carboxylic acids: Anti-trypanosomal compounds. Med. Chem. Res. 2014, 23, 1170–1179. [Google Scholar] [CrossRef]
- Scott, F.J.; Khalaf, A.I.; Giordani, F.; Wong, P.E.; Duffy, S.; Barrett, M.; Avery, V.M.; Suckling, C.J. An evaluation of Minor Groove Binders as anti-Trypanosoma brucei brucei therapeutics. Eur. J. Med. Chem. 2016, 116, 116–125. [Google Scholar] [CrossRef]
- Rodríguez Arce, E.; Mosquillo, M.F.; Pérez-Díaz, L.; Echeverría, G.A.; Piro, O.E.; Merlino, A.; Coitiño, E.L.; Maríngolo Ribeiro, C.; Leite, C.Q.F.; Pavan, F.R.; et al. Aromatic amine N-oxide organometallic compounds: Searching for prospective agents against infectious diseases. Dalton Trans. 2015, 44, 14453–14464. [Google Scholar] [CrossRef]
- Mosquillo, M.F.; Bilbao, L.; Hernández, F.; Tissot, F.; Gambino, D.; Garat, B.; Pérez-Díaz, L. Trypanosoma cruzi biochemical changes and cell death induced by an organometallic platinum-based compound. Chem. Biol. Drug Des. 2018, 92, 1657–1669. [Google Scholar] [CrossRef]
- Biot, C.; Dive, D. Bioorganometallic Chemistry and Malaria. In Medicinal Organometallic Chemistry; Jaouen, G., Metzler-Nolte, N., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 155–193. ISBN 978-3-642-13185-1. [Google Scholar]
- Dubar, F.; Egan, T.J.; Pradines, B.; Kuter, D.; Ncokazi, K.K.; Forge, D.; Paul, J.-F.; Pierrot, C.; Kalamou, H.; Khalife, J.; et al. The Antimalarial Ferroquine: Role of the Metal and Intramolecular Hydrogen Bond in Activity and Resistance. ACS Chem. Biol. 2011, 6, 275–287. [Google Scholar] [CrossRef]
- Aguirre, G.; Boiani, L.; Cerecetto, H.; Fernández, M.; González, M.; Denicola, A.; Otero, L.; Gambino, D.; Rigol, C.; Olea-Azar, C.; et al. In vitro activity and mechanism of action against the protozoan parasite Trypanosoma cruzi of 5-nitrofuryl containing thiosemicarbazones. Bioorganic Med. Chem. 2004, 12, 4885–4893. [Google Scholar] [CrossRef]
- Krauth-Siegel, L.R.; Comini, M.A.; Schlecker, T. The Trypanothione System. In Peroxiredoxin Systems: Structures and Functions; Flohé, L., Harris, J.R., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 231–251. ISBN 978-1-4020-6051-9. [Google Scholar]
- Moutiez, M.; Meziane-cherif, D.; Aumercier, M.; Sergheraert, C.; Tartar, A. Compared Reactivities of Trypanothione and Glutathione in Conjugation Reactions. Chem. Pharm. Bull. 1994, 42, 2641–2644. [Google Scholar] [CrossRef]
- Wilkinson, S.R.; Horn, D.; Prathalingam, S.R.; Kelly, J.M. RNA Interference Identifies Two Hydroperoxide Metabolizing Enzymes That Are Essential to the Bloodstream Form of the African Trypanosome. J. Biol. Chem. 2003, 278, 31640–31646. [Google Scholar] [CrossRef]
- Budde, H.; Flohé, L.; Hecht, H.-J.; Hofmann, B.; Stehr, M.; Wissing, J.; Lünsdorf, H. Kinetics and redox-sensitive oligomerisation reveal negative subunit cooperativity in tryparedoxin peroxidase of Trypanosoma brucei brucei. Biol. Chem. 2003, 384, 619–633. [Google Scholar] [CrossRef]
- Schlecker, T.; Schmidt, A.; Dirdjaja, N.; Voncken, F.; Clayton, C.; Krauth-Siegel, R.L. Substrate Specificity, Localization, and Essential Role of the Glutathione Peroxidase-type Tryparedoxin Peroxidases in Trypanosoma brucei. J. Biol. Chem. 2005, 280, 14385–14394. [Google Scholar] [CrossRef]
- Fairlamb, A.H.; Henderson, G.B.; Bacchi, C.J.; Cerami, A. In vivo effects of difluoromethylornithine on trypanothione and polyamine levels in bloodstream forms of Trypanosoma brucei. Mol. Biochem. Parasitol. 1987, 24, 185–191. [Google Scholar] [CrossRef]
- Zíková, A.; Verner, Z.; Nenarokova, A.; Michels, P.A.M.; Lukeš, J. A paradigm shift: The mitoproteomes of procyclic and bloodstream Trypanosoma brucei are comparably complex. PLoS Pathog. 2017, 13, e1006679. [Google Scholar] [CrossRef]
- Seung, K.J.; Keshavjee, S.; Rich, M.L. Multidrug-Resistant Tuberculosis and Extensively Drug-Resistant Tuberculosis. Cold Spring Harb. Perspect. Med. 2015, 5, a017863. [Google Scholar] [CrossRef] [PubMed]
- Lange, C.; Abubakar, I.; Alffenaar, J.-W.C.; Bothamley, G.; Caminero, J.A.; Carvalho, A.C.C.; Chang, K.-C.; Codecasa, L.; Correia, A.; Crudu, V.; et al. Management of patients with multidrug-resistant/extensively drug-resistant tuberculosis in Europe: A TBNET consensus statement. Eur. Respir. J. 2014, 44, 23–63. [Google Scholar] [CrossRef] [PubMed]
- Günther, G.; Gomez, G.B.; Lange, C.; Rupert, S.; van Leth, F. Availability, price and affordability of anti-tuberculosis drugs in Europe: A TBNET survey. Eur. Respir. J. 2015, 45, 1081. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control/WHO Regional Office for Europe Tuberculosis Surveillance and Monitoring in Europe 2018–2016 Data. Available online: www.ecdc.europa.eu/en/publications-data/tuberculosis-surveillance-and-monitoring-europe-2018 (accessed on 22 August 2020).
- Rowland, J.L.; Niederweis, M. Resistance mechanisms of Mycobacterium tuberculosis against phagosomal copper overload. Tuberculosis 2012, 92, 202–210. [Google Scholar] [CrossRef] [PubMed]
- White, C.; Lee, J.; Kambe, T.; Fritsche, K.; Petris, M.J. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J. Biol. Chem. 2009, 284, 33949–33956. [Google Scholar] [CrossRef]
- Jin, Y.; Lewis, M.A.; Gokhale, N.H.; Long, E.C.; Cowan, J.A. Influence of Stereochemistry and Redox Potentials on the Single- and Double-Strand DNA Cleavage Efficiency of Cu(II) and Ni(II)·Lys-Gly-His-Derived ATCUN Metallopeptides. J. Am. Chem. Soc. 2007, 129, 8353–8361. [Google Scholar] [CrossRef]
- Russell, D.G. Mycobacterium tuberculosis: Here today, and here tomorrow. Nat. Rev. Mol. Cell Biol. 2001, 2, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.Q.; Helaine, S.; Lewis, K.; Ackermann, M.; Aldridge, B.; Andersson, D.I.; Brynildsen, M.P.; Bumann, D.; Camilli, A.; Collins, J.J.; et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 2019, 17, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Gomez, J.E.; McKinney, J.D. M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis 2004, 84, 29–44. [Google Scholar] [CrossRef]
- Stewart, G.R.; Robertson, B.D.; Young, D.B. Tuberculosis: A problem with persistence. Nat. Rev. Microbiol. 2003, 1, 97–105. [Google Scholar] [CrossRef]
- Vijay, S.; Mukkayyan, N.; Ajitkumar, P. Highly Deviated Asymmetric Division in Very Low Proportion of Mycobacterial Mid-log Phase Cells. Open Microbiol. J. 2014, 8, 40–50. [Google Scholar] [CrossRef]
- Vijay, S.; Nagaraja, M.; Sebastian, J.; Ajitkumar, P. Asymmetric cell division in Mycobacterium tuberculosis and its unique features. Arch. Microbiol. 2014, 196, 157–168. [Google Scholar] [CrossRef]
- Vijay, S.; Nair, R.R.; Sharan, D.; Jakkala, K.; Mukkayyan, N.; Swaminath, S.; Pradhan, A.; Joshi, N.V.; Ajitkumar, P. Mycobacterial Cultures Contain Cell Size and Density Specific Sub-populations of Cells with Significant Differential Susceptibility to Antibiotics, Oxidative and Nitrite Stress. Front. Microbiol. 2017, 8, 463. [Google Scholar] [CrossRef]
- Roca, F.J.; Ramakrishnan, L. TNF dually mediates resistance and susceptibility to mycobacteria via mitochondrial reactive oxygen species. Cell 2013, 153, 521–534. [Google Scholar] [CrossRef] [PubMed]
- Butler, R.E.; Brodin, P.; Jang, J.; Jang, M.-S.; Robertson, B.D.; Gicquel, B.; Stewart, G.R. The balance of apoptotic and necrotic cell death in Mycobacterium tuberculosis infected macrophages is not dependent on bacterial virulence. PLoS ONE 2012, 7, e47573. [Google Scholar] [CrossRef]
- Espinosa-Cueto, P.; Magallanes-Puebla, A.; Castellanos, C.; Mancilla, R. Dendritic cells that phagocytose apoptotic macrophages loaded with mycobacterial antigens activate CD8 T cells via cross-presentation. PLoS ONE 2017, 12, e0182126. [Google Scholar] [CrossRef]
- Winau, F.; Weber, S.; Sad, S.; de Diego, J.; Hoops, S.L.; Breiden, B.; Sandhoff, K.; Brinkmann, V.; Kaufmann, S.H.E.; Schaible, U.E. Apoptotic Vesicles Crossprime CD8 T Cells and Protect against Tuberculosis. Immunity 2006, 24, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, Z.; Kandasamy, V.; Lee, S.Y.; Solem, C.; Jensen, P.R. Harnessing the respiration machinery for high-yield production of chemicals in metabolically engineered Lactococcus lactis. Metab. Eng. 2017, 44, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Fang, H.; Groom, L.; Cheng, A.; Zhang, W.; Liu, J.; Wang, X.; Li, K.; Han, P.; Zheng, M.; et al. Superoxide Flashes in Single Mitochondria. Cell 2008, 134, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Shen, E.-Z.; Song, C.-Q.; Lin, Y.; Zhang, W.-H.; Su, P.-F.; Liu, W.-Y.; Zhang, P.; Xu, J.; Lin, N.; Zhan, C.; et al. Mitoflash frequency in early adulthood predicts lifespan in Caenorhabditis elegans. Nature 2014, 508, 128–132. [Google Scholar] [CrossRef]
- Schwarzländer, M.; Logan, D.C.; Fricker, M.D.; Sweetlove, L.J. The circularly permuted yellow fluorescent protein cpYFP that has been used as a superoxide probe is highly responsive to pH but not superoxide in mitochondria: Implications for the existence of superoxide ‘flashes. ’ Biochem. J. 2011, 437, 381–387. [Google Scholar] [CrossRef]
- Schwarzländer, M.; Wagner, S.; Ermakova, Y.G.; Belousov, V.V.; Radi, R.; Beckman, J.S.; Buettner, G.R.; Demaurex, N.; Duchen, M.R.; Forman, H.J.; et al. The “mitoflash” probe cpYFP does not respond to superoxide. Nature 2014, 514, E12–E14. [Google Scholar] [CrossRef]
- White, R.M.; Sessa, A.; Burke, C.; Bowman, T.; LeBlanc, J.; Ceol, C.; Bourque, C.; Dovey, M.; Goessling, W.; Burns, C.E.; et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2008, 2, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.Y.; Aramburu, J.; Douglas, P.S.; Izumo, S. Transgenic expression of green fluorescence protein can cause dilated cardiomyopathy. Nat. Med. 2000, 6, 482–483. [Google Scholar] [CrossRef]
- Agbulut, O.; Coirault, C.; Niederländer, N.; Huet, A.; Vicart, P.; Hagège, A.; Puceat, M.; Menasché, P. GFP expression in muscle cells impairs actin-myosin interactions: Implications for cell therapy. Nat. Methods 2006, 3, 331. [Google Scholar] [CrossRef] [PubMed]
- Baens, M.; Noels, H.; Broeckx, V.; Hagens, S.; Fevery, S.; Billiau, A.D.; Vankelecom, H.; Marynen, P. The Dark Side of EGFP: Defective Polyubiquitination. PLoS ONE 2006, 1, e54. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Ren, W.; Wright, Q.E.; Ai, H. Genetically Encoded Fluorescent Probe for the Selective Detection of Peroxynitrite. J. Am. Chem. Soc. 2013, 135, 14940–14943. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, S.; Li, X.; Ai, H.-W. A High-Performance Genetically Encoded Fluorescent Biosensor for Imaging Physiological Peroxynitrite. Biochemistry 2020. [Google Scholar] [CrossRef]
- Chen, S.; Chen, Z.; Ren, W.; Ai, H. Reaction-Based Genetically Encoded Fluorescent Hydrogen Sulfide Sensors. J. Am. Chem. Soc. 2012, 134, 9589–9592. [Google Scholar] [CrossRef]
- Youssef, S.; Zhang, S.; Ai, H. A Genetically Encoded, Ratiometric Fluorescent Biosensor for Hydrogen Sulfide. ACS Sens. 2019, 4, 1626–1632. [Google Scholar] [CrossRef]
- Chen, Y.; Song, X.; Ye, S.; Miao, L.; Zhu, Y.; Zhang, R.-G.; Ji, G. Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement. Protein Cell 2013, 4, 299–309. [Google Scholar] [CrossRef]
- Dana, H.; Mohar, B.; Sun, Y.; Narayan, S.; Gordus, A.; Hasseman, J.P.; Tsegaye, G.; Holt, G.T.; Hu, A.; Walpita, D.; et al. Sensitive red protein calcium indicators for imaging neural activity. elife 2016, 5, e12727. [Google Scholar] [CrossRef]
- Akerboom, J.; Chen, T.-W.; Wardill, T.J.; Tian, L.; Marvin, J.S.; Mutlu, S.; Calderon, N.C.; Esposti, F.; Borghuis, B.G.; Sun, X.R.; et al. Optimization of a GCaMP Calcium Indicator for Neural Activity Imaging. J. Neurosci. 2012, 32, 13819–13840. [Google Scholar] [CrossRef] [PubMed]
- Dana, H.; Sun, Y.; Mohar, B.; Hulse, B.K.; Kerlin, A.M.; Hasseman, J.P.; Tsegaye, G.; Tsang, A.; Wong, A.; Patel, R.; et al. High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat. Methods 2019, 16, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Berezin, M.Y.; Achilefu, S. Fluorescence Lifetime Measurements and Biological Imaging. Chem. Rev. 2010, 110, 2641–2684. [Google Scholar] [CrossRef] [PubMed]
- Kwok, S.; Lee, C.; Sánchez, S.A.; Hazlett, T.L.; Gratton, E.; Hayashi, Y. Genetically encoded probe for fluorescence lifetime imaging of CaMKII activity. Biochem. Biophys. Res. Commun. 2008, 369, 519–525. [Google Scholar] [CrossRef]
- Yasuda, R.; Harvey, C.D.; Zhong, H.; Sobczyk, A.; van Aelst, L.; Svoboda, K. Supersensitive Ras activation in dendrites and spines revealed by two-photon fluorescence lifetime imaging. Nat. Neurosci. 2006, 9, 283–291. [Google Scholar] [CrossRef]
- Tantama, M.; Hung, Y.P.; Yellen, G. Imaging intracellular pH in live cells with a genetically encoded red fluorescent protein sensor. J. Am. Chem. Soc. 2011, 133, 10034–10037. [Google Scholar] [CrossRef]
- Nakabayashi, T.; Wang, H.-P.; Kinjo, M.; Ohta, N. Application of fluorescence lifetime imaging of enhanced green fluorescent protein to intracellular pH measurements. Photochem. Photobiol. Sci. 2008, 7, 668. [Google Scholar] [CrossRef]
- Zhuo, Y.; Solntsev, K.M.; Reddish, F.; Tang, S.; Yang, J.J. Effect of Ca 2+ on the Steady-State and Time-Resolved Emission Properties of the Genetically Encoded Fluorescent Sensor CatchER. J. Phys. Chem. B 2015, 119, 2103–2111. [Google Scholar] [CrossRef]
- Weller, J.; Kizina, K.M.; Can, K.; Bao, G.; Müller, M. Response properties of the genetically encoded optical H2O2 sensor HyPer. Free Radic. Biol. Med. 2014, 76, 227–241. [Google Scholar] [CrossRef]
- Ravotto, L.; Duffet, L.; Zhou, X.; Weber, B.; Patriarchi, T. A Bright and Colorful Future for G-Protein Coupled Receptor Sensors. Front. Cell. Neurosci. 2020, 14, 67. [Google Scholar] [CrossRef]
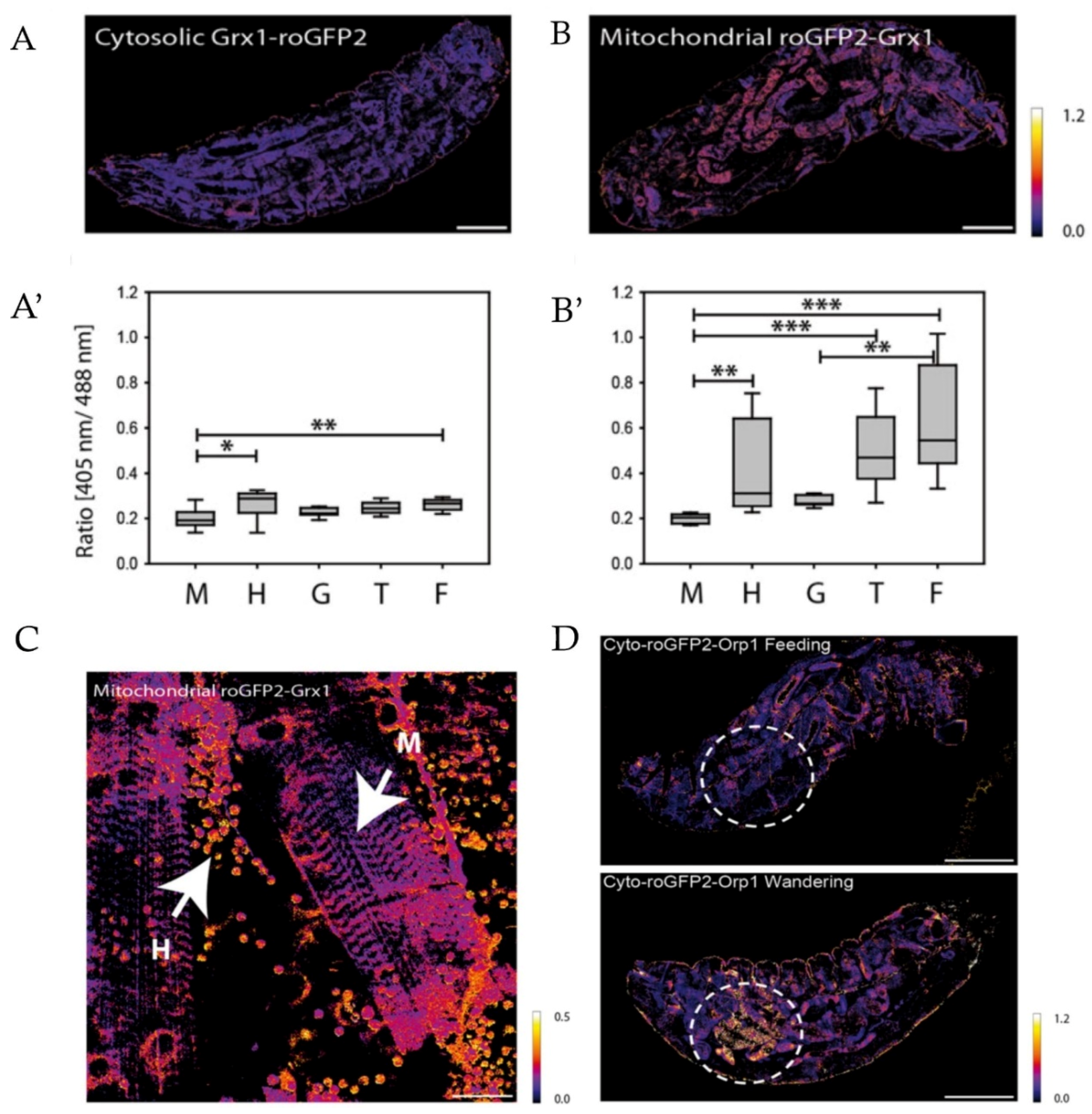
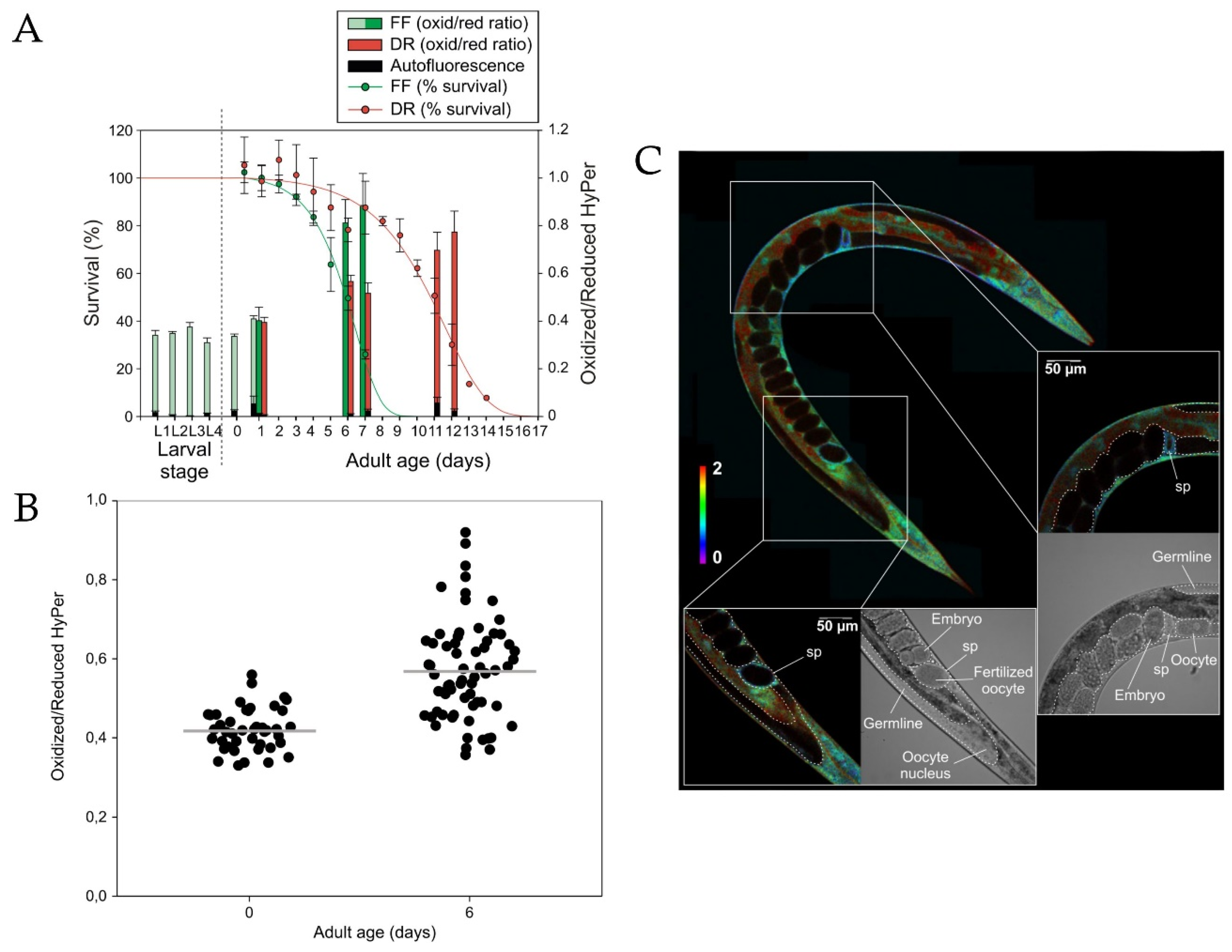

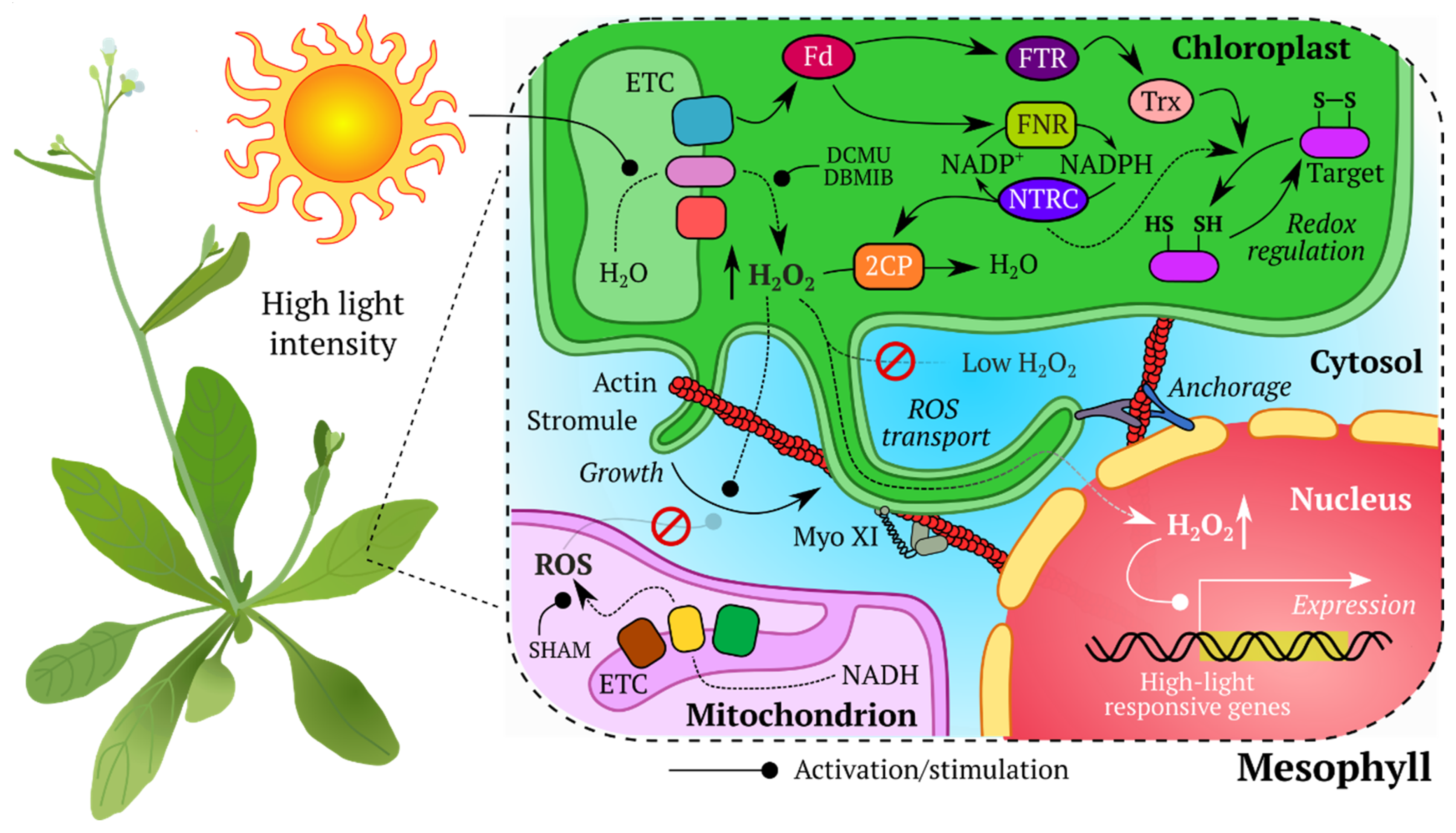
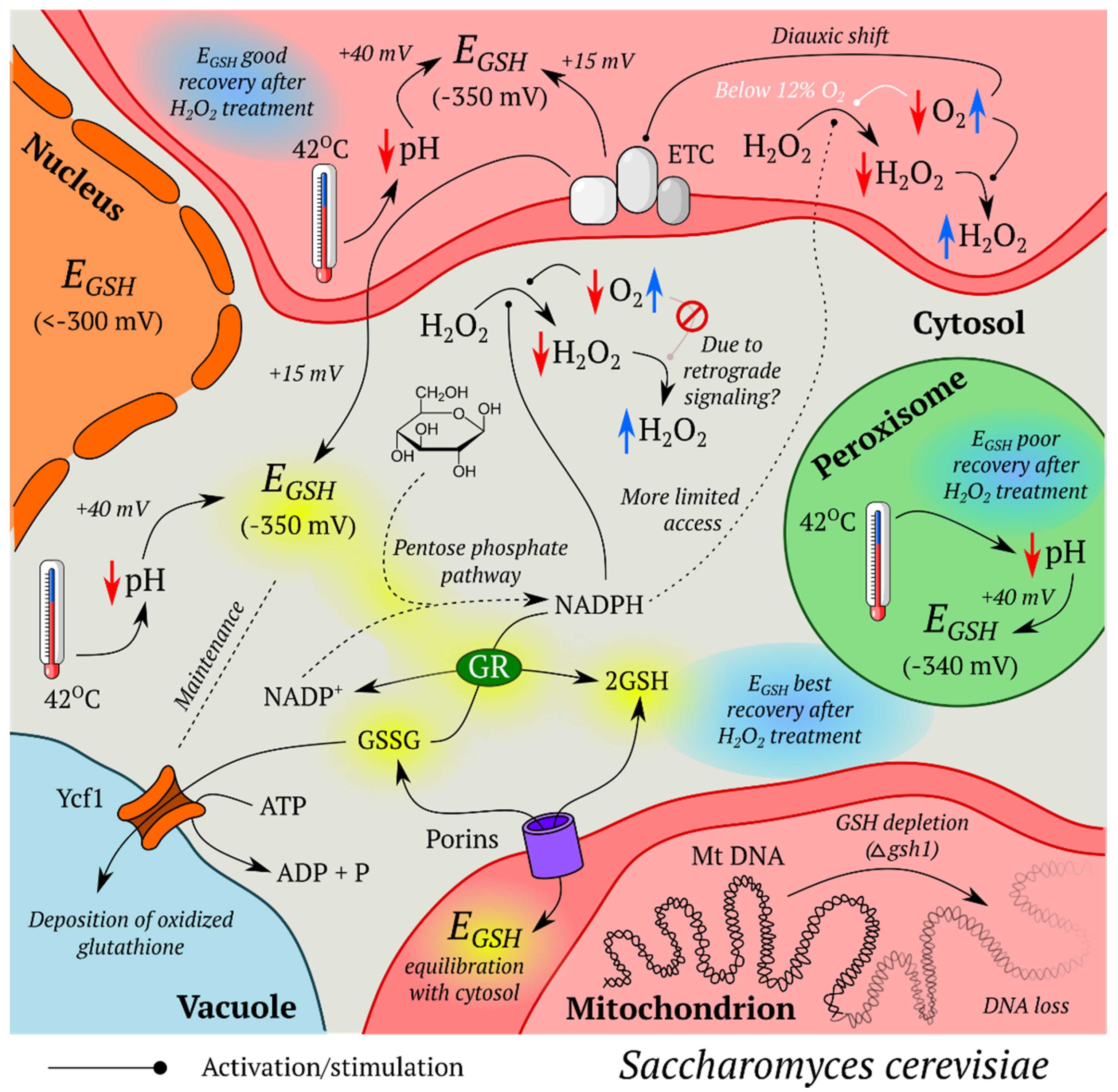
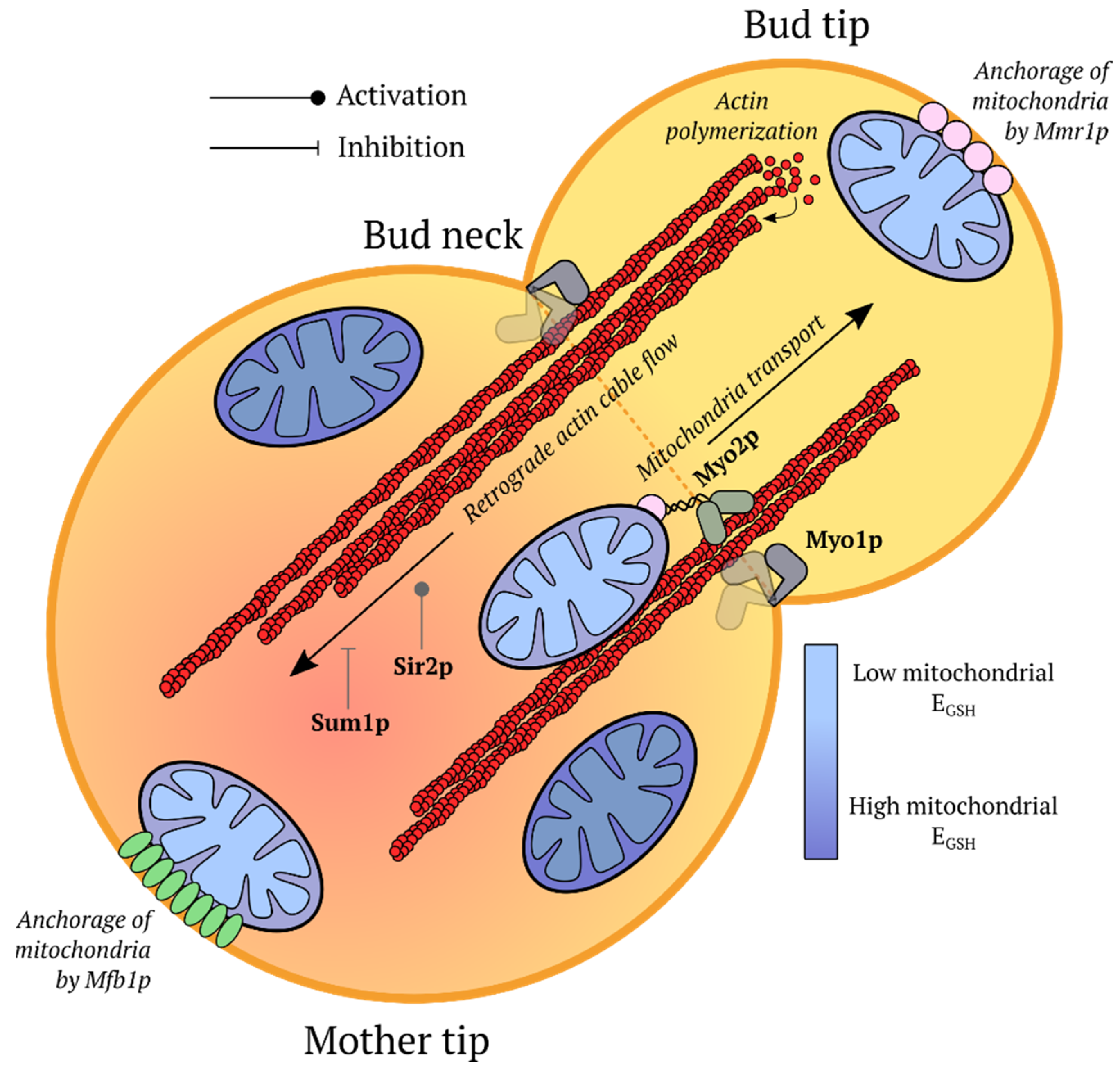

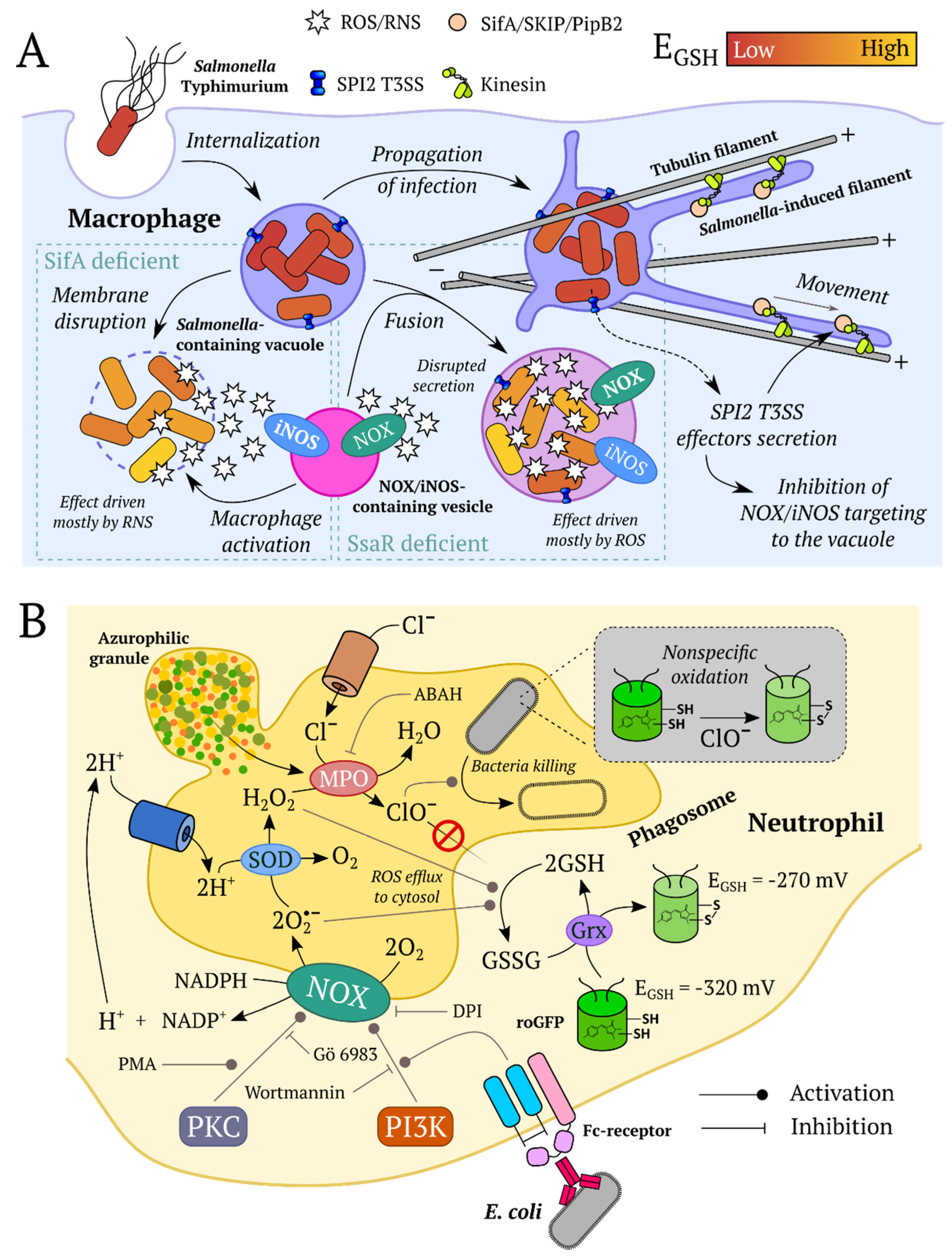
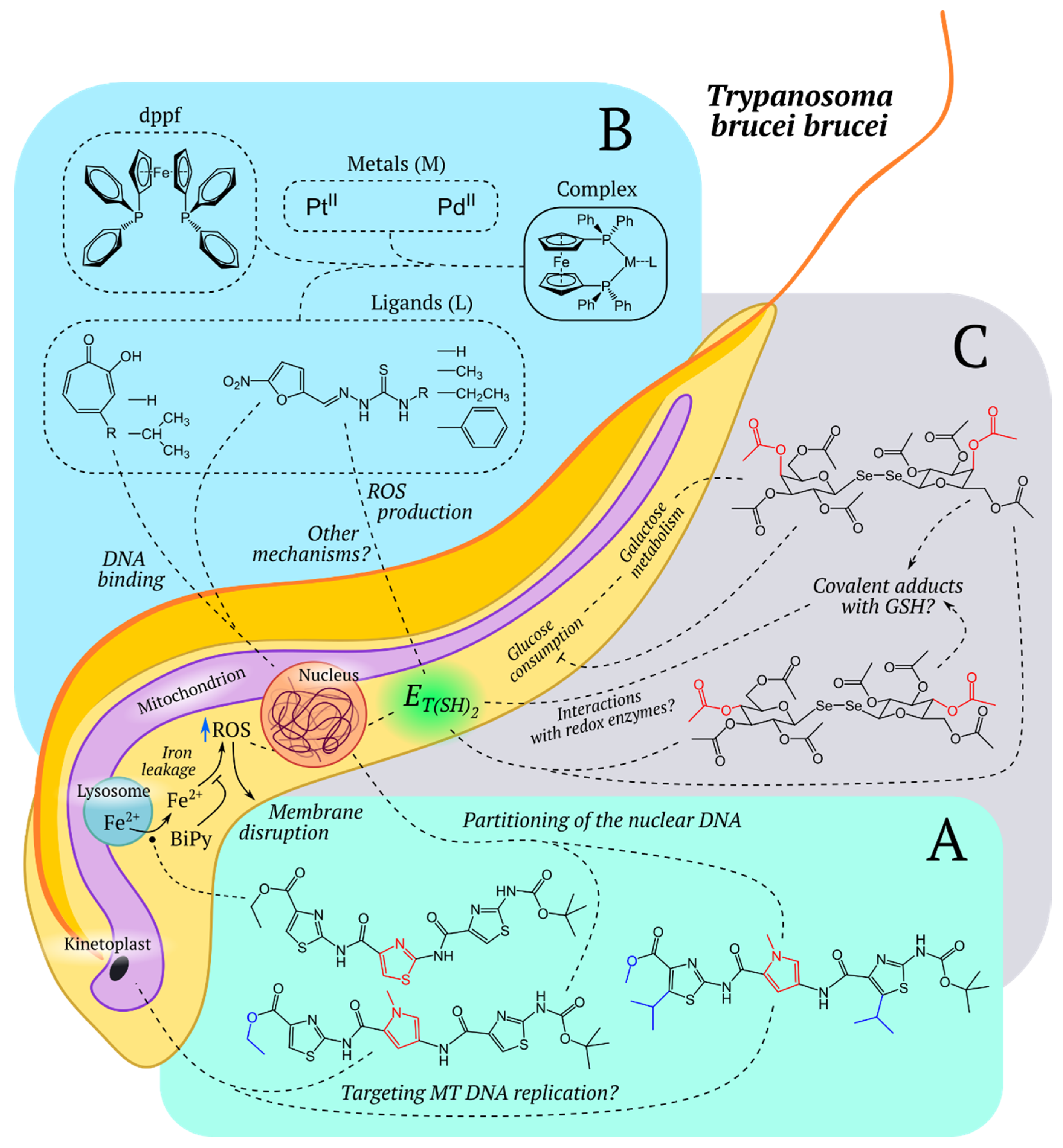
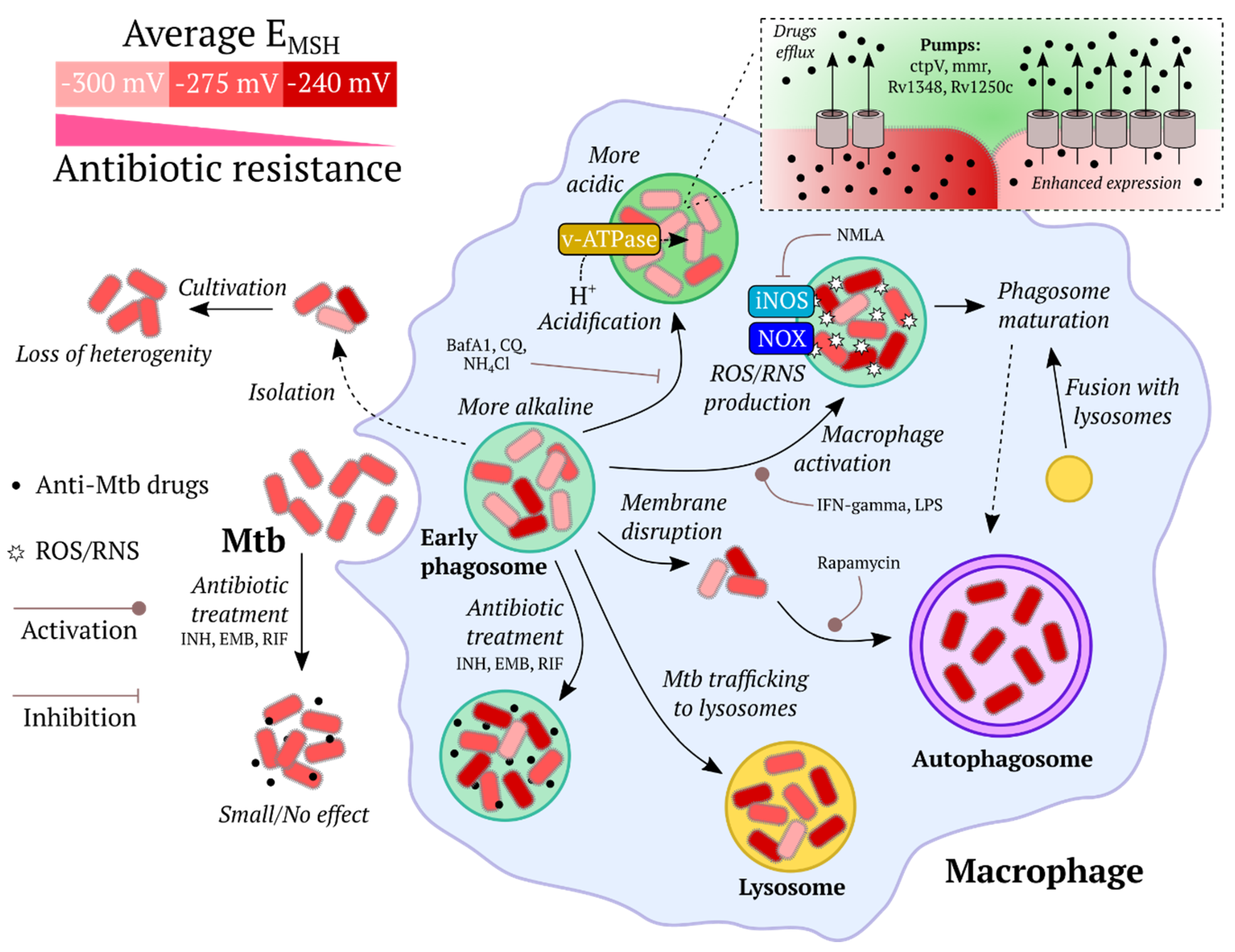

| Analyte | Name | λex | λem | Response Amplitude | Midpoint Potential/EC50/Kd/Ks | Reference |
|---|---|---|---|---|---|---|
| general redox state | Oba-Qc | 430 | 480 | ~2 | MP = −249 mV | [95] |
| NADH/NAD+ | Peredox | 400/587 | 510/610 | 2.5 | Kd to NADH <5 nM for initial P0 construct | [107] |
| H2O2 | NeonOxIrr | 508 | 520 | 2.8 (in vitro) | NM | [105] |
| NADH | Frex | 420/500 | 518 | 9 (fl. increase) | Kd~3.7 μM at pH 7.4 | [109] |
| NADH | FrexH | 420/500 | 518 | 3 (fl. decrease) | Kd~40 nM | [109] |
| NADH/NAD+ | RexYFP | 490 | 516 | ~2 (in vitro) | K’(NADH) = 180 nM, K’(NADPH) = 6.2 μM | [110] |
| NADH/NAD+ | SoNar | 420/485 | 528 | 15 | Kd(NADH) ~0.2 μM, Kd(NAD+) ~5.0 μM at pH 7.4 | [111] |
| NAD+/AXP (AXP = ATP + ADP) | FiNad | 485 | ~520 | ~7 | Kd(NAD+) shifts from ~14 μM to ~1.3 mM in the presence of ATP or ADP | [112] |
| NADPH | iNaps | 420/500 | 515 | 10 | Kd = 2.0 ÷ 120 μM | [75] |
| EGSH | rxYFP | 512 | 523 | 2.2 (in vitro) | MP = −261 mV | [93] |
| EGSH | roGFP1 | 400/475 | 508 | ~6 (in vitro) | MP = −294 mV | [85,117] |
| EGSH | roGFP1-Rx family | 395/475 | NM | 5.4–7.5 | MP = −263 mV to −284 mV | [86] |
| EGSH | roGFP1-iX | 395/465 | 505 | 2.4–7.2 | MP = −229 mV to −246 mV | [87] |
| EGSH | roGFP2 | 400/490 | 511 | ~6 (in vitro) | MP = −287 mV | [85,117] |
| general redox state | roUnaG | 498 | 527 | ~9 (in vitro) | MP = −275 mV | [99] |
| EGSH | Grx1-roGFP2 | 400/490 * | 511 | ~4.4 (in living cells) | MP = −280 mV | [88] |
| EMSH | Mrx1-roGFP2 | 390/490 | 510 | ~8 (in vitro) | MP = −280 mV | [52] |
| EBSH | Brx-roGFP2 | 405/488 | NM | ~4 (in vitro) | NM | [89] |
| ET(SH)2 | Tpx-roGFP2 | 400/490 * | 511 | NM | NM | [90] |
| H2O2 | TriPer | 405/488 | NM | NM | NM | [104] |
| H2O2 | HyPer | 420/500 | 516 | 3.3 (in vitro) | Ks = 5 × 105 M−1 s−1 | [66,100] |
| H2O2 | HyPer-2 | 420/500 | 516 | ~6 (HeLa cells) | Ks = 1.2 × 105 M−1 s−1 | [66,101] |
| H2O2 | HyPer-3 | 420/500 | 516 | ~6 (HeLa cells) | Ks = 2.5 × 105 M−1 s−1 | [66] |
| H2O2 | HyPer7 | 400/499 | 516 | ~10 (in vitro) | i.v. = 26.9 ± 0.28 a.u./s (for HyPer3 it is 0.315 ± 0.007 a.u./s) | [102] |
| H2O2 | roGFP2-Orp1 | 400/490 * | 511 | 4.8 (HeLa cells) | responds to low micromolar concentrations of exogenously applied H2O2 (HeLa cells) | [91] |
| H2O2 | roGFP2-Tsa2ΔCR | 400/490 * | 511 | ~6 (in vitro) | low-nanomolar or high-picomolar endogenous H2O2 concentrations | [92] |
| S-MetO | MetSOx | 425/505 | 510–516 | ~6 | K0.5 = 0.5 μM (in vitro) | [113] |
| R-MetO | MetROx | 410/500 | 510–516 | ~6 | K0.5 = 177 μM (in vitro) | [113] |
| ROOH | OHSer | 519 | 526 | 2 | NM | [114] |
| general redox state | rxRFP | 576 | 600 | 4 | MP = −290 mV | [96] |
| EGSH | Grx1-roCherry | 589 | 610 | 1.5 | MP = −311 mV (pH = 7.0) | [97] |
| H2O2 | HyPerRed | 575 | 605 | ~2 | Ks = 3 × 105 M−1 s−1 | [103] |
| general redox state | Redoxfluor | NM | 476/527 | ~1.5 (H2O2 in vitro) | MP = −213 mV (pH = 7.0) | [44] |
| Trx redox state | CROST | NM | 480/530 | ~3 | MP (CROST1) = −266 mV (pH = 7.5) MP(CROST2) = −296 mV (pH = 7.5) | [116] |
| NADP+ | NADPsor | NM | 478/526 | ~1.4 | Kd = 2 mM, detection limit = 1 μM (for NADPsor-K1-K72W version) | [115] |
| Model organism | Parameter | GEFI | References |
|---|---|---|---|
| Animals | |||
| Caenorhabditis elegans | EGSH | roGFP1 | [22,23,34] |
| roGFP2 | [118] | ||
| roGFP | [119] | ||
| roGFP1-R12 | [50] | ||
| Grx1-roGFP2 | [120,121,122] | ||
| H2O2 | HyPer | [34,70,120,123,124,125,126] | |
| HyPer-2 | [127] | ||
| roGFP2-Orp1 | [128,129,130] | ||
| roGFP2-Tsa2ΔCR | [129] | ||
| NADH/NAD+ | Peredox | [56,131] | |
| Drosophila melanogaster | EGSH | roGFP2 | [61] |
| Grx1-roGFP2 | [18] | ||
| H2O2 | roGFP2-Orp1 | [18,132] | |
| Danio rerio | EGSH | roGFP2 | [133] |
| Grx1-roGFP2 | [134,135,136,137,138] | ||
| Grx1-roCherry | [97] | ||
| NADH/NAD+ | SoNar | [139] | |
| RexYFP | [64] | ||
| NAD+/AXP, AXP = ATP + ADP | FiNad | [112] | |
| NADPH | iNap | [75,139] | |
| H2O2 | HyPer | [57,58,63,65,67,68,69,140,141,142] | |
| HyPer-3 | [66] | ||
| HyPerRed | [75,139] | ||
| HyPer7 | [102] | ||
| roGFP2-Orp1 | [137,138] | ||
| Mus musculus | EGSH | roGFP1 | [53,54,143,144] |
| roGFP2 | [60] | ||
| roGFP | [21,145,146,147] | ||
| Grx1-roGFP2 | [55,62] | ||
| NADH/NAD+ | Peredox | [76] | |
| SoNar | [111,148,149] | ||
| NAD+/AXP | FiNad | [112] | |
| H2O2 | roGFP2-Orp1 | [150] | |
| NeonOxIrr | [105] | ||
| Xenopus laevis | H2O2 | HyPer | [71,72,151] |
| Plants | |||
| Arabidopsis thaliana | EGSH | roGFP | [152] |
| roGFP1 | [153,154,155,156,157,158] | ||
| roGFP2 | [153,159,160,161,162,163,164,165,166,167] | ||
| Grx1-roGFP2 | [168,169,170,171,172,173,174,175,176,177] | ||
| roGFP2-iL | [178] | ||
| GRX1-roGFP2-iL | [178] | ||
| Trx redox state | CROST | [116] | |
| H2O2 | roGFP2-Orp1 | [174,179] | |
| HyPer | [180,181,182,183,184,185] | ||
| Medicago truncatula | H2O2 | HyPer | [186] |
| Nicotiana benthamiana | H2O2 | HyPer | [187] |
| HyPer-2 | [188] | ||
| Nicotiana tabacum | EGSH | roGFP1 | [153] |
| roGFP2 | [153,189] | ||
| Solanum lycopersicum | EGSH | roGFP1 | [190] |
| Fungi | |||
| Botrytis cinerea | EGSH | roGFP2 | [191,192] |
| Grx1-roGFP2 | [191] | ||
| Cochliobolus heterostrophus | H2O2 | HyPer | [193] |
| Fusarium graminearum | H2O2 | HyPer-2 | [194] |
| Magnaporthe oryzae | EGSH | Grx1-roGFP2 | [195] |
| H2O2 | MoHyPer | [196] | |
| Pichia pastoris | EGSH | roGFP1 | [37,197] |
| roGFP1-iE | [37,197,198] | ||
| roGFP1-iL | [198] | ||
| general redox state | Redoxfluor | [44] | |
| Saccharomyces cerevisiae | EGSH | rxYFP | [31,43,46,199,200,201] |
| roGFP1 | [25,26,27,28,29,202,203] | ||
| roGFP2 | [12,19,33,204,205,206] | ||
| roGFP | [207] | ||
| eroGFP | [35,36,38,39,40,41,42,208] | ||
| Grx1-roGFP2 | [15,24,32,47,48,209,210,211,212,213,214] | ||
| general redox state | rxRFP | [30] | |
| Redoxfluor | [44,215] | ||
| H2O2 | HyPer | [92] | |
| HyPerRed | [216] | ||
| roGFP2-Orp1 | [13,214,217] | ||
| roGFP2-Tsa2ΔCR | [14,15,92,206] | ||
| roGFP2-Tsa2ΔCRΔCP | [92] | ||
| Schizosaccharomyces pombe | H2O2 | HyPer | [218] |
| roGFP2-Tpx1.C169S * | [218] | ||
| Ustilago maydis | NADH/NAD+ | Peredox | [219] |
| Eukaryotic unicellular organisms | |||
| Chlamydomonas reinhardtii | EGSH | ObaQc | [220] |
| Phaeodactylum tricornutum | EGSH | roGFP | [221,222] |
| Plasmosium falciparum | EGSH | Grx1-roGFP2 | [16,73,223,224,225] |
| sfroGFP2 | [73] | ||
| H2O2 | HyPer-3 | [226] | |
| roGFP2-Orp1 | [17,226] | ||
| Toxoplasma gondii | EGSH | roGFP1, roGFP-iL | [45] |
| Trypanosoma brucei brucei | EGSH/ET(SH)2 | Grx1-roGFP2 | [90,227,228,229,230,231] |
| roGFP2 | [90] | ||
| ET(SH)2 | Tpx-roGFP2 | [90,232] | |
| Bacteria | |||
| Caulobacter crescentus | EGSH | roGFP2 | [233] |
| Chlamydia trachomatis | EGSH | roGFP2 | [234] |
| Citrobacter rodentium | EGSH | roGFP2 | [235] |
| Corynebacterium glutamicum | EMSH | Mrx1-roGFP2 | [74] |
| Escherichia coli | EGSH | rxYFP | [93] |
| roGFP1 | [236] | ||
| roGFP2 | [235,237,238,239] | ||
| Grx1-roGFP2 | [239,240] | ||
| general redox state | roUnaG | [99] | |
| H2O2 | roGFP2-Orp1 | [239,240] | |
| S- and R-MetO | MetSOx, MetROx | [113] | |
| Lactobacillus paracasei | NADH | Frex | [241] |
| Lactococcus lactis | EGSH | roGFP1-R12 | [242] |
| Methylococcus capsulatus | NADH/NAD+ | Peredox | [243] |
| Mycobacterium marinum | NADH/NAD+ | Peredox | [244] |
| Mycobacterium smegmatis | EMSH | Mrx1-roGFP2 | [52,245,246,247,248,249] |
| NADH/NAD+ | Peredox | [250] | |
| Mycobacterium tuberculosis | EMSH | Mrx1-roGFP2 | [49,52,245,246,251,252,253] |
| roGFP1-R12 | [254,255] | ||
| NADH/NAD+ | Peredox | [250] | |
| Pantoe eucalypti | EGSH | roGFP2 | [256] |
| Ralstonia eutropha | NADH/NAD+ | Peredox | [108] |
| NADH | Frex | [257] | |
| Salmonella Typhi | EGSH | roGFP2 | [235] |
| Salmonella Typhimurium | EGSH | roGFP2 | [235,258,259] |
| Staphylococcus aureus | EBSH | Brx-roGFP2 | [89,260,261,262] |
| H2O2 | Tpx-roGFP2 * | [260] | |
| Streptococcus oligofermentans | H2O2 | HyPer | [263] |
| Synechococcus elongatus | EGSH | roGFP1 | [264] |
| Yersinia pseudotuberculosis | EGSH | roGFP2 | [235] |
| Transgene | Species | Redox Sensor | Target | References |
|---|---|---|---|---|
| Thy1-mito-Grx1-roGFP2 | Mus musculus | Grx1-roGFP2 | mitochondria of neurons in the CNS and the PNS | [62] |
| Thy1-roGFP1c | Mus musculus | roGFP1 | cytosol of neurons in the CNS and the PNS | [59] |
| Thy1-roGFP1m | Mus musculus | roGFP1 | mitochondria of neurons in the CNS and the PNS | [59] |
| TH-mito-roGFP2 | Mus musculus | roGFP2 | mitochondria of dopaminergic neurons | [60] |
| elav-Gal4; UAS-MTSroGFP2 | Drosophila melanogaster | roGFP2 | mitochondria of neurons | [61] |
| ubi-HyPer | Danio rerio | HyPer | ubiquitous expression | [63] |
| myo6b-REX-YFP | Danio rerio | REX-YFP | hair cells of lateral-line system | [64] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kostyuk, A.I.; Panova, A.S.; Kokova, A.D.; Kotova, D.A.; Maltsev, D.I.; Podgorny, O.V.; Belousov, V.V.; Bilan, D.S. In Vivo Imaging with Genetically Encoded Redox Biosensors. Int. J. Mol. Sci. 2020, 21, 8164. https://doi.org/10.3390/ijms21218164
Kostyuk AI, Panova AS, Kokova AD, Kotova DA, Maltsev DI, Podgorny OV, Belousov VV, Bilan DS. In Vivo Imaging with Genetically Encoded Redox Biosensors. International Journal of Molecular Sciences. 2020; 21(21):8164. https://doi.org/10.3390/ijms21218164
Chicago/Turabian StyleKostyuk, Alexander I., Anastasiya S. Panova, Aleksandra D. Kokova, Daria A. Kotova, Dmitry I. Maltsev, Oleg V. Podgorny, Vsevolod V. Belousov, and Dmitry S. Bilan. 2020. "In Vivo Imaging with Genetically Encoded Redox Biosensors" International Journal of Molecular Sciences 21, no. 21: 8164. https://doi.org/10.3390/ijms21218164
APA StyleKostyuk, A. I., Panova, A. S., Kokova, A. D., Kotova, D. A., Maltsev, D. I., Podgorny, O. V., Belousov, V. V., & Bilan, D. S. (2020). In Vivo Imaging with Genetically Encoded Redox Biosensors. International Journal of Molecular Sciences, 21(21), 8164. https://doi.org/10.3390/ijms21218164