Cooperative Interaction of Janthinobacterium sp. SLB01 and Flavobacterium sp. SLB02 in the Diseased Sponge Lubomirskia baicalensis
Abstract
:1. Introduction
2. Results
2.1. Cultivation Strains, Genome Assembly, Scaffolding and Features
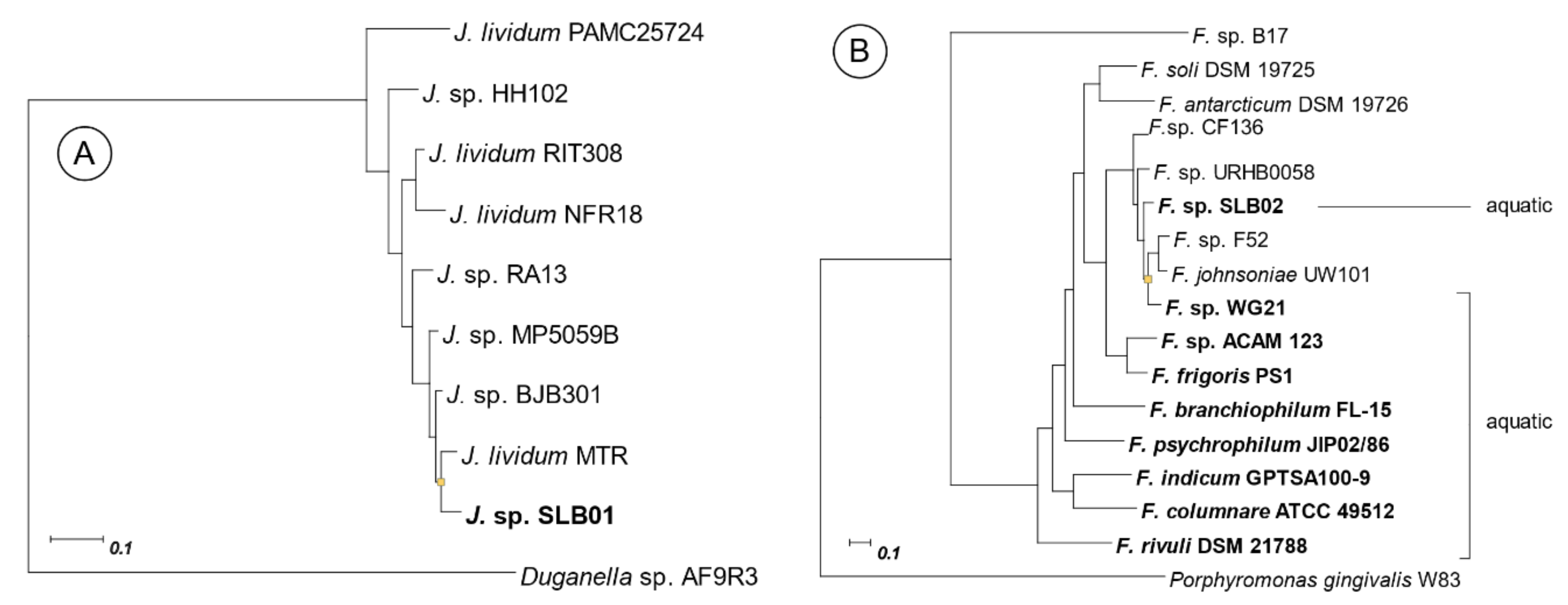
2.2. Phylogenetic Relationship with Closer Species
2.3. Violacein Synthesis by Janthinobacterium sp. SLB01
2.4. Type VI Secretion System Genes Identification
2.5. Quorum Sensing in Janthinobacterium sp. SLB01
2.6. Floc Formation by Janthinobacterium sp. SLB01
2.7. Comparative Genomics of Janthinobacterium Species
2.8. Polysaccharides Utilization Loci Analysis in Flavobacterium sp. SLB02
2.9. Carbon Sources Metabolism
3. Discussion
3.1. The Role of Each Species in the Joint Action
3.2. Probable Scenario of Strains Interaction
4. Materials and Methods
4.1. Bacterial Strains and Growth Media
4.2. Genome Assembly, Annotation and Phylogenetic Relationship
4.3. In Silico Analysis of Type VI Secretion System Loci
4.4. Genome Subsystems and Comparative Genomics
4.5. Polysaccharides Utilization Loci Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BUSCO | Benchmarking universal single-copy orthologs |
| CTERM | C-terminal |
| EPS | Exopolysaccharides |
| PGAP | Prokaryotic genome annotation pipeline |
| PEP | Pro-Glu-Pro |
| PUL | Polysaccharides utilization loci |
| T6SS | Type VI secretion system |
References
- Pile, A.J.; Patterson, M.R.; Savarese, M.; Chernykh, V.I.; Fialkov, V.A. Trophic effects of sponge feeding within Lake Baikal’s littoral zone. 2. Sponge abundance, diet, feeding efficiency, and carbon flux. Limnol. Oceanogr. 1997, 42, 178–184. [Google Scholar] [CrossRef]
- Jensen, K.S.; Pedersen, M.F. Photosynthesis by symbiotic algae in the freshwater sponge, Spongilla lacustris. Limnol. Oceanogr. 1994, 39, 551–561. [Google Scholar] [CrossRef]
- Chernogor, L.; Denikina, N.; Kondratov, I.; Solovarov, I.; Khanaev, I.; Belikov, S.; Ehrlich, H. Isolation and identification of the microalgal symbiont from primmorphs of the endemic freshwater sponge Lubomirskia baicalensis (Lubomirskiidae, Porifera). Eur. J. Phycol. 2013, 48, 497–508. [Google Scholar] [CrossRef]
- Bil, K.; Titlyanov, E.; Berner, T.; Fomina, I.; Muscatine, L. Some aspects of the physiology and biochemistry of Lubomirska baikalensis, a sponge from Lake Baikal containing symbiotic algae. Symbiosis 1999, 26, 179–191. [Google Scholar]
- Kravtsova, L.S.; Izhboldina, L.A.; Khanaev, I.V.; Pomazkina, G.V.; Rodionova, E.V.; Domysheva, V.M.; Sakirko, M.V.; Tomberg, I.V.; Kostornova, T.Y.; Kravchenko, O.S.; et al. Nearshore benthic blooms of filamentous green algae in Lake Baikal. J. Gt. Lakes Res. 2014, 40, 441–448. [Google Scholar] [CrossRef]
- Khanaev, I.V.; Kravtsova, L.S.; Maikova, O.O.; Bukshuk, N.A.; Sakirko, M.V.; Kulakova, N.V.; Butina, T.V.; Nebesnykh, I.A.; Belikov, S.I. Current state of the sponge fauna (Porifera: Lubomirskiidae) of Lake Baikal: Sponge disease and the problem of conservation of diversity. J. Gt. Lakes Res. 2018, 44, 77–85. [Google Scholar] [CrossRef]
- Belikov, S.; Belkova, N.; Butina, T.; Chernogor, L.; Van Kley, A.M.; Nalian, A.; Rorex, C.; Khanaev, I.; Maikova, O.; Feranchuk, S. Diversity and shifts of the bacterial community associated with Baikal sponge mass mortalities. PLoS ONE 2019, 14, e9080. [Google Scholar] [CrossRef] [PubMed]
- Chernogor, L.; Klimenko, E.; Khanaev, I.; Belikov, S. Microbiome analysis of healthy and diseased sponges Lubomirskia baicalensis by using cell cultures of primmorphs. PeerJ 2020, 8, e9080. [Google Scholar] [CrossRef] [PubMed]
- Petrushin, I.S.; Belikov, S.I.; Chernogor, L.I. Draft Genome Sequence of Janthinobacterium sp. Strain SLB01, Isolated from the Diseased Sponge Lubomirskia baicalensis. Microbiol. Resour. Announc. 2019, 8, e01108-19. [Google Scholar] [CrossRef] [Green Version]
- Petrushin, I.S.; Belikov, S.I.; Chernogor, L.I. Draft Genome Sequence of Flavobacterium sp. Strain SLB02, Isolated from the Diseased Sponge Lubomirskia baicalensis. Microbiol. Resour. Announc. 2020, 9, e00530-20. [Google Scholar] [CrossRef] [PubMed]
- Durán, N.; Justo, G.Z.; Ferreira, C.V.; Melo, P.S.; Cordi, L.; Martins, D. Violacein: Properties and biological activities. Biotechnol. Appl. Biochem. 2007, 48, 127. [Google Scholar]
- Hornung, C.; Poehlein, A.; Haack, F.S.; Schmidt, M.; Dierking, K.; Pohlen, A.; Schulenburg, H.; Blokesch, M.; Plener, L.; Jung, K.; et al. The Janthinobacterium sp. HH01 Genome Encodes a Homologue of the V. cholerae CqsA and L. pneumophila LqsA Autoinducer Synthases. PLoS ONE 2013, 8, e55045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pantanella, F.; Berlutti, F.; Passariello, C.; Sarli, S.; Morea, C.; Schippa, S. Violacein and biofilm production in Janthinobacterium lividum. J. Appl. Microbiol. 2007, 102, 992–999. [Google Scholar] [CrossRef]
- Aqeel, H.; Basuvaraj, M.; Hall, M.; Neufeld, J.D.; Liss, S.N. Microbial dynamics and properties of aerobic granules developed in a laboratory-scale sequencing batch reactor with an intermediate filamentous bulking stage. Appl. Microbiol. Biotechnol. 2016, 100, 447–460. [Google Scholar] [CrossRef]
- Gao, N.; Xia, M.; Dai, J.; Yu, D.; An, W.; Li, S.; Liu, S.; He, P.; Zhang, L.; Wu, Z.; et al. Both widespread PEP-CTERM proteins and exopolysaccharides are required for floc formation of Zoogloea resiniphila and other activated sludge bacteria. Environ. Microbiol. 2018, 20, 1677–1692. [Google Scholar] [CrossRef] [PubMed]
- Haft, D.H.; Paulsen, I.T.; Ward, N.; Selengut, J.D. Exopolysaccharide-associated protein sorting in environmental organisms: The PEP-CTERM/SpsH system. Application of a novel phylogenetic profiling heuristic. BMC Biol. 2006, 4, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Blom, J.; Loch, T.P.; Faisal, M.; Walker, E.D. The emerging fish pathogen Flavobacterium spartansii isolated from Chinook salmon: Comparative genome analysis and molecular manipulation. Front. Microbiol. 2017, 8, 2339. [Google Scholar] [CrossRef] [PubMed]
- Kinnula, H.; Mappes, J.; Sundberg, L.R. Coinfection outcome in an opportunistic pathogen depends on the inter-strain interactions. BMC Evol. Biol. 2017, 17, 77. [Google Scholar] [CrossRef] [Green Version]
- Kolton, M.; Sela, N.; Elad, Y.; Cytryn, E. Comparative Genomic Analysis Indicates that Niche Adaptation of Terrestrial Flavobacteria Is Strongly Linked to Plant Glycan Metabolism. PLoS ONE 2013, 8, e76704. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, H.; Tanaka, K.; Teramura, N.; Hattori, S. Expression of collagenase in Flavobacterium psychrophilum isolated from cold-water disease-affected ayu (Plecoglossus altivelis). Biosci. Biotechnol. Biochem. 2016, 80, 135–144. [Google Scholar] [CrossRef] [Green Version]
- Kharade, S.S.; McBride, M.J. Flavobacterium johnsoniae chitinase ChiA is required for chitin utilization and is secreted by the type IX secretion system. J. Bacteriol. 2014, 196, 961–970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuqua, C.; Parsek, M.R.; Greenberg, E.P. Regulation of gene expression by cell-to-cell communication: Acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 2001, 35, 439–468. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Mishra, A.; Jha, B. Anti-quorum sensing and anti-biofilm activity of Delftia tsuruhatensis extract by attenuating the quorum sensing-controlled virulence factor production in Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 2017, 7, 337. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Kolmogorov, M.; Armstrong, J.; Raney, B.J.; Streeter, I.; Dunn, M.; Yang, F.; Odom, D.; Flicek, P.; Keane, T.M.; Thybert, D.; et al. Chromosome assembly of large and complex genomes using multiple references. Genome Res. 2018, 28, 1720–1732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waterhouse, R.M.; Seppey, M.; Simao, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 2018, 35, 543–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haack, F.S.; Poehlein, A.; Kröger, C.; Voigt, C.A.; Piepenbring, M.; Bode, H.B.; Daniel, R.; Schäfer, W.; Streit, W.R. Molecular Keys to the Janthinobacterium and Duganella spp. Interaction with the Plant Pathogen Fusarium graminearum. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, W.; Guo, F.; Song, Y.; Gao, N.; Bai, S.; Dai, J.; Wei, H.; Zhang, L.; Yu, D.; Xia, M.; et al. Comparative genomics analyses on EPS biosynthesis genes required for floc formation of Zoogloea resiniphila and other activated sludge bacteria. Water Res. 2016, 102, 494–504. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Terrapon, N.; Lombard, V.; Drula, É.; Lapébie, P.; Al-Masaudi, S.; Gilbert, H.J.; Henrissat, B. PULDB: The expanded database of Polysaccharide Utilization Loci. Nucleic Acids Res. 2018, 46, D677–D683. [Google Scholar] [CrossRef] [PubMed]
- Basson, A.; Flemming, L.A.; Chenia, H.Y. Evaluation of adherence, hydrophobicity, aggregation, and biofilm development of Flavobacterium johnsoniae-like isolates. Microb. Ecol. 2008, 55, 1–14. [Google Scholar] [CrossRef]
- Lee, J.; Cho, D.H.; Ramanan, R.; Kim, B.H.; Oh, H.M.; Kim, H.S. Microalgae-associated bacteria play a key role in the flocculation of Chlorella vulgaris. Bioresour. Technol. 2013, 131, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Alexander, B.E.; Mueller, B.; Vermeij, M.J.A.; van der Geest, H.H.G.; de Goeij, J.M. Biofouling of inlet pipes affects water quality in running seawater aquaria and compromises sponge cell proliferation. PeerJ 2015, 3, e1430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silverman, J.M.; Brunet, Y.R.; Cascales, E.; Mougous, J.D. Structure and regulation of the type VI secretion system. Annu. Rev. Microbiol. 2012, 66, 453–472. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Brodmann, M.; Basler, M. Assembly and subcellular localization of bacterial type VI secretion systems. Annu. Rev. Microbiol. 2019, 73, 621–638. [Google Scholar] [CrossRef] [Green Version]
- Larsbrink, J.; Zhu, Y.; Kharade, S.S.; Kwiatkowski, K.J.; Eijsink, V.G.H.; Koropatkin, N.M.; McBride, M.J.; Pope, P.B. A polysaccharide utilization locus from Flavobacterium johnsoniae enables conversion of recalcitrant chitin. Biotechnol. Biofuels 2016, 9. [Google Scholar] [CrossRef] [Green Version]
- McBride, M.J.; Xie, G.; Martens, E.C.; Lapidus, A.; Henrissat, B.; Rhodes, R.G.; Goltsman, E.; Wang, W.; Xu, J.; Hunnicutt, D.W.; et al. Novel features of the polysaccharide-digesting gliding bacterium Flavobacterium johnsoniae as revealed by genome sequence analysis. Appl. Environ. Microbiol. 2009, 75, 6864–6875. [Google Scholar] [CrossRef] [Green Version]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42. [Google Scholar] [CrossRef]
- Dieser, M.; Smith, H.J.; Ramaraj, T.; Foreman, C.M. Janthinobacterium CG23_2: Comparative Genome Analysis Reveals Enhanced Environmental Sensing and Transcriptional Regulation for Adaptation to Life in an Antarctic Supraglacial Stream. Microorganisms 2019, 7, 454. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Lu, D.; Qin, B.; Liu, Q.; Zhao, Y.; Liu, H.; Ma, J. Highly efficient nitrogen removal of a coldness-resistant and low nutrient needed bacterium, Janthinobacterium sp. M-11. Bioresour. Technol. 2018, 256, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Allen, H.K.; Klimowicz, A.K.; Mlot, C.; Gross, J.A.; Savengsuksa, S.; McEllin, J.; Clardy, J.; Ruess, R.W.; Handelsman, J. Psychrotrophic Strain of Janthinobacterium lividum from a Cold Alaskan Soil Produces Prodigiosin. DNA Cell Biol. 2010, 29, 533–541. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Skrivergaard, S.; Korsgaard, B.S.; Schreiber, L.; Marshall, I.P.G.; Finster, K.; Schramm, A. High quality draft genome sequence of Janthinobacterium psychrotolerans sp. nov., isolated from a frozen freshwater pond. Stand. Genom. Sci. 2017, 12, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tezuka, Y. Cation-dependent flocculation in a Flavobacterium species predominant in activated sludge. Appl. Microbiol. 1969, 17, 222–226. [Google Scholar] [CrossRef] [Green Version]
- Park, M.; Ryu, S.H.; Vu, T.H.T.; Ro, H.S.; Yun, P.Y.; Jeon, C.O. Flavobacterium defluvii sp. nov., isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 2007, 57, 233–237. [Google Scholar] [CrossRef]
- Hantula, J.; Bamford, D.H. The efficiency of the protein-dependent flocculation of Flavobacterium sp. is sensitive to the composition of growth medium. Appl. Microbiol. Biotechnol. 1991, 36, 100–104. [Google Scholar] [CrossRef]
- Tatusova, T.; Dicuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Asnicar, F.; Thomas, A.M.; Beghini, F.; Mengoni, C.; Manara, S.; Manghi, P.; Zhu, Q.; Bolzan, M.; Cumbo, F.; May, U.; et al. Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef]
- Li, J.; Yao, Y.; Xu, H.H.; Hao, L.; Deng, Z.; Rajakumar, K.; Ou, H.Y. SecReT6: A web-based resource for type VI secretion systems found in bacteria. Environ. Microbiol. 2015, 17, 2196–2202. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; Varlamov, A.; Vaskin, Y.; Efremov, I.; German Grehov, O.G.; Kandrov, D.; Rasputin, K.; Syabro, M.; et al. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [Green Version]
- Terrapon, N.; Lombard, V.; Gilbert, H.J.; Henrissat, B. Automatic prediction of polysaccharide utilization loci in Bacteroidetes species. Bioinformatics 2015, 31, 647–655. [Google Scholar] [CrossRef] [Green Version]





| Property | Janthinobacterium sp. SLB01 | Flavobacterium sp. SLB02 |
|---|---|---|
| Raw reads | 12,099,942 * | 17,921,744 * |
| GenBank accession number | VZAB00000000 ** | CP045928 *** |
| Genome size, bp | 6,467,981 | 6,363,829 |
| Number of contigs | 2 | 1 |
| GC content | 62.63% | 35.50% |
| Number of genes | 6023 | 4964 |
| Protein-coding sequences | 5863 | 4901 |
| tRNAs | 65 | 56 |
| Noncoding RNAs | 4 | 3 |
| Pseudogenes | 78 | 73 |
| Reference to genome report | [9] | [10] |
| Janthinobacterium sp. SLB01 | Janthinobacterium sp. HH01 | |||
|---|---|---|---|---|
| Locus Tag | Annotation | Locus Tag | % Ident ** | % Simi-larity ** |
| F3B38_RS23475 | quorum-sensing autoinducer synthase | Jab_2c24330 * | 68.6 | 81.0 |
| F3B38_RS23480 | HAMP domain-containing histidine kinase | Jab_2c24340 | 60.6 | 73.1 |
| F3B38_RS23485 | response regulator | Jab_2c24350 | 68.2 | 79.8 |
| Carbon Source Group | Subsystem * Name | SLB01 ** | SLB02 *** |
|---|---|---|---|
| Central carbohydrate Metabolism | TCA Cycle | 15 | 0 |
| Pentose phosphate pathway | 0 | 9 | |
| Di- and oligosaccharides | Sucrose utilization | 0 | 2 |
| Organic acids | Methylcitrate cycle | 7 | 0 |
| Propionate-CoA to succinate module | 6 | 0 | |
| Lactose and galactose uptake and utilization | 0 | 8 | |
| Fermentation | Mixed acid | 0 | 7 |
| Polysaccharides | Glycogen metabolism | 0 | 4 |
| Monosaccharides | 2-Ketogluconate utilization | 4 | 0 |
| L-Arabinose utilization | 0 | 9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrushin, I.; Belikov, S.; Chernogor, L. Cooperative Interaction of Janthinobacterium sp. SLB01 and Flavobacterium sp. SLB02 in the Diseased Sponge Lubomirskia baicalensis. Int. J. Mol. Sci. 2020, 21, 8128. https://doi.org/10.3390/ijms21218128
Petrushin I, Belikov S, Chernogor L. Cooperative Interaction of Janthinobacterium sp. SLB01 and Flavobacterium sp. SLB02 in the Diseased Sponge Lubomirskia baicalensis. International Journal of Molecular Sciences. 2020; 21(21):8128. https://doi.org/10.3390/ijms21218128
Chicago/Turabian StylePetrushin, Ivan, Sergei Belikov, and Lubov Chernogor. 2020. "Cooperative Interaction of Janthinobacterium sp. SLB01 and Flavobacterium sp. SLB02 in the Diseased Sponge Lubomirskia baicalensis" International Journal of Molecular Sciences 21, no. 21: 8128. https://doi.org/10.3390/ijms21218128






