Whole-Transcriptome Sequencing-Based Analysis of DAZL and Its Interacting Genes during Germ Cells Specification and Zygotic Genome Activation in Chickens
Abstract
:1. Introduction
2. Results and Discussion
2.1. Prediction of DAZL Interacting Genes and Motifs Analysis
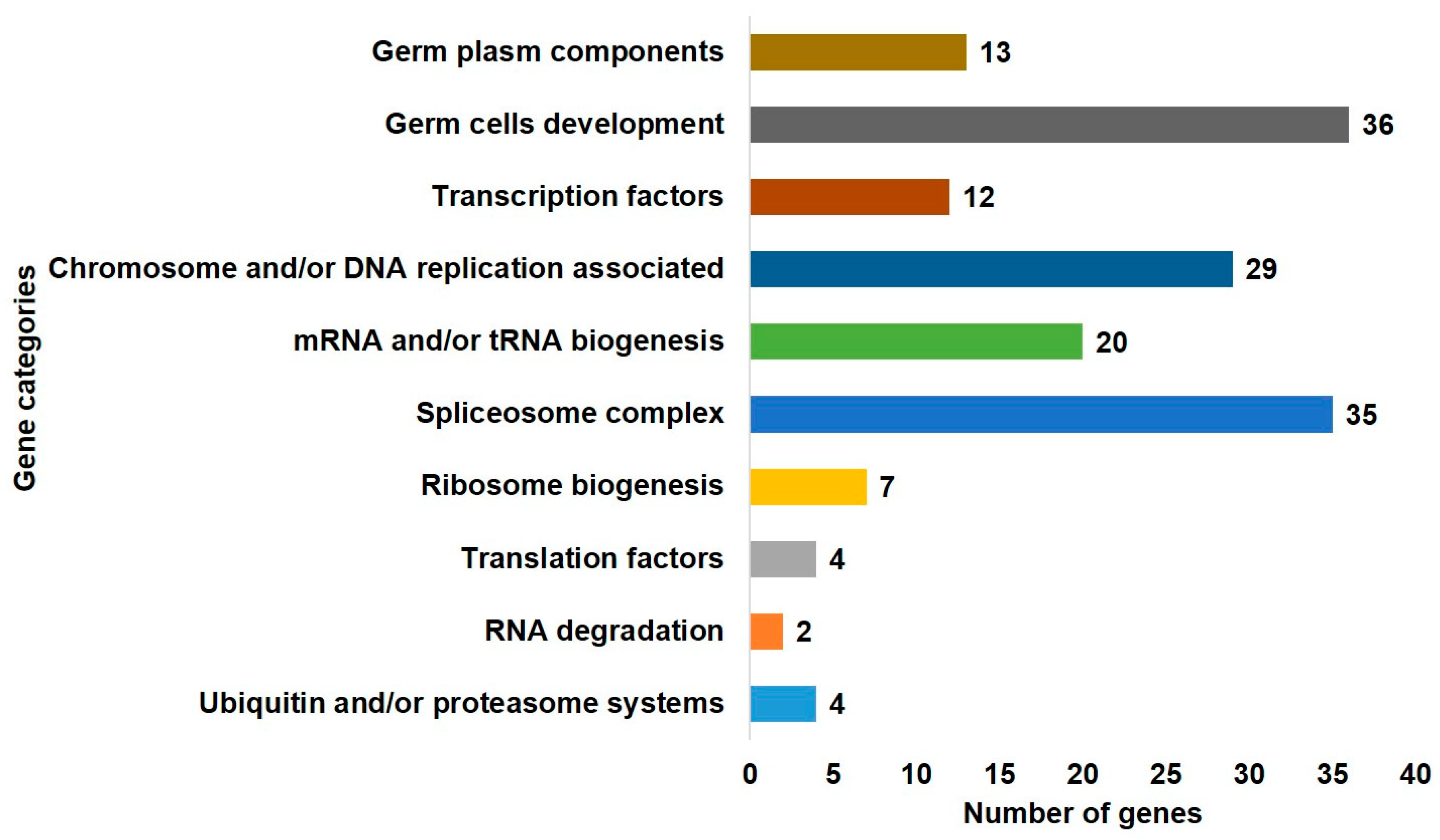
2.2. Gene Ontology Enrichment of DAZL and Its Interacting Genes
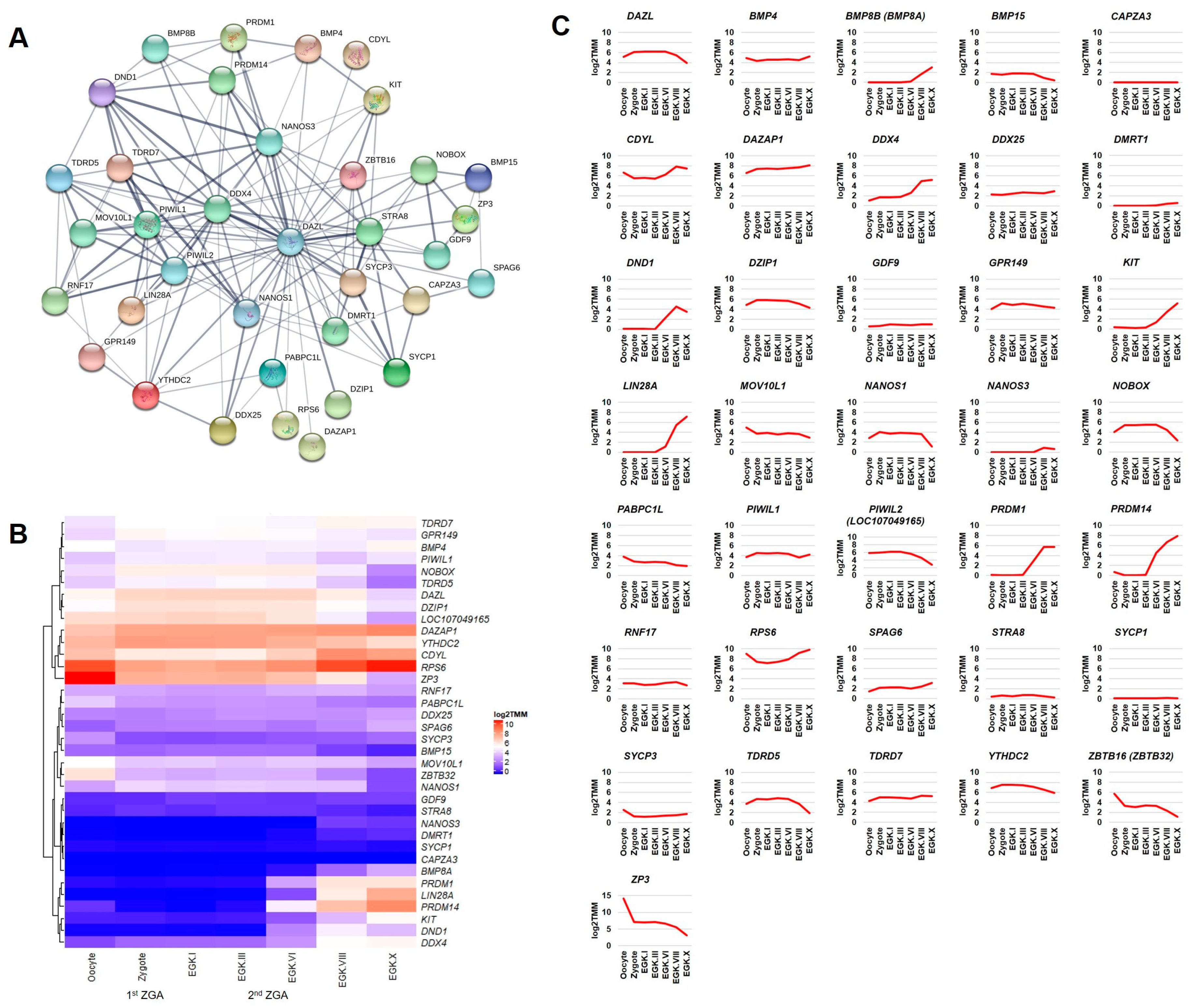
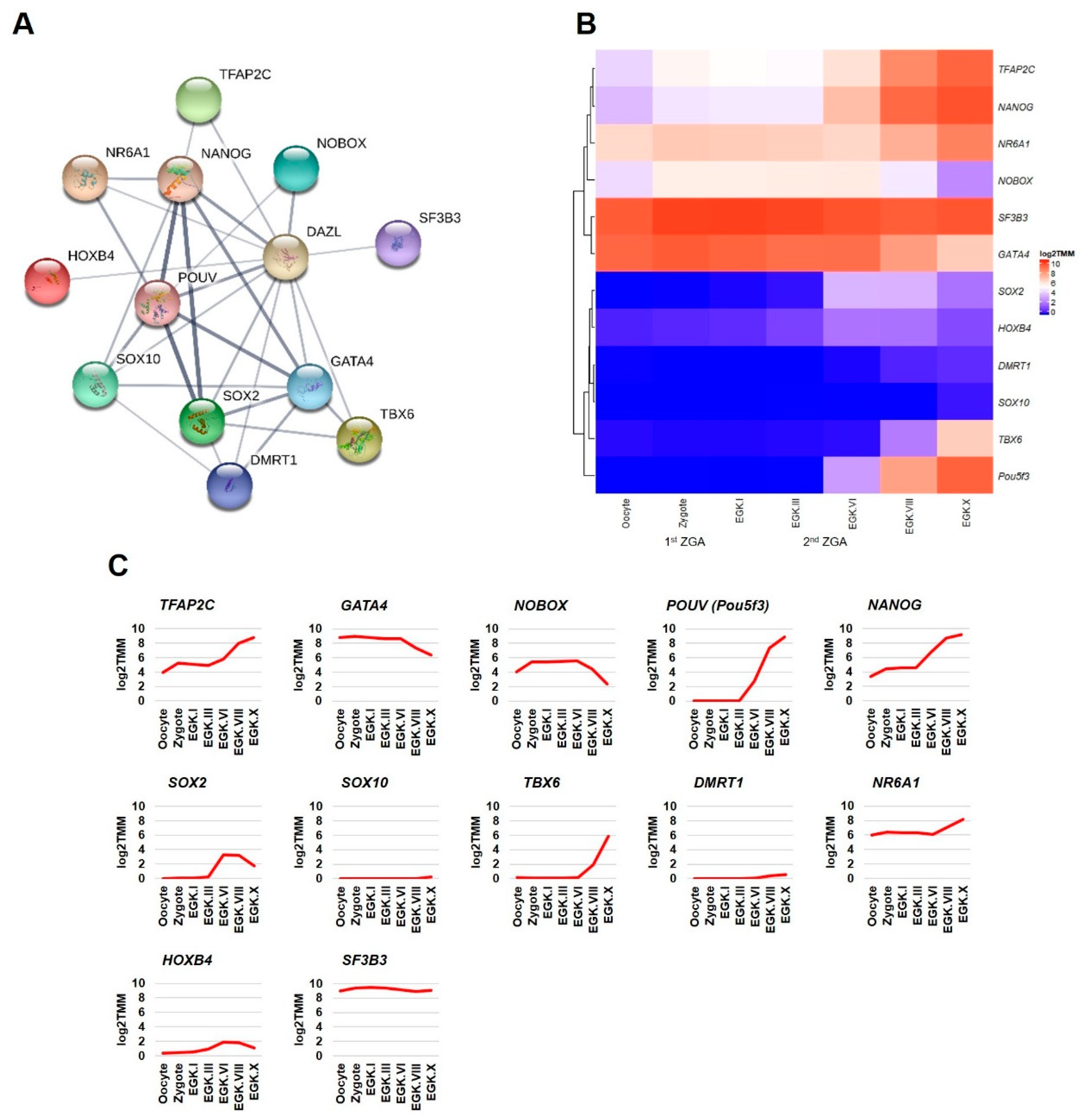
2.3. Detection of DAZL and Its Interacting Genes in the Chicken Intrauterine Embryos
2.3.1. Germ Plasm and Germ Cells Development Categories
2.3.2. Transcription Factors Category
2.3.3. Other Categories of Processes Related to ZGA/MZT
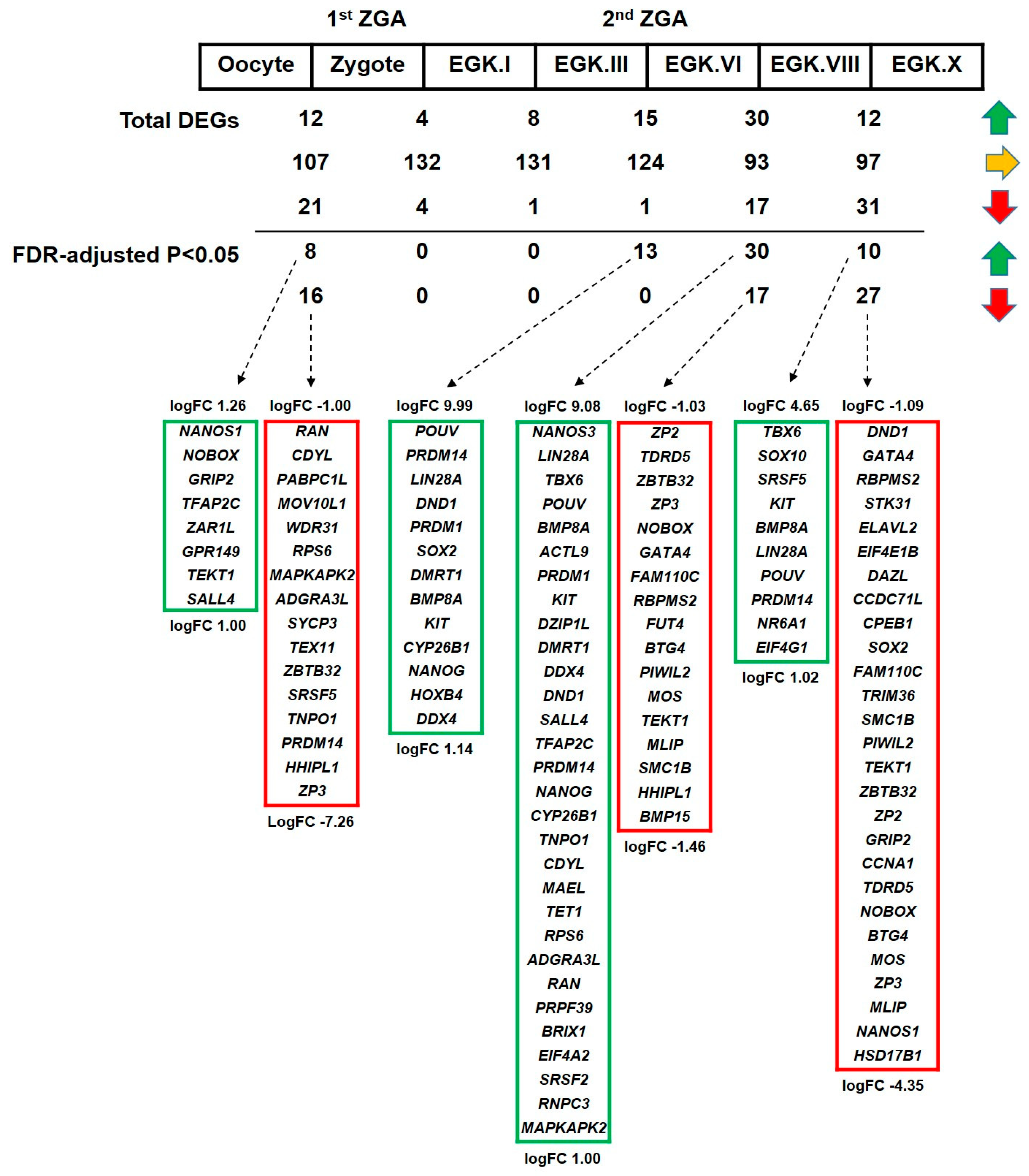
2.3.4. Differential Expression of DAZL and Its Interacting Genes between Consecutive Stages
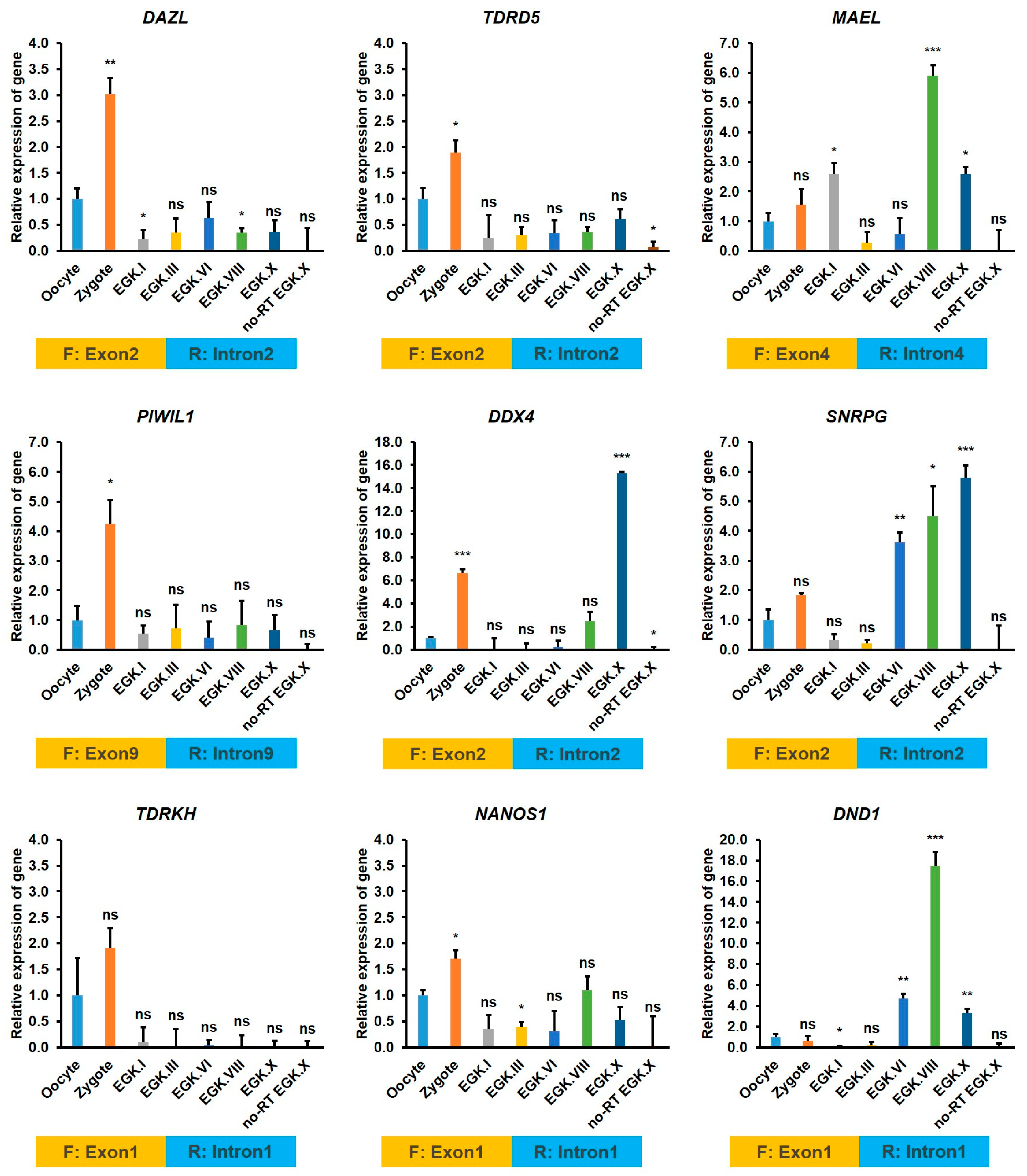
2.3.5. Exon–Intron Specific RT-qPCR Validation of DAZL and Candidate Interacting Genes
3. Materials and Methods
3.1. Prediction of DAZL Interacting Genes and Motifs Analysis
3.2. Gene Ontology Enrichment, and Sub-Interaction Analysis of Predicted Genes with DAZL
3.3. Preprocessing of WTS Data from the Chicken Intrauterine Embryos
3.4. Alignment and Quantification of Mapped Reads
3.5. Experimental Animals and Animal Care
3.6. Sample Collection and Exon–Intron Specific RT-qPCR Validation of DAZL and Its Interacting Genes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mitalipov, S.; Wolf, D. Totipotency, pluripotency and nuclear reprogramming. Adv. Biochem. Eng. Biotechnol. 2009, 114, 185–199. [Google Scholar] [CrossRef] [Green Version]
- Jamieson-Lucy, A.; Mullins, M.C. The vertebrate Balbiani body, germ plasm, and oocyte polarity. In Current Topics in Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2019; Volume 135, pp. 1–34. [Google Scholar] [CrossRef]
- Tang, W.W.; Kobayashi, T.; Irie, N.; Dietmann, S.; Surani, M.A. Specification and epigenetic programming of the human germ line. Nat. Rev. Genet. 2016, 17, 585–600. [Google Scholar] [CrossRef]
- Hird, S.N.; Paulsen, J.E.; Strome, S. Segregation of germ granules in living Caenorhabditis elegans embryos: Cell-type-specific mechanisms for cytoplasmic localisation. Development 1996, 122, 1303–1312. [Google Scholar]
- Lehmann, R. Germ-plasm formation and germ-cell determination in Drosophila. Curr. Opin. Genet. Dev. 1992, 2, 543–549. [Google Scholar] [CrossRef]
- Kloc, M.; Bilinski, S.; Chan, A.P.; Allen, L.H.; Zearfoss, N.R.; Etkin, L.D. RNA localization and germ cell determination in Xenopus. Int. Rev. Cytol. 2001, 203, 63–91. [Google Scholar] [CrossRef]
- Dosch, R. Next generation mothers: Maternal control of germline development in zebrafish. Crit. Rev. Biochem. Mol. Biol. 2015, 50, 54–68. [Google Scholar] [CrossRef]
- Haston, K.M.; Tung, J.Y.; Reijo Pera, R.A. Dazl functions in maintenance of pluripotency and genetic and epigenetic programs of differentiation in mouse primordial germ cells in vivo and in vitro. PLoS ONE 2009, 4, e5654. [Google Scholar] [CrossRef]
- Rengaraj, D.; Zheng, Y.H.; Kang, K.S.; Park, K.J.; Lee, B.R.; Lee, S.I.; Choi, J.W.; Han, J.Y. Conserved expression pattern of chicken DAZL in primordial germ cells and germ-line cells. Theriogenology 2010, 74, 765–776. [Google Scholar] [CrossRef]
- Ruggiu, M.; Speed, R.; Taggart, M.; McKay, S.J.; Kilanowski, F.; Saunders, P.; Dorin, J.; Cooke, H.J. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature 1997, 389, 73–77. [Google Scholar] [CrossRef]
- Gill, M.E.; Hu, Y.C.; Lin, Y.; Page, D.C. Licensing of gametogenesis, dependent on RNA binding protein DAZL, as a gateway to sexual differentiation of fetal germ cells. Proc. Natl. Acad. Sci. USA 2011, 108, 7443–7448. [Google Scholar] [CrossRef] [Green Version]
- Nicholls, P.K.; Schorle, H.; Naqvi, S.; Hu, Y.C.; Fan, Y.; Carmell, M.A.; Dobrinski, I.; Watson, A.L.; Carlson, D.F.; Fahrenkrug, S.C.; et al. Mammalian germ cells are determined after PGC colonization of the nascent gonad. Proc. Natl. Acad. Sci. USA 2019, 116, 25677–25687. [Google Scholar] [CrossRef]
- Zagore, L.L.; Sweet, T.J.; Hannigan, M.M.; Weyn-Vanhentenryck, S.M.; Jobava, R.; Hatzoglou, M.; Zhang, C.; Licatalosi, D.D. DAZL regulates germ cell survival through a network of polyA-proximal mRNA interactions. Cell Rep. 2018, 25, 1225–1240.e6. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.H.; Welling, M.; Bloch, D.B.; Munoz, J.; Mientjes, E.; Chen, X.; Tramp, C.; Wu, J.; Yabuuchi, A.; Chou, Y.F.; et al. DAZL limits pluripotency, differentiation, and apoptosis in developing primordial germ cells. Stem Cell Rep. 2014, 3, 892–904. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Melton, C.; Suh, N.; Oh, J.S.; Horner, K.; Xie, F.; Sette, C.; Blelloch, R.; Conti, M. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev. 2011, 25, 755–766. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Liang, Z.; Yang, J.; Wang, D.; Wang, H.; Zhu, M.; Geng, B.; Xu, E.Y. DAZL is a master translational regulator of murine spermatogenesis. Natl. Sci. Rev. 2019, 6, 455–468. [Google Scholar] [CrossRef] [Green Version]
- Mikedis, M.M.; Fan, Y.; Nicholls, P.K.; Endo, T.; Jackson, E.K.; Cobb, S.A.; de Rooij, D.G.; Page, D.C. DAZL mediates a broad translational program regulating expansion and differentiation of spermatogonial progenitors. eLife 2020, 9, e56523. [Google Scholar] [CrossRef]
- Lee, H.C.; Choi, H.J.; Lee, H.G.; Lim, J.M.; Ono, T.; Han, J.Y. DAZL expression explains origin and central formation of primordial germ cells in chickens. Stem Cells Dev. 2016, 25, 68–79. [Google Scholar] [CrossRef]
- Eyal-Giladi, H.; Kochav, S. From cleavage to primitive streak formation: A complementary normal table and a new look at the first stages of the development of the chick. I. General morphology. Dev. Biol. 1976, 49, 321–337. [Google Scholar] [CrossRef]
- Kochav, S.; Ginsburg, M.; Eyal-Giladi, H. From cleavage to primitive streak formation: A complementary normal table and a new look at the first stages of the development of the chick. II. Microscopic anatomy and cell population dynamics. Dev. Biol. 1980, 79, 296–308. [Google Scholar] [CrossRef]
- Rengaraj, D.; Hwang, Y.S.; Lee, H.C.; Han, J.Y. Zygotic genome activation in the chicken: A comparative review. Cell. Mol. Life Sci. 2020, 77, 1879–1891. [Google Scholar] [CrossRef]
- Hwang, Y.S.; Seo, M.; Lee, B.R.; Lee, H.J.; Park, Y.H.; Kim, S.K.; Lee, H.C.; Choi, H.J.; Yoon, J.; Kim, H.; et al. The transcriptome of early chicken embryos reveals signaling pathways governing rapid asymmetric cellularization and lineage segregation. Development 2018, 145, dev157453. [Google Scholar] [CrossRef] [Green Version]
- Hwang, Y.S.; Seo, M.; Bang, S.; Kim, H.; Han, J.Y. Transcriptional and translational dynamics during maternal-to-zygotic transition in early chicken development. FASEB J. 2018, 32, 2004–2011. [Google Scholar] [CrossRef] [Green Version]
- Hwang, Y.S.; Seo, M.; Kim, S.K.; Bang, S.; Kim, H.; Han, J.Y. Zygotic gene activation in the chicken occurs in two waves, the first involving only maternally derived genes. eLife 2018, 7, e39381. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Maegawa, S.; Yamashita, M.; Yasuda, K.; Inoue, K. Zebrafish DAZ-like protein controls translation via the sequence ‘GUUC’. Genes Cells 2002, 7, 971–984. [Google Scholar] [CrossRef]
- Venables, J.P.; Ruggiu, M.; Cooke, H.J. The RNA-binding specificity of the mouse Dazl protein. Nucleic Acids Res. 2001, 29, 2479–2483. [Google Scholar] [CrossRef] [Green Version]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.C.; Choi, H.J.; Park, T.S.; Lee, S.I.; Kim, Y.M.; Rengaraj, D.; Nagai, H.; Sheng, G.; Lim, J.M.; Han, J.Y. Cleavage events and sperm dynamics in chick intrauterine embryos. PLoS ONE 2013, 8, e80631. [Google Scholar] [CrossRef] [Green Version]
- Hwang, Y.S.; Seo, M.; Choi, H.J.; Kim, S.K.; Kim, H.; Han, J.Y. The first whole transcriptomic exploration of pre-oviposited early chicken embryos using single and bulked embryonic RNA-sequencing. Gigascience 2018, 7, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Tsunekawa, N.; Naito, M.; Sakai, Y.; Nishida, T.; Noce, T. Isolation of chicken vasa homolog gene and tracing the origin of primordial germ cells. Development 2000, 127, 2741–2750. [Google Scholar]
- Wan, Z.; Rui, L.; Li, Z. Expression patterns of prdm1 during chicken embryonic and germline development. Cell Tissue Res. 2014, 356, 341–356. [Google Scholar] [CrossRef] [Green Version]
- Schulz, K.N.; Harrison, M.M. Mechanisms regulating zygotic genome activation. Nat. Rev. Genet. 2019, 20, 221–234. [Google Scholar] [CrossRef]
- Han, J.Y.; Lee, H.G.; Park, Y.H.; Hwang, Y.S.; Kim, S.K.; Rengaraj, D.; Cho, B.W.; Lim, J.M. Acquisition of pluripotency in the chick embryo occurs during intrauterine embryonic development via a unique transcriptional network. J. Anim. Sci. Biotechnol. 2018, 9, 31. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.T.; Bonneau, A.R.; Giraldez, A.J. Zygotic genome activation during the maternal-to-zygotic transition. Annu. Rev. Cell Dev. Biol. 2014, 30, 581–613. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Huang, B.; Chen, H.; Yin, Q.; Liu, Y.; Xiang, Y.; Zhang, B.; Liu, B.; Wang, Q.; Xia, W.; et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 2016, 534, 652–657. [Google Scholar] [CrossRef]
- Will, C.L.; Luhrmann, R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011, 3, a003707. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Ma, Y.; Shang, Y.; Huo, R.; Li, W. Post-translational regulation of the maternal-to-zygotic transition. Cell. Mol. Life Sci. 2018, 75, 1707–1722. [Google Scholar] [CrossRef]
- Braude, P.; Bolton, V.; Moore, S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature 1988, 332, 459–461. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Huang, K.; Cai, C.; Cai, L.; Jiang, C.Y.; Feng, Y.; Liu, Z.; Zeng, Q.; Cheng, L.; Sun, Y.E.; et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 2013, 500, 593–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sayers, E.W.; Agarwala, R.; Bolton, E.E.; Brister, J.R.; Canese, K.; Clark, K.; Connor, R.; Fiorini, N.; Funk, K.; Hefferon, T.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2019, 47, D23–D28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]

| Samples | Raw Reads | QC Passed Reads | QC Passed Ratio | Mapped Reads | Mapped Ratio | Uniquely Mapped Reads | Uniquely Mapped Ratio |
|---|---|---|---|---|---|---|---|
| Oocyte_S1 | 118,184,502 | 112,049,150 | 94.81% | 100,138,495 | 89.37% | 88,202,016 | 88.08% |
| Oocyte_S2 | 119,066,268 | 112,087,560 | 94.14% | 100,319,697 | 89.50% | 84,762,286 | 84.49% |
| Oocyte_S3 | 124,554,030 | 118,997,350 | 95.54% | 105,789,456 | 88.90% | 92,455,076 | 87.40% |
| Zygote_S4 | 110,348,652 | 106,756,296 | 96.74% | 94,855,747 | 88.85% | 84,724,358 | 89.32% |
| Zygote_S5 | 111,604,036 | 107,999,168 | 96.77% | 96,008,462 | 88.90% | 84,931,128 | 88.46% |
| Zygote_S6 | 102,075,504 | 100,055,858 | 98.02% | 87,757,716 | 87.71% | 78,370,790 | 89.30% |
| EGK.I_S1 | 116,899,114 | 113,818,628 | 97.36% | 94,434,196 | 82.97% | 80,459,294 | 85.20% |
| EGK.I_S2 | 125,479,340 | 122,894,028 | 97.94% | 100,660,269 | 81.91% | 85,257,994 | 84.70% |
| EGK.I_S4 | 103,691,686 | 100,377,694 | 96.80% | 88,604,778 | 88.27% | 77,917,230 | 87.94% |
| EGK.III_S3 | 125,126,674 | 121,753,362 | 97.30% | 102,957,681 | 84.56% | 87,289,704 | 84.78% |
| EGK.III_S4 | 115,130,736 | 112,715,380 | 97.90% | 94,110,789 | 83.49% | 80,524,216 | 85.56% |
| EGK.III_S5 | 93,275,856 | 91,430,970 | 98.02% | 76,574,666 | 83.75% | 65,587,730 | 85.65% |
| EGK.VI_S1 | 127,298,262 | 124,150,076 | 97.53% | 100,420,929 | 80.89% | 84,066,832 | 83.71% |
| EGK.VI_S5 | 133,416,206 | 130,446,328 | 97.77% | 115,208,886 | 88.32% | 100,992,302 | 87.66% |
| EGK.VI_S6 | 101,065,710 | 99,208,584 | 98.16% | 83,024,395 | 83.69% | 70,955,576 | 85.46% |
| EGK.VIII_S2 | 138,471,542 | 134,802,776 | 97.35% | 107,760,966 | 79.94% | 90,024,204 | 83.54% |
| EGK.VIII_S3 | 116,500,772 | 112,792,536 | 96.82% | 87,374,002 | 77.46% | 71,465,540 | 81.79% |
| EGK.VIII_S4 | 146,364,730 | 142,618,126 | 97.44% | 114,447,799 | 80.25% | 96,284,262 | 84.13% |
| EGK.X_S5 | 141,549,042 | 135,461,004 | 95.70% | 110,279,481 | 81.41% | 92,973,012 | 84.31% |
| EGK.X_S6 | 155,994,076 | 148,219,000 | 95.02% | 118,622,560 | 80.03% | 99,114,152 | 83.55% |
| EGK.X_S7 | 133,595,886 | 126,451,838 | 94.65% | 101,943,745 | 80.62% | 85,789,668 | 84.15% |
| Average | 121,890,125 | 117,861,224 | 96.75% | 99,109,272 | 84.32% | 84,864,160 | 85.68% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rengaraj, D.; Won, S.; Han, J.W.; Yoo, D.; Kim, H.; Han, J.Y. Whole-Transcriptome Sequencing-Based Analysis of DAZL and Its Interacting Genes during Germ Cells Specification and Zygotic Genome Activation in Chickens. Int. J. Mol. Sci. 2020, 21, 8170. https://doi.org/10.3390/ijms21218170
Rengaraj D, Won S, Han JW, Yoo D, Kim H, Han JY. Whole-Transcriptome Sequencing-Based Analysis of DAZL and Its Interacting Genes during Germ Cells Specification and Zygotic Genome Activation in Chickens. International Journal of Molecular Sciences. 2020; 21(21):8170. https://doi.org/10.3390/ijms21218170
Chicago/Turabian StyleRengaraj, Deivendran, Sohyoung Won, Jong Won Han, DongAhn Yoo, Heebal Kim, and Jae Yong Han. 2020. "Whole-Transcriptome Sequencing-Based Analysis of DAZL and Its Interacting Genes during Germ Cells Specification and Zygotic Genome Activation in Chickens" International Journal of Molecular Sciences 21, no. 21: 8170. https://doi.org/10.3390/ijms21218170
APA StyleRengaraj, D., Won, S., Han, J. W., Yoo, D., Kim, H., & Han, J. Y. (2020). Whole-Transcriptome Sequencing-Based Analysis of DAZL and Its Interacting Genes during Germ Cells Specification and Zygotic Genome Activation in Chickens. International Journal of Molecular Sciences, 21(21), 8170. https://doi.org/10.3390/ijms21218170





