Potent and Broad but not Unselective Cleavage of Cytokines and Chemokines by Human Neutrophil Elastase and Proteinase 3
Abstract
1. Introduction
2. Results
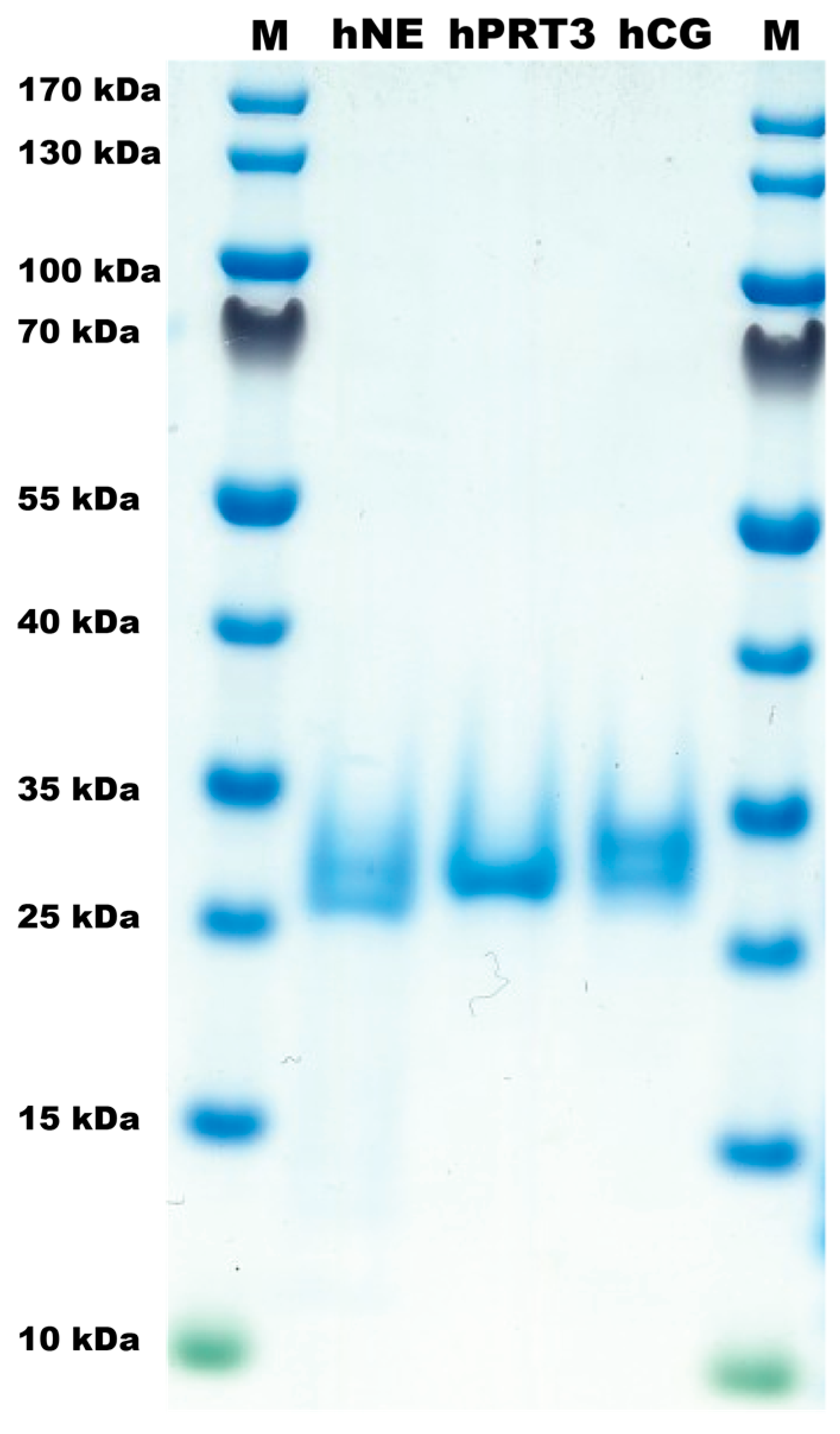
2.1. The Proteases
2.2. Analysis of Sensitivity to Cleavage by hNE and hPR3
2.3. Simultaneous Cleavage by Three Neutrophil Proteases, hNE, hPR3 and hCG
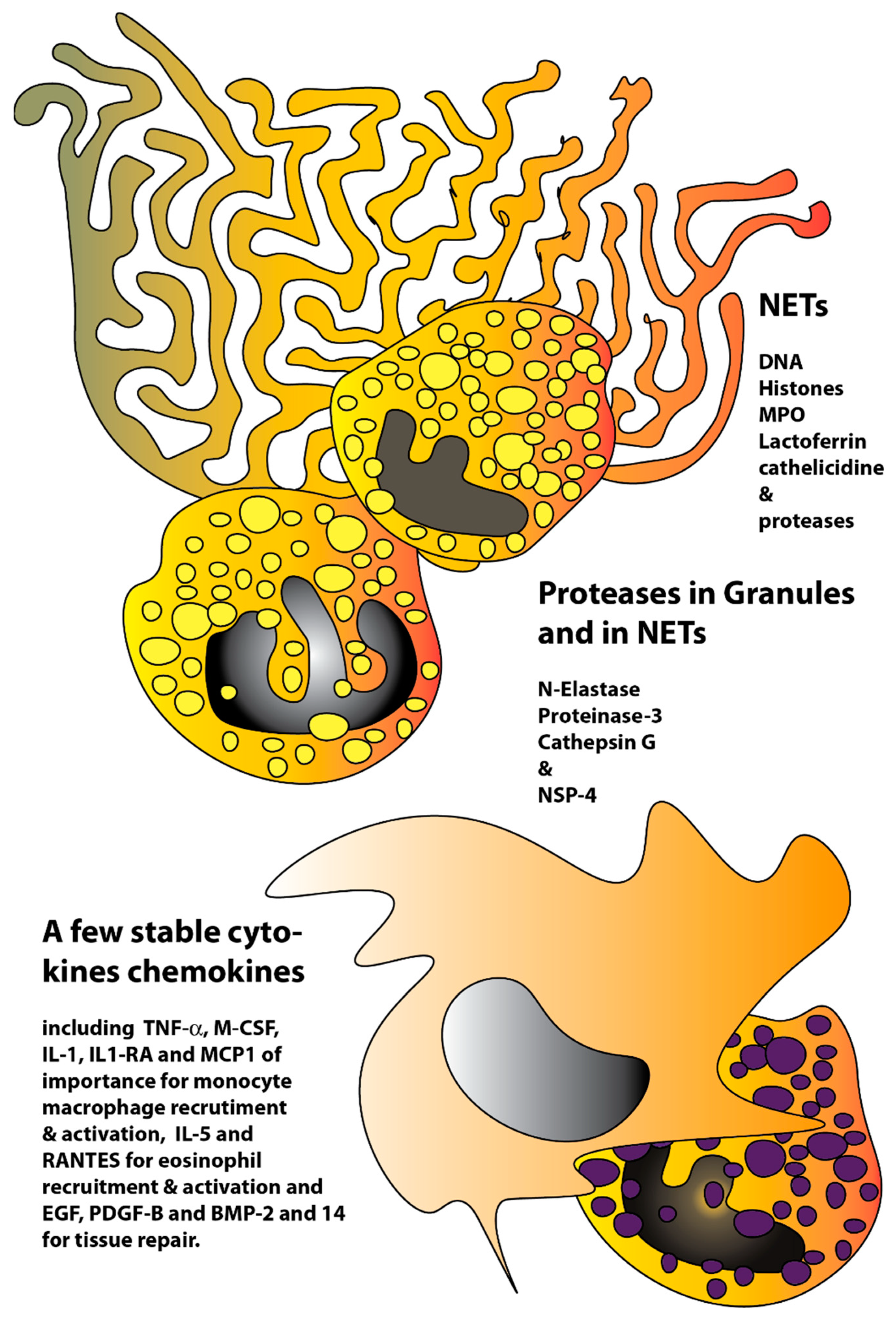
3. Discussion
4. Materials and Methods
4.1. Enzymes
4.2. Recombinant Cytokines and Chemokines
4.3. Analysis of the Sensitivity to Cleavage by hNE and hPR3
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| hNE | human N-elastase |
| hPR-3 | human proteinase 3 |
| HC | human chymase |
| hCG | human cathepsin G |
References
- Korkmaz, B.; Moreau, T.; Gauthier, F. Neutrophil elastase, proteinase 3 and cathepsin G: Physicochemical properties, activity and physiopathological functions. Biochimie 2008, 90, 227–242. [Google Scholar] [CrossRef] [PubMed]
- Pejler, G.; Ronnberg, E.; Waern, I.; Wernersson, S. Mast cell proteases: Multifaceted regulators of inflammatory disease. Blood 2010, 115, 4981–4990. [Google Scholar] [CrossRef] [PubMed]
- Caughey, G.H. Mast cell proteases as protective and inflammatory mediators. Adv. Exp. Med. Biol. 2011, 716, 212–234. [Google Scholar] [CrossRef] [PubMed]
- Hellman, L.; Thorpe, M. Granule proteases of hematopoietic cells, a family of versatile inflammatory mediators—An update on their cleavage specificity, in vivo substrates, and evolution. Biol. Chem. 2014, 395, 15–49. [Google Scholar] [CrossRef] [PubMed]
- Akula, S.; Thorpe, M.; Boinapally, V.; Hellman, L. Granule Associated Serine Proteases of Hematopoietic Cells—An Analysis of Their Appearance and Diversification during Vertebrate Evolution. PLoS ONE 2015, 10, e0143091. [Google Scholar] [CrossRef]
- Schwartz, L.B.; Irani, A.M.; Roller, K.; Castells, M.C.; Schechter, N.M. Quantitation of histamine, tryptase, and chymase in dispersed human T and TC mast cells. J. Immunol. 1987, 138, 2611–2615. [Google Scholar]
- Pereira, H.A.; Spitznagel, J.K.; Pohl, J.; Wilson, D.E.; Morgan, J.; Palings, I.; Larrick, J.W. CAP 37, a 37 kD human neutrophil granule cationic protein shares homology with inflammatory proteinases. Life Sci. 1990, 46, 189–196. [Google Scholar] [CrossRef]
- O’Donoghue, A.J.; Jin, Y.; Knudsen, G.M.; Perera, N.C.; Jenne, D.E.; Murphy, J.E.; Craik, C.S.; Hermiston, T.W. Global substrate profiling of proteases in human neutrophil extracellular traps reveals consensus motif predominantly contributed by elastase. PLoS ONE 2013, 8, e75141. [Google Scholar] [CrossRef]
- Thorpe, M.; Fu, Z.; Chahal, G.; Akula, S.; Kervinen, J.; de Garavilla, L.; Hellman, L. Extended cleavage specificity of human neutrophil cathepsin G: A low activity protease with dual chymase and tryptase-type specificities. PLoS ONE 2018, 13, e0195077. [Google Scholar] [CrossRef]
- Fu, Z.; Thorpe, M.; Alemayehu, R.; Roy, A.; Kervinen, J.; de Garavilla, L.; Abrink, M.; Hellman, L. Highly Selective Cleavage of Cytokines and Chemokines by the Human Mast Cell Chymase and Neutrophil Cathepsin, G. J. Immunol. 2017, 198, 1474–1483. [Google Scholar] [CrossRef]
- Fu, Z.; Akula, S.; Thorpe, M.; Hellman, L. Highly Selective Cleavage of TH2-Promoting Cytokines by the Human and the Mouse Mast Cell Tryptases, Indicating a Potent Negative Feedback Loop on TH2 Immunity. Int. J. Mol. Sci. 2019, 20, 5174. [Google Scholar] [CrossRef] [PubMed]
- Laurell, C.B. Is emphysema in alpha 1-antitrypsin deficiency a result of autodigestion? Scand. J. Clin. Lab. Investig. 1971, 28, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Kawabata, K.; Hagio, T.; Matsuoka, S. The role of neutrophil elastase in acute lung injury. Eur. J. Pharmacol. 2002, 451, 1–10. [Google Scholar] [CrossRef]
- Moraes, T.J.; Chow, C.W.; Downey, G.P. Proteases and lung injury. Crit. Care Med. 2003, 31, S189–S194. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Thorpe, M.; Akula, S.; Chahal, G.; Hellman, L. Extended cleavage specificity of human neutrophil elastase, human proteinase 3 and their distant orthologue clawed frog PR3—three elastases with similar primary but different extended specificities and stability. Front. Immunol. 2018, 9, 2387. [Google Scholar] [CrossRef]
- Raymond, W.W.; Trivedi, N.N.; Makarova, A.; Ray, M.; Craik, C.S.; Caughey, G.H. How immune peptidases change specificity: Cathepsin G gained tryptic function but lost efficiency during primate evolution. J. Immunol. 2010, 185, 5360–5368. [Google Scholar] [CrossRef]
- Novick, D.; Rubinstein, M.; Azam, T.; Rabinkov, A.; Dinarello, C.A.; Kim, S.H. Proteinase 3 is an IL-32 binding protein. Proc. Natl. Acad. Sci. USA 2006, 103, 3316–3321. [Google Scholar] [CrossRef]
- Pham, C.T. Neutrophil serine proteases fine-tune the inflammatory response. Int. J. Biochem. Cell Biol. 2008, 40, 1317–1333. [Google Scholar] [CrossRef]
- Schauer, C.; Janko, C.; Munoz, L.E.; Zhao, Y.; Kienhofer, D.; Frey, B.; Lell, M.; Manger, B.; Rech, J.; Naschberger, E.; et al. Aggregated neutrophil extracellular traps limit inflammation by degrading cytokines and chemokines. Nat. Med. 2014, 20, 511–517. [Google Scholar] [CrossRef]
- Hahn, J.; Schauer, C.; Czegley, C.; Kling, L.; Petru, L.; Schmid, B.; Weidner, D.; Reinwald, C.; Biermann, M.H.C.; Blunder, S.; et al. Aggregated neutrophil extracellular traps resolve inflammation by proteolysis of cytokines and chemokines and protection from antiproteases. FASEB J. 2019, 33, 1401–1414. [Google Scholar] [CrossRef]
- Urban, C.F.; Ermert, D.; Schmid, M.; Abu-Abed, U.; Goosmann, C.; Nacken, W.; Brinkmann, V.; Jungblut, P.R.; Zychlinsky, A. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog. 2009, 5, e1000639. [Google Scholar] [CrossRef] [PubMed]
- Henry, C.M.; Sullivan, G.P.; Clancy, D.M.; Afonina, I.S.; Kulms, D.; Martin, S.J. Neutrophil-Derived Proteases Escalate Inflammation through Activation of IL-36 Family Cytokines. Cell Rep. 2016, 14, 708–722. [Google Scholar] [CrossRef] [PubMed]
- Clancy, D.M.; Henry, C.M.; Sullivan, G.P.; Martin, S.J. Neutrophil extracellular traps can serve as platforms for processing and activation of IL-1 family cytokines. FEBS J. 2017, 284, 1712–1725. [Google Scholar] [CrossRef] [PubMed]
- Clancy, D.M.; Sullivan, G.P.; Moran, H.B.T.; Henry, C.M.; Reeves, E.P.; McElvaney, N.G.; Lavelle, E.C.; Martin, S.J. Extracellular Neutrophil Proteases Are Efficient Regulators of IL-1, IL-33, and IL-36 Cytokine Activity but Poor Effectors of Microbial Killing. Cell Rep. 2018, 22, 2937–2950. [Google Scholar] [CrossRef] [PubMed]
- Lefrancais, E.; Duval, A.; Mirey, E.; Roga, S.; Espinosa, E.; Cayrol, C.; Girard, J.P. Central domain of IL-33 is cleaved by mast cell proteases for potent activation of group-2 innate lymphoid cells. Proc. Natl. Acad. Sci. USA 2014, 111, 15502–15507. [Google Scholar] [CrossRef]
- Padrines, M.; Wolf, M.; Walz, A.; Baggiolini, M. Interleukin-8 processing by neutrophil elastase, cathepsin G and proteinase-3. FEBS Lett. 1994, 352, 231–235. [Google Scholar] [CrossRef]
- Perera, N.C.; Jenne, D.E. Perspectives and potential roles for the newly discovered NSP4 in the immune system. Expert Rev. Clin. Immunol. 2012, 8, 501–503. [Google Scholar] [CrossRef]
- Perera, N.C.; Schilling, O.; Kittel, H.; Back, W.; Kremmer, E.; Jenne, D.E. NSP4, an elastase-related protease in human neutrophils with arginine specificity. Proc. Natl. Acad. Sci. USA 2012, 109, 6229–6234. [Google Scholar] [CrossRef]
- Neuhoff, V.; Arold, N.; Taube, D.; Ehrhardt, W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 1988, 9, 255–262. [Google Scholar] [CrossRef]
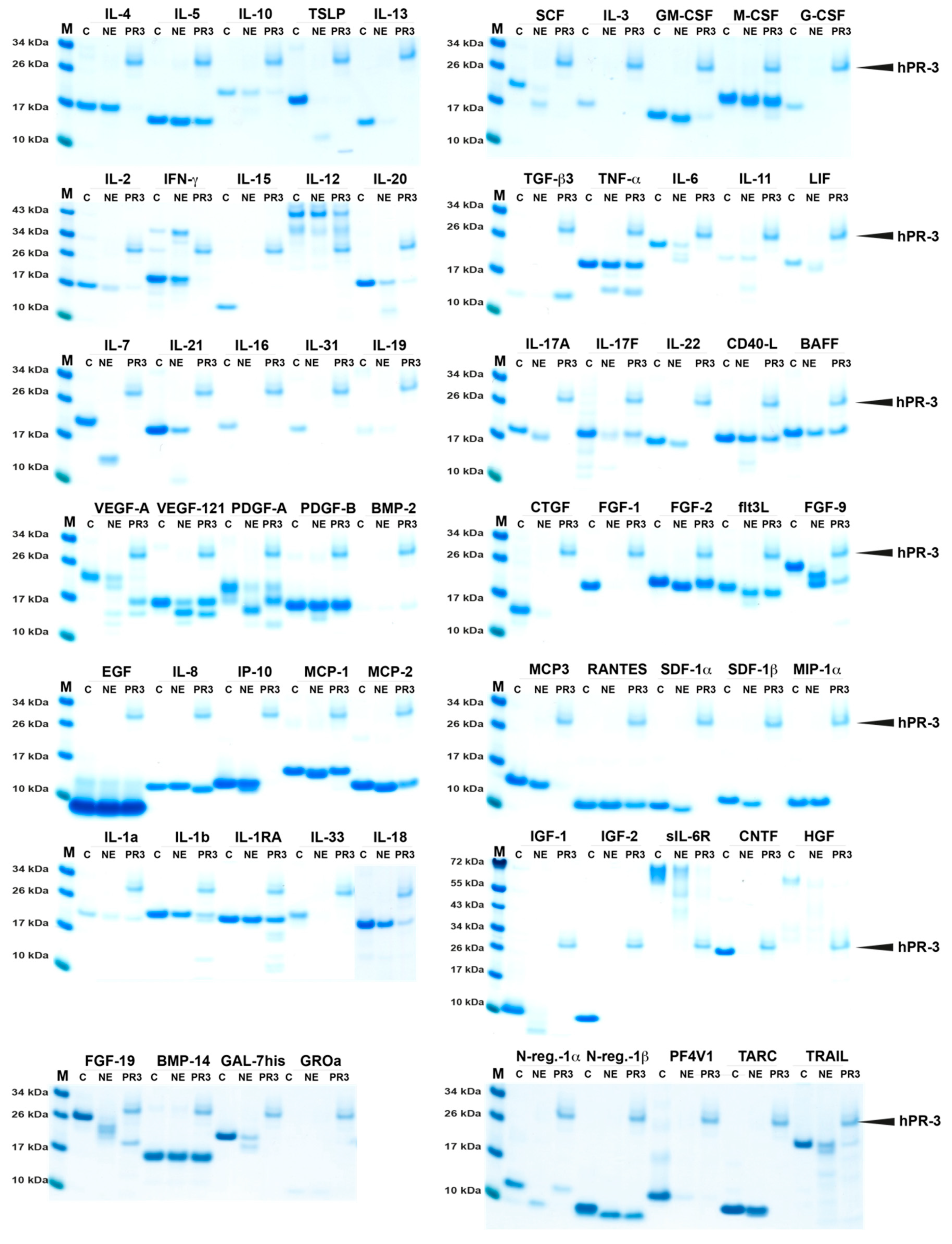
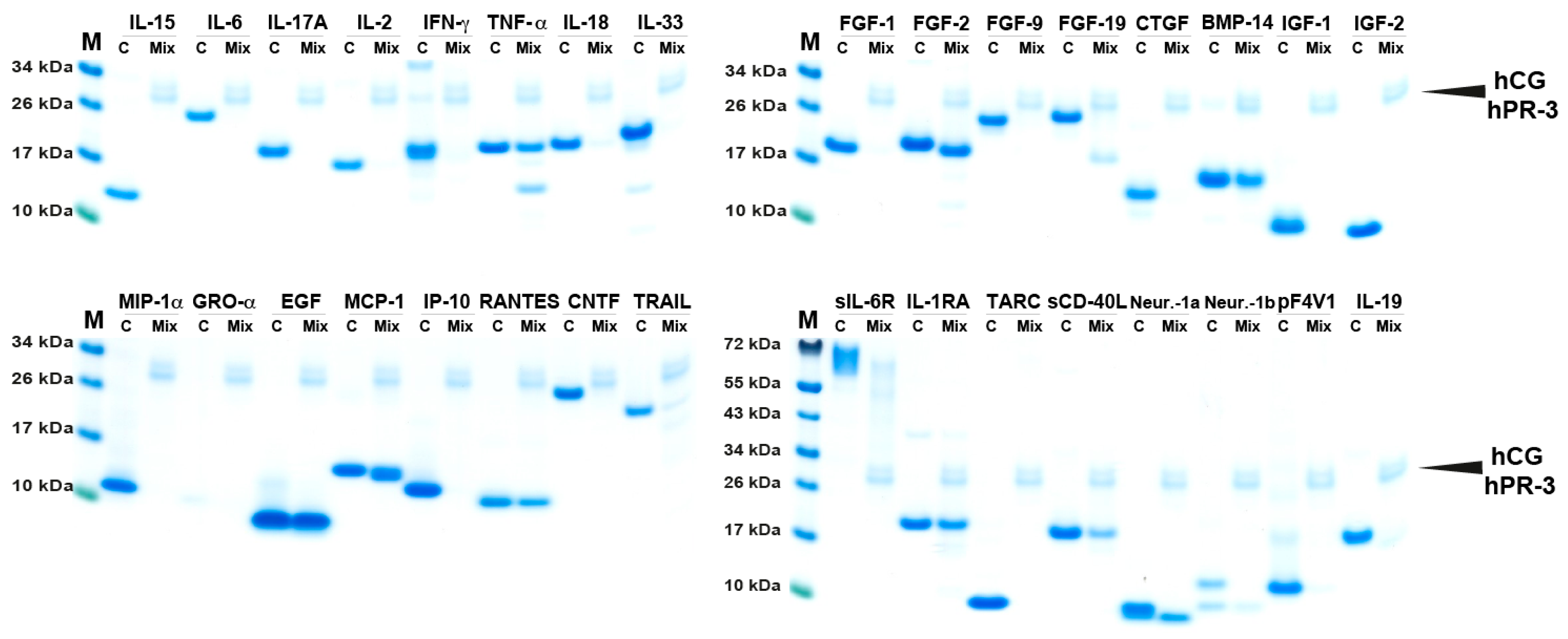
| Cytokine | HC | hCG | hNE | hPR3 | Mix | Cytokine | hNE | hPR3 | Mix |
|---|---|---|---|---|---|---|---|---|---|
| IL-4 | - | - | - | ++++ | M-CSF | ¨+¨ | ¨+¨ | ||
| IL-5 | ¨-¨ | ¨-¨ | ¨-¨ | ¨+¨ | VEGF-121 | ++ | + | ||
| IL-10 | - | - | + | +++ | IL-16 | ++++ | ++++ | ||
| TSLP | + | - | ++++ | ++++ | FGF-9 | ++++ | ++++ | ++++ | |
| IL-13 | + | - | +++ | ++++ | IL-1α | ¨-¨ | ¨-¨ | ||
| SCF | - | ++++ | ++++ | ++++ | IL-1RA | ¨-¨ | ¨+¨ | ¨+¨ | |
| IL-3 | + | ++++ | ++++ | ++++ | IGF-1 | ++++ | ++++ | ++++ | |
| GM-CSF | - | + | + | +++ | IGF-2 | ++++ | ++++ | ++++ | |
| IL-9 | - | - | sIL-6R | + | +++ | +++ | |||
| G-CSF | - | - | ++++ | ++++ | CNTF | ++++ | ++++ | ++++ | |
| IL-2 | - | + | ++ | +++ | ++++ | HGF | ++ | +++ | |
| IFN-γ | + | + | + | ++++ | ++++ | FGF-19 | ++++ | ++++ | ++++ |
| IL-15 | ++++ | ++++ | ++++ | ++++ | ++++ | BMP-14 | ¨-¨ | ¨-¨ | ¨+¨ |
| IL-12 | - | - | - | ++ | GAL-7H | +++ | ++++ | ||
| IL-20 | - | - | ++ | +++ | Neuregulin-1a | ++++ | + | ++ | |
| IL-1β | - | - | + | ++ | Neuregulin-1b | ++++ | ++++ | ++++ | |
| TNF-α | ¨-¨ | ¨+¨ | ¨+¨ | ¨+¨ | ¨+¨ | PF4V1 | ++++ | ++++ | ++++ |
| IL-6 | ++ | ++++ | +++ | ++++ | ++++ | TARC | + | ++++ | ++++ |
| IL-11 | + | + | + | ++++ | TRAIL | ++ | +++ | +++ | |
| LIF | + | + | ++ | ++++ | BMP-2 | ¨-¨ | ¨-¨ | ||
| IL-7 | - | +++ | ++++ | ++++ | TGF-β3 | ++++ | - | ||
| IL-21 | - | ++ | +++ | ++++ | GROα | - | ++++ | ++++ | |
| IL-24 | - | + | |||||||
| IL-31 | - | ++++ | ++++ | ++++ | |||||
| IL-19 | - | - | ++ | ++++ | ++++ | ||||
| IL-17A | - | ++ | ++++ | ++++ | ++++ | ||||
| IL-17F | - | - | +++ | ++ | |||||
| IL-22 | - | + | ++ | ++++ | |||||
| sCD40L | - | + | + | ++ | ++ | ||||
| BAFF | + | + | ++ | ++ | |||||
| VEGF-A | - | +++ | ++ | ++++ | |||||
| BMP-7 | - | + | |||||||
| PDGF-A | - | - | +++ | ++ | |||||
| PDGF-B | ¨-¨ | ¨-¨ | ¨+¨ | ¨-¨ | |||||
| IL-33 | ++++ | ++++ | ++++ | ++++ | ++++ | ||||
| CTGF | + | +++ | ++++ | ++++ | ++++ | ||||
| FGF-1 | + | ++++ | ++++ | ++++ | ++++ | ||||
| FGF-2 | + | + | ++ | + | ++ | ||||
| flt3L | ++ | + | ++++ | ++++ | |||||
| IL-18 | +++ | ++++ | + | +++ | ++++ | ||||
| EGF | ¨-¨ | ¨-¨ | ¨-¨ | ¨-¨ | ¨-¨ | ||||
| IL-8 | ¨-¨ | ¨-¨ | ¨-¨ | ¨++¨ | |||||
| IP-10 | - | - | - | ++++ | ++++ | ||||
| MCP-1 | ¨-¨ | ¨-¨ | + | ¨-¨ | ¨-¨ | ||||
| MCP-2 | - | - | - | ++ | |||||
| MCP-3 | - | - | + | ++++ | |||||
| RANTES | ¨-¨ | ¨-¨ | ¨-¨ | ¨+¨ | ¨+¨ | ||||
| SDF-1α | - | - | ++ | ++++ | |||||
| SDF-1β | - | + | ++ | ++++ | |||||
| MIP-1α | - | - | - | ++++ | ++++ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, Z.; Akula, S.; Thorpe, M.; Hellman, L. Potent and Broad but not Unselective Cleavage of Cytokines and Chemokines by Human Neutrophil Elastase and Proteinase 3. Int. J. Mol. Sci. 2020, 21, 651. https://doi.org/10.3390/ijms21020651
Fu Z, Akula S, Thorpe M, Hellman L. Potent and Broad but not Unselective Cleavage of Cytokines and Chemokines by Human Neutrophil Elastase and Proteinase 3. International Journal of Molecular Sciences. 2020; 21(2):651. https://doi.org/10.3390/ijms21020651
Chicago/Turabian StyleFu, Zhirong, Srinivas Akula, Michael Thorpe, and Lars Hellman. 2020. "Potent and Broad but not Unselective Cleavage of Cytokines and Chemokines by Human Neutrophil Elastase and Proteinase 3" International Journal of Molecular Sciences 21, no. 2: 651. https://doi.org/10.3390/ijms21020651
APA StyleFu, Z., Akula, S., Thorpe, M., & Hellman, L. (2020). Potent and Broad but not Unselective Cleavage of Cytokines and Chemokines by Human Neutrophil Elastase and Proteinase 3. International Journal of Molecular Sciences, 21(2), 651. https://doi.org/10.3390/ijms21020651






