Extended Cleavage Specificities of Two Mast Cell Chymase-Related Proteases and One Granzyme B-Like Protease from the Platypus, a Monotreme
Abstract
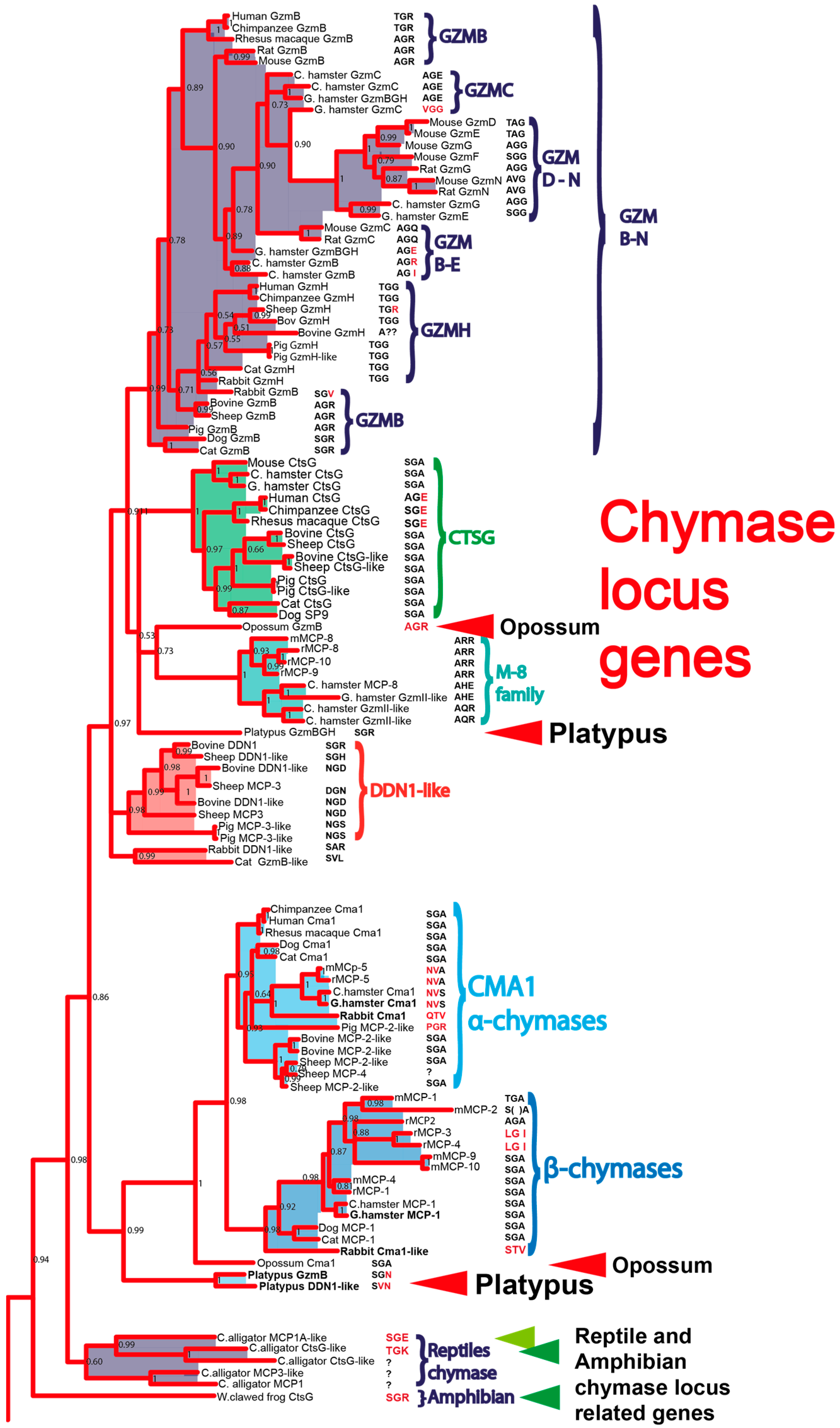
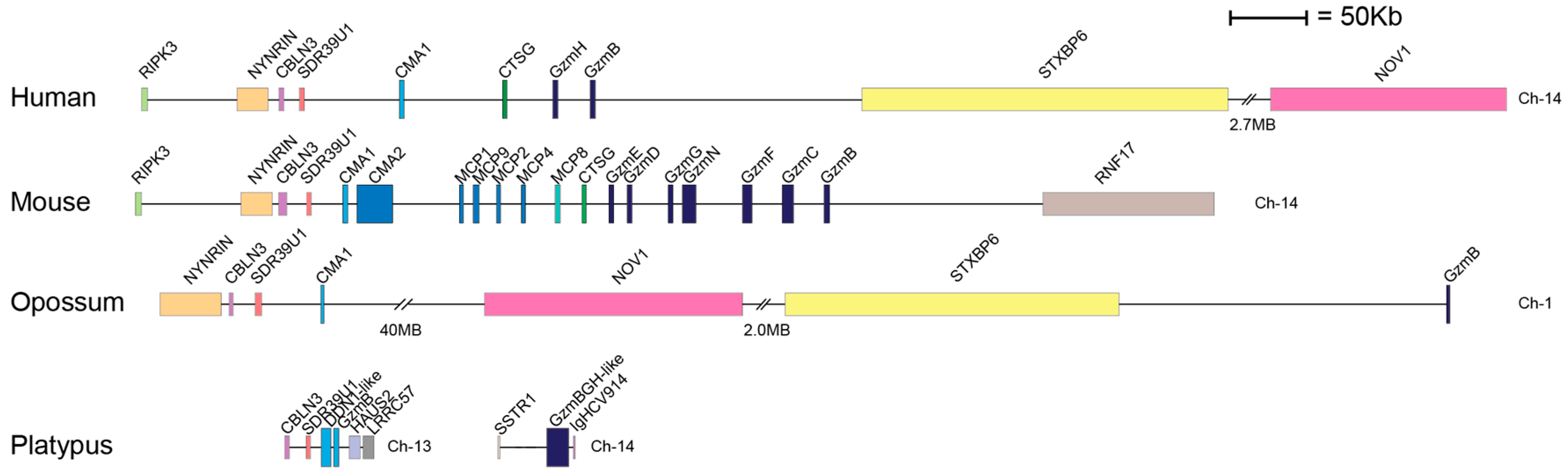
1. Introduction
2. Results
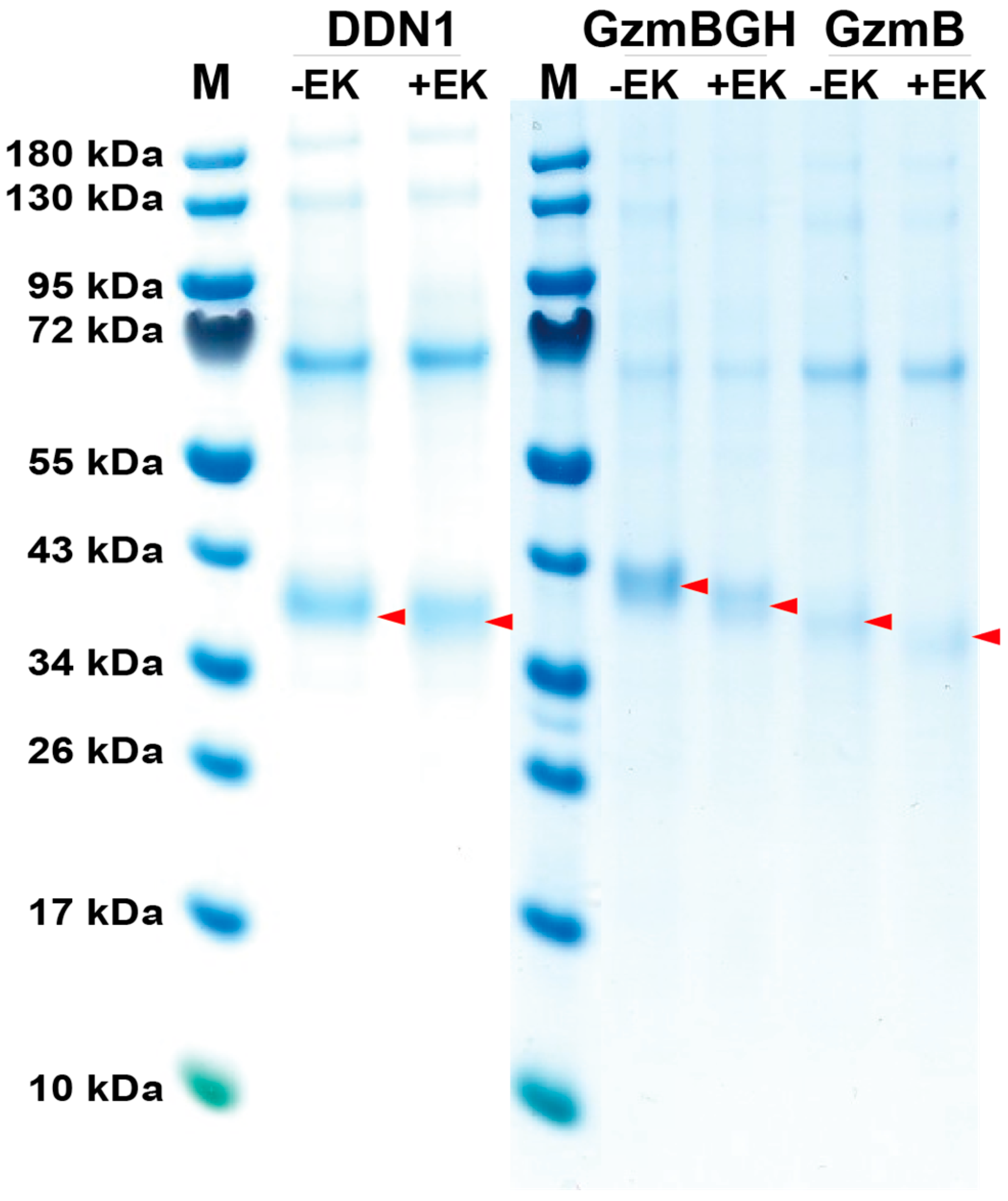
2.1. Production, Activation and Purification of Recombinant Platypus Enzymes
2.2. Chromogenic Substrate Assay and Phage Display
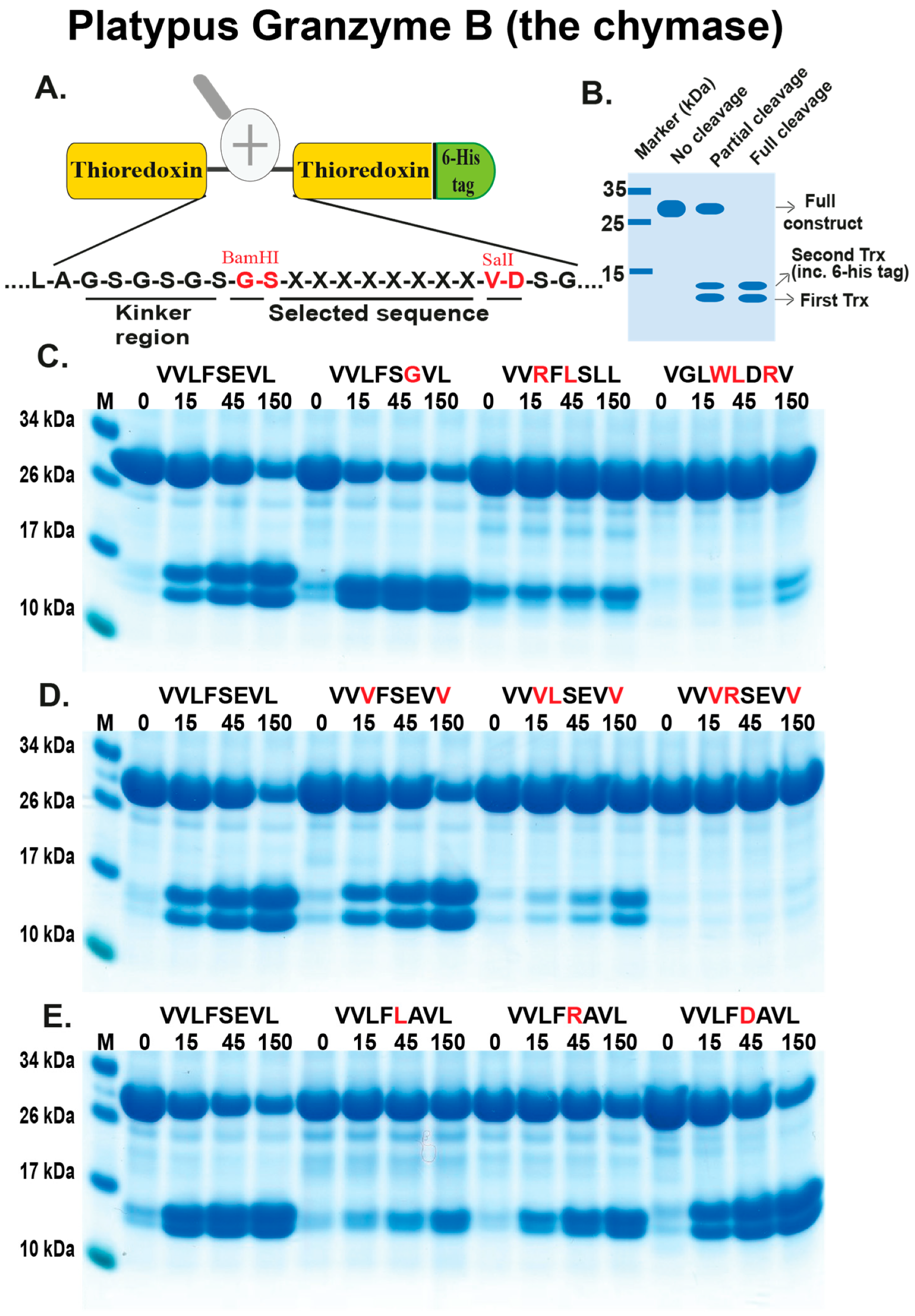
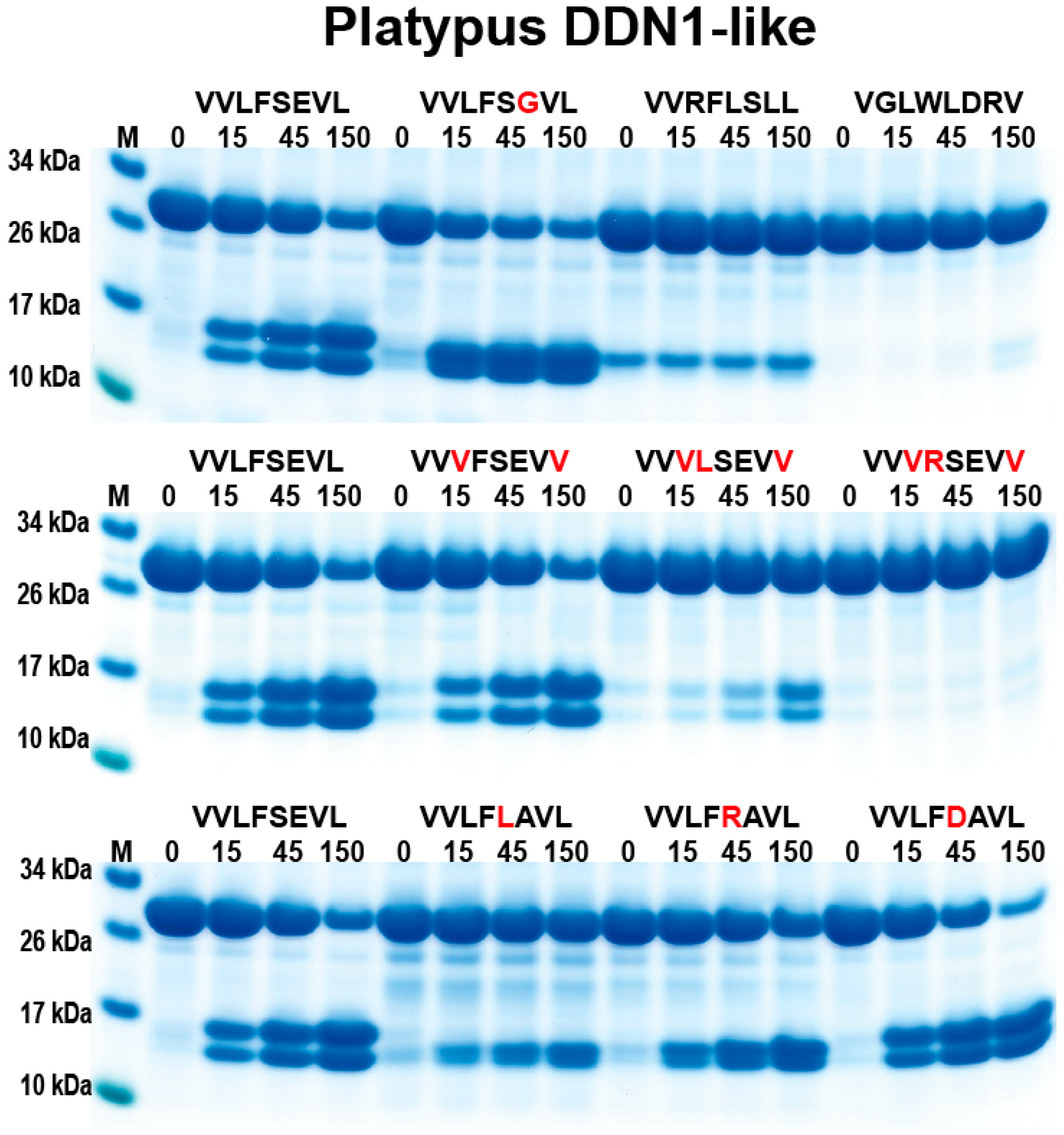
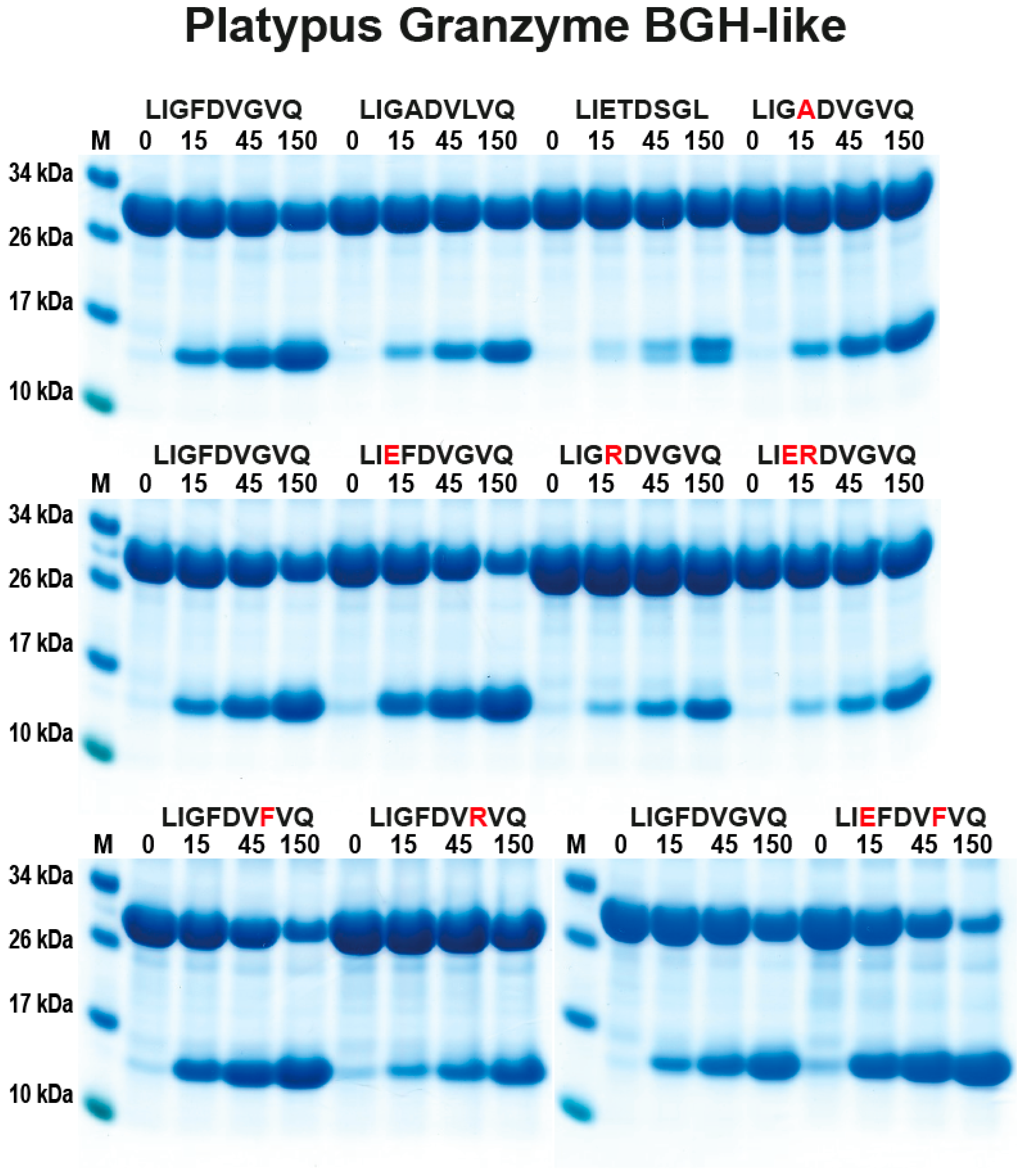
2.3. Analysis of the Extended Cleavage Specificity by the Use of Recombinant Protein Substrates
3. Discussion
4. Materials and Methods
4.1. Production and Purification of Recombinant Platypus Enzymes
4.2. Generation of Recombinant Substrates for the Analysis of the Cleavage Specificity
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| MC | mast cell |
| aa | amino acid(s) |
| mMCP | mouse mast cell protease |
| LD | linear dichroism |
| rMCP | rat mast cell protease |
| HC | human chymase |
| Ang | angiotensin |
| Ni-NTA | nickel-nitrilotriacetic acid |
| EK | enterokinase |
References
- Galli, S.J. Mast cells and basophils. Curr. Opin. Hematol. 2000, 7, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Galli, S.J.; Grimbaldeston, M.; Tsai, M. Immunomodulatory mast cells: Negative, as well as positive, regulators of immunity. Nat. Rev. Immunol. 2008, 8, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Hellman, L.T.; Akula, S.; Thorpe, M.; Fu, Z. Tracing the Origins of IgE, Mast Cells, and Allergies by Studies of Wild Animals. Front. Immunol. 2017, 8, 1749. [Google Scholar] [CrossRef] [PubMed]
- Abe, T.; Swieter, M.; Imai, T.; Hollander, N.D.; Befus, A.D. Mast cell heterogeneity: Two-dimensional gel electrophoretic analyses of rat peritoneal and intestinal mucosal mast cell. Eur. J. Immunol. 1990, 20, 1941–1947. [Google Scholar] [CrossRef] [PubMed]
- Lützelschwab, C.; Aveskogh, M.; Pejler, G.; Hellman, L. Secretory Granule Proteases in Rat Mast Cells. Cloning of 10 Different Serine Proteases and a Carboxypeptidase A from Various Rat Mast Cell Populations. J. Exp. Med. 1997, 185, 13–30. [Google Scholar] [CrossRef] [PubMed]
- Pejler, G.; Rönnberg, E.; Waern, I.; Wernersson, S. Mast cell proteases: Multifaceted regulators of inflammatory disease. Blood 2010, 115, 4981–4990. [Google Scholar] [CrossRef] [PubMed]
- Caughey, G.H. Mast cell tryptases and chymases in inflammation and host defense. Immunol. Rev. 2007, 217, 141–154. [Google Scholar] [CrossRef]
- Caughey, G.H. Mast cell proteases as protective and inflammatory mediators. Single Mol. Single Cell Seq. 2011, 716, 212–234. [Google Scholar]
- Chandrasekharan, U.M.; Sanker, S.; Glynias, M.J.; Karnik, S.S.; Husain, A. Angiotensin II-Forming Activity in a Reconstructed Ancestral Chymase. Science 1996, 271, 502–505. [Google Scholar] [CrossRef]
- Huang, R.; Blom, T.; Hellman, L. Cloning and structural analysis of MMCP-1, MMCP-4 and MMCP-5, three mouse mast cell-specific serine proteases. Eur. J. Immunol. 1991, 21, 1611–1621. [Google Scholar] [CrossRef]
- Hellman, L.; Thorpe, M. Granule proteases of hematopoietic cells, a family of versatile inflammatory mediators—An update on their cleavage specificity, in vivo substrates, and evolution. Boil. Chem. 2014, 395, 15–49. [Google Scholar] [CrossRef] [PubMed]
- Akula, S.; Thorpe, M.; Boinapally, V.; Hellman, L. Granule Associated Serine Proteases of Hematopoietic Cells—An Analysis of Their Appearance and Diversification during Vertebrate Evolution. PLoS ONE 2015, 10, e0143091. [Google Scholar]
- Gallwitz, M.; Reimer, J.M.; Hellman, L. Expansion of the mast cell chymase locus over the past 200 million years of mammalian evolution. Immunogenetics 2006, 58, 655–669. [Google Scholar] [CrossRef] [PubMed]
- Kunori, Y.; Koizumi, M.; Masegi, T.; Kasai, H.; Kawabata, H.; Yamazaki, Y.; Fukamizu, A. Rodent alpha-chymases are elastase-like proteases. JBIC J. Boil. Inorg. Chem. 2002, 269. [Google Scholar] [CrossRef]
- Karlson, U.; Pejler, G.; Tomasini-Johansson, B.; Hellman, L. Extended substrate specificity of rat mast cell protease 5, a rodent alpha-chymase with elastase-like primary specificity. J. Biol. Chem. 2003, 278, 39625–39631. [Google Scholar] [CrossRef]
- Kervinen, J.; Abad, M.; Crysler, C.; Kolpak, M.; Mahan, A.D.; Masucci, J.A.; Bayoumy, S.; Cummings, M.D.; Yao, X.; Olson, M.; et al. Structural basis for elastolytic substrate specificity in rodent alpha-chymases. J. Biol. Chem. 2008, 283, 427–436. [Google Scholar] [CrossRef]
- Thorpe, M.; Fu, Z.; Albat, E.; Akula, S.; De Garavilla, L.; Kervinen, J.; Hellman, L. Extended cleavage specificities of mast cell proteases 1 and 2 from golden hamster: Classical chymase and an elastolytic protease comparable to rat and mouse MCP-5. PLoS ONE 2018, 13, e0207826. [Google Scholar] [CrossRef]
- Caughey, G.H.; Beauchamp, J.; Schlatter, D.; Raymond, W.W.; Trivedi, N.N.; Banner, D.; Mauser, H.; Fingerle, J. Guinea pig chymase is leucine-specific: A novel example of functional plasticity in the chymase/granzyme family of serine peptidases. J. Boil. Chem. 2008, 283, 13943–13951. [Google Scholar] [CrossRef]
- Yuan, Z.W.; Akula, S.; Fu, Z.; de Garavilla, L.; Kervinen, J.; Thorpe, M.; Hellman, L. Extended Cleavage Specificities of Rabbit and Guinea Pig Mast Cell Chymases: Two Highly Specific Leu-Ases. Int. J. Mol. Sci. 2019, 20, 6340. [Google Scholar] [CrossRef]
- Vernersson, M. Generation of therapeutic antibody responses against IgE through vaccination. FASEB J. 2002, 16, 875–877. [Google Scholar] [CrossRef]
- Gallwitz, M.; Enoksson, M.; Thorpe, M.; Ge, X.; Hellman, L. The extended substrate recognition profile of the dog mast cell chymase reveals similarities and differences to the human chymase. Int. Immunol. 2010, 22, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.K.; Thorpe, M.; Hellman, L. Arg143 and Lys192 of the human mast cell chymase mediate the preference for acidic amino acids in position P2′ of substrates. FEBS J. 2010, 277, 2255–2267. [Google Scholar] [CrossRef] [PubMed]
- Thorpe, M.; Yu, J.; Boinapally, V.; Ahooghalandari, P.; Kervinen, J.; De Garavilla, L.; Hellman, L. Extended cleavage specificity of the mast cell chymase from the crab-eating macaque (Macaca fascicularis): An interesting animal model for the analysis of the function of the human mast cell chymase. Int. Immunol. 2012, 24, 771–782. [Google Scholar] [CrossRef] [PubMed]
- Gallwitz, M.; Enoksson, M.; Thorpe, M.; Hellman, L. The Extended Cleavage Specificity of Human Thrombin. PLoS ONE 2012, 7, e31756. [Google Scholar] [CrossRef]
- Chahal, G.; Thorpe, M.; Hellman, L. The Importance of Exosite Interactions for Substrate Cleavage by Human Thrombin. PLoS ONE 2015, 10, e0129511. [Google Scholar] [CrossRef]
- Fu, Z.; Thorpe, M.; Hellman, L. rMCP-2, the Major Rat Mucosal Mast Cell Protease, an Analysis of Its Extended Cleavage Specificity and Its Potential Role in Regulating Intestinal Permeability by the Cleavage of Cell Adhesion and Junction Proteins. PLoS ONE 2015, 10, e0131720. [Google Scholar] [CrossRef]
- Thorpe, M.; Akula, S.; Hellman, L. Channel catfish granzyme-like I is a highly specific serine protease with metase activity that is expressed by fish NK-like cells. Dev. Comp. Immunol. 2016, 63, 84–95. [Google Scholar] [CrossRef]
- Fu, Z.; Thorpe, M.; Akula, S.; Hellman, L. Asp-ase Activity of the Opossum Granzyme B Supports the Role of Granzyme B as Part of Anti-Viral Immunity Already during Early Mammalian Evolution. PLoS ONE 2016, 11, e0154886. [Google Scholar] [CrossRef][Green Version]
- Thorpe, M.; Fu, Z.; Chahal, G.; Akula, S.; Kervinen, J.; De Garavilla, L.; Hellman, L. Extended cleavage specificity of human neutrophil cathepsin G: A low activity protease with dual chymase and tryptase-type specificities. PLoS ONE 2018, 13, e0195077. [Google Scholar] [CrossRef]
- Fu, Z.; Thorpe, M.; Akula, S.; Chahal, G.; Hellman, L. Extended cleavage specificity of human neutrophil elastase, human proteinase 3 and their distant orthologue clawed frog PR3—three elastases with similar primary but different extended specificities and stability. Front. Immunol. 2018, 9, 1–19. [Google Scholar] [CrossRef]
- Zamolodchikova, T.S.; Scherbakov, I.T.; Khrennikov, B.N.; Svirshchevskaya, E.V. Expression of duodenase-like protein in epitheliocytes of Brunner’s glands in human duodenal mucosa. Biochemistry (Moscow) 2013, 78, 954–957. [Google Scholar] [CrossRef] [PubMed]
- Zamolodchikova, T.S.; Vorotyntseva, T.I.; Antonov, V.K. Duodenase, a new serine protease of unusual specificity from bovine duodenal mucosa. Purification and properties. Eur. J. Biochem. 1995, 227, 866–872. [Google Scholar] [CrossRef] [PubMed]
- Pejler, G.; Abrink, M.; Ringvall, M.; Wernersson, S. Mast Cell Proteases. Adv. Immunol. 2007, 95, 167–255. [Google Scholar] [PubMed]
- Guo, C.; Ju, H.; Leung, D.; Massaeli, H.; Shi, M.; Rabinovitch, M. A novel vascular smooth muscle chymase is upregulated in hypertensive rats. J. Clin. Investig. 2001, 107, 703–715. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ju, H.; Gros, R.; You, X.; Tsang, S.; Husain, M.; Rabinovitch, M. Conditional and targeted overexpression of vascular chymase causes hypertension in transgenic mice. Proc. Natl. Acad. Sci. USA 2001, 98, 7469–7474. [Google Scholar] [CrossRef] [PubMed]
- Neuhoff, V.; Arold, N.; Taube, D.; Ehrhardt, W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 1988, 9, 255–262. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, Z.; Akula, S.; Thorpe, M.; Hellman, L. Extended Cleavage Specificities of Two Mast Cell Chymase-Related Proteases and One Granzyme B-Like Protease from the Platypus, a Monotreme. Int. J. Mol. Sci. 2020, 21, 319. https://doi.org/10.3390/ijms21010319
Fu Z, Akula S, Thorpe M, Hellman L. Extended Cleavage Specificities of Two Mast Cell Chymase-Related Proteases and One Granzyme B-Like Protease from the Platypus, a Monotreme. International Journal of Molecular Sciences. 2020; 21(1):319. https://doi.org/10.3390/ijms21010319
Chicago/Turabian StyleFu, Zhirong, Srinivas Akula, Michael Thorpe, and Lars Hellman. 2020. "Extended Cleavage Specificities of Two Mast Cell Chymase-Related Proteases and One Granzyme B-Like Protease from the Platypus, a Monotreme" International Journal of Molecular Sciences 21, no. 1: 319. https://doi.org/10.3390/ijms21010319
APA StyleFu, Z., Akula, S., Thorpe, M., & Hellman, L. (2020). Extended Cleavage Specificities of Two Mast Cell Chymase-Related Proteases and One Granzyme B-Like Protease from the Platypus, a Monotreme. International Journal of Molecular Sciences, 21(1), 319. https://doi.org/10.3390/ijms21010319





