The Effect of Dysfunctional Ubiquitin Enzymes in the Pathogenesis of Most Common Diseases
Abstract
1. Introduction
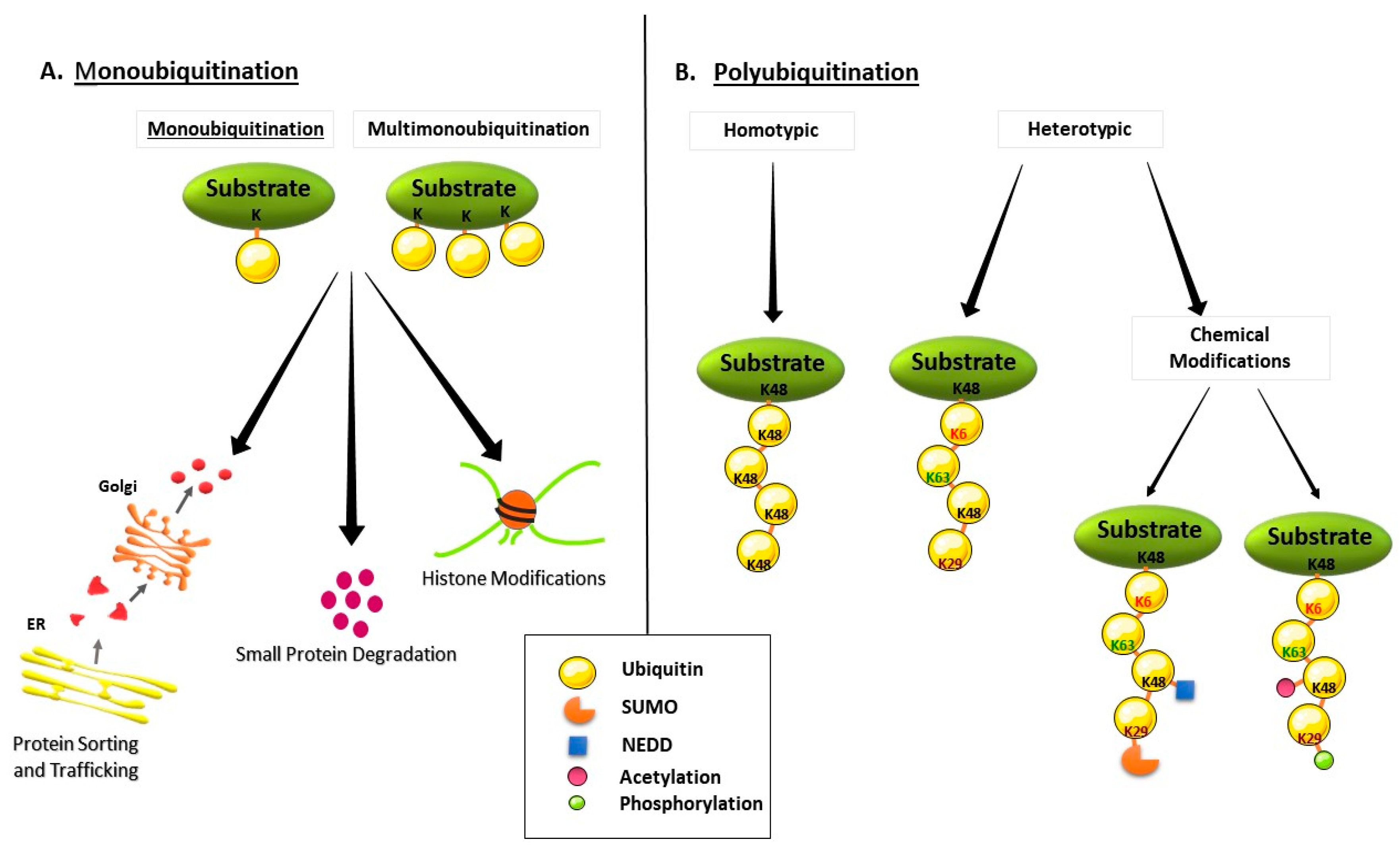
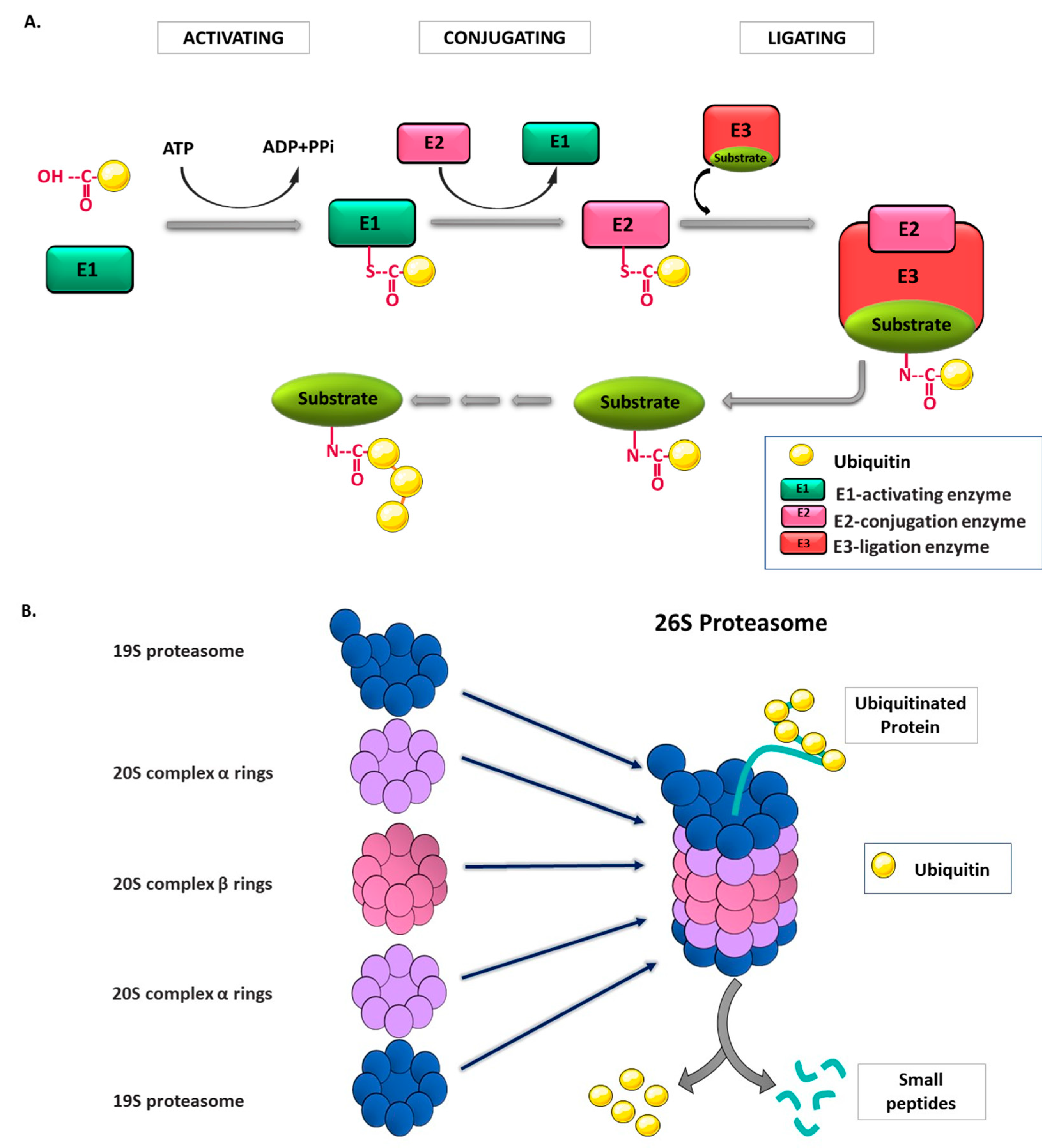
2. Ubiquitin and Ubiquitin Proteasome System (UPS)
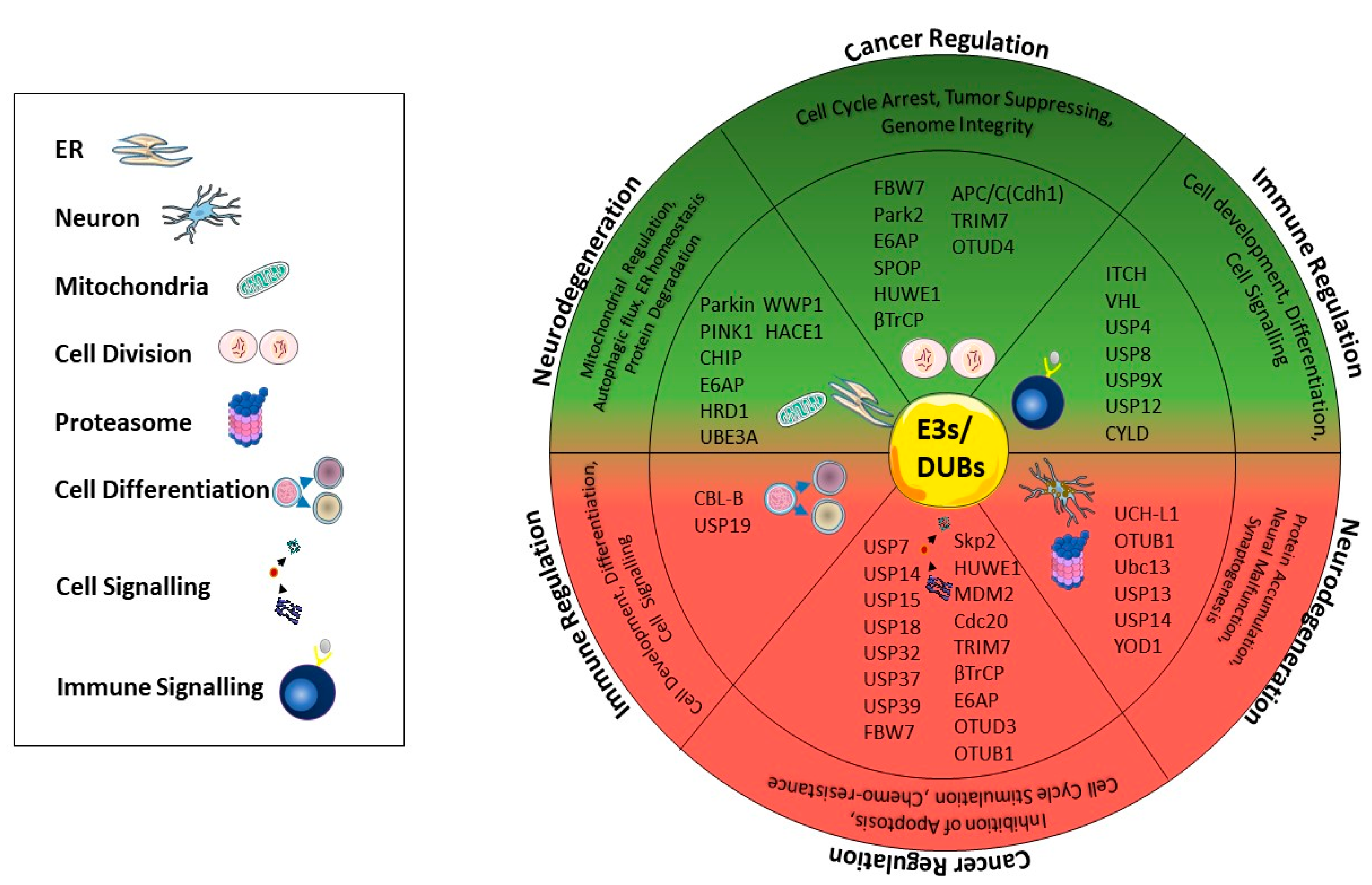
3. Ubiquitination-Mediated Regulation in Cancer
4. Ubiquitination-Mediated Regulation in Neurodegeneration and Neurodegenerative Diseases
5. Ubiquitination in Immune-Related Diseases
6. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Goldstein, G.; Scheid, M.; Hammerling, U.; Schlesinger, D.H.; Niall, H.D.; Boyse, E.A. Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc. Natl. Acad. Sci. USA 1975, 72, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Kocaturk, N.M.; Gozuacik, D. Crosstalk Between Mammalian Autophagy and the Ubiquitin-Proteasome System. Front. Cell Dev. Biol. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Gilberto, S.; Peter, M. Dynamic ubiquitin signaling in cell cycle regulation. J. Cell Biol. 2017, 216, 2259–2271. [Google Scholar] [CrossRef] [PubMed]
- Vucic, D.; Dixit, V.M.; Wertz, I.E. Ubiquitylation in apoptosis: A post-translational modification at the edge of life and death. Nat. Rev. Mol. Cell Biol. 2011, 12, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.L.; Urbe, S. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 2007, 8, 355–368. [Google Scholar] [CrossRef]
- Cao, J.; Yan, Q. Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front. Oncol. 2012, 2, 26. [Google Scholar] [CrossRef]
- Scheuring, D.; Künzl, F.; Viotti, C.; Yan, M.S.; Jiang, L.; Schellmann, S.; Robinson, D.G.; Pimpl, P. Ubiquitin initiates sorting of Golgi and plasma membrane proteins into the vacuolar degradation pathway. BMC Plant Biol. 2012, 12, 164. [Google Scholar] [CrossRef]
- Ronai, Z.e.A. Monoubiquitination in proteasomal degradation. Proc. Natl. Acad. Sci. USA 2016, 113, 8894–8896. [Google Scholar] [CrossRef]
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef]
- Chen, Z.J.; Sun, L.J. Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 2009, 33, 275–286. [Google Scholar] [CrossRef]
- Zhao, B.; Bhuripanyo, K.; Schneider, J.; Zhang, K.; Schindelin, H.; Boone, D.; Yin, J. Specificity of the E1-E2-E3 enzymatic cascade for ubiquitin C-terminal sequences identified by phage display. ACS Chem. Biol. 2012, 7, 2027–2035. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.; Shabek, N. Ubiquitin Ligases: Structure, Function, and Regulation. Annu. Rev. Biochem. 2017, 86, 129–157. [Google Scholar] [CrossRef] [PubMed]
- George, A.J.; Hoffiz, Y.C.; Charles, A.J.; Zhu, Y.; Mabb, A.M. A Comprehensive Atlas of E3 Ubiquitin Ligase Mutations in Neurological Disorders. Front. Genet. 2018, 9. [Google Scholar] [CrossRef]
- Stewart, M.D.; Ritterhoff, T.; Klevit, R.E.; Brzovic, P.S. E2 enzymes: More than just middle men. Cell Res. 2016, 26, 423–440. [Google Scholar] [CrossRef]
- Lecker, S.H.; Goldberg, A.L.; Mitch, W.E. Protein Degradation by the Ubiquitin–Proteasome Pathway in Normal and Disease States. J. Am. Soc. Nephrol. 2006, 17, 1807–1819. [Google Scholar] [CrossRef]
- Benaroudj, N.; Zwickl, P.; Seemuller, E.; Baumeister, W.; Goldberg, A.L. ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol. Cell 2003, 11, 69–78. [Google Scholar] [CrossRef]
- Tanaka, K. The proteasome: Overview of structure and functions. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009, 85, 12–36. [Google Scholar] [CrossRef]
- Abdul Rehman, S.A.; Kristariyanto, Y.A.; Choi, S.Y.; Nkosi, P.J.; Weidlich, S.; Labib, K.; Hofmann, K.; Kulathu, Y. MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes. Mol. Cell 2016, 63, 146–155. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Zhou, Z.; Shah, A.A.; Zou, H.; Tao, J.; Chen, Q.; Wan, Y. The emerging role of deubiquitinating enzymes in genomic integrity, diseases, and therapeutics. Cell Biosci. 2016, 6, 62. [Google Scholar] [CrossRef]
- Zou, Q.; Jin, J.; Hu, H.; Li, H.S.; Romano, S.; Xiao, Y.; Nakaya, M.; Zhou, X.; Cheng, X.; Yang, P.; et al. USP15 stabilizes MDM2 to mediate cancer-cell survival and inhibit antitumor T cell responses. Nat. Immunol. 2014, 15, 562–570. [Google Scholar] [CrossRef]
- Li, L.; Martinez, S.S.; Hu, W.; Liu, Z.; Tjian, R. A specific E3 ligase/deubiquitinase pair modulates TBP protein levels during muscle differentiation. Elife 2015, 4, e08536. [Google Scholar] [CrossRef] [PubMed]
- Grou, C.P.; Pinto, M.P.; Mendes, A.V.; Domingues, P.; Azevedo, J.E. The de novo synthesis of ubiquitin: Identification of deubiquitinases acting on ubiquitin precursors. Sci. Rep. 2015, 5, 12836. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Kitagaki, J.; Dai, R.M.; Tsai, Y.C.; Lorick, K.L.; Ludwig, R.L.; Pierre, S.A.; Jensen, J.P.; Davydov, I.V.; Oberoi, P.; et al. Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Res. 2007, 67, 9472–9481. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Shaik, S.; Dai, X.; Wu, Q.; Zhou, X.; Wang, Z.; Wei, W. Targeting the ubiquitin pathway for cancer treatment. Biochim. Biophys. Acta 2015, 1855, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, S.M.; Okoye, I.; Chaleshtari, M.G.; Hazhirkarzar, B.; Mohamadnejad, J.; Azizi, G.; Hojjat-Farsangi, M.; Mohammadi, H.; Shotorbani, S.S.; Jadidi-Niaragh, F. E2 ubiquitin-conjugating enzymes in cancer: Implications for immunotherapeutic interventions. Clin. Chim. Acta 2019, 498, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Ceccarelli, D.F.; Tang, X.; Pelletier, B.; Orlicky, S.; Xie, W.; Plantevin, V.; Neculai, D.; Chou, Y.C.; Ogunjimi, A.; Al-Hakim, A.; et al. An allosteric inhibitor of the human Cdc34 ubiquitin-conjugating enzyme. Cell 2011, 145, 1075–1087. [Google Scholar] [CrossRef]
- Scheper, J.; Guerra-Rebollo, M.; Sanclimens, G.; Moure, A.; Masip, I.; González-Ruiz, D.; Rubio, N.; Crosas, B.; Meca-Cortés, O.; Loukili, N.; et al. Protein-protein interaction antagonists as novel inhibitors of non-canonical polyubiquitylation. PLoS ONE 2010, 5, e11403. [Google Scholar] [CrossRef]
- Ushiyama, S.; Umaoka, H.; Kato, H.; Suwa, Y.; Morioka, H.; Rotinsulu, H.; Losung, F.; Mangindaan, R.E.; de Voogd, N.J.; Yokosawa, H.; et al. Manadosterols A and B, sulfonated sterol dimers inhibiting the Ubc13-Uev1A interaction, isolated from the marine sponge Lissodendryx fibrosa. J. Nat. Prod. 2012, 75, 1495–1499. [Google Scholar] [CrossRef]
- Hodge, C.D.; Edwards, R.A.; Markin, C.J.; McDonald, D.; Pulvino, M.; Huen, M.S.; Zhao, J.; Spyracopoulos, L.; Hendzel, M.J.; Glover, J.N. Covalent Inhibition of Ubc13 Affects Ubiquitin Signaling and Reveals Active Site Elements Important for Targeting. ACS Chem. Biol. 2015, 10, 1718–1728. [Google Scholar] [CrossRef]
- Huang, D.T.; Miller, D.W.; Mathew, R.; Cassell, R.; Holton, J.M.; Roussel, M.F.; Schulman, B.A. A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8. Nat. Struct. Mol. Biol. 2004, 11, 927–935. [Google Scholar] [CrossRef]
- Kumar, A.; Ito, A.; Hirohama, M.; Yoshida, M.; Zhang, K.Y.J. Identification of Sumoylation Inhibitors Targeting a Predicted Pocket in Ubc9. J. Chem. Inf. Model. 2014, 54, 2784–2793. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Chong, R.A.; Yu, Q.; Bai, J.; Spratt, D.E.; Ching, K.; Lee, C.; Miao, H.; Tappin, I.; Hurwitz, J.; et al. Suramin inhibits cullin-RING E3 ubiquitin ligases. Proc. Natl. Acad. Sci. USA 2016, 113, E2011–E2018. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Li, H.; Xing, C.; Ye, H.; Feng, J.; Wu, J.; Lu, Z.; Fang, J.; Gao, S. Honokiol induces proteasomal degradation of AML1-ETO oncoprotein via increasing ubiquitin conjugase UbcH8 expression in leukemia. Biochem. Pharm. 2017, 128, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Kothayer, H.; Spencer, S.M.; Tripathi, K.; Westwell, A.D.; Palle, K. Synthesis and in vitro anticancer evaluation of some 4,6-diamino-1,3,5-triazine-2-carbohydrazides as Rad6 ubiquitin conjugating enzyme inhibitors. Bioorg. Med. Chem. Lett. 2016, 26, 2030–2034. [Google Scholar] [CrossRef]
- Yada, M.; Hatakeyama, S.; Kamura, T.; Nishiyama, M.; Tsunematsu, R.; Imaki, H.; Ishida, N.; Okumura, F.; Nakayama, K.; Nakayama, K.I. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004, 23, 2116–2125. [Google Scholar] [CrossRef]
- Yumimoto, K.; Nakayama, K.I. Recent insight into the role of FBXW7 as a tumor suppressor. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef]
- Xu, W.; Taranets, L.; Popov, N. Regulating Fbw7 on the road to cancer. Semin. Cancer Biol. 2016, 36, 62–70. [Google Scholar] [CrossRef]
- Bahram, F.; von der Lehr, N.; Cetinkaya, C.; Larsson, L.G. c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood 2000, 95, 2104–2110. [Google Scholar] [CrossRef]
- Yeh, C.-H.; Bellon, M.; Nicot, C. FBXW7: A critical tumor suppressor of human cancers. Mol. Cancer 2018, 17, 115. [Google Scholar] [CrossRef]
- Reavie, L.; Buckley, S.M.; Loizou, E.; Takeishi, S.; Aranda-Orgilles, B.; Ndiaye-Lobry, D.; Abdel-Wahab, O.; Ibrahim, S.; Nakayama, K.I.; Aifantis, I. Regulation of c-Myc ubiquitination controls chronic myelogenous leukemia initiation and progression. Cancer Cell 2013, 23, 362–375. [Google Scholar] [CrossRef]
- Jin, X.; Yang, C.; Fan, P.; Xiao, J.; Zhang, W.; Zhan, S.; Liu, T.; Wang, D.; Wu, H. CDK5/FBW7-dependent ubiquitination and degradation of EZH2 inhibits pancreatic cancer cell migration and invasion. J. Biol. Chem. 2017, 292, 6269–6280. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Oh, Y.T.; Yue, P.; Khuri, F.R.; Sun, S.Y. Inhibition of mTOR complex 2 induces GSK3/FBXW7-dependent degradation of sterol regulatory element-binding protein 1 (SREBP1) and suppresses lipogenesis in cancer cells. Oncogene 2016, 35, 642–650. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.-H.; Bellon, M.; Pancewicz-Wojtkiewicz, J.; Nicot, C. Oncogenic mutations in the FBXW7 gene of adult T-cell leukemia patients. Proc. Natl. Acad. Sci. USA 2016, 113, 6731–6736. [Google Scholar] [CrossRef] [PubMed]
- Imura, S.; Tovuu, L.O.; Utsunomiya, T.; Morine, Y.; Ikemoto, T.; Arakawa, Y.; Kanamoto, M.; Iwahashi, S.; Saito, Y.; Takasu, C.; et al. Role of Fbxw7 expression in hepatocellular carcinoma and adjacent non-tumor liver tissue. J. Gastroenterol. Hepatol. 2014, 29, 1822–1829. [Google Scholar] [CrossRef] [PubMed]
- Iwatsuki, M.; Mimori, K.; Ishii, H.; Yokobori, T.; Takatsuno, Y.; Sato, T.; Toh, H.; Onoyama, I.; Nakayama, K.I.; Baba, H.; et al. Loss of FBXW7, a cell cycle regulating gene, in colorectal cancer: Clinical significance. Int. J. Cancer 2010, 126, 1828–1837. [Google Scholar] [CrossRef] [PubMed]
- Yokobori, T.; Mimori, K.; Iwatsuki, M.; Ishii, H.; Tanaka, F.; Sato, T.; Toh, H.; Sudo, T.; Iwaya, T.; Tanaka, Y.; et al. Copy number loss of FBXW7 is related to gene expression and poor prognosis in esophageal squamous cell carcinoma. Int. J. Oncol. 2012, 41, 253–259. [Google Scholar] [CrossRef]
- Wertz, I.E.; Kusam, S.; Lam, C.; Okamoto, T.; Sandoval, W.; Anderson, D.J.; Helgason, E.; Ernst, J.A.; Eby, M.; Liu, J.; et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature 2011, 471, 110–114. [Google Scholar] [CrossRef]
- Devine, T.; Dai, M.S. Targeting the ubiquitin-mediated proteasome degradation of p53 for cancer therapy. Curr. Pharm. Des. 2013, 19, 3248–3262. [Google Scholar] [CrossRef]
- Wade, M.; Li, Y.C.; Wahl, G.M. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat. Rev. Cancer 2013, 13, 83–96. [Google Scholar] [CrossRef]
- Wang, Z.; Kang, W.; You, Y.; Pang, J.; Ren, H.; Suo, Z.; Liu, H.; Zheng, Y. USP7: Novel Drug Target in Cancer Therapy. Front. Pharmacol. 2019, 10. [Google Scholar] [CrossRef]
- Shaikh, M.F.; Morano, W.F.; Lee, J.; Gleeson, E.; Babcock, B.D.; Michl, J.; Sarafraz-Yazdi, E.; Pincus, M.R.; Bowne, W.B. Emerging Role of MDM2 as Target for Anti-Cancer Therapy: A Review. Ann. Clin. Lab. Sci. 2016, 46, 627–634. [Google Scholar]
- Alfarsi, L.H.; Ansari, R.E.; Craze, M.L.; Toss, M.S.; Masisi, B.; Ellis, I.O.; Rakha, E.A.; Green, A.R. CDC20 expression in oestrogen receptor positive breast cancer predicts poor prognosis and lack of response to endocrine therapy. Breast Cancer Res. Treat. 2019, 178, 535–544. [Google Scholar] [CrossRef]
- Cheng, S.; Castillo, V.; Sliva, D. CDC20 associated with cancer metastasis and novel mushroom-derived CDC20 inhibitors with antimetastatic activity. Int. J. Oncol. 2019, 54, 2250–2256. [Google Scholar] [CrossRef] [PubMed]
- Han, T.; Jiang, S.; Zheng, H.; Yin, Q.; Xie, M.; Little, M.R.; Yin, X.; Chen, M.; Song, S.J.; Beg, A.A.; et al. Interplay between c-Src and the APC/C co-activator Cdh1 regulates mammary tumorigenesis. Nat. Commun. 2019, 10, 3716. [Google Scholar] [CrossRef] [PubMed]
- Schrock, M.S.; Stromberg, B.R.; Scarberry, L.; Summers, M.K. APC/C ubiquitin ligase: Functions and mechanisms in tumorigenesis. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Shaik, S.; Wan, L.; Tron, A.E.; Wang, Z.; Sun, L.; Inuzuka, H.; Wei, W. SCF β-TRCP targets MTSS1 for ubiquitination-mediated destruction to regulate cancer cell proliferation and migration. Oncotarget 2013, 4, 2339–2353. [Google Scholar] [CrossRef]
- Ma, J.; Lu, Y.; Zhang, S.; Li, Y.; Huang, J.; Yin, Z.; Ren, J.; Huang, K.; Liu, L.; Yang, K.; et al. β-Trcp ubiquitin ligase and RSK2 kinase-mediated degradation of FOXN2 promotes tumorigenesis and radioresistance in lung cancer. Cell Death Differ. 2018, 25, 1473–1485. [Google Scholar] [CrossRef]
- Shaik, S.; Nucera, C.; Inuzuka, H.; Gao, D.; Garnaas, M.; Frechette, G.; Harris, L.; Wan, L.; Fukushima, H.; Husain, A.; et al. SCF(β-TRCP) suppresses angiogenesis and thyroid cancer cell migration by promoting ubiquitination and destruction of VEGF receptor 2. J. Exp. Med. 2012, 209, 1289–1307. [Google Scholar] [CrossRef]
- Li, C.; Du, L.; Ren, Y.; Liu, X.; Jiao, Q.; Cui, D.; Wen, M.; Wang, C.; Wei, G.; Wang, Y.; et al. SKP2 promotes breast cancer tumorigenesis and radiation tolerance through PDCD4 ubiquitination. J. Exp. Clin. Cancer Res. 2019, 38, 76. [Google Scholar] [CrossRef]
- Ma, J.; Chang, K.; Peng, J.; Shi, Q.; Gan, H.; Gao, K.; Feng, K.; Xu, F.; Zhang, H.; Dai, B.; et al. SPOP promotes ATF2 ubiquitination and degradation to suppress prostate cancer progression. J. Exp. Clin. Cancer Res. 2018, 37, 145. [Google Scholar] [CrossRef]
- Fong, K.-w.; Zhao, J.C.; Song, B.; Zheng, B.; Yu, J. TRIM28 protects TRIM24 from SPOP-mediated degradation and promotes prostate cancer progression. Nat. Commun. 2018, 9, 5007. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.J.; Raghu, D.; Chan, A.L.; Gulati, T.; Lambeth, L.; Takano, E.; Herold, M.J.; Hagekyriakou, J.; Vessella, R.L.; Fedele, C.; et al. Restoration of tumor suppression in prostate cancer by targeting the E3 ligase E6AP. Oncogene 2016, 35, 6235–6245. [Google Scholar] [CrossRef] [PubMed]
- Raghu, D.; Paul, P.J.; Gulati, T.; Deb, S.; Khoo, C.; Russo, A.; Gallo, E.; Blandino, G.; Chan, A.-L.; Takano, E.; et al. E6AP promotes prostate cancer by reducing p27 expression. Oncotarget 2017, 8, 42939–42948. [Google Scholar] [CrossRef] [PubMed]
- Gamell, C.; Bandilovska, I.; Gulati, T.; Kogan, A.; Lim, S.C.; Kovacevic, Z.; Takano, E.A.; Timpone, C.; Agupitan, A.D.; Litchfield, C.; et al. E6AP Promotes a Metastatic Phenotype in Prostate Cancer. iScience 2019, 22, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Venuto, S.; Merla, G. E3 Ubiquitin Ligase TRIM Proteins, Cell Cycle and Mitosis. Cells 2019, 8, 510. [Google Scholar] [CrossRef]
- Zhu, L.; Qin, C.; Li, T.; Ma, X.; Qiu, Y.; Lin, Y.; Ma, D.; Qin, Z.; Sun, C.; Shen, X.; et al. The E3 ubiquitin ligase TRIM7 suppressed hepatocellular carcinoma progression by directly targeting Src protein. Cell Death Differ. 2019. [Google Scholar] [CrossRef]
- Jin, J.; Lu, Z.; Wang, X.; Liu, Y.; Han, T.; Wang, Y.; Wang, T.; Gan, M.; Xie, C.; Wang, J.; et al. E3 ubiquitin ligase TRIM7 negatively regulates NF-kappa B signaling pathway by degrading p65 in lung cancer. Cell. Signal. 2020, 69. [Google Scholar] [CrossRef]
- Chakraborty, A.; Diefenbacher, M.E.; Mylona, A.; Kassel, O.; Behrens, A. The E3 ubiquitin ligase Trim7 mediates c-Jun/AP-1 activation by Ras signalling. Nat. Commun. 2015, 6, 6782. [Google Scholar] [CrossRef]
- Yang, D.; Cheng, D.; Tu, Q.; Yang, H.; Sun, B.; Yan, L.; Dai, H.; Luo, J.; Mao, B.; Cao, Y.; et al. HUWE1 controls the development of non-small cell lung cancer through down-regulation of p53. Theranostics 2018, 8, 3517–3529. [Google Scholar] [CrossRef]
- Gong, X.; Du, D.; Deng, Y.; Zhou, Y.; Sun, L.; Yuan, S. The structure and regulation of the E3 ubiquitin ligase HUWE1 and its biological functions in cancer. Investig. New Drugs 2020, 38, 515–524. [Google Scholar] [CrossRef]
- Fujiwara, M.; Marusawa, H.; Wang, H.Q.; Iwai, A.; Ikeuchi, K.; Imai, Y.; Kataoka, A.; Nukina, N.; Takahashi, R.; Chiba, T. Parkin as a tumor suppressor gene for hepatocellular carcinoma. Oncogene 2008, 27, 6002–6011. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.C.; Xu, L.; Chen, Y.; Yan, H.; Hazawa, M.; Doan, N.; Said, J.W.; Ding, L.W.; Liu, L.Z.; Yang, H.; et al. Genomic and Functional Analysis of the E3 Ligase PARK2 in Glioma. Cancer Res. 2015, 75, 1815–1827. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Ghosh, M.K. Cell Death and Deubiquitinases: Perspectives in Cancer. Biomed. Res. Int. 2014, 2014, 435197. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chen, X.; Xu, D.; Yang, L.; Yang, Z.; Yang, Q.; Yan, D.; Zhang, P.; Feng, D.; Liu, J. Autophagy Induced by Proteasomal DUB Inhibitor NiPT Restricts NiPT-Mediated Cancer Cell Death. Front. Oncol. 2020, 10, 348. [Google Scholar] [CrossRef] [PubMed]
- Fraile, J.M.; Quesada, V.; Rodriguez, D.; Freije, J.M.; Lopez-Otin, C. Deubiquitinases in cancer: New functions and therapeutic options. Oncogene 2012, 31, 2373–2388. [Google Scholar] [CrossRef]
- Akhavantabasi, S.; Akman, H.B.; Sapmaz, A.; Keller, J.; Petty, E.M.; Erson, A.E. USP32 is an active, membrane-bound ubiquitin protease overexpressed in breast cancers. Mamm. Genome 2010, 21, 388–397. [Google Scholar] [CrossRef]
- Wei, R.; Liu, X.; Yu, W.; Yang, T.; Cai, W.; Liu, J.; Huang, X.; Xu, G.-t.; Zhao, S.; Yang, J.; et al. Deubiquitinases in cancer. Oncotarget 2015, 6, 12872–12889. [Google Scholar] [CrossRef]
- Lee, J.E.; Park, C.M.; Kim, J.H. USP7 deubiquitinates and stabilizes EZH2 in prostate cancer cells. Genet. Mol. Biol. 2020, 43, e20190338. [Google Scholar] [CrossRef]
- Tan, Y.; Zhou, G.; Wang, X.; Chen, W.; Gao, H. USP18 promotes breast cancer growth by upregulating EGFR and activating the AKT/Skp2 pathway. Int. J. Oncol. 2018, 53, 371–383. [Google Scholar] [CrossRef]
- Pan, J.; Deng, Q.; Jiang, C.; Wang, X.; Niu, T.; Li, H.; Chen, T.; Jin, J.; Pan, W.; Cai, X.; et al. USP37 directly deubiquitinates and stabilizes c-Myc in lung cancer. Oncogene 2015, 34, 3957–3967. [Google Scholar] [CrossRef]
- Diefenbacher, M.E.; Chakraborty, A.; Blake, S.M.; Mitter, R.; Popov, N.; Eilers, M.; Behrens, A. Usp28 counteracts Fbw7 in intestinal homeostasis and cancer. Cancer Res. 2015, 75, 1181–1186. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Ramakrishna, S.; Kim, K.S. Critical Roles of Deubiquitinating Enzymes in the Nervous System and Neurodegenerative Disorders. Mol. Cells 2020. [Google Scholar] [CrossRef]
- Yi, L.; Cui, Y.; Xu, Q.; Jiang, Y. Stabilization of LSD1 by deubiquitinating enzyme USP7 promotes glioblastoma cell tumorigenesis and metastasis through suppression of the p53 signaling pathway. Oncol. Rep. 2016, 36, 2935–2945. [Google Scholar] [CrossRef] [PubMed]
- Ding, K.; Ji, J.; Zhang, X.; Huang, B.; Chen, A.; Zhang, D.; Li, X.; Wang, X.; Wang, J. RNA splicing factor USP39 promotes glioma progression by inducing TAZ mRNA maturation. Oncogene 2019, 38, 6414–6428. [Google Scholar] [CrossRef] [PubMed]
- Sha, B.; Chen, X.; Wu, H.; Li, M.; Shi, J.; Wang, L.; Liu, X.; Chen, P.; Hu, T.; Li, P. Deubiquitylatinase inhibitor b-AP15 induces c-Myc-Noxa-mediated apoptosis in esophageal squamous cell carcinoma. Apoptosis 2019, 24, 826–836. [Google Scholar] [CrossRef]
- Narayanan, S.; Cai, C.Y.; Assaraf, Y.G.; Guo, H.Q.; Cui, Q.; Wei, L.; Huang, J.J.; Ashby, C.R., Jr.; Chen, Z.S. Targeting the ubiquitin-proteasome pathway to overcome anti-cancer drug resistance. Drug Resist. Updat 2020, 48, 100663. [Google Scholar] [CrossRef]
- Fu, Y.; Ma, G.; Liu, G.; Li, B.; Li, H.; Hao, X.; Liu, L. USP14 as a novel prognostic marker promotes cisplatin resistance via Akt/ERK signaling pathways in gastric cancer. Cancer Med. 2018, 7, 5577–5588. [Google Scholar] [CrossRef]
- Zhao, C.; Chen, X.; Zang, D.; Lan, X.; Liao, S.; Yang, C.; Zhang, P.; Wu, J.; Li, X.; Liu, N.; et al. A novel nickel complex works as a proteasomal deubiquitinase inhibitor for cancer therapy. Oncogene 2016, 35, 5916–5927. [Google Scholar] [CrossRef]
- Jin, Y.; Cheng, H.; Cao, J.; Shen, W. MicroRNA 32 promotes cell proliferation, migration, and suppresses apoptosis in colon cancer cells by targeting OTU domain containing 3. J. Cell Biochem. 2019, 120, 18629–18639. [Google Scholar] [CrossRef]
- Zhou, Y.; Wu, J.; Fu, X.; Du, W.; Zhou, L.; Meng, X.; Yu, H.; Lin, J.; Ye, W.; Liu, J.; et al. OTUB1 promotes metastasis and serves as a marker of poor prognosis in colorectal cancer. Mol. Cancer 2014, 13, 258. [Google Scholar] [CrossRef]
- Zhang, P.; Li, C.; Li, H.; Yuan, L.; Dai, H.; Peng, Z.; Deng, Z.; Chang, Z.; Cui, C.P.; Zhang, L. Ubiquitin ligase CHIP regulates OTUD3 stability and suppresses tumour metastasis in lung cancer. Cell Death Differ. 2020. [Google Scholar] [CrossRef]
- Zhao, X.; Su, X.; Cao, L.; Xie, T.; Chen, Q.; Li, J.; Xu, R.; Jiang, C. OTUD4: A Potential Prognosis Biomarker for Multiple Human Cancers. Cancer Manag. Res. 2020, 12, 1503–1512. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, A.A.; Zuvanich, E.G.; Khan, D.A.; Mushtaq, S.; Silswal, N.; Qureshi, N. Proteasome inhibitors modulate anticancer and anti-proliferative properties via NF-kB signaling, and ubiquitin-proteasome pathways in cancer cell lines of different organs. Lipids Health Dis. 2018, 17, 62. [Google Scholar] [CrossRef]
- Wang, Z.; Wan, L.; Zhong, J.; Inuzuka, H.; Liu, P.; Sarkar, F.H.; Wei, W. Cdc20: A potential novel therapeutic target for cancer treatment. Curr. Pharm. Des. 2013, 19, 3210–3214. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Higuera, I.; Manchado, E.; Dubus, P.; Canamero, M.; Mendez, J.; Moreno, S.; Malumbres, M. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat. Cell Biol. 2008, 10, 802–811. [Google Scholar] [CrossRef] [PubMed]
- Lehman, N.L.; Tibshirani, R.; Hsu, J.Y.; Natkunam, Y.; Harris, B.T.; West, R.B.; Masek, M.A.; Montgomery, K.; van de Rijn, M.; Jackson, P.K. Oncogenic regulators and substrates of the anaphase promoting complex/cyclosome are frequently overexpressed in malignant tumors. Am. J. Pathol. 2007, 170, 1793–1805. [Google Scholar] [CrossRef][Green Version]
- Belaïdouni, N.; Peuchmaur, M.; Perret, C.; Florentin, A.; Benarous, R.; Besnard-Guérin, C. Overexpression of human βTrCP1 deleted of its F box induces tumorigenesis in transgenic mice. Oncogene 2005, 24, 2271–2276. [Google Scholar] [CrossRef][Green Version]
- Frescas, D.; Pagano, M. Deregulated proteolysis by the F-box proteins SKP2 and β-TrCP: Tipping the scales of cancer. Nat. Rev. Cancer 2008, 8, 438–449. [Google Scholar] [CrossRef]
- Chan, C.H.; Morrow, J.K.; Li, C.F.; Gao, Y.; Jin, G.; Moten, A.; Stagg, L.J.; Ladbury, J.E.; Cai, Z.; Xu, D.; et al. Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell 2013, 154, 556–568. [Google Scholar] [CrossRef]
- Wang, G.; Chan, C.H.; Gao, Y.; Lin, H.K. Novel roles of Skp2 E3 ligase in cellular senescence, cancer progression, and metastasis. Chin. J. Cancer 2012, 31, 169–177. [Google Scholar] [CrossRef]
- Geng, C.; He, B.; Xu, L.; Barbieri, C.E.; Eedunuri, V.K.; Chew, S.A.; Zimmermann, M.; Bond, R.; Shou, J.; Li, C.; et al. Prostate cancer-associated mutations in speckle-type POZ protein (SPOP) regulate steroid receptor coactivator 3 protein turnover. Proc. Natl. Acad. Sci. USA 2019, 116, 14386–14387. [Google Scholar] [CrossRef] [PubMed]
- Geng, C.; Kaochar, S.; Li, M.; Rajapakshe, K.; Fiskus, W.; Dong, J.; Foley, C.; Dong, B.; Zhang, L.; Kwon, O.J.; et al. SPOP regulates prostate epithelial cell proliferation and promotes ubiquitination and turnover of c-MYC oncoprotein. Oncogene 2017, 36, 4767–4777. [Google Scholar] [CrossRef] [PubMed]
- Gamell, C.; Gulati, T.; Levav-Cohen, Y.; Young, R.J.; Do, H.; Pilling, P.; Takano, E.; Watkins, N.; Fox, S.B.; Russell, P.; et al. Reduced abundance of the E3 ubiquitin ligase E6AP contributes to decreased expression of the INK4/ARF locus in non-small cell lung cancer. Sci. Signal. 2017, 10. [Google Scholar] [CrossRef]
- Mortensen, F.; Schneider, D.; Barbic, T.; Sladewska-Marquardt, A.; Kuhnle, S.; Marx, A.; Scheffner, M. Role of ubiquitin and the HPV E6 oncoprotein in E6AP-mediated ubiquitination. Proc. Natl. Acad. Sci. USA 2015, 112, 9872–9877. [Google Scholar] [CrossRef]
- Senft, D.; Qi, J.; Ronai, Z.A. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat. Rev. Cancer 2018, 18, 69–88. [Google Scholar] [CrossRef] [PubMed]
- Adhikary, S.; Marinoni, F.; Hock, A.; Hulleman, E.; Popov, N.; Beier, R.; Bernard, S.; Quarto, M.; Capra, M.; Goettig, S.; et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell 2005, 123, 409–421. [Google Scholar] [CrossRef]
- Su, C.; Wang, T.; Zhao, J.; Cheng, J.; Hou, J. Meta-analysis of gene expression alterations and clinical significance of the HECT domain-containing ubiquitin ligase HUWE1 in cancer. Oncol. Lett. 2019, 18, 2292–2303. [Google Scholar] [CrossRef]
- Xu, L.; Lin, D.C.; Yin, D.; Koeffler, H.P. An emerging role of PARK2 in cancer. J. Mol. Med. (Berl) 2014, 92, 31–42. [Google Scholar] [CrossRef]
- Eichhorn, P.J.; Rodón, L.; Gonzàlez-Juncà, A.; Dirac, A.; Gili, M.; Martínez-Sáez, E.; Aura, C.; Barba, I.; Peg, V.; Prat, A.; et al. USP15 stabilizes TGF-β receptor I and promotes oncogenesis through the activation of TGF-β signaling in glioblastoma. Nat. Med. 2012, 18, 429–435. [Google Scholar] [CrossRef]
- Qin, T.; Li, B.; Feng, X.; Fan, S.; Liu, L.; Liu, D.; Mao, J.; Lu, Y.; Yang, J.; Yu, X.; et al. Abnormally elevated USP37 expression in breast cancer stem cells regulates stemness, epithelial-mesenchymal transition and cisplatin sensitivity. J. Exp. Clin. Cancer Res. 2018, 37, 287. [Google Scholar] [CrossRef]
- Du, T.; Li, H.; Fan, Y.; Yuan, L.; Guo, X.; Zhu, Q.; Yao, Y.; Li, X.; Liu, C.; Yu, X.; et al. The deubiquitylase OTUD3 stabilizes GRP78 and promotes lung tumorigenesis. Nat. Commun. 2019, 10, 2914. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Pozo, A.; Frosch, M.P.; Masliah, E.; Hyman, B.T. Neuropathological alterations in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2011, 1, a006189. [Google Scholar] [CrossRef] [PubMed]
- Dickson, D.W. Parkinson’s disease and parkinsonism: Neuropathology. Cold Spring Harb. Perspect Med. 2012, 2. [Google Scholar] [CrossRef] [PubMed]
- Stancu, I.C.; Ferraiolo, M.; Terwel, D.; Dewachter, I. Tau Interacting Proteins: Gaining Insight into the Roles of Tau in Health and Disease. Adv. Exp. Med. Biol. 2019, 1184, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Huang, T.; Zhang, L.; Zhou, Y.; Luo, H.; Xu, H.; Wang, X. Dysregulation of Ubiquitin-Proteasome System in Neurodegenerative Diseases. Front. Aging Neurosci. 2016, 8, 303. [Google Scholar] [CrossRef] [PubMed]
- Rosen, K.M.; Moussa, C.E.; Lee, H.K.; Kumar, P.; Kitada, T.; Qin, G.; Fu, Q.; Querfurth, H.W. Parkin reverses intracellular beta-amyloid accumulation and its negative effects on proteasome function. J. Neurosci. Res. 2010, 88, 167–178. [Google Scholar] [CrossRef]
- Martinez, A.; Ramirez, J.; Osinalde, N.; Arizmendi, J.M.; Mayor, U. Neuronal proteomic analysis of the ubiquitinated substrates of the disease-linked E3 ligases parkin and Ube3a. BioMed. Res. Int. 2018, 2018, 1–14. [Google Scholar] [CrossRef]
- Kaneko, M.; Koike, H.; Saito, R.; Kitamura, Y.; Okuma, Y.; Nomura, Y. Loss of HRD1-mediated protein degradation causes amyloid precursor protein accumulation and amyloid-beta generation. J. Neurosci. 2010, 30, 3924–3932. [Google Scholar] [CrossRef]
- Saito, R.; Kaneko, M.; Kitamura, Y.; Takata, K.; Kawada, K.; Okuma, Y.; Nomura, Y. Effects of oxidative stress on the solubility of HRD1, a ubiquitin ligase implicated in Alzheimer’s disease. PLoS ONE 2014, 9, e94576. [Google Scholar] [CrossRef]
- Sahara, N.; Murayama, M.; Mizoroki, T.; Urushitani, M.; Imai, Y.; Takahashi, R.; Murata, S.; Tanaka, K.; Takashima, A. In vivo evidence of CHIP up-regulation attenuating tau aggregation. J. Neurochem. 2005, 94, 1254–1263. [Google Scholar] [CrossRef]
- Shimura, H.; Schwartz, D.; Gygi, S.P.; Kosik, K.S. CHIP-Hsc70 complex ubiquitinates phosphorylated tau and enhances cell survival. J. Biol. Chem. 2004, 279, 4869–4876. [Google Scholar] [CrossRef] [PubMed]
- Dickey, C.A.; Yue, M.; Lin, W.L.; Dickson, D.W.; Dunmore, J.H.; Lee, W.C.; Zehr, C.; West, G.; Cao, S.; Clark, A.M.; et al. Deletion of the ubiquitin ligase CHIP leads to the accumulation, but not the aggregation, of both endogenous phospho- and caspase-3-cleaved tau species. J. Neurosci. 2006, 26, 6985–6996. [Google Scholar] [CrossRef] [PubMed]
- Evgen’ev, M.; Bobkova, N.; Krasnov, G.; Garbuz, D.; Funikov, S.; Kudryavtseva, A.; Kulikov, A.; Samokhin, A.; Maltsev, A.; Nesterova, I. The Effect of Human HSP70 Administration on a Mouse Model of Alzheimer’s Disease Strongly Depends on Transgenicity and Age. J. Alzheimer’s Dis. 2019, 67, 1391–1404. [Google Scholar] [CrossRef]
- Lee, S.; Choi, B.R.; Kim, J.; LaFerla, F.M.; Park, J.H.Y.; Han, J.S.; Lee, K.W.; Kim, J. Sulforaphane Upregulates the Heat Shock Protein Co-Chaperone CHIP and Clears Amyloid-β and Tau in a Mouse Model of Alzheimer’s Disease. Mol. Nutr. Food Res. 2018, 62. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Cai, F.; Zhang, S.; Zhang, S.; Song, W. Overexpression of ubiquitin carboxyl-terminal hydrolase L1 (UCHL1) delays Alzheimer’s progression in vivo. Sci. Rep. 2014, 4, 7298. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.-J.; Kim, J.S.; Park, S.M. Ubiquitin C-terminal Hydrolase L1 Regulates Lipid Raft-dependent Endocytosis. Exp. Neurobiol. 2018, 27, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Kumar, R.; Kumar, S.; Singh, A.K.; Hanpude, P.; Jangir, D.; Maiti, T.K. Amyloid aggregates of the deubiquitinase OTUB1 are neurotoxic, suggesting that they contribute to the development of Parkinson’s disease. J. Biol. Chem. 2020, 295, 3466–3484. [Google Scholar] [CrossRef]
- Ortuno, D.; Carlisle, H.J.; Miller, S. Does inactivation of USP14 enhance degradation of proteasomal substrates that are associated with neurodegenerative diseases? F1000Res 2016, 5, 137. [Google Scholar] [CrossRef]
- Zeng, X.S.; Geng, W.S.; Jia, J.J.; Chen, L.; Zhang, P.P. Cellular and Molecular Basis of Neurodegeneration in Parkinson Disease. Front. Aging Neurosci. 2018, 10, 109. [Google Scholar] [CrossRef]
- Kondapalli, C.; Kazlauskaite, A.; Zhang, N.; Woodroof, H.I.; Campbell, D.G.; Gourlay, R.; Burchell, L.; Walden, H.; Macartney, T.J.; Deak, M.; et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012, 2, 120080. [Google Scholar] [CrossRef]
- Narendra, D.; Tanaka, A.; Suen, D.F.; Youle, R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008, 183, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.Y.; Kishinevsky, S.; Mazzulli, J.R.; Graziotto, J.; Mrejeru, A.; Mosharov, E.V.; Puspita, L.; Valiulahi, P.; Sulzer, D.; Milner, T.A.; et al. Parkin and PINK1 Patient iPSC-Derived Midbrain Dopamine Neurons Exhibit Mitochondrial Dysfunction and alpha-Synuclein Accumulation. Stem Cell Rep. 2016, 7, 664–677. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.Y.; Park, H.R.; Lee, S.J.; Lee, S.H.; Kim, J.S.; Jung, Y.S.; Hwang, S.H.; Ha, N.C.; Seol, W.G.; Lee, J.; et al. Elevated TRAF2/6 expression in Parkinson’s disease is caused by the loss of Parkin E3 ligase activity. Lab. Invest. 2013, 93, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Murata, H.; Sakaguchi, M.; Kataoka, K.; Huh, N.H. SARM1 and TRAF6 bind to and stabilize PINK1 on depolarized mitochondria. Mol. Biol. Cell 2013, 24, 2772–2784. [Google Scholar] [CrossRef] [PubMed]
- Matheoud, D.; Sugiura, A.; Bellemare-Pelletier, A.; Laplante, A.; Rondeau, C.; Chemali, M.; Fazel, A.; Bergeron, J.J.; Trudeau, L.E.; Burelle, Y.; et al. Parkinson’s Disease-Related Proteins PINK1 and Parkin Repress Mitochondrial Antigen Presentation. Cell 2016, 166, 314–327. [Google Scholar] [CrossRef] [PubMed]
- Matheoud, D.; Cannon, T.; Voisin, A.; Penttinen, A.M.; Ramet, L.; Fahmy, A.M.; Ducrot, C.; Laplante, A.; Bourque, M.J.; Zhu, L.; et al. Intestinal infection triggers Parkinson’s disease-like symptoms in Pink1 −/− mice. Nature 2019, 571, 565–569. [Google Scholar] [CrossRef]
- Mulherkar, S.A.; Sharma, J.; Jana, N.R. The ubiquitin ligase E6-AP promotes degradation of alpha-synuclein. J. Neurochem. 2009, 110, 1955–1964. [Google Scholar] [CrossRef]
- Liu, X.; Hebron, M.; Shi, W.; Lonskaya, I.; Moussa, C.E.-H. Ubiquitin specific protease-13 independently regulates parkin ubiquitination and alpha-synuclein clearance in alpha-synucleinopathies. Hum. Mol. Genet. 2018, 28, 548–560. [Google Scholar] [CrossRef]
- Bishop, P.; Rubin, P.; Thomson, A.R.; Rocca, D.; Henley, J.M. The ubiquitin C-terminal hydrolase L1 (UCH-L1) C terminus plays a key role in protein stability, but its farnesylation is not required for membrane association in primary neurons. J. Biol. Chem. 2014, 289, 36140–36149. [Google Scholar] [CrossRef]
- Cai, C.-Z.; Zhou, H.-F.; Yuan, N.-N.; Wu, M.-Y.; Lee, S.M.-Y.; Ren, J.-Y.; Su, H.-X.; Lu, J.-J.; Chen, X.-P.; Li, M.; et al. Natural alkaloid harmine promotes degradation of alpha-synuclein via PKA-mediated ubiquitin-proteasome system activation. Phytomedicine 2019, 61, 152842. [Google Scholar] [CrossRef]
- Myöhänen, T.T.; Norrbacka, S.; Savolainen, M.H. Prolyl oligopeptidase inhibition attenuates the toxicity of a proteasomal inhibitor, lactacystin, in the alpha-synuclein overexpressing cell culture. Neurosci. Lett. 2017, 636, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Ortega, Z.; Lucas, J.J. Ubiquitin-proteasome system involvement in Huntington’s disease. Front. Mol. Neurosci. 2014, 7, 77. [Google Scholar] [CrossRef]
- Miller, V.M.; Nelson, R.F.; Gouvion, C.M.; Williams, A.; Rodriguez-Lebron, E.; Harper, S.Q.; Davidson, B.L.; Rebagliati, M.R.; Paulson, H.L. CHIP suppresses polyglutamine aggregation and toxicity in vitro and in vivo. J. Neurosci. 2005, 25, 9152–9161. [Google Scholar] [CrossRef] [PubMed]
- Bhat, K.P.; Yan, S.; Wang, C.E.; Li, S.; Li, X.J. Differential ubiquitination and degradation of huntingtin fragments modulated by ubiquitin-protein ligase E3A. Proc. Natl. Acad. Sci. USA 2014, 111, 5706–5711. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Jin, Z.; Tan, H.; Xu, Q.; Peng, T.; Li, H. Atypical ubiquitination by E3 ligase WWP1 inhibits the proteasome-mediated degradation of mutant huntingtin. Brain Res. 2016, 1643, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Rubio, I.; Rodríguez, J.; Tomas-Zapico, C.; Ruiz, C.; Casarejos, M.; Perucho, J.; Gómez, A.; Rodal, I.; Lucas, J.; Mena, M.; et al. Effects of partial suppression of parkin on huntingtin mutant R6/1 mice. Brain Res. 2009, 1281, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhong, X.; Ballar, P.; Luo, S.; Shen, Y.; Rubinsztein, D.C.; Monteiro, M.J.; Fang, S. Ubiquitin ligase Hrd1 enhances the degradation and suppresses the toxicity of polyglutamine-expanded huntingtin. Exp. Cell Res. 2007, 313, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Rotblat, B.; Southwell, A.L.; Ehrnhoefer, D.E.; Skotte, N.H.; Metzler, M.; Franciosi, S.; Leprivier, G.; Somasekharan, S.P.; Barokas, A.; Deng, Y.; et al. HACE1 reduces oxidative stress and mutant Huntingtin toxicity by promoting the NRF2 response. Proc. Natl. Acad. Sci. USA 2014, 111, 3032–3037. [Google Scholar] [CrossRef]
- Tanji, K.; Mori, F.; Miki, Y.; Utsumi, J.; Sasaki, H.; Kakita, A.; Takahashi, H.; Wakabayashi, K. YOD1 attenuates neurogenic proteotoxicity through its deubiquitinating activity. Neurobiol. Dis. 2018, 112, 14–23. [Google Scholar] [CrossRef]
- Lee, B.H.; Lee, M.J.; Park, S.; Oh, D.C.; Elsasser, S.; Chen, P.C.; Gartner, C.; Dimova, N.; Hanna, J.; Gygi, S.P.; et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 2010, 467, 179–184. [Google Scholar] [CrossRef]
- Hyrskyluoto, A.; Bruelle, C.; Lundh, S.H.; Do, H.T.; Kivinen, J.; Rappou, E.; Reijonen, S.; Waltimo, T.; Petersén, Å.; Lindholm, D.; et al. Ubiquitin-specific protease-14 reduces cellular aggregates and protects against mutant huntingtin-induced cell degeneration: Involvement of the proteasome and ER stress-activated kinase IRE1α. Hum. Mol. Genet. 2014, 23, 5928–5939. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.N.; Her, L.S. The Ubiquitin Receptor ADRM1 Modulates HAP40-Induced Proteasome Activity. Mol. Neurobiol. 2017, 54, 7382–7400. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Hernandez, M.; Valera, A.G.; Moran, M.A.; Gomez-Ramos, P.; Alvarez-Castelao, B.; Castano, J.G.; Hernandez, F.; Lucas, J.J. Inhibition of 26S proteasome activity by huntingtin filaments but not inclusion bodies isolated from mouse and human brain. J. Neurochem. 2006, 98, 1585–1596. [Google Scholar] [CrossRef] [PubMed]
- Schipper-Krom, S.; Juenemann, K.; Jansen, A.H.; Wiemhoefer, A.; van den Nieuwendijk, R.; Smith, D.L.; Hink, M.A.; Bates, G.P.; Overkleeft, H.; Ovaa, H.; et al. Dynamic recruitment of active proteasomes into polyglutamine initiated inclusion bodies. FEBS Lett. 2014, 588, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Aki, D.; Li, Q.; Li, H.; Liu, Y.C.; Lee, J.H. Immune regulation by protein ubiquitination: Roles of the E3 ligases VHL and Itch. Protein Cell 2019, 10, 395–404. [Google Scholar] [CrossRef]
- Wang, A.; Zhu, F.; Liang, R.; Li, D.; Li, B. Regulation of T cell differentiation and function by ubiquitin-specific proteases. Cell Immunol. 2019, 340, 103922. [Google Scholar] [CrossRef]
- Wu, Y.; Kang, J.; Zhang, L.; Liang, Z.; Tang, X.; Yan, Y.; Qian, H.; Zhang, X.; Xu, W.; Mao, F. Ubiquitination regulation of inflammatory responses through NF-κB pathway. Am. J. Transl. Res. 2018, 10, 881–891. [Google Scholar]
- Thien, C.B.; Langdon, W.Y. c-Cbl and Cbl-b ubiquitin ligases: Substrate diversity and the negative regulation of signalling responses. Biochem. J. 2005, 391, 153–166. [Google Scholar] [CrossRef]
- Dou, H.; Buetow, L.; Hock, A.; Sibbet, G.J.; Vousden, K.H.; Huang, D.T. Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl. Nat. Struct. Mol. Biol. 2012, 19, 184–192. [Google Scholar] [CrossRef]
- Huang, F.; Kitaura, Y.; Jang, I.; Naramura, M.; Kole, H.H.; Liu, L.; Qin, H.; Schlissel, M.S.; Gu, H. Establishment of the major compatibility complex-dependent development of CD4+ and CD8+ T cells by the Cbl family proteins. Immunity 2006, 25, 571–581. [Google Scholar] [CrossRef]
- Kitaura, Y.; Jang, I.K.; Wang, Y.; Han, Y.C.; Inazu, T.; Cadera, E.J.; Schlissel, M.; Hardy, R.R.; Gu, H. Control of the B cell-intrinsic tolerance programs by ubiquitin ligases Cbl and Cbl-b. Immunity 2007, 26, 567–578. [Google Scholar] [CrossRef] [PubMed]
- Lyle, C.; Richards, S.; Yasuda, K.; Napoleon, M.A.; Walker, J.; Arinze, N.; Belghasem, M.; Vellard, I.; Yin, W.; Ravid, J.D.; et al. c-Cbl targets PD-1 in immune cells for proteasomal degradation and modulates colorectal tumor growth. Sci. Rep. 2019, 9, 20257. [Google Scholar] [CrossRef] [PubMed]
- Abe, T.; Hirasaka, K.; Kohno, S.; Ochi, A.; Yamagishi, N.; Ohno, A.; Teshima-Kondo, S.; Nikawa, T. Ubiquitin ligase Cbl-b and obesity-induced insulin resistance. Endocr. J. 2014, 61, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Abe, T.; Hirasaka, K.; Nikawa, T. Involvement of Cbl-b-mediated macrophage inactivation in insulin resistance. World J. Diabetes 2017, 8, 97. [Google Scholar] [CrossRef]
- Huang, H.; Jeon, M.S.; Liao, L.; Yang, C.; Elly, C.; Yates, J.R., III; Liu, Y.C. K33-linked polyubiquitination of T cell receptor-zeta regulates proteolysis-independent T cell signaling. Immunity 2010, 33, 60–70. [Google Scholar] [CrossRef]
- O’Connor, H.F.; Lyon, N.; Leung, J.W.; Agarwal, P.; Swaim, C.D.; Miller, K.M.; Huibregtse, J.M. Ubiquitin-Activated Interaction Traps (UBAITs) identify E3 ligase binding partners. EMBO Rep. 2015, 16, 1699–1712. [Google Scholar] [CrossRef]
- Maxwell, P.H.; Wiesener, M.S.; Chang, G.W.; Clifford, S.C.; Vaux, E.C.; Cockman, M.E.; Wykoff, C.C.; Pugh, C.W.; Maher, E.R.; Ratcliffe, P.J. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 1999, 399, 271–275. [Google Scholar] [CrossRef]
- Dang, E.V.; Barbi, J.; Yang, H.Y.; Jinasena, D.; Yu, H.; Zheng, Y.; Bordman, Z.; Fu, J.; Kim, Y.; Yen, H.R.; et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell 2011, 146, 772–784. [Google Scholar] [CrossRef]
- Lee, J.H.; Elly, C.; Park, Y.; Liu, Y.C. E3 Ubiquitin Ligase VHL Regulates Hypoxia-Inducible Factor-1α to Maintain Regulatory T Cell Stability and Suppressive Capacity. Immunity 2015, 42, 1062–1074. [Google Scholar] [CrossRef]
- Izquierdo, H.M.; Brandi, P.; Gómez, M.J.; Conde-Garrosa, R.; Priego, E.; Enamorado, M.; Martínez-Cano, S.; Sánchez, I.; Conejero, L.; Jimenez-Carretero, D.; et al. Von Hippel-Lindau Protein Is Required for Optimal Alveolar Macrophage Terminal Differentiation, Self-Renewal, and Function. Cell Rep. 2018, 24, 1738–1746. [Google Scholar] [CrossRef]
- Palazon, A.; Goldrath, A.W.; Nizet, V.; Johnson, R.S. HIF transcription factors, inflammation, and immunity. Immunity 2014, 41, 518–528. [Google Scholar] [CrossRef] [PubMed]
- Labrousse-Arias, D.; Martínez-Alonso, E.; Corral-Escariz, M.; Bienes-Martínez, R.; Berridy, J.; Serrano-Oviedo, L.; Conde, E.; García-Bermejo, M.L.; Giménez-Bachs, J.M.; Salinas-Sánchez, A.S.; et al. VHL promotes immune response against renal cell carcinoma via NF-κB-dependent regulation of VCAM-1. J. Cell Biol. 2017, 216, 835–847. [Google Scholar] [CrossRef] [PubMed]
- Trotta, A.M.; Santagata, S.; Zanotta, S.; D’Alterio, C.; Napolitano, M.; Rea, G.; Camerlingo, R.; Esposito, F.; Lamantia, E.; Anniciello, A.; et al. Mutated von Hippel-Lindau-renal cell carcinoma (RCC) promotes patients specific natural killer (NK) cytotoxicity. J. Exp. Clin. Cancer Res. 2018, 37, 297. [Google Scholar] [CrossRef] [PubMed]
- Quesada, V.; Diaz-Perales, A.; Gutierrez-Fernandez, A.; Garabaya, C.; Cal, S.; Lopez-Otin, C. Cloning and enzymatic analysis of 22 novel human ubiquitin-specific proteases. Biochem. Biophys. Res. Commun. 2004, 314, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Xu, P.; Han, L.; Guo, Z.; Wang, X.; Chen, Z.; Nie, J.; Yin, S.; Piccioni, M.; Tsun, A.; et al. Cutting Edge: Ubiquitin-Specific Protease 4 Promotes Th17 Cell Function under Inflammation by Deubiquitinating and Stabilizing RORγt. J. Immunol. 2015, 194, 4094–4097. [Google Scholar] [CrossRef] [PubMed]
- Dufner, A.; Kisser, A.; Niendorf, S.; Basters, A.; Reissig, S.; Schonle, A.; Aichem, A.; Kurz, T.; Schlosser, A.; Yablonski, D.; et al. The ubiquitin-specific protease USP8 is critical for the development and homeostasis of T cells. Nat. Immunol. 2015, 16, 950–960. [Google Scholar] [CrossRef]
- Huang, C.; Shi, Y.; Zhao, Y. USP8 mutation in Cushing’s disease. Oncotarget 2015, 6, 18240–18241. [Google Scholar] [CrossRef]
- Park, Y.; Jin, H.S.; Liu, Y.C. Regulation of T cell function by the ubiquitin-specific protease USP9X via modulating the Carma1-Bcl10-Malt1 complex. Proc. Natl. Acad. Sci. USA 2013, 110, 9433–9438. [Google Scholar] [CrossRef]
- Jahan, A.S.; Lestra, M.; Swee, L.K.; Fan, Y.; Lamers, M.M.; Tafesse, F.G.; Theile, C.S.; Spooner, E.; Bruzzone, R.; Ploegh, H.L.; et al. Usp12 stabilizes the T-cell receptor complex at the cell surface during signaling. Proc. Natl. Acad. Sci. USA 2016, 113, E705–E714. [Google Scholar] [CrossRef]
- Wu, X.; Lei, C.; Xia, T.; Zhong, X.; Yang, Q.; Shu, H.B. Regulation of TRIF-mediated innate immune response by K27-linked polyubiquitination and deubiquitination. Nat. Commun. 2019, 10, 4115. [Google Scholar] [CrossRef]
- Bignell, G.R.; Warren, W.; Seal, S.; Takahashi, M.; Rapley, E.; Barfoot, R.; Green, H.; Brown, C.; Biggs, P.J.; Lakhani, S.R.; et al. Identification of the familial cylindromatosis tumour-suppressor gene. Nat. Genet. 2000, 25, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Tsagaratou, A.; Trompouki, E.; Grammenoudi, S.; Kontoyiannis, D.L.; Mosialos, G. Thymocyte-specific truncation of the deubiquitinating domain of CYLD impairs positive selection in a NF-kappaB essential modulator-dependent manner. J. Immunol. 2010, 185, 2032–2043. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Stirling, B.; Temmerman, S.T.; Ma, C.A.; Fuss, I.J.; Derry, J.M.; Jain, A. Impaired regulation of NF-kappaB and increased susceptibility to colitis-associated tumorigenesis in CYLD-deficient mice. J. Clin. Investig. 2006, 116, 3042–3049. [Google Scholar] [CrossRef] [PubMed]
- Reiley, W.W.; Jin, W.; Lee, A.J.; Wright, A.; Wu, X.; Tewalt, E.F.; Leonard, T.O.; Norbury, C.C.; Fitzpatrick, L.; Zhang, M.; et al. Deubiquitinating enzyme CYLD negatively regulates the ubiquitin-dependent kinase Tak1 and prevents abnormal T cell responses. J. Exp. Med. 2007, 204, 1475–1485. [Google Scholar] [CrossRef]
- Lork, M.; Verhelst, K.; Beyaert, R. CYLD, A20 and OTULIN deubiquitinases in NF-κB signaling and cell death: So similar, yet so different. Cell Death Differ. 2017, 24, 1172–1183. [Google Scholar] [CrossRef]
- Jin, Y.J.; Wang, S.; Cho, J.; Selim, M.A.; Wright, T.; Mosialos, G.; Zhang, J.Y. Epidermal CYLD inactivation sensitizes mice to the development of sebaceous and basaloid skin tumors. JCI Insight 2016, 1. [Google Scholar] [CrossRef]
- Nikolaou, K.; Tsagaratou, A.; Eftychi, C.; Kollias, G.; Mosialos, G.; Talianidis, I. Inactivation of the Deubiquitinase CYLD in Hepatocytes Causes Apoptosis, Inflammation, Fibrosis, and Cancer. Cancer Cell 2012, 21, 738–750. [Google Scholar] [CrossRef]
- Cleynen, I.; Vazeille, E.; Artieda, M.; Verspaget, H.W.; Szczypiorska, M.; Bringer, M.A.; Lakatos, P.L.; Seibold, F.; Parnell, K.; Weersma, R.K.; et al. Genetic and microbial factors modulating the ubiquitin proteasome system in inflammatory bowel disease. Gut 2014, 63, 1265–1274. [Google Scholar] [CrossRef]
- Cascio, P.; Hilton, C.; Kisselev, A.F.; Rock, K.L.; Goldberg, A.L. 26S proteasomes and immunoproteasomes produce mainly N-extended versions of an antigenic peptide. EMBO J. 2001, 20, 2357–2366. [Google Scholar] [CrossRef]
- Lin, H.; Li, S.; Shu, H.-B. The Membrane-Associated MARCH E3 Ligase Family: Emerging Roles in Immune Regulation. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef]
- Egerer, K.; Kuckelkorn, U.; Rudolph, P.E.; Rückert, J.C.; Dörner, T.; Burmester, G.R.; Kloetzel, P.M.; Feist, E. Circulating proteasomes are markers of cell damage and immunologic activity in autoimmune diseases. J. Rheumatol. 2002, 29, 2045–2052. [Google Scholar] [PubMed]
- Xiao, F.; Lin, X.; Tian, J.; Wang, X.; Chen, Q.; Rui, K.; Ma, J.; Wang, S.; Wang, Q.; Wang, X.; et al. Proteasome inhibition suppresses Th17 cell generation and ameliorates autoimmune development in experimental Sjögren’s syndrome. Cell Mol. Immunol. 2017, 14, 924–934. [Google Scholar] [CrossRef] [PubMed]
- Landré, V.; Rotblat, B.; Melino, S.; Bernassola, F.; Melino, G. Screening for E3-ubiquitin ligase inhibitors: Challenges and opportunities. Oncotarget 2014, 5, 7988–8013. [Google Scholar] [CrossRef] [PubMed]
- Harrigan, J.A.; Jacq, X.; Martin, N.M.; Jackson, S.P. Deubiquitylating enzymes and drug discovery: Emerging opportunities. Nat. Rev. Drug Discov. 2018, 17, 57–78. [Google Scholar] [CrossRef] [PubMed]
- Thibaudeau, T.A.; Smith, D.M. A Practical Review of Proteasome Pharmacology. Pharmacol. Rev. 2019, 71, 170–197. [Google Scholar] [CrossRef]
| Compounds | Targeted E2 Enzyme | References |
|---|---|---|
| CC0651 | Ube2R1 | [26] |
| Leucettamol A | Ubc13–Uev1A interaction | [27] |
| Manadosterols A and B | Ubc13–Uev1A interaction | [28] |
| BAY 11-7082 | Ubc13 | [29] |
| UBC12N26 | UBC12–NAE binding | [30] |
| Sumoylation Inhibitors | Ubc9 | [31] |
| Suramin | Cdc34–CRL interaction | [32] |
| NSC697923 | Ube2N | [29] |
| Honokiol | UbcH8 | [33] |
| Triazines Compounds | Rad6B | [34] |
| E3s and DUBs | Disease Association | Related Gene Expression | Ref |
|---|---|---|---|
| FBW7 | Hepatocellular carcinoma, colorectal cancer, esophageal squamous cell carcinoma | Underexpressed | [44,45] |
| MDM2 | Breast cancer | Overexpressed | [50] |
| Cdc20 | Breast, pancreatic cancer | Overexpressed | [94] |
| APC/C(Cdh1) | Breast cancer, melanoma | Downregulated | [95,96] |
| βTrCP | Breast, prostate, lung cancer | Expression depends on tissue type | [97,98] |
| Skp2 | Breast cancer, prostate cancer | Overexpressed | [99,100] |
| SPOP | Prostate cancer | Downregulated | [101,102] |
| E6AP | Prostate cancer, cervical and lung cancer | Expression depends on tissue type | [103,104] |
| TRIM7 | Lung cancer | Expression depends on tissue type | [67,68] |
| HUWE1 | Lung, breast, and colorectal carcinoma | Expression depends on tissue type | [105,106,107] |
| Park2 | Glioma, hepatocellular carcinoma, and lymphoma | Underexpressed | [71,108] |
| USP7 | Prostate cancer, brain tumors | Overexpressed | [78,82] |
| USP14 | Gastric cancer | Overexpressed | [87] |
| USP15 | Breast Cancer | Overexpressed | [109] |
| USP18 | Breast cancer | Overexpressed | [79] |
| USP32 | Breast cancer | Overexpressed | [76] |
| USP37 | Breast and lung cancer | Overexpressed | [80,110] |
| USP39 | Glioma | Overexpressed | [84] |
| OTUD3 | Colorectal and lung cancer | Expression depends on tissue type | [89,91,111] |
| OTUB1 | Colorectal cancer | Overexpressed | [90] |
| OTUD4 | Breast, liver, and lung cancers | Overexpressed | [92] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Celebi, G.; Kesim, H.; Ozer, E.; Kutlu, O. The Effect of Dysfunctional Ubiquitin Enzymes in the Pathogenesis of Most Common Diseases. Int. J. Mol. Sci. 2020, 21, 6335. https://doi.org/10.3390/ijms21176335
Celebi G, Kesim H, Ozer E, Kutlu O. The Effect of Dysfunctional Ubiquitin Enzymes in the Pathogenesis of Most Common Diseases. International Journal of Molecular Sciences. 2020; 21(17):6335. https://doi.org/10.3390/ijms21176335
Chicago/Turabian StyleCelebi, Gizem, Hale Kesim, Ebru Ozer, and Ozlem Kutlu. 2020. "The Effect of Dysfunctional Ubiquitin Enzymes in the Pathogenesis of Most Common Diseases" International Journal of Molecular Sciences 21, no. 17: 6335. https://doi.org/10.3390/ijms21176335
APA StyleCelebi, G., Kesim, H., Ozer, E., & Kutlu, O. (2020). The Effect of Dysfunctional Ubiquitin Enzymes in the Pathogenesis of Most Common Diseases. International Journal of Molecular Sciences, 21(17), 6335. https://doi.org/10.3390/ijms21176335





