The Cruciality of Single Amino Acid Replacement for the Spectral Tuning of Biliverdin-Binding Cyanobacteriochromes
Abstract
1. Introduction
2. Results
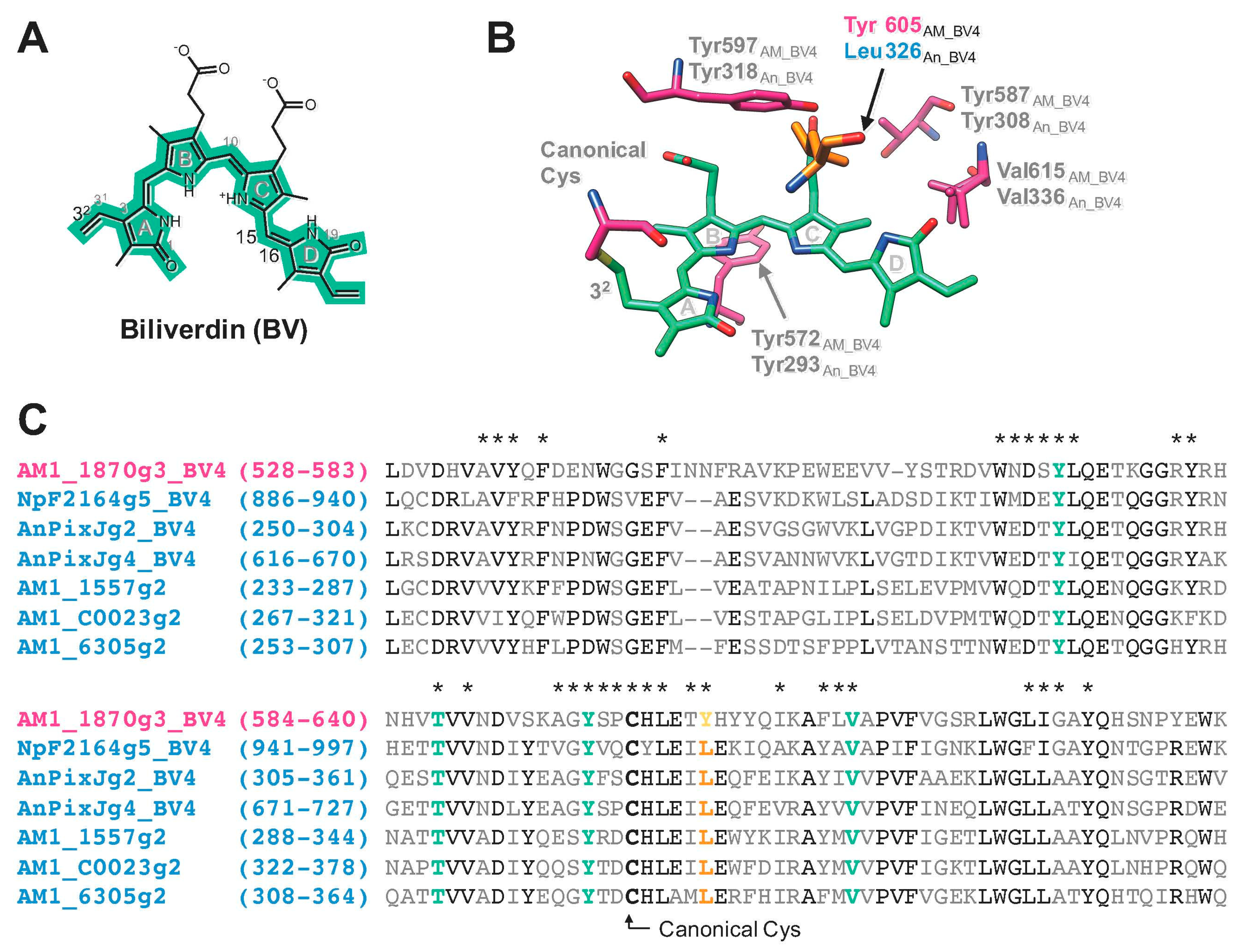
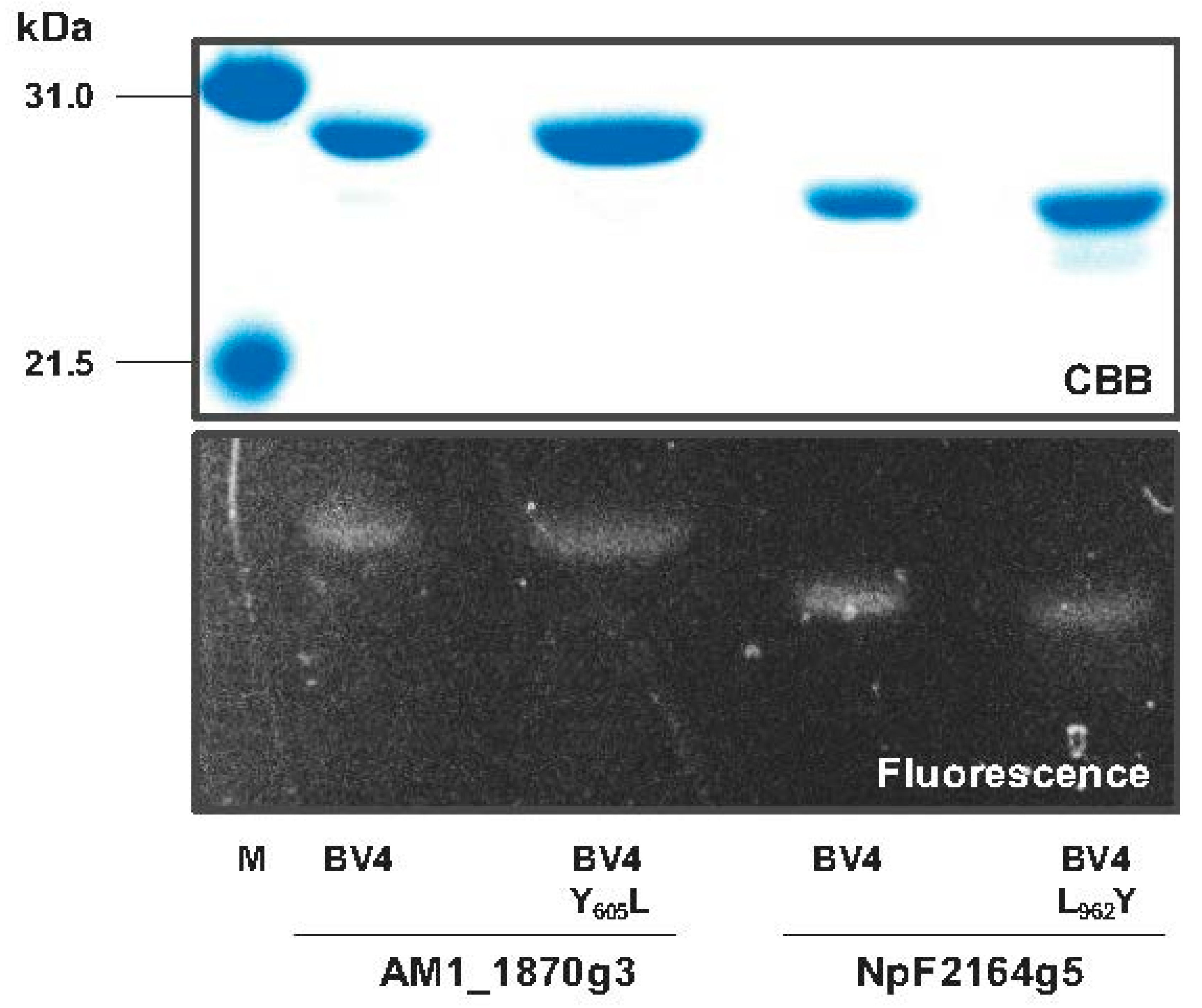
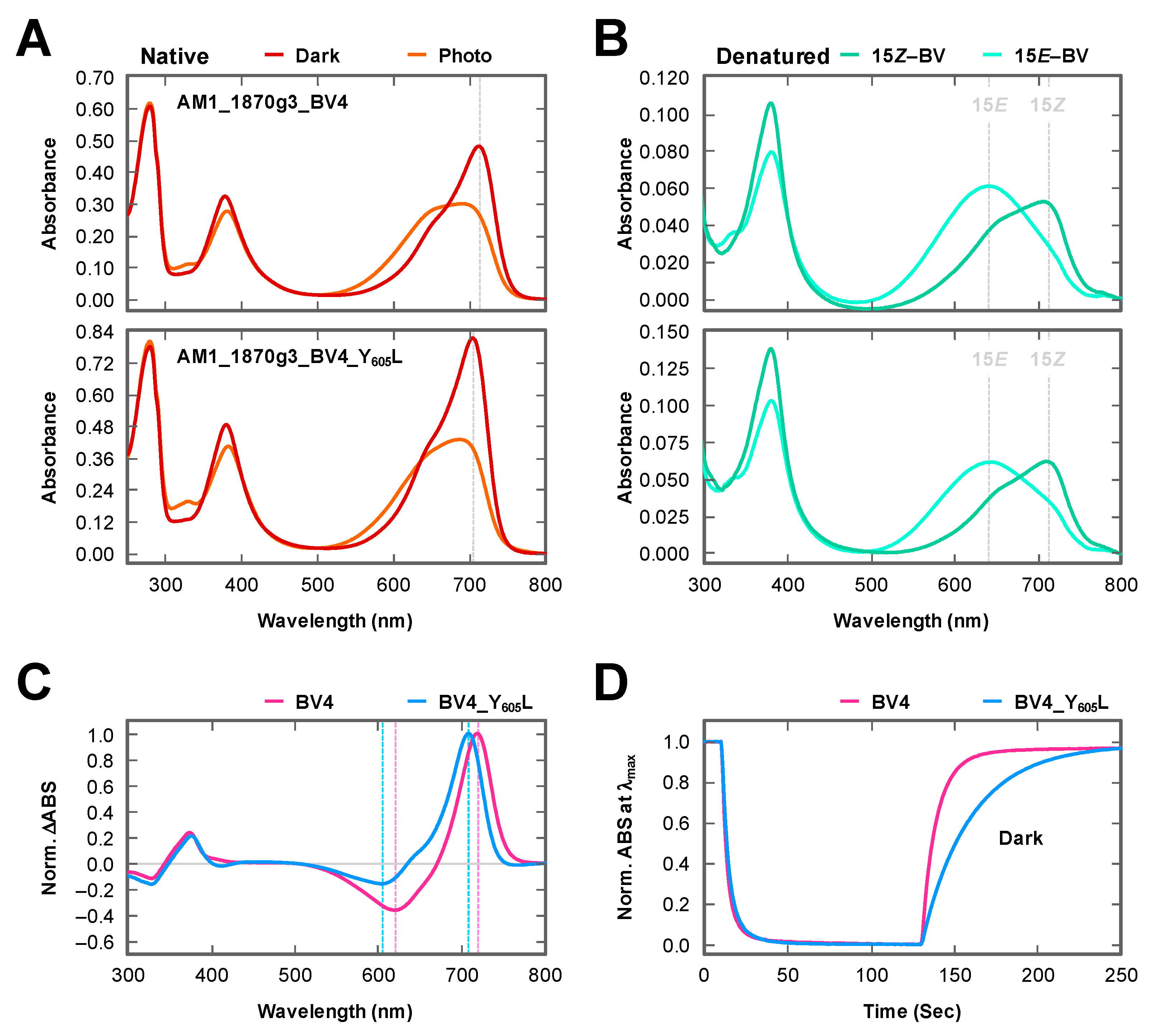
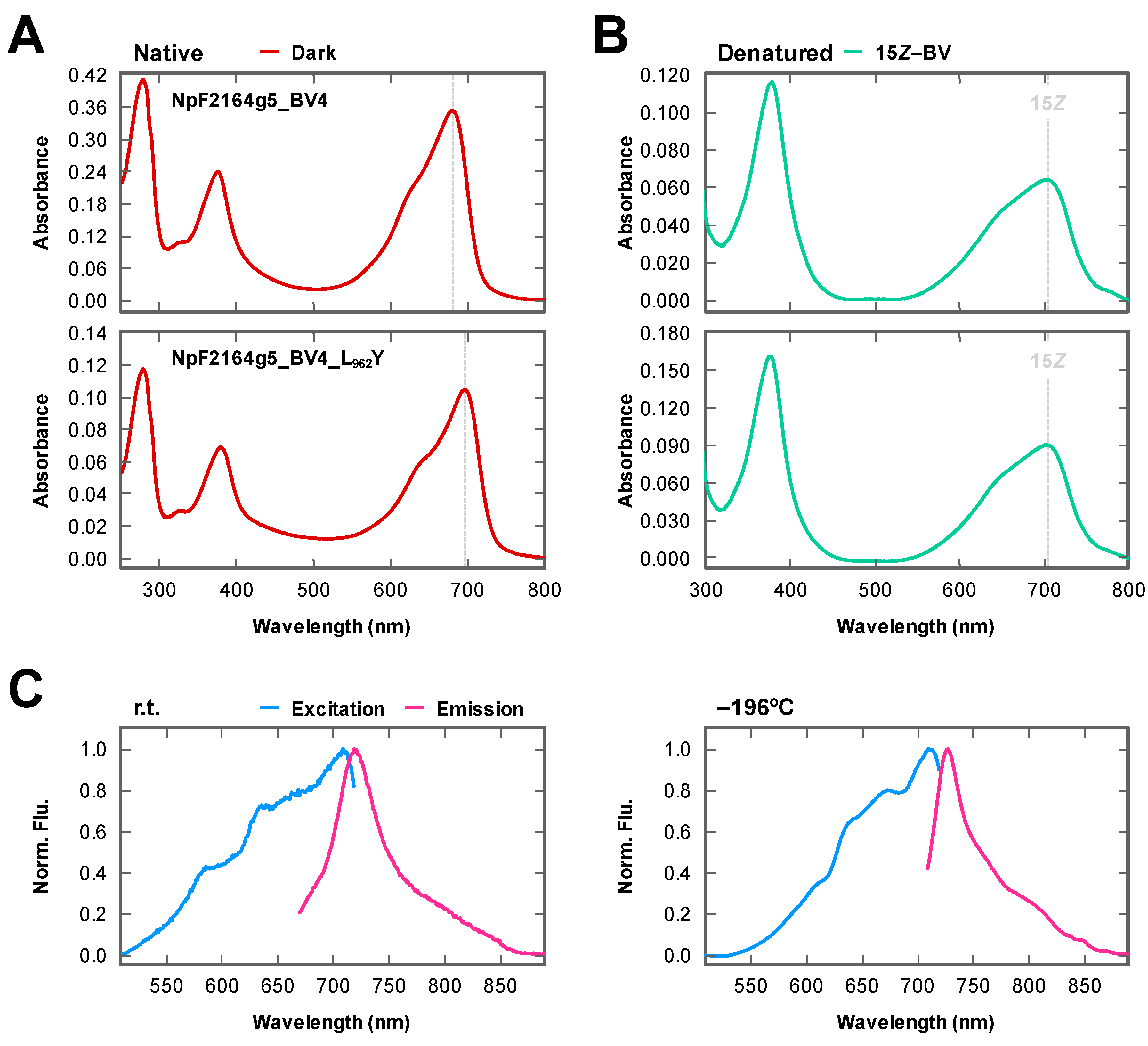
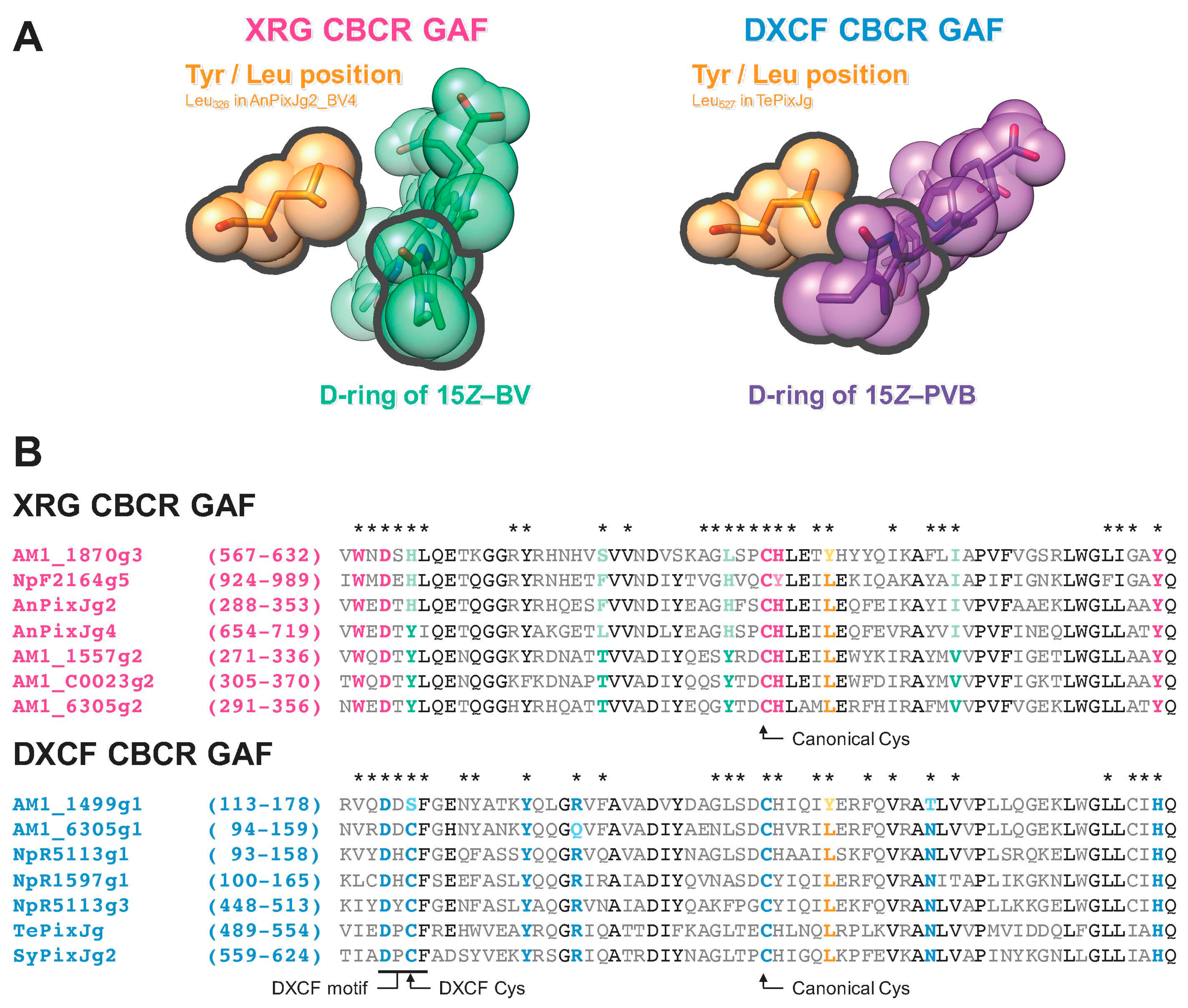
2.1. Rational Site-Directed Mutagenesis for Spectral Tuning
2.2. Dark Reversion
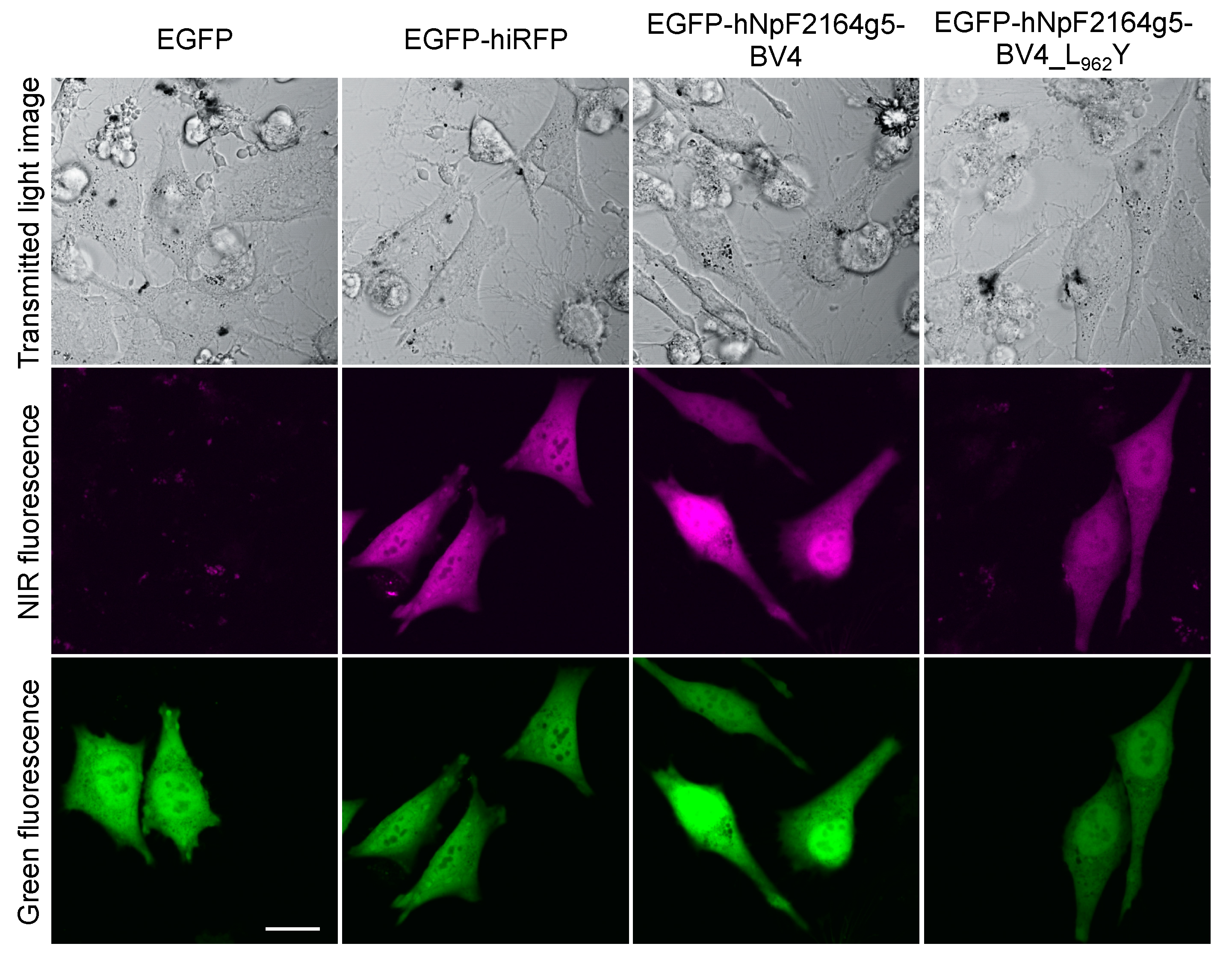
2.3. Fluorescence Imaging in Living Mammalian Cells Using NpF2164g5_BV4_L962Y
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Media
4.2. Bioinformatics
4.3. Plasmid Construction
4.4. Protein Expression and Purification
4.5. Electrophoresis and Zinc-Induced Fluorescence Assay
4.6. Spectroscopic Analysis
4.7. Biochemical and Photochemical Characterization of Cyanobacteriochromes
4.8. Mammalian Cell Culture and Transfection
4.9. Confocal Fluorescence Imaging
Author Contributions
Funding
Conflicts of Interest
References
- Fushimi, K.; Narikawa, R. Cyanobacteriochromes: Photoreceptors covering the entire UV-to-visible spectrum. Curr. Opin. Struct. Biol. 2019, 57, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Fushimi, K.; Ikeuchi, M.; Narikawa, R. The expanded red/green cyanobacteriochrome lineage: An evolutionary hot spot. Photochem. Photobiol. 2017, 93, 903–906. [Google Scholar] [CrossRef] [PubMed]
- Narikawa, R.; Fukushima, Y.; Ishizuka, T.; Itoh, S.; Ikeuchi, M. A novel photoactive GAF domain of cyanobacteriochrome AnPixJ that shows reversible green/red photoconversion. J. Mol. Biol. 2008, 380, 844–855. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Martin, S.S.; Lagarias, J.C. Red/green cyanobacteriochromes: Sensors of color and power. Biochemistry 2012, 51, 9667–9677. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, J.; Luo, J.; Tu, J.-M.; Zeng, X.-L.; Xie, J.; Zhou, M.; Zhao, J.-Q.; Scheer, H.; Zhao, K.-H. Photophysical diversity of two novel cyanobacteriochromes with phycocyanobilin chromophores: Photochemistry and dark reversion kinetics. FEBS J. 2012, 279, 40–54. [Google Scholar] [CrossRef]
- Fukushima, Y.; Iwaki, M.; Narikawa, R.; Ikeuchi, M.; Tomita, Y.; Itoh, S. Photoconversion mechanism of a green/red photosensory cyanobacteriochrome AnPixJ: Time-resolved optical spectroscopy and FTIR analysis of the AnPixJ-GAF2 domain. Biochemistry 2011, 50, 6328–6339. [Google Scholar] [CrossRef]
- Velazquez Escobar, F.; Utesch, T.; Narikawa, R.; Ikeuchi, M.; Mroginski, M.A.; Gärtner, W.; Hildebrandt, P. Photoconversion mechanism of the second GAF domain of cyanobacteriochrome AnPixJ and the cofactor structure of its green-absorbing state. Biochemistry 2013, 52, 4871–4880. [Google Scholar] [CrossRef]
- Scarbath-Evers, L.K.; Jähnigen, S.; Elgabarty, H.; Song, C.; Narikawa, R.; Matysik, J.; Sebastiani, D. Structural heterogeneity in a parent ground-state structure of AnPixJg2 revealed by theory and spectroscopy. Phys. Chem. Chem. Phys. 2017, 19, 13882–13894. [Google Scholar] [CrossRef]
- Song, C.; Velazquez Escobar, F.; Xu, X.-L.; Narikawa, R.; Ikeuchi, M.; Siebert, F.; Gärtner, W.; Matysik, J.; Hildebrandt, P. A red/green cyanobacteriochrome sustains its color despite a change in the bilin chromophore’s protonation state. Biochemistry 2015, 54, 5839–5848. [Google Scholar] [CrossRef]
- Song, C.; Narikawa, R.; Ikeuchi, M.; Gärtner, W.; Matysik, J. Color tuning in red/green cyanobacteriochrome AnPixJ: Photoisomerization at C15 causes an excited-state destabilization. J. Phys. Chem. B 2015, 119, 9688–9695. [Google Scholar] [CrossRef]
- Slavov, C.; Xu, X.; Zhao, K.-H.; Gärtner, W.; Wachtveitl, J. Detailed insight into the ultrafast photoconversion of the cyanobacteriochrome Slr1393 from Synechocystis sp. Biochim. Biophys. Acta 2015, 1847, 1335–1344. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.W.; Freer, L.H.; Rockwell, N.C.; Martin, S.S.; Lagarias, J.C.; Larsen, D.S. Second-chance forward isomerization dynamics of the red/green cyanobacteriochrome NpR6012g4 from Nostoc punctiforme. J. Am. Chem. Soc. 2012, 134, 130–133. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kim, P.W.; Freer, L.H.; Rockwell, N.C.; Martin, S.S.; Lagarias, J.C.; Larsen, D.S. Femtosecond photodynamics of the red/green cyanobacteriochrome NpR6012g4 from Nostoc punctiforme. 1. Forward dynamics. Biochemistry 2012, 51, 608–618. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.W.; Freer, L.H.; Rockwell, N.C.; Martin, S.S.; Lagarias, J.C.; Larsen, D.S. Femtosecond photodynamics of the red/green cyanobacteriochrome NpR6012g4 from Nostoc punctiforme. 2. Reverse dynamics. Biochemistry 2012, 51, 619–630. [Google Scholar] [CrossRef]
- Chang, C.-W.; Gottlieb, S.M.; Kim, P.W.; Rockwell, N.C.; Lagarias, J.C.; Larsen, D.S. Reactive ground-state pathways are not ubiquitous in red/green cyanobacteriochromes. J. Phys. Chem. B 2013, 117, 11229–11238. [Google Scholar] [CrossRef]
- Rockwell, N.C.; Martin, S.S.; Gulevich, A.G.; Lagarias, J.C. Conserved phenylalanine residues are required for blue-shifting of cyanobacteriochrome photoproducts. Biochemistry 2014, 53, 3118–3130. [Google Scholar] [CrossRef]
- Rockwell, N.C.; Martin, S.S.; Lim, S.; Lagarias, J.C.; Ames, J.B. Characterization of red/green cyanobacteriochrome NpR6012g4 by solution nuclear magnetic resonance spectroscopy: A protonated bilin ring system in both photostates. Biochemistry 2015, 54, 2581–2600. [Google Scholar] [CrossRef]
- Rockwell, N.C.; Martin, S.S.; Lim, S.; Lagarias, J.C.; Ames, J.B. Characterization of red/green cyanobacteriochrome NpR6012g4 by solution nuclear magnetic resonance spectroscopy: A hydrophobic pocket for the C15-E,anti chromophore in the photoproduct. Biochemistry 2015, 54, 3772–3783. [Google Scholar] [CrossRef]
- Gottlieb, S.M.; Kim, P.W.; Chang, C.-W.; Hanke, S.J.; Hayer, R.J.; Rockwell, N.C.; Martin, S.S.; Lagarias, J.C.; Larsen, D.S. Conservation and diversity in the primary forward photodynamics of red/green cyanobacteriochromes. Biochemistry 2015, 54, 1028–1042. [Google Scholar] [CrossRef]
- Narikawa, R.; Ishizuka, T.; Muraki, N.; Shiba, T.; Kurisu, G.; Ikeuchi, M. Structures of cyanobacteriochromes from phototaxis regulators AnPixJ and TePixJ reveal general and specific photoconversion mechanism. Proc. Natl. Acad. Sci. USA 2013, 110, 918–923. [Google Scholar] [CrossRef]
- Lim, S.; Yu, Q.; Gottlieb, S.M.; Chang, C.-W.; Rockwell, N.C.; Martin, S.S.; Madsen, D.; Lagarias, J.C.; Larsen, D.S.; Ames, J.B. Correlating structural and photochemical heterogeneity in cyanobacteriochrome NpR6012g4. Proc. Natl. Acad. Sci. USA 2018, 115, 4387–4392. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Port, A.; Wiebeler, C.; Zhao, K.-H.; Schapiro, I.; Gärtner, W. Structural elements regulating the photochromicity in a cyanobacteriochrome. Proc. Natl. Acad. Sci. USA 2020, 117, 2432–2440. [Google Scholar] [CrossRef] [PubMed]
- Fushimi, K.; Nakajima, T.; Aono, Y.; Yamamoto, T.; Ikeuchi, M.; Sato, M.; Narikawa, R. Photoconversion and fluorescence properties of a red/green-type cyanobacteriochrome AM1_C0023g2 that binds not only phycocyanobilin but also biliverdin. Front. Microbiol. 2016, 7, 588. [Google Scholar] [CrossRef] [PubMed]
- Narikawa, R.; Nakajima, T.; Aono, Y.; Fushimi, K.; Enomoto, G.; Itoh, S.; Sato, M.; Ikeuchi, M. A biliverdin-binding cyanobacteriochrome from the chlorophyll d-bearing cyanobacterium Acaryochloris marina. Sci. Rep. 2015, 5, 7950. [Google Scholar] [CrossRef]
- Fushimi, K.; Miyazaki, T.; Kuwasaki, Y.; Nakajima, T.; Yamamoto, T.; Suzuki, K.; Ueda, Y.; Miyake, K.; Takeda, Y.; Choi, J.-H.; et al. Rational conversion of chromophore selectivity of cyanobacteriochromes to accept mammalian intrinsic biliverdin. Proc. Natl. Acad. Sci. USA 2019, 116, 8301–8309. [Google Scholar] [CrossRef]
- Filonov, G.S.; Piatkevich, K.D.; Ting, L.-M.; Zhang, J.; Kim, K.; Verkhusha, V.V. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat. Biotechnol. 2011, 29, 757–761. [Google Scholar] [CrossRef]
- Shu, X.; Royant, A.; Lin, M.Z.; Aguilera, T.A.; Lev-Ram, V.; Steinbach, P.A.; Tsien, R.Y. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science 2009, 324, 804–807. [Google Scholar] [CrossRef]
- Shcherbakova, D.M.; Stepanenko, O.V.; Turoverov, K.K.; Verkhusha, V.V. Near-infrared fluorescent proteins: Multiplexing and optogenetics across scales. Trends Biotechnol. 2018, 36, 1230–1243. [Google Scholar] [CrossRef]
- Redchuk, T.A.; Omelina, E.S.; Chernov, K.G.; Verkhusha, V.V. Near-infrared optogenetic pair for protein regulation and spectral multiplexing. Nat. Chem. Biol. 2017, 13, 633–639. [Google Scholar] [CrossRef]
- Rockwell, N.C.; Martin, S.S.; Lagarias, J.C. Mechanistic insight into the photosensory versatility of DXCF cyanobacteriochromes. Biochemistry 2012, 51, 3576–3585. [Google Scholar] [CrossRef]
- Rockwell, N.C.; Martin, S.S.; Gulevich, A.G.; Lagarias, J.C. Phycoviolobilin formation and spectral tuning in the DXCF cyanobacteriochrome subfamily. Biochemistry 2012, 51, 1449–1463. [Google Scholar] [CrossRef] [PubMed]
- Fushimi, K.; Hasegawa, M.; Ito, T.; Rockwell, N.C.; Enomoto, G.; Lagarias, J.C.; Ikeuchi, M.; Narikawa, R. Evolution-inspired design of multicolored photoswitches from a single cyanobacteriochrome scaffold. Proc. Natl. Acad. Sci. USA 2020, 117, 15573–15580. [Google Scholar] [CrossRef] [PubMed]
- Velazquez Escobar, F.; Lang, C.; Takiden, A.; Schneider, C.; Balke, J.; Hughes, J.; Alexiev, U.; Hildebrandt, P.; Mroginski, M.A. Protonation-dependent structural heterogeneity in the chromophore binding site of cyanobacterial phytochrome Cph1. J. Phys. Chem. B 2017, 121, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Burgie, E.S.; Walker, J.M.; Phillips, G.N., Jr.; Vierstra, R.D. A photo-labile thioether linkage to phycoviolobilin provides the foundation for the blue/green photocycles in DXCF-cyanobacteriochromes. Structure 2013, 21, 88–97. [Google Scholar] [CrossRef]
- Miyake, K.; Fushimi, K.; Kashimoto, T.; Maeda, K.; Kimura, H.; Sugishima, M.; Ikeuchi, M.; Narikawa, R. Functional diversification of two bilin reductases for light perception and harvesting in unique cyanobacterium Acaryochloris marina MBIC 11017. FEBS J. 2020. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
| Absorption Maximum (Dark State) a | Dark State–Photoproduct State | Dark Reversion | |||
| λmax (nm) | SAR b | Positive (nm) | Negative (nm) | Half Life (sec) c | |
| AM1_1870g3 | |||||
| BV4 | 713 | 0.79 | 718 | 619 | 7.3 ± 0.03 |
| BV4_Y605L | 704 | 0.85 | 708 | 605 | 20.6 ± 0.11 |
| Absorption Maximum (Dark State) d | Fluorescence Maximum e | Fluorescence | |||
| λmax (nm) | SAR b | Excitation (nm) | Emission (nm) | Quantum Yield (%) f | |
| NpF2164g5 | |||||
| BV4 | 680 | 0.86 | 696 | 707 | 4 |
| BV4_L962Y | 697 | 0.89 | 711 | 728 | 2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fushimi, K.; Hoshino, H.; Shinozaki-Narikawa, N.; Kuwasaki, Y.; Miyake, K.; Nakajima, T.; Sato, M.; Kano, F.; Narikawa, R. The Cruciality of Single Amino Acid Replacement for the Spectral Tuning of Biliverdin-Binding Cyanobacteriochromes. Int. J. Mol. Sci. 2020, 21, 6278. https://doi.org/10.3390/ijms21176278
Fushimi K, Hoshino H, Shinozaki-Narikawa N, Kuwasaki Y, Miyake K, Nakajima T, Sato M, Kano F, Narikawa R. The Cruciality of Single Amino Acid Replacement for the Spectral Tuning of Biliverdin-Binding Cyanobacteriochromes. International Journal of Molecular Sciences. 2020; 21(17):6278. https://doi.org/10.3390/ijms21176278
Chicago/Turabian StyleFushimi, Keiji, Hiroki Hoshino, Naeko Shinozaki-Narikawa, Yuto Kuwasaki, Keita Miyake, Takahiro Nakajima, Moritoshi Sato, Fumi Kano, and Rei Narikawa. 2020. "The Cruciality of Single Amino Acid Replacement for the Spectral Tuning of Biliverdin-Binding Cyanobacteriochromes" International Journal of Molecular Sciences 21, no. 17: 6278. https://doi.org/10.3390/ijms21176278
APA StyleFushimi, K., Hoshino, H., Shinozaki-Narikawa, N., Kuwasaki, Y., Miyake, K., Nakajima, T., Sato, M., Kano, F., & Narikawa, R. (2020). The Cruciality of Single Amino Acid Replacement for the Spectral Tuning of Biliverdin-Binding Cyanobacteriochromes. International Journal of Molecular Sciences, 21(17), 6278. https://doi.org/10.3390/ijms21176278





