Abstract
DExD (DDX)- and DExH (DHX)-box RNA helicases, named after their Asp-Glu-x-Asp/His motifs, are integral to almost all RNA metabolic processes in eukaryotic cells. They play myriad roles in processes ranging from transcription and mRNA-protein complex remodeling, to RNA decay and translation. This last facet, translation, is an intricate process that involves DDX/DHX helicases and presents a regulatory node that is highly targetable. Studies aimed at better understanding this family of conserved proteins have revealed insights into their structures, catalytic mechanisms, and biological roles. They have also led to the development of chemical modulators that seek to exploit their essential roles in diseases. Herein, we review the most recent insights on several general and target-specific DDX/DHX helicases in eukaryotic translation initiation.
1. Introduction
RNA helicases play a role in all facets of RNA metabolism, ranging from transcription and translation, to processing and decay. These highly conserved enzymes use ATP to bind, unwind, and disrupt RNA structures and RNA-protein complexes [1]. Found in all domains of life, helicases are classified into six superfamilies (SFs), which are further sub-divided into families [1]. All eukaryotic RNA helicases belong to SFs 1 and 2, which comprise non-ring-forming structures [2]. Within SF2, the two largest families are the DExD-box (DDX) and DExH-box (DHX) helicases, named after their Asp-Glu-x-Asp/His signatures, which share evolutionarily conserved motifs within their core.
Eukaryotic translation initiation is an intensively studied process and is subject to much regulation—either through the modulation of Met-tRNAiMet•GTP•eukaryotic initiation factor (eIF) 2 ternary complex availability or the flow of ribosome recruitment to mRNA templates by eIF4F [3]. This latter avenue requires the translation apparatus to successfully negotiate intricate mRNA secondary structures and stably bound proteins in order to establish a bona fide 80S complex at the initiation codon that is competent for protein synthesis. It is here that RNA helicases play a central role in the regulation of gene expression. Dubbed the “godfather” of DEAD-box helicases, eIF4A is a minimal DDX protein that consists of only the conserved ATP-binding and RNA-binding sites and short N- and C- terminal extensions (~50 and ~30 aa, respectively) [4]. It is the best-characterized RNA helicase in translation initiation and represents a prototypical DDX protein. Studies surrounding eIF4A have illuminated much of our understanding of the roles that DDX family members play, and many multi-faceted RNA helicases have since been explored.
In this review, we seek to outline the different RNA helicases that have been implicated in eukaryotic translation initiation (Table 1). We refer the reader to a previous excellent review on this topic [5], which this current piece seeks to update. First, we introduce the DDX helicase family and then provide an up-to-date summary on the biophysical and biochemical information surrounding its archetypal member, eIF4A. Then, we review recent information on other helicases that have been implicated in translation, some of which have been attributed a general role, and others a more mRNA- or structure-specific function. Due to space limitations, we restrict, for the most part, our discussions to the mammalian setting and to those helicases with a putatively direct role in translation initiation.

Table 1.
RNA Helicases Implicated in Translational Control.
1.1. Translation Initiation and the Role of RNA Helicases
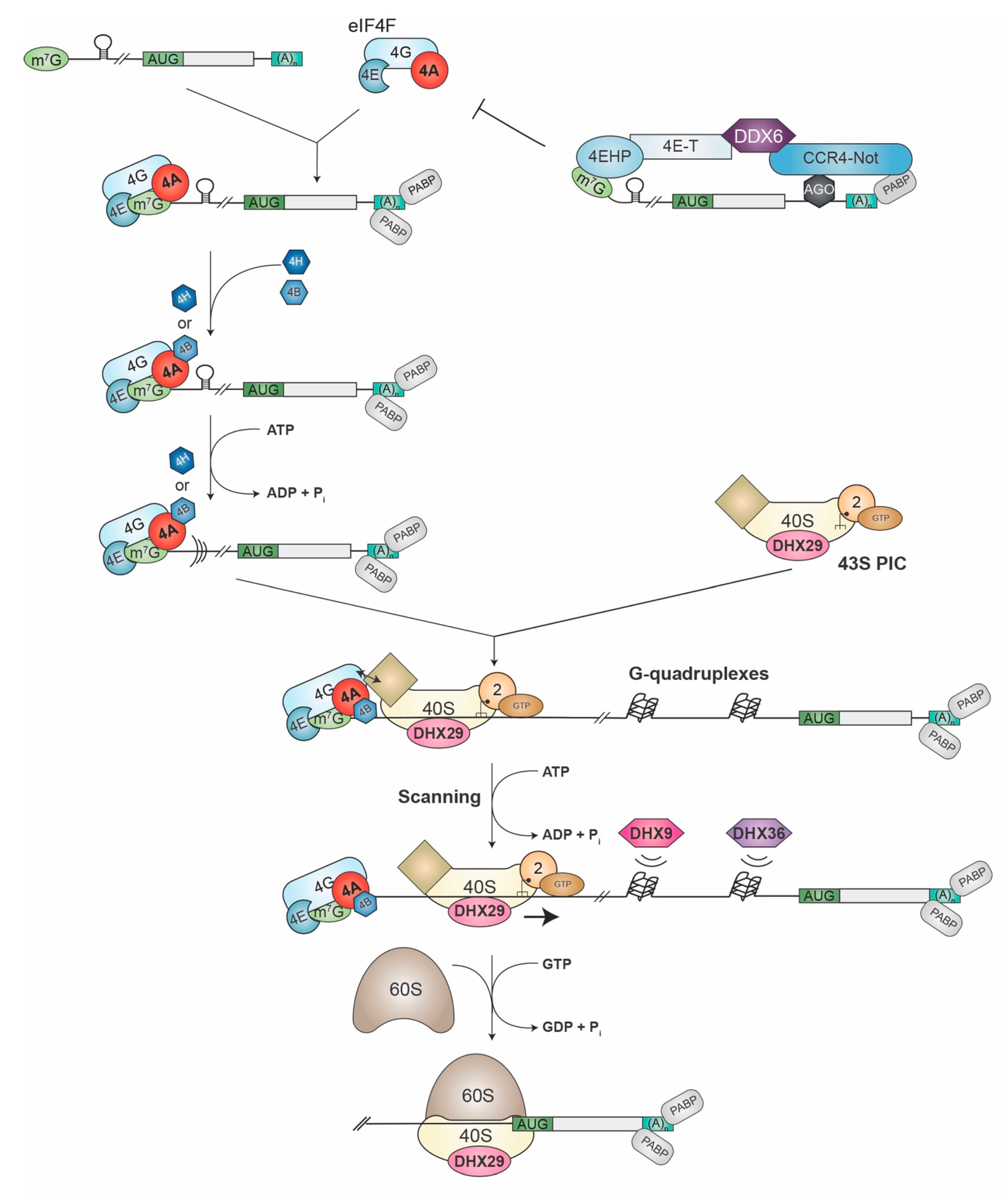
The recruitment of ribosomes to cytoplasmic mRNAs occurs by one of two mechanisms—cap-dependent and cap-independent processes. Cap-dependent translation is facilitated by the 5′-terminal m7GpppN (where N is any nucleotide) cap structure and is mediated by the eIF4F complex, which consists of eIF4E (the cap-binding subunit), eIF4G (a large scaffolding protein), and eIF4A (the ATP-dependent RNA helicase) (Figure 1). The RNA chaperones, eIF4B and eIF4H, interact with eIF4A, stimulate its activity (see below), and facilitate ribosome recruitment. An array of factors (the multi-subunit eIF3, eIF1, eIF1A, eIF2•GTP•Met-tRNAiMet, eIF5) bind to the 40S ribosomal subunit and convert it into the 43S pre-initiation complex (PIC). Subsequent interactions between eIF4G and eIF3 are critical for recruitment of the small ribosomal subunit to mRNAs. Once bound, the 43S PIC is thought to scan the 5′ leader region until an appropriate initiation codon is encountered, at which point initiation factors are evicted from the 48S complex, an additional molecule of GTP is hydrolyzed by eIF5B, and a 60S subunit is joined to the 40S to form an 80S complex now poised for elongation. This process is stimulated by the circularization of the mRNA, mediated by interactions between eIF4G and the 3′-end-bound poly (A) binding protein (PABP) [6]. Although the binding of eIF4E to the cap does not require ATP hydrolysis, the subsequent binding of eIF4A and eIF4B to the mRNA [7], as well as ribosome scanning [8], are ATP-dependent—a reliance that is thought to reflect the involvement of helicase activity in cap-dependent initiation.
Figure 1.
Schematic diagram of mammalian cap-dependent translation initiation pathway. See text for details. Please note for ease of viewing, the PABP•eIF4G bridging interactions are not shown. The diamond shape on the 40S ribosomes symbolizes eIF3, eIF1, eIF1A, and eIF5.
Cap-independent translation, mediated by internal ribosome entry sites (IRESes), on the other hand, have been stratified into four classes (types), with Types 1 and 2 requiring eIF4G and eIF4A, and Types 3 and 4 capable of recruiting ribosomes without the aid of any eIF4F subunits [3,9]. Moreover, initiation on some IRESes bypasses the requirement for scanning by directly situating the ribosomes at the initiation codon [3]. Helicase requirements for initiation are thus not equivalent among all IRES-containing mRNAs.
A determinant of eIF4F activity is mRNA secondary structure. The secondary structure within 5′ leader regions has long been known to inhibit translation and, when located near the cap, to negatively impact the interaction of eIF4F and eIF4B with the mRNA [10,11,12,13,14]. Similarly, stable cap-proximal mRNA-protein complexes are also effective at suppressing translation [15]. The degree of secondary structure within an mRNA 5′ leader region links initiation of that mRNA to eIF4A activity. The greater the amount of structure, the more dependent that mRNA appears to be on eIF4A (and hence eIF4F) activity for initiation [14]. Unstructured synthetic mRNA templates are able to recruit ribosomes in the absence of ATP. However, this process is stimulated by eIF4A, and introducing even a weak stem-loop structure (ΔG = −6.7 kcal/mol) renders the reaction eIF4A- and ATP-dependent [16]. A class of cellular mRNAs containing very short 5′ leaders known as TISU (translation initiator of short 5′ UTR) elements are capable of initiating translation in an eIF4E-dependent, but eIF4A-independent manner [17,18]. TISU elements are common in mRNAs encoding mitochondrial proteins, which may have evolved to be less dependent on eIF4A [18]. Given the link between mitochondrial outer membrane integrity and apoptosis, this may be a way to buffer against large changes in eIF4A availability while remaining tuned to the eIF4E activity. Structure within the 5′ leader, therefore, is a critical determinant of translational efficiency—a notion that has been supported by large-scale ribosome footprinting studies, wherein eIF4A activity has been perturbed by small molecule inhibitors [19,20,21].
1.2. Structural Basis of Helicase Activity
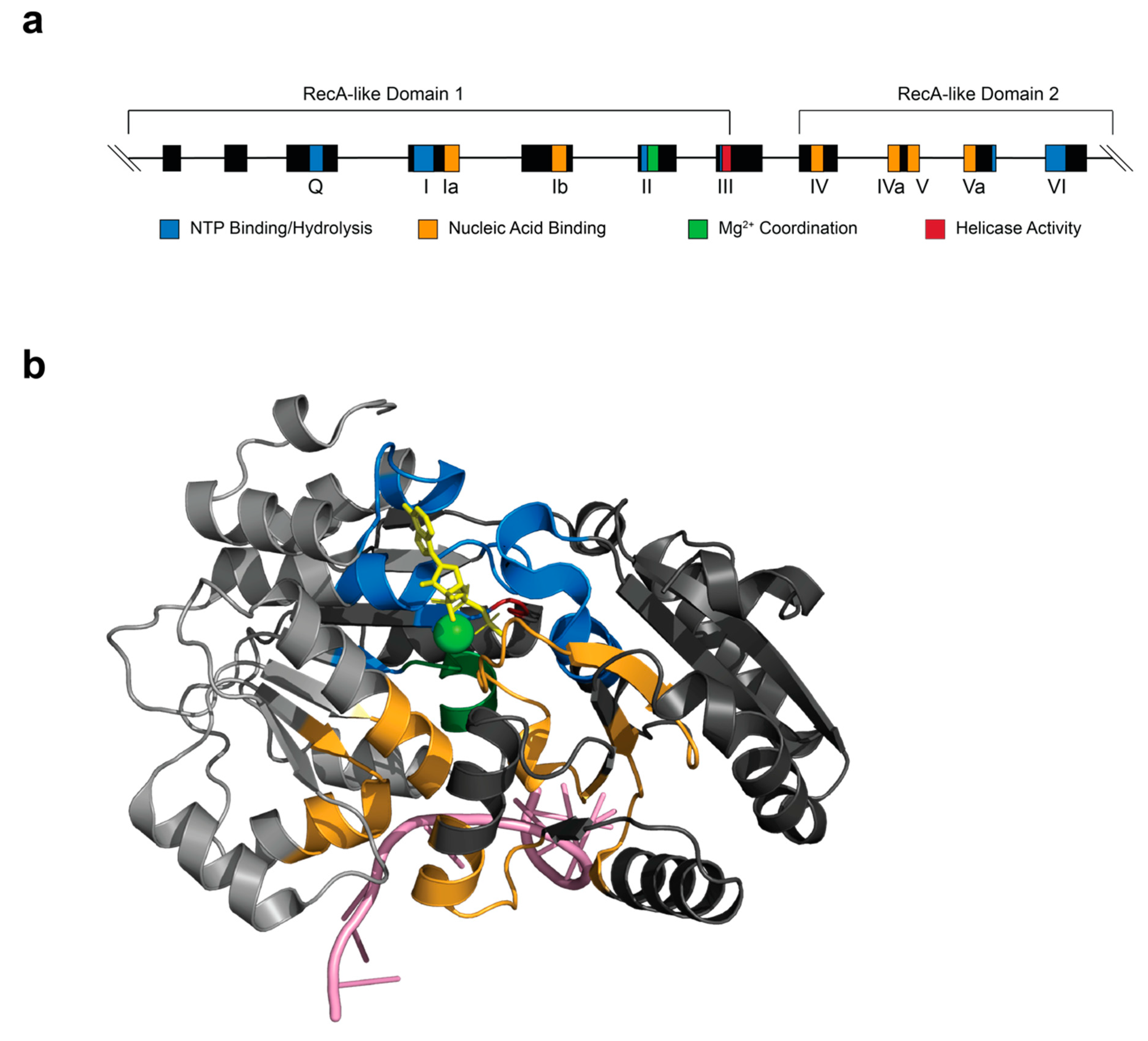
DDX/DHX proteins harbor two recombinase A (RecA)-like domains tethered together by a short, flexible linker at their core (Figure 2) [2]. The helicase core has been further characterized to contain at least 12 conserved motifs, with motif II housing the DExD/H sequence. These motifs are evenly spaced and line the cleft of the “dumbbell”-shaped protein [22] (Figure 2). Between the two RecA-like domains lie the ATP- and RNA-binding sites whose functional significance has been extensively reviewed [22] (Figure 2a). Structural and biochemical studies have shown that the ATP-binding cleft in DDX proteins must be in a closed conformation to efficiently bind and hydrolyze ATP [23].
Figure 2.
Structure of a conserved DDX/DHX helicase core and the archetypal eIF4A. (a) Exon-intron organization of the eIF4A1 gene with the location of the conserved domains highlighted. The functional role of each domain is denoted by a color code. (b) Three-dimensional structure of eIF4A complexed with AMP-PNP (yellow) and RNA (pink). The relative spatial location of the different functional domains is shown and color-coded as in (a). A magnesium ion is shown in green. The structure is from Protein Data Bank (PDB, 5ZC9).
DDX helicases act as molecular switches that normally adopt an open conformation in the absence of RNA or ATP and close upon cooperative binding of the two molecules, which occurs via a two-step process [24,25]. This new, more rigid structure induces contacts between ATP and residues within motifs I, II, and VI (Figure 2b). Nucleotide specificity for adenine is conferred by the Q-motif through a conserved glutamine residue [26,27,28] (Figure 2). RNA-binding involves motifs Ia, Ib, IV, IVa, V, and Va (Figure 2). Communication between the RNA- and ATP-binding sites is thought to occur through motifs III, IV, and IVa [29] (Figure 2). In the closed conformation, a single-stranded (ss) RNA is bent such that duplex formation is unfavorable [25,30]. The sugar phosphate backbone of the RNA makes contact with residues of the helicase core, resulting in the kinking of the molecule [26]. The unwound RNA is then released, an event which may be coupled to ATP-binding in some cases and ATP hydrolysis in others [30,31]. In this manner, RNA and ATP stabilize the helicase core and, in the case of eIF4A, ATP stimulates RNA-binding and unwinding, and RNA stimulates ATP-binding and hydrolysis [24,26,27,32]. The unwinding mechanism of DDX helicases, however, remains distinct from those of other families. Rather than operating by a translocation or “threading” mechanism, DDX proteins facilitate local duplex unwinding by clamping onto the duplex region and pulling the strands apart [33,34]. Therefore, this unusual mechanism limits unwinding to only short RNA duplexes, 10–12 nucleotides in length (for eIF4A), at any given time [35].
Beyond the helicase core, most DDX/DHX helicases also contain N- and C-terminal extensions. EIF4A is an exception in that it has very short extensions. For the other DDX/DHX helicases, however, these extensions impart functional diversity to the family. Among some of the activities gained are modified RNA/DNA/protein-binding, oligomerization, and nuclease capabilities [1].
It is important to note that DHX proteins, despite being overall quite similar to DDX family members, exhibit significant differences in key residues within their conserved motifs [36]. DDX family members are generally ATP-specific, whereas DHX members can hydrolyze different NTPs—a feature attributed to the presence of the Q-motif in DDX family members, and its absence in their DHX counterparts.
2. General DExD/H-Box Helicases in Translation
2.1. eIF4A: A Minimal DDX RNA Helicase
EIF4A was one of the first DDX proteins discovered and represents the quintessential DEAD-box helicase due to its minimal structure that contains all the activities characteristic of the family but not much more (Figure 2) [4]. EIF4A is present in mammals as three paralogs: Human eIF4A1 (DDX2A) and eIF4A2 (DDX2B), which share 90% identity at the amino acid level, and the distant cousin, eIF4A3 (DDX48), which shares 66% identity to eIF4A1 [3]. While eIF4A1 is essential and plays an active role within translation initiation, eIF4A2 is generally less abundant, non-essential, and less well-characterized in its role [3]. In fact, suppression of eIF4A1 leads to an increase in eIF4A2 that, on a molar basis, should rescue the eIF4A1-deficient phenotype but does not [37]. Furthermore, the two proteins are differentially expressed, with eIF4A1 levels being higher in proliferating cells, and eIF4A2 expression being elevated in quiescent cells [38,39]. The third paralog, eIF4A3, is largely a misnomer, as it does not participate in translation initiation but rather in exon-junction complexes (EJCs) [40].
Similar to all other DDX helicases, eIF4A does not exhibit directional or processive helicase activity alone [41]. It unwinds ~10–12 base pairs/hydrolyzed ATP, with a turnover rate (kcat) of 0.01–0.05 per second [42]. The RNA-binding site has further been mapped to span 10–15 nts in length [43], and although RNA binding to eIF4A1 occurs with low affinity and generally independently of sequence, ssRNA-binding substantially stimulates ATPase activity in vitro [44,45]. In contrast, dsRNA binds weakly to eIF4A and does not significantly stimulate the ATPase activity [32,44]. In terms of substrates, eIF4A alone has been shown to unwind RNA/RNA, RNA/DNA, and DNA/RNA, but not DNA/DNA duplexes in vitro [46]. While the unwinding rate depends on the thermodynamic stability of the duplex substrate and not its length nor sequence, ssRNA tails on either end increase the rate by ~30% [46]. Furthermore, eIF4A1 and eIF4A2 have slight, inherent biases for binding polypurine-rich RNA sequences [47].
While it has been demonstrated that dependence on the eIF4A unwinding activity is directly proportional to the amount of secondary structure within the 5′ leader, whether all structural barriers play equivalent roles in determining eIF4A-dependency is not clear. Certain studies, such as that of Wolfe et al. [19] suggest G-quadruplexes (G4) to be key determinants of eIF4A-mediated translation. However, the very existence of these G4 structures is debated, as they have proven elusive in more recent studies [48,49]. Both Guo et al. [48] and Waldron et al. [49] used a combination of transcription stalling assays, dimethyl sulphate treatment, SHAPE analysis, and 7-deazaguanine incorporation to show that G4s appear to be largely unfolded in eukaryotic cells and dictate eIF4A-dependence to a lesser extent than classical stable hairpins. A subsequent study by Waldron et al. [21], using hippuristanol (an inhibitor of eIF4A that will be discussed further below), suggests that the secondary structure closest to the AUG start codon is most important in dictating eIF4A dependencies, but this result awaits biochemical validation. RNA pseudoknots are another example of complex intramolecular RNA structures and consist of two stem-loops in which half of one stem is intercalated between the half of the other. A pseudoknot structure is present in the 5’ leader region of human interferon gamma (IFNG) mRNA for example and thought to be a component of its regulation by the interferon-inducible protein kinase, PKR [50]. Whether this structure can be resolved by eIF4A, implicating eIF4A in a PKR regulatory circuitry, has yet to be determined and could present a novel regulatory loop.
EIF4A2 has been implicated in translational repression and microRNA (miRNA) regulation through its interactions with the Ccr4-Not complex [51,52]. This complex is recruited to mRNAs via PABP, miRNAs, and RNA-binding proteins (RBPs), among others, when an mRNA is targeted for deadenylation and decay [53,54,55,56]. Meijer et al. [57] first demonstrated that eIF4A2 activity is required for miRNA-mediated mRNA destabilization. Then, the same lab reported that eIF4A2 may exert this function by becoming incorporated into the Ccr4-Not complex and inhibiting the CNOT7 deadenylation activity or repressing translation by binding to purine-rich sequences proximal to the AUG start codon [51,52]. These experiments implicating a repressive function for eIF4A2 are at odds with eIF4A2 being able to incorporate into the eIF4F complex, which plays a stimulatory role in translation [58]. Moreover, eIF4A2 is not essential for cell viability, and miRNA regulation is not impaired in its absence [59].
EIF4A does not function alone in translation initiation. As indicated above, eIF4B and eIF4H act as eIF4A cofactors to stimulate protein synthesis [60]. EIF4B stimulates the ATPase and RNA-unwinding activities of eIF4A [61], increases the coupling of ATP hydrolysis and helicase activity [62], confers directionality to eIF4A [63], helps recruit the 43S PIC to mRNA [64], and has even been reported to stimulate translation of mRNAs with complex 5′ leaders independently of eIF4A [65]. EIF4H and eIF4B share a common binding site on eIF4A, thus making the binding of these two cofactors mutually exclusive, but eIF4H exhibits many of the same effects as eIF4B when complexed with eIF4A [60].
Only ~5% of eIF4A is thought to reside within the eIF4F complex at one time, while the majority exists as the free form in cells [66]. However, eIF4F-bound eIF4A exhibits drastically altered biochemical properties. Upon recruitment of free eIF4A into eIF4F, eIF4A gains 5′→3′ directionality, for example [41]. Introduction of either human eIF4G682-1105 (middle domain of eIF4G sufficient to bind to eIF43, RNA, and eIF4A), eIF4H, or eIF4B alone enhance the ability of eIF4A to unwind by just under 2-fold [63]. However, addition of both eIF4G682-1105 and eIF4H, or eIF4G682-1105 and eIF4B, stimulated activity synergistically, with a greater increase being observed for the 4A/4G/4B combination (~100-fold) [63]. The resulting complex also became processive, with translocations occurring in discrete steps of 11 +/− 2 base pairs (roughly equal to one turn of an RNA double helix), as assessed by a single-molecule optical trapping assay [63]. Similar findings have been reported by others using gel-shift unwinding [60], real-time fluorescence [62], RNA pull-down [67], and single-molecule fluorescence resonance energy transfer (smFRET) assays [68].
The mechanism by which the ATP turnover rate increased was found to be through a joint stimulation of eIF4A by eIF4G and eIF4B of the open-closed conformational cycle of the helicase [68]. Both eIF4B and eIF4H increase the affinity of eIF4A or eIF4F for nucleotides and RNA, as well as the coupling of ATP hydrolysis with RNA unwinding [69]. Additionally, the presence of eIF4H or eIF4B conferred greater substrate specificity to eIF4F—namely, only substrates with at least a 20-nt ssRNA tail adjacent to the duplex and only RNA/RNA duplexes [70].
2.2. Regulation of eIF4A and Additional Roles
Beyond its interactions with other initiation factors, the eIF4A availability itself is dictated by factors such as the tumor suppressor programmed cell death 4 (PDCD4) protein. Structural studies have revealed that PDCD4 binds eIF4A and prevents the formation of the closed conformation, thus blocking RNA binding [71]. Under normal circumstances, the interaction between PDCD4 and eIF4A is subject to S6 kinase 1 (S6K1)- and S6K2-mediated phosphorylation, both of which are under control of the mammalian target of rapamycin complex 1 (mTORC1) [72,73]. PDCD4 phosphorylation leads to its rapid degradation, thus linking mTORC1 regulation to the eIF4A:PDCD4 interaction [72,73]. Additionally, mitogenic signals may be transduced via eIF4G (e.g., through the phosphorylation of its HEAT-1/2 interdomain linker) and eIF4E (e.g., via eIF4E-binding proteins [4E-BPs]), which are also under mTOR regulation) to modulate eIF4A helicase activity indirectly [3].
The eIF4A activity is also regulated by the long non-coding (lnc) RNA, human brain cytoplasmic RNA 1 (aka BC200), a 200-nt, predominantly cytoplasmic RNA linked to neurodegeneration and cancer initiation and progression [74]. BC200 is overexpressed in a number of cancer types yet is undetectable in corresponding normal tissues [74]. A number of initiation factors have been found to interact with BC200 and its mouse homolog BC1, including PABP, eIF4A, and eIF4B [74]. Binding of PABP and eIF4B to BC200/BC1 is thought to lead to suppression of translation by sequestering these factors, thereby preventing their participation in initiation [74]. Interaction of BC200 with eIF4A uncouples ATP hydrolysis from the helicase activity, leading to a reduction in cap-dependent translation [75,76]. BC1 also colocalizes with dendritic mRNAs and polysomes, raising a potential role in localized inhibition of translation [77].
With regards to modified or additional roles of eIF4A, a recent study by Sokabe and Fraser [78] suggests the possibility of eIF4A participating in mRNA recruitment to the ribosome in a helicase activity-independent manner. Through fluorescence-based anisotropy assays, they showed that eIF3j, which binds anti-cooperatively with RNA to the A-site of the 40S ribosomal subunit, can have its affinity reduced by the addition of eIF4A and ATP alone [78]. This reduction was achieved in a helicase-independent manner, and the authors showed that eIF4A can use its ATPase activity to control the conformation of the 43S PIC [78]. This study is consistent with a subsequent investigation by Yourik et al. [79] using yeast eIF4A, which showed mRNA recruitment to the ribosome to be independent of the 5′ leader structural complexity.
The functional diversity of eIF4A was recently expanded by Tauber et al. [80], who described the modulation of RNA condensation in stress granules by the DDX protein. Specifically, Tauber et al. showed that eIF4A limits stress granule formation by acting as an RNA chaperone to prevent aggregation [80]. DDX proteins have previously been described to localize to stress granules in mammals, yeast, and bacteria [81,82,83]. The authors showed that eIF4A prevents intermolecular RNA-RNA interactions and condensation within stress granules via its RNA-binding activity [80]. They also demonstrate that although the RNA-binding activity of the DDX protein alone is enough to decrease RNA condensation, its ATP hydrolysis activity increases the effect by allowing multiple cycles of RNA binding [80].
2.3. DDX3X
DDX3, or Ded1p in yeast, is ubiquitously expressed and is involved in a plethora of biological processes. DDX3 can be considered the next-most-investigated DDX family member, after eIF4A [84]. Ded1p is essential in yeast and shares a high degree of homology with human DDX3 [85]. There are two DDX3 subfamily members: DDX3X, which is X-linked (Xp11.3-11.23), and DDX3Y, which is found on the Y chromosome and expressed predominantly in the testes [86,87]. Both proteins have longer N-and C-terminal extensions than eIF4A [84]. DDX3 members maintain RNA-dependent ATPase activity and ATP-dependent RNA helicase activity, but the RecA-like domains 1 and 2 alone are insufficient for full ATPase activity, suggesting that the sequences flanking the helicase core contribute to this function [88,89]. A crystal structure of a DDX3X-AMP complex and subsequent functional characterization have revealed that DDX3 harbors a unique sequence (10 nts: Residues 250–259) between motifs I and Ia, which is involved in nucleic acid binding, hydrolysis, and unwinding [88,89]. In contrast to many other DDX family members, DDX3 has weak specificity for NTP and sugar binding, yet many of the conserved interactions, including the Q-motif with nucleotides, are maintained [90]. The roles of DDX3/Ded1p are myriad and have been comprehensively reviewed [84,85]. Beyond translation, DDX3/Ded1p appear to function in nucleocytoplasmic shuttling [91], as a host factor in viral infection [92], in chromosome segregation during mitosis [93], and even in EJC and spliceosome association [94]. DDX3/Ded1p subfamily members exhibit stunningly diverse functions.
In translation, conflicting reports have found that DDX3X in HeLa and Huh7 reporter cell assays stimulate cap-dependent translation [95,96] but can also inhibit translation upon ectopic expression in Huh7 cells [97]. This dual role as a stimulator and repressor of translation has been suggested by Hilliker et al. [98] to be due to Ded1p/DDX3’s ability to form a Ded1p-mRNA-eIF4F complex (via Ded1p•eIF4G1 interactions) that is sequestered to stress granules but is then released following ATP hydrolysis by Ded1p. Microscopy experiments showed the formation of a stalled mRNP that is resolved by Ded1p’s ATPase activity [98]. This was also supported by the finding that Ded1p overexpression leads to an increased processing body (P-body) formation, where Ded1p further accumulates [99]. Several groups have also reported that DDX3X promotes the translation of a subset of mRNAs, including cyclins and transcripts with longer, more structured 5′ leaders [100,101,102]. Ribosome footprinting experiments with temperature-sensitive mutants of Ded1p and eIF4A showed that Ded1p-dependent transcripts harbor more cap-distal stem loops in their 5′ leaders and greater overall structure than eIF4A-dependent mRNAs [102]. The emerging picture from yeast is that eIF4A is required for translation initiation on all mRNAs, whereas Ded1p is needed to resolve 5’ leader structural barriers [102]. Furthermore, Ded1p also plays a key role in near-cognate initiation codon recognition—when Ded1p activity is repressed or diminished, the scanning ribosome initiates at near-cognate initiation codons that are within the 5’ leader and immediately upstream of the secondary structure [103]. A recent report has documented a role for Ded1p as a stress sensor that responds to environmental changes [104]. At elevated temperatures (>39 °C), Ded1p condenses/aggregates, an event that is associated with a repression in translation of mRNAs encoding housekeeping functions [104]. A role for DDX3X and DDX3Y in mammalian translation is less clearly defined and necessitates further investigation.
2.4. DHX29
DHX29 binds to 40S ribosomes through its N-terminal domain [105]. Its NTPase activity is strongly stimulated by 40S ribosomes, is required for efficient 48S complex formation, and plays a role in promoting translation initiation on mRNAs with elevated 5′ leader secondary structure [106]. It is present in 48S complexes but not polysomes, indicating that it recycles during the initiation of translation [107]. CryoEM studies have placed DHX29 at the tip of helix H16 of the 18S ribosomal RNA, with its distal part extending along the mRNA entry channel [108]. DHX29 promotes unwinding of stable stems and ensures a linear nucleotide-by-nucleotide scanning during initiation; in its absence, stem-loops enter the mRNA-binding channel but are not threaded through the exit channel [109]. It has been suggested that DHX29 could be responsible for closing the mRNA entry channel latch following slotting of the mRNA into the mRNA-binding cleft. This would trap the mRNA inside the channel and contribute to an increased efficiency of 43S scanning [108]. On its own, DHX29 does not possess processive helicase activity [106]; rather, recent data have shown that it makes contact with 40S-bound eIF3 subunits, which may implicate it in the rearrangement of the ribosomal complex and higher processivity in mRNA unwinding during scanning [110,111]. A consequence of this remodeling may explain the ability of DHX29 to suppress leaky scanning through upstream AUG codons (but not through non-cognate CUG codons), where DHX29 would rearrange the positioning of eIF1A and eIF2α (indirectly), thereby impacting AUG recognition [112].
Depleting DHX29 by RNAi disrupts polysomes, suppresses translation, and inhibits cancer cell proliferation [107]. Whether DHX29 is a viable anti-cancer target whose inhibition could achieve a workable therapeutic index remains to be explored. However, CRISPR/Cas9-mediated knockout of DHX29 in human and mouse non-transformed, primary cells is also lethal, indicating that DHX29 is an essential gene [113]. A role for DHX29 beyond translation has recently been reported, where it was shown to recognize double-stranded RNA and specifically interact with MDA5 to enhance innate antiviral immunity [113].
3. Target- or Structure-Specific Helicases
A number of less well-studied DDX/DHX-box helicases have also been implicated in translation. Although the evidence for these playing a role in the initiation process is mounting, biochemical and mechanistic insights are required to consolidate their role in this process and to properly situate where they stand in the initiation pathway (Table 1). Here, we review the DDX/DHX helicases currently implicated in the translation of specific mRNA species.
3.1. DHX36
DHX36 (aka RHAU or G4R1) binds DNA and RNA G4 structures with a high affinity in vitro and in vivo and is capable of resolving such structures [114,115,116]. It has been implicated in transcriptional and post-transcriptional regulation of gene expression, with essential roles in heart development, hematopoiesis, and embryogenesis [117,118,119]. A role for DHX36 (RHAU and G4R1) in the selective translation of mRNAs encoding mixed lineage leukemia (MLL) 1 and MLL4—pivotal leukemogenic transcriptional regulators—has been documented, whereby the Aven RNA-binding protein recruits DHX36 to G4 sequences [120]. Depletion of DHX36 in HEK293 cells does not impair global protein synthesis, but does result in a shift of both MLL1 and MLL4 mRNAs into lighter polysomes [120]. A specialized role for DHX36 in regulating expression of G4-containing mRNAs was obtained from genome-wide studies undertaken in HeLa cells, which showed that: (i) mRNAs with G4 structures in their 5′ leader regions are inefficiently translated, (ii) stable G4s can promote the translation of upstream open reading frames (uORFs) when positioned downstream of the uORF initiation codon, (iii) DHX36 (and DHX9) are polysome-associated, and (iv) suppression of DHX36 and DHX9 results in an impairment of translation of mRNAs harboring G4s in their 5′ leader region. Overall structure within the 5′ leader region was not a determinant of DHX36-responsiveness, although length was [121]. The consequences of DHX36 knockdown in HEK293T cells were also analyzed at the proteomic level, where only 1.9% of the identified proteins (1837 sampled total) were downregulated by at least 70% [122]. Among the suppressed targets, 33% were encoded by mRNAs containing G4s in their 5′ leader regions [122].
Co-crystallization of DHX36 with a DNA G4 structure has revealed that DHX36 interacts with both the top face of the G4 structure and the adjacent single-stranded nucleic acid segment [123]. DHX36 does not discriminate between DNA and RNA substrates, as its contacts with the nucleic acid backbone are primarily with phosphates [123]. The drosophila DHX36 homolog has been shown to stabilize G4s in the absence of nucleotide or when bound to AMP-PNP or ADP, but resolves said structures in the presence of ATP [124]. The domains interacting with both the G4 structure and 3′ tail are necessary for this destabilization [124].
DHX36 can also associate with the BC200 lncRNA [125]. This interaction is not G4-mediated but rather occurs through a 3′-end, adenosine-rich region [125]. Given the interaction between eIF4A and BC200, it would be interesting to investigate whether BC200 co-regulates the activity of eIF4A and DHX36. Whether and how the activity of DHX36 is coupled to core translation initiation factors and if it can be regulated in an mRNA-specific manner remains open to questions.
3.2. DHX9
DHX9 has been implicated in a number of cellular processes ranging from DNA replication, transcription, translation, RNA processing and transport, miRNA processing, and maintenance of genomic stability [126]. A role for DHX9 in mRNA-specific translation has been documented, with implicated roles in the expression of viral and JunD mRNAs harboring a 5′ post-transcription control element (PCE)—a complex structural feature consisting of two stem-loops [127]. The NTPase/helicase activity of DHX9 is required for this function [128], and association with PCE-containing mRNAs occurs in the nucleus and cytoplasm, which may reflect an early post-transcriptional mark that serves to ensure efficient routing of the tagged mRNA into polysomes [128,129]. JunD mRNA translation is cap-dependent [130], and the possible interplay between eIF4F and DHX9 would be intriguing to explore. Bioinformatics analysis has identified ~200 human genes containing PCEs [131].
DHX9 facilitates the translation of α1 and α2 type I collagen, but in this case, via a different mechanism. The 5′ leader regions of the α1(I) and α2(I) mRNAs both harbor a stem-loop to which the La ribonucleoprotein (LARP) 6 family member binds and recruits DHX9, stimulating ribosome loading and translation initiation [132]. Knockdown of DHX9 in human lung fibroblasts prevents collagen mRNAs from loading into polysomes without affecting mRNA levels [133]. Similar to PCE-containing mRNAs, the binding of transcripts by LARP6/DHX9 occurs in the nucleus, prior to the resulting stimulation in translation [133].
DHX9 has also been implicated in IRES-mediated translation—specifically, of an IRES in the 5′ leader region of the p53 mRNA that is responsive to DNA damage. DHX9 and translational control protein (TCP) 80 cooperatively bind to the p53 IRES in response to exposure of cells to DNA-damaging agents [134,135]. The presence of DHX9 is thought to stimulate an IRES-mediated initiation event, leading to an increased translation of the p53 mRNA.
In vitro, DHX9 has also been reported to bind and resolve RNA G4 structures [136]. Similar to DHX36, DHX9 is also found to be polysome- and G4-associated, with the translation of mRNAs harboring G4s being most sensitive to DHX9 knockdown [121]. Depletion of DHX9 in HeLa cells does not lead to a global inhibition of translation, and the spectrum of mRNAs inhibited somewhat overlapped with those responsive to DHX36 suppression (Pearson correlation = 0.56) [121]. Individual-nucleotide resolution UV crosslinking and immunoprecipitation (iCLIP) experiments revealed G4 sequences to be enriched for, and located ~40 nts upstream of, the DHX9 peak centers, possibly reflecting the pre-resolution of the G4 sequence with the helicase loading downstream of the structure [121]. Taken together, it appears that G4s are key determinants of DHX9 (and DHX36)-responsiveness.
3.3. DHX33
DHX33 is not well-studied and was initially implicated in rRNA biogenesis [137]. However, its suppression in several cell lines led to a reduction in polysomes and a concomitant increase in monosomes—not what would be expected for an inhibitor of rRNA synthesis, but rather more akin to what one would expect for a block in initiation or disruption of polysomes [138]. Consistent with this, DHX33 knockdown reduces translation as assessed in 35S-Met metabolic labelling and luciferase reporter assays [138]. Sub-cellular fractionation studies have shown that although the majority of DHX33 is nuclear, a significant proportion is cytoplasmic [137,139]. DHX33 co-sediments with monosomes, but not polysomes, in sucrose gradients and can be found in pull-down experiments to be associated with eIF3g and several ribosomal proteins (rpL27, rpL26, rpL7 (but not rpS2)) [137,139]. DHX33 helicase activity was not required for this association, nor were the associations RNA-mediated (since they were not affected by RNAse treatment) [139]. In vitro, DHX33 failed to stimulate translation of a firefly luciferase reporter mRNA, indicating it may not be limiting in reticulocyte lysates, or that its role in translation in cellula is indirect [139].
DHX33 is highly expressed in several cancer types, including glioblastoma [140]. Knockdown in the U118-MG glioblastoma line resulted in reduced cell proliferation, cell migration, and/or reduced anchorage-independent growth [140], a result that is interesting, since it appears that DHX33 is not essential in most cell lines (https://oncologynibr.shinyapps.io/drive/). Overexpression of DHX33 conferred resistance to the mTOR inhibitors, rapamycin, and Torin1 [140].
3.4. DDX1
DDX1 was found to be an insulin mRNA-binding protein in RNA pull-down assays [141]. Prolonged exposure to free fatty acids impairs insulin secretion from pancreatic cells, and DDX1 is thought to mediate the palmitate-induced inhibition of insulin mRNA translation [141]. Palmitate treatment of cells causes DDX1 to re-localize from the cytoplasm to the nucleus. Knockdown of DDX1 results in a significant reduction in proinsulin levels with no change in insulin mRNA levels, whereas ectopic overexpression of DDX1 increases proinsulin protein levels (with mRNA levels remaining unchanged) [141]. DDX1 suppression causes insulin mRNA to shift into lighter polysomes, while palmitate treatment of cells leads to phosphorylation of DDX1 at S295, a modification that affects its association with insulin mRNA and its ability to rescue shDDX1-mediated suppression [141]. Immunoprecipitation experiments to identify interacting partners indicated an association with the core translation factors, eIF4B, eIF3A, eIF3B, eIF3M, eIF4G1, and eIF4G2 [141]. DDX1 was also found in a complex with hCLE, HSPC117, and FAM98B, in which hCLE was reported to have cap-binding activity [142]. Suppression of hCLE reduced the incorporation of 35S-Met into cellular proteins without impacting global mRNA levels. Whether and how hCLE specifically and directly recognizes cap structures awaits structural resolution. In pull-down experiments, 144 mRNAs were found to interact with hCLE in HEK293 cells [142]. Whether DDX1 plays a role in selecting target mRNAs remains to be established.
3.5. DDX41
DDX41 was postulated to have a role in translation following its identification in RNA pull-down assays using the in vitro synthesized 3′ leader region of p21 (cyclin-dependent kinase inhibitor) mRNA [143]. Suppression of DDX41 led to elevated p21 protein levels with no change in mRNA levels, indicating that DDX41 negatively regulates p21 expression [143]. The helicase activity of DDX41 is essential to its ability to suppress p21 expression [143]. What is now needed are experiments providing mechanistic insight into DDX41-mediated translational control.
3.6. DDX6
DDX6 has been implicated in mRNA storage, decay, and the repression of translation [144]. It has been shown to contribute to the translational repression exerted by miRNAs. The silencing activity of miRNAs is mediated by the Ccr4-Not deadenylase complex, with DDX6 and 4EHP (aka eIF4E2) also present as components of the miRNA-induced silencing complex (miRISC) [145]. 4EHP is a cap-binding protein that unlike eIF4E, does not interact with eIF4G, and its binding to the cap has been associated with a repressive function [3]. The C-terminal RecA-like domain of DDX6 interacts with the CNOT1 MIF4G domain in a manner reminiscent of the eIF4G-eIF4A complex [146]. DDX6 tethers 4E-T (eIF4E transporter) [147], which in turn can interact with eIF4E or 4EHP [148,149]. When 4E-T is bound to eIF4E, this enhances the decay of mRNAs targeted by the Ccr4-Not complex, including miRNAs [149]. The binding of 4E-T to 4EHP increases the affinity of 4EHP for the cap and engenders a closed-loop complex with 4EHP bound to the cap and DDX6 bound to Ccr4-Not residing at a miRNA-binding site (Figure 1). Since 4EHP cannot interact with eIF4G, this complex cannot recruit ribosomes and thus prevents eIF4F from initiating on the cyclized mRNA template [148]. In this manner, DDX6 is able to coordinate the repression of specific mRNA templates.
DDX6 has also been implicated in the stimulation of translation of specific mRNAs. In this case, ectopic expression of DDX6 resulted in elevated levels of c-Myc protein in COS-7 and SW480 human colorectal cells [150]. Translation of the c-Myc mRNA is cap-dependent and eIF4E-responsive [151], but it may also harbor an IRES [152]. DDX6 is thought to be an IRES-trans-acting factor (ITAF), where its helicase activity might resolve structural elements within the c-Myc IRES—an event that would be required for initiation of c-Myc [153]. In contrast, DDX6 has also been implicated as a repressor of the vascular endothelial growth factor (VEGF) IRES [154]. The VEGF IRES confers sustained translation under conditions of hypoxia. Recombinant DDX6 inhibited translation of VEGF mRNA in vitro, DDX6 levels decline during hypoxia, and DDX6 depletion stimulated DDX6 expression under hypoxia [154]. In this scenario, DDX6 would rearrange the VEGF IRES and render it non-functional under normoxic conditions. These contrasting activities of DDX6 might depend on a select suite of binding partner(s) that intimately regulate DDX6 activity while imparting mRNA binding specificity.
4. Targeting DDX/DHX-Box Helicases in Translation Initiation
Protein synthesis is the most energetically expensive process in cells, and consequently, its regulation or dysregulation will have profound implications on cellular physiology. Ranging from cancer biology to viral replication, perturbation of translation is found in many diseases. In particular, cancer (aberrant proliferation, angiogenesis, changes in the immune system, etc.) has been tied to altered translation initiation, the rate-limiting step of the entire process (reviewed in [155,156]). EIF4E overexpression, for example, is sufficient for neoplastic transformation and tumorigenesis both in vitro and in vivo [157,158,159]. Additionally, viruses, a subject of topical interest, also rely on the translation machinery and DDX/DHX helicases for their replication [160,161,162]. Beyond perturbing the translation process, however, diseases such as cancer and viral infection often develop a dependence on altered mRNA translation, raising the possibility that by targeting protein synthesis, one could drug this vulnerability [155,156]. For example, many oncogenes harbor long, highly structured 5′ leaders that exhibit a greater dependence on eIF4A helicase activity [19]. It has also been demonstrated that the translation of these mRNAs with complex 5′ leaders is directly proportional to the amount of eIF4A activity [14]. By targeting this therapeutic window, wherein translation of highly structured mRNAs (many of which encode for oncogenic functions) will be disproportionately affected compared to normal, non-oncogenic transcripts, one might be able to treat the disease without harming the individual [155,156]. The same can be said of viral infections, where the viruses’ replication depends more on host translation than do most of our regular homeostatic mechanisms.
To date, several small molecules have been identified that selectively target DDX/DHX-box helicase family members, and these have been extensively reviewed in [163] (summarized in Table 2). In brief, helicase inhibitors can be stratified into two groups: (i) interfacial inhibitors, in which the compounds interact with both the protein target and the RNA, and (ii) compounds that inhibit RNA binding. Rocaglates and Pateamine A (and analogs) are interfacial inhibitors that cause eIF4A to clamp onto RNA, significantly stabilizing the resulting complex [164,165]. This gain-of-function produces a number of effects on translation initiation that inhibits the pathway through several modes: (i) clamped eIF4A molecules on 5′ leader regions inhibit scanning, (ii) eIF4F becomes clamped at the cap resulting in a reduced 43S pre-initiation complex loading, and (iii) stabilization of eIF4F onto RNA diminishes the pool of eIF4F and exerts an effect in trans on initiation [166]. Furthermore, eIF4A appears to also become clamped to ribosomes, the functional consequence of which remains to be investigated [167]. Interfacial inhibitors represent a powerful approach through which to inhibit a biological process such as translation, since only a small fraction of eIF4A molecules (enough to impose a steric blockade) needs to be engaged by the small molecule. Specificity of binding, at least for rocaglates where this has been defined through structural studies [164], is conferred by interacting amino acids (F163 and Q195) only found at these locations in eIF4A [164].

Table 2.
Current Selective Inhibitors of DDX/DHX Family Members Implicated in Translation.
On the other hand, hippuristanol and 1,4-diacylpiperazine derivatives, respectively, target eIF4A1/2 and eIF4A3 by binding to the CTD and inhibiting RNA binding [168,169]. It appears from NMR studies that both compounds share fairly conserved binding pockets. Variomics screens undertaken with both compounds have clearly indicated that these are not pan-helicase inhibitors and have linked their biological effects to target engagement [170] (J. Pelletier, unpublished data).
Two inhibitors of DDX3X have recently been reported [171,172]. RK-33 is a ring-expanded nucleoside that directly interacts with DDX3X and inhibits its ATPase and helicase activity. Given that DDX3X has been implicated in the replication of a number of viruses, RK-33 also shows broad-spectrum anti-viral activity [67]. Takeda also recently identified a new inhibitor of DDX3X helicase activity, C1 [172]. We envision that if these compounds can be shown to be selective, they will be useful in better defining the role of DDX3X in translation and its other associated processes.
Twenty-six compounds emerged from a high-throughput screen undertaken by Takeda for DDX41 ATPase inhibitors [173]. Their counter-screens probed for activity against DDX48 and DnaK, and they ensured that primary hits did not have RNA intercalation activity (which could non-specifically inhibit helicase activity). Two compounds that emerged from this screen (Compounds #1 and 2) are thought to inhibit nucleic acid substrate binding and act as non-competitive inhibitors of ATP binding [173].
5. Summary and Outlook
There is emerging interest in the study of RNA helicases, given their fundamental role in a number of biological processes. Even with the best-studied DDX helicase, eIF4A, there is much that remains to be uncovered with respect to its role in translation alone. Why is only ~5% of eIF4A present in the eIF4F complex? What role, if any, does the remaining 95% free eIF4A play in or outside of translation? It is becoming evident that one key to understanding helicase specificity of action is defining their interactomes, as their associated factors often impact function and modulate activity in drastic manners. Helicases also exhibit diversity in activity—from resolving RNA secondary structure, to affecting the processivity of the scanning ribosome, to remodeling protein complexes. How these activities are regulated is another aspect that remains to be explored.
Lastly, the characterization of small molecules that can selectively interfere with helicase activity has opened up the possibility of targeting these for drug development and provided a roadmap for effectively inhibiting downstream gene expression. Due to their central role in many disease processes, RNA helicases represent a biological target with significant, unexplored potential. Whether it be targeting host helicases in cancer or viral helicases in infection, small molecules and therapeutics can only be further developed if we continue to expand on our knowledge.
Author Contributions
Writing—original draft preparation, L.S.; writing—review and editing, L.S. and J.P.; visualization, L.S. and J.P.; supervision, J.P.; fund administration and acquisition, J.P. All authors have read and agreed to the published version of the manuscript.
Funding
Work in J. Pelletier’s lab on RNA helicases is funded by The Canadian Institutes of Health Research (FDN-148366).
Acknowledgments
We apologize to those authors whose work we did not cite due to space constraints. We are grateful to Francis Robert and Marc Fabian for their insight and constructive comments.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| 4EHP | eIF4E Homology Protein |
| 4E-T | eIF4E Transporter |
| CCR4-Not | Carbon Catabolite Repression—Negative On TATA-less |
| DDX | DExD-box |
| DHX | DExH-box |
| eIF | Eukaryotic Initiation Factor |
| IFNG | Human Interferon Gamma |
| IRES | Internal Ribosome Entry Site |
| G4 | G-quadruplexes |
| HEAT | Huntington, Elongation Factor 3, PR65/A, TOR |
| iCLIP | Individual-nucleotide resolution UV crosslinking and immunoprecipitation |
| ITAF | IRES Trans-Acting Factor |
| LARP | La Ribonucleoprotein |
| Lnc | Long Non-coding |
| MIF4G | Middle domain of eIF4G |
| miRISC | miRNA-Induced Silencing Complex |
| mTORC1 | Mammalian Target of Rapamycin Complex 1 |
| MLL | Mixed Lineage Leukemia |
| PABP | Poly (A) Binding Protein |
| PCE | Post-transcription Control Element |
| PIC | Pre-initiation Complex |
| PDCD4 | Programmed Cell Death 4 |
| PKR | Protein Kinase R |
| RBP | RNA Binding Protein |
| RecA | Recombinase A |
| S6K | Ribosomal Protein S6 kinase |
| SF | Superfamily |
| smFRET | single-molecule Fluorescence Resonance Energy Transfer |
| TCP | Translational Control Protein |
| TISU | Translation Initiator of Short 5’ UTR |
| uORFs | Upstream Open Reading Frames |
References
- Jankowsky, E. RNA helicases at work: Binding and rearranging. Trends Biochem. Sci. 2011, 36, 19–29. [Google Scholar] [CrossRef]
- Singleton, M.R.; Dillingham, M.S.; Wigley, D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007, 76, 23–50. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, J.; Sonenberg, N. The organizing principles of eukaryotic ribosome recruitment. Annu. Rev. Biochem. 2019, 88, 307–335. [Google Scholar] [CrossRef]
- Rogers, G.W., Jr.; Komar, A.A.; Merrick, W.C. eIF4A: The godfather of the DEAD box helicases. Prog. Nucleic Acid Res. Mol. Biol. 2002, 72, 307–331. [Google Scholar]
- Parsyan, A.; Svitkin, Y.; Shahbazian, D.; Gkogkas, C.; Lasko, P.; Merrick, W.C.; Sonenberg, N. mRNA helicases: The tacticians of translational control. Nat. Rev. Mol. Cell Biol. 2011, 12, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Gallie, D.R. A tale of two termini: A functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene 1998, 216, 1–11. [Google Scholar] [CrossRef]
- Sonenberg, N. ATP/Mg++-dependent cross-linking of cap binding proteins to the 5’ end of eukaryotic mRNA. Nucleic Acids Res. 1981, 9, 1643–1656. [Google Scholar] [CrossRef]
- Kozak, M. Role of ATP in binding and migration of 40S ribosomal subunits. Cell 1980, 22, 459–467. [Google Scholar] [CrossRef]
- Pestova, T.V.; Shatsky, I.N.; Fletcher, S.P.; Jackson, R.J.; Hellen, C.U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998, 12, 67–83. [Google Scholar] [CrossRef]
- Pelletier, J.; Sonenberg, N. Insertion mutagenesis to increase secondary structure within the 5’ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell 1985, 40, 515–526. [Google Scholar] [CrossRef]
- Pelletier, J.; Sonenberg, N. Photochemical cross-linking of cap binding proteins to eucaryotic mRNAs: Effect of mRNA 5’ secondary structure. Mol. Cell. Biol. 1985, 5, 3222–3230. [Google Scholar] [CrossRef] [PubMed]
- Parkin, N.T.; Cohen, E.A.; Darveau, A.; Rosen, C.; Haseltine, W.; Sonenberg, N. Mutational analysis of the 5’ non-coding region of human immunodeficiency virus type 1: Effects of secondary structure on translation. EMBO J. 1988, 7, 2831–2837. [Google Scholar] [CrossRef] [PubMed]
- Babendure, J.R.; Babendure, J.L.; Ding, J.H.; Tsien, R.Y. Control of mammalian translation by mRNA structure near caps. RNA 2006, 12, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Svitkin, Y.V.; Pause, A.; Haghighat, A.; Pyronnet, S.; Witherell, G.; Belsham, G.J.; Sonenberg, N. The requirement for eukaryotic initiation factor 4A (eIF4A) in translation is in direct proportion to the degree of mRNA 5’ secondary structure. RNA 2001, 7, 382–394. [Google Scholar] [CrossRef] [PubMed]
- Muckenthaler, M.; Gray, N.K.; Hentze, M.W. IRP-1 binding to ferritin mRNA prevents the recruitment of the small ribosomal subunit by the cap-binding complex eIF4F. Mol. Cell 1998, 2, 383–388. [Google Scholar] [CrossRef]
- Pestova, T.V.; Kolupaeva, V.G. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002, 16, 2906–2922. [Google Scholar] [CrossRef] [PubMed]
- Elfakess, R.; Dikstein, R. A translation initiation element specific to mRNAs with very short 5’UTR that also regulates transcription. PLoS ONE 2008, 3, e3094. [Google Scholar] [CrossRef] [PubMed]
- Gandin, V.; Masvidal, L.; Hulea, L.; Gravel, S.-P.; Cargnello, M.; McLaughlan, S.; Cai, Y.; Balanathan, P.; Morita, M.; Rajakumar, A. NanoCAGE reveals 5’ UTR features that define specific modes of translation of functionally related MTOR-sensitive mRNAs. Genome Res. 2016, 26, 636–648. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, A.L.; Singh, K.; Zhong, Y.; Drewe, P.; Rajasekhar, V.K.; Sanghvi, V.R.; Mavrakis, K.J.; Jiang, M.; Roderick, J.E.; Van der Meulen, J.; et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature 2014, 513, 65–70. [Google Scholar] [CrossRef]
- Rubio, C.A.; Weisburd, B.; Holderfield, M.; Arias, C.; Fang, E.; DeRisi, J.L.; Fanidi, A. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol. 2014, 15, 476. [Google Scholar] [CrossRef]
- Waldron, J.A.; Tack, D.C.; Ritchey, L.E.; Gillen, S.L.; Wilczynska, A.; Turro, E.; Bevilacqua, P.C.; Assmann, S.M.; Bushell, M.; Le Quesne, J. mRNA structural elements immediately upstream of the start codon dictate dependence upon eIF4A helicase activity. Genome Biol. 2019, 20, 300. [Google Scholar] [CrossRef] [PubMed]
- Linder, P.; Jankowsky, E. From unwinding to clamping—The DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 2011, 12, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Hilbert, M.; Karow, A.R.; Klostermeier, D. The mechanism of ATP-dependent RNA unwinding by DEAD box proteins. Biol. Chem. 2009, 390, 1237–1250. [Google Scholar] [CrossRef] [PubMed]
- Theissen, B.; Karow, A.R.; Kohler, J.; Gubaev, A.; Klostermeier, D. Cooperative binding of ATP and RNA induces a closed conformation in a DEAD box RNA helicase. Proc. Natl. Acad. Sci. USA 2008, 105, 548–553. [Google Scholar] [CrossRef] [PubMed]
- Henn, A.; Cao, W.; Hackney, D.D.; De La Cruz, E.M. The ATPase cycle mechanism of the DEAD-box rRNA helicase, DbpA. J. Mol. Biol. 2008, 377, 193–205. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sengoku, T.; Nureki, O.; Nakamura, A.; Kobayashi, S.; Yokoyama, S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell 2006, 125, 287–300. [Google Scholar] [CrossRef]
- Bono, F.; Ebert, J.; Lorentzen, E.; Conti, E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell 2006, 126, 713–725. [Google Scholar] [CrossRef]
- Tanner, N.K. The newly identified Q motif of DEAD box helicases is involved in adenine recognition. Cell Cycle 2003, 2, 18–19. [Google Scholar] [CrossRef]
- Banroques, J.; Doere, M.; Dreyfus, M.; Linder, P.; Tanner, N.K. Motif III in superfamily 2 “helicases” helps convert the binding energy of ATP into a high-affinity RNA binding site in the yeast DEAD-box protein Ded1. J. Mol. Biol. 2010, 396, 949–966. [Google Scholar] [CrossRef]
- Henn, A.; Cao, W.; Licciardello, N.; Heitkamp, S.E.; Hackney, D.D.; Enrique, M. Pathway of ATP utilization and duplex rRNA unwinding by the DEAD-box helicase, DbpA. Proc. Natl. Acad. Sci. USA 2010, 107, 4046–4050. [Google Scholar] [CrossRef]
- Liu, F.; Putnam, A.; Jankowsky, E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc. Natl. Acad. Sci. USA 2008, 105, 20209–20214. [Google Scholar] [CrossRef] [PubMed]
- Lorsch, J.R.; Herschlag, D. The DEAD box protein eIF4A. 1. A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry 1998, 37, 2180–2193. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Jankowsky, E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat. Struct. Mol. Biol. 2006, 13, 981–986. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Del Campo, M.; Lambowitz, A.M.; Jankowsky, E. DEAD-box proteins unwind duplexes by local strand separation. Mol. Cell 2007, 28, 253–263. [Google Scholar] [CrossRef]
- Rogers, G.W., Jr.; Richter, N.J.; Merrick, W.C. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem. 1999, 274, 12236–12244. [Google Scholar] [CrossRef]
- Fairman-Williams, M.E.; Guenther, U.-P.; Jankowsky, E. SF1 and SF2 helicases: Family matters. Curr. Opin. Struct. Biol. 2010, 20, 313–324. [Google Scholar] [CrossRef]
- Galicia-Vazquez, G.; Cencic, R.; Robert, F.; Agenor, A.Q.; Pelletier, J. A cellular response linking eIF4AI activity to eIF4AII transcription. RNA 2012, 18, 1373–1384. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, P.J.; Trachsel, H. The mouse protein synthesis initiation factor 4A gene family includes two related functional genes which are differentially expressed. EMBO J. 1988, 7, 2097–2105. [Google Scholar] [CrossRef]
- Williams-Hill, D.M.; Duncan, R.F.; Nielsen, P.J.; Tahara, S.M. Differential expression of the murine eukaryotic translation initiation factor isogenes eIF4A(I) and eIF4A(II) is dependent upon cellular growth status. Arch. Biochem. Biophys. 1997, 338, 111–120. [Google Scholar] [CrossRef]
- Chan, C.C.; Dostie, J.; Diem, M.D.; Feng, W.; Mann, M.; Rappsilber, J.; Dreyfuss, G. eIF4A3 is a novel component of the exon junction complex. RNA 2004, 10, 200–209. [Google Scholar] [CrossRef]
- Rozen, F.; Edery, I.; Meerovitch, K.; Dever, T.E.; Merrick, W.C.; Sonenberg, N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol. Cell. Biol. 1990, 10, 1134–1144. [Google Scholar] [CrossRef] [PubMed]
- Hilbert, M.; Kebbel, F.; Gubaev, A.; Klostermeier, D. eIF4G stimulates the activity of the DEAD box protein eIF4A by a conformational guidance mechanism. Nucleic Acids Res. 2011, 39, 2260–2270. [Google Scholar] [CrossRef] [PubMed]
- Peck, M.L.; Herschlag, D. Effects of oligonucleotide length and atomic composition on stimulation of the ATPase activity of translation initiation factor elF4A. RNA 1999, 5, 1210–1221. [Google Scholar] [CrossRef] [PubMed]
- Abramson, R.D.; Dever, T.E.; Lawson, T.G.; Ray, B.K.; Thach, R.E.; Merrick, W.C. The ATP-dependent interaction of eukaryotic initiation factors with mRNA. J. Biol. Chem. 1987, 262, 3826–3832. [Google Scholar]
- Pause, A.; Methot, N.; Sonenberg, N. The HRIGRXXR region of the DEAD box RNA helicase eukaryotic translation initiation factor 4A is required for RNA binding and ATP hydrolysis. Mol. Cell. Biol. 1993, 13, 6789–6798. [Google Scholar] [CrossRef][Green Version]
- Rogers, G.W., Jr.; Lima, W.F.; Merrick, W.C. Further characterization of the helicase activity of eIF4A. Substrate specificity. J. Biol. Chem. 2001, 276, 12598–12608. [Google Scholar] [CrossRef]
- Chu, J.; Zhang, W.; Cencic, R.; Devine, W.G.; Beglov, D.; Henkel, T.; Brown, L.E.; Vajda, S.; Porco, J.A.; Pelletier, J. Amidino-rocaglates: A potent class of eIF4A inhibitors. Cell Chem. Biol. 2019, 26, 1586–1593. [Google Scholar] [CrossRef]
- Guo, J.U.; Bartel, D.P. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science 2016, 353, aaf5371. [Google Scholar] [CrossRef]
- Waldron, J.A.; Raza, F.; Le Quesne, J. eIF4A alleviates the translational repression mediated by classical secondary structures more than by G-quadruplexes. Nucleic Acids Res. 2018, 46, 3075–3087. [Google Scholar] [CrossRef]
- Ben-Asouli, Y.; Banai, Y.; Pel-Or, Y.; Shir, A.; Kaempfer, R. Human interferon-γ mRNA autoregulates its translation through a pseudoknot that activates the interferon-inducible protein kinase PKR. Cell 2002, 108, 221–232. [Google Scholar] [CrossRef]
- Meijer, H.A.; Schmidt, T.; Gillen, S.L.; Langlais, C.; Jukes-Jones, R.; de Moor, C.H.; Cain, K.; Wilczynska, A.; Bushell, M. DEAD-box helicase eIF4A2 inhibits CNOT7 deadenylation activity. Nucleic Acids Res. 2019, 47, 8224–8238. [Google Scholar] [CrossRef] [PubMed]
- Wilczynska, A.; Gillen, S.L.; Schmidt, T.; Meijer, H.A.; Jukes-Jones, R.; Langlais, C.; Kopra, K.; Lu, W.-T.; Godfrey, J.D.; Hawley, B.R.; et al. eIF4A2 drives repression of translation at initiation by Ccr4-Not through purine-rich motifs in the 5’UTR. Genome Biol. 2019, 20, 262. [Google Scholar] [CrossRef] [PubMed]
- Webster, M.W.; Chen, Y.-H.; Stowell, J.A.; Alhusaini, N.; Sweet, T.; Graveley, B.R.; Coller, J.; Passmore, L.A. mRNA deadenylation is coupled to translation rates by the differential activities of Ccr4-Not nucleases. Mol. Cell 2018, 70, 1089–1100.e1088. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Park, J.; Ha, M.; Lim, J.; Chang, H.; Kim, V.N. PABP cooperates with the CCR4-NOT complex to promote mRNA deadenylation and block precocious decay. Mol. Cell 2018, 70, 1081–1088. [Google Scholar] [CrossRef]
- Braun, J.E.; Huntzinger, E.; Fauser, M.; Izaurralde, E. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol. Cell 2011, 44, 120–133. [Google Scholar] [CrossRef]
- Yamaji, M.; Jishage, M.; Meyer, C.; Suryawanshi, H.; Der, E.; Yamaji, M.; Garzia, A.; Morozov, P.; Manickavel, S.; McFarland, H.L. DND1 maintains germline stem cells via recruitment of the CCR4–NOT complex to target mRNAs. Nature 2017, 543, 568–572. [Google Scholar] [CrossRef]
- Meijer, H.A.; Kong, Y.W.; Lu, W.T.; Wilczynska, A.; Spriggs, R.V.; Robinson, S.W.; Godfrey, J.D.; Willis, A.E.; Bushell, M. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science 2013, 340, 82–85. [Google Scholar] [CrossRef]
- Conroy, S.C.; Dever, T.E.; Owens, C.L.; Merrick, W.C. Characterization of the 46,000-dalton subunit of eIF-4F. Arch. Biochem. Biophys. 1990, 282, 363–371. [Google Scholar] [CrossRef]
- Galicia-Vázquez, G.; Chu, J.; Pelletier, J. eIF4AII is dispensable for miRNA-mediated gene silencing. RNA 2015, 21, 1826–1833. [Google Scholar] [CrossRef]
- Rogers, G.W., Jr.; Richter, N.J.; Lima, W.F.; Merrick, W.C. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 2001, 276, 30914–30922. [Google Scholar] [CrossRef]
- Altmann, M.; Müller, P.; Wittmer, B.; Ruchti, F.; Lanker, S.; Trachsel, H. A Saccharomyces cerevisiae homologue of mammalian translation initiation factor 4B contributes to RNA helicase activity. EMBO J. 1993, 12, 3997–4003. [Google Scholar] [CrossRef] [PubMed]
- Özeş, A.R.; Feoktistova, K.; Avanzino, B.C.; Fraser, C.S. Duplex unwinding and ATPase activities of the DEAD-Box helicase eIF4A are coupled by eIF4G and eIF4B. J. Mol. Biol. 2011, 412, 674–687. [Google Scholar] [PubMed]
- García-García, C.; Frieda, K.L.; Feoktistova, K.; Fraser, C.S.; Block, S.M. RNA biochemistry. Factor-dependent processivity in human eIF4A DEAD-box helicase. Science 2015, 348, 1486–1488. [Google Scholar] [CrossRef] [PubMed]
- Pestova, T.V.; Lorsch, J.R.; Hellen, C.U. The mechanism of translation initiation in eukaryotes. Cold Spring Harb. Monogr. Ser. 2007, 48, 87. [Google Scholar]
- Sen, N.D.; Zhou, F.; Harris, M.S.; Ingolia, N.T.; Hinnebusch, A.G. eIF4B stimulates translation of long mRNAs with structured 5’ UTRs and low closed-loop potential but weak dependence on eIF4G. Proc. Natl. Acad. Sci. USA 2016, 113, 10464–10472. [Google Scholar] [CrossRef]
- Kulak, N.A.; Pichler, G.; Paron, I.; Nagaraj, N.; Mann, M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 2014, 11, 319–324. [Google Scholar] [CrossRef]
- Nielsen, K.H.; Behrens, M.A.; He, Y.; Oliveira, C.L.P.; Sottrup Jensen, L.; Hoffmann, S.V.; Pedersen, J.S.; Andersen, G.R. Synergistic activation of eIF4A by eIF4B and eIF4G. Nucleic Acids Res. 2010, 39, 2678–2689. [Google Scholar] [CrossRef]
- Andreou, A.Z.; Klostermeier, D. eIF4B and eIF4G jointly stimulate eIF4A ATPase and unwinding activities by modulation of the eIF4A conformational cycle. J. Mol. Biol. 2014, 426, 51–61. [Google Scholar] [CrossRef]
- Marintchev, A.; Edmonds, K.A.; Marintcheva, B.; Hendrickson, E.; Oberer, M.; Suzuki, C.; Herdy, B.; Sonenberg, N.; Wagner, G. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell 2009, 136, 447–460. [Google Scholar] [CrossRef]
- Richter, N.J.; Rogers, G.W., Jr.; Hensold, J.O.; Merrick, W.C. Further biochemical and kinetic characterization of human eukaryotic initiation factor 4H. J. Biol. Chem. 1999, 274, 35415–35424. [Google Scholar] [CrossRef]
- Chang, J.H.; Cho, Y.H.; Sohn, S.Y.; Choi, J.M.; Kim, A.; Kim, Y.C.; Jang, S.K.; Cho, Y. Crystal structure of the eIF4A-PDCD4 complex. Proc. Natl. Acad. Sci. USA 2009, 106, 3148–3153. [Google Scholar] [CrossRef] [PubMed]
- Dorrello, N.V.; Peschiaroli, A.; Guardavaccaro, D.; Colburn, N.H.; Sherman, N.E.; Pagano, M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science 2006, 314, 467–471. [Google Scholar] [CrossRef]
- Liwak, U.; Thakor, N.; Jordan, L.E.; Roy, R.; Lewis, S.M.; Pardo, O.E.; Seckl, M.; Holcik, M. Tumor suppressor PDCD4 represses internal ribosome entry site-mediated translation of antiapoptotic proteins and is regulated by S6 kinase 2. Mol. Cell. Biol. 2012, 32, 1818–1829. [Google Scholar] [CrossRef] [PubMed]
- Samson, J.; Cronin, S.; Dean, K. BC200 (BCYRN1)—The shortest, long, non-coding RNA associated with cancer. Non-Coding RNA Res. 2018, 3, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Iacoangeli, A.; Popp, S.; Muslimov, I.A.; Imataka, H.; Sonenberg, N.; Lomakin, I.B.; Tiedge, H. Dendritic BC1 RNA: Functional role in regulation of translation initiation. J. Neurosci. 2002, 22, 10232–10241. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Pestova, T.V.; Hellen, C.U.; Tiedge, H. Translational control by a small RNA: Dendritic BC1 RNA targets the eukaryotic initiation factor 4A helicase mechanism. Mol. Cell. Biol. 2008, 28, 3008–3019. [Google Scholar] [CrossRef] [PubMed]
- Tiedge, H.; Fremeau, R.T.; Weinstock, P.H.; Arancio, O.; Brosius, J. Dendritic location of neural BC1 RNA. Proc. Natl. Acad. Sci. USA 1991, 88, 2093–2097. [Google Scholar] [CrossRef] [PubMed]
- Sokabe, M.; Fraser, C.S. A helicase-independent activity of eIF4A in promoting mRNA recruitment to the human ribosome. Proc. Natl. Acad. Sci. USA 2017, 114, 6304–6309. [Google Scholar] [CrossRef]
- Yourik, P.; Aitken, C.E.; Zhou, F.; Gupta, N.; Hinnebusch, A.G.; Lorsch, J.R. Yeast eIF4A enhances recruitment of mRNAs regardless of their structural complexity. ELife 2017, 6, e31476. [Google Scholar] [CrossRef]
- Tauber, D.; Tauber, G.; Khong, A.; Van Treeck, B.; Pelletier, J.; Parker, R. Modulation of RNA condensation by the DEAD-Box protein eIF4A. Cell 2020, 180, 411–426. [Google Scholar] [CrossRef]
- Jain, S.; Wheeler, J.R.; Walters, R.W.; Agrawal, A.; Barsic, A.; Parker, R. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 2016, 164, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Al-Husini, N.; Tomares, D.T.; Bitar, O.; Childers, W.S.; Schrader, J.M. α-Proteobacterial RNA degradosomes assemble liquid-liquid phase-separated RNP bodies. Mol. Cell 2018, 71, 1027–1039. [Google Scholar] [CrossRef] [PubMed]
- Hondele, M.; Sachdev, R.; Heinrich, S.; Wang, J.; Vallotton, P.; Fontoura, B.M.; Weis, K. DEAD-box ATPases are global regulators of phase-separated organelles. Nature 2019, 573, 144–148. [Google Scholar] [CrossRef]
- Sharma, D.; Jankowsky, E. The Ded1/DDX3 subfamily of DEAD-box RNA helicases. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 343–360. [Google Scholar] [CrossRef]
- Iost, I.; Dreyfus, M.; Linder, P. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J. Biol. Chem. 1999, 274, 17677–17683. [Google Scholar] [CrossRef] [PubMed]
- Soto-Rifo, R.; Ohlmann, T. The role of the DEAD-box RNA helicase DDX3 in mRNA metabolism. Wires RNA 2013, 4, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Ditton, H.; Zimmer, J.; Kamp, C.; Rajpert-De Meyts, E.; Vogt, P. The AZFa gene DBY (DDX3Y) is widely transcribed but the protein is limited to the male germ cells by translation control. Hum. Mol. Genet. 2004, 13, 2333–2341. [Google Scholar] [CrossRef]
- Högbom, M.; Collins, R.; van den Berg, S.; Jenvert, R.-M.; Karlberg, T.; Kotenyova, T.; Flores, A.; Hedestam, G.B.K.; Schiavone, L.H. Crystal structure of conserved domains 1 and 2 of the human DEAD-box helicase DDX3X in complex with the mononucleotide AMP. J. Mol. Biol. 2007, 372, 150–159. [Google Scholar] [CrossRef]
- Garbelli, A.; Beermann, S.; Di Cicco, G.; Dietrich, U.; Maga, G. A motif unique to the human Dead-box protein DDX3 is important for nucleic acid binding, ATP hydrolysis, RNA/DNA unwinding and HIV-1 replication. PLoS ONE 2011, 6, e19810. [Google Scholar] [CrossRef]
- Franca, R.; Belfiore, A.; Spadari, S.; Maga, G. Human DEAD-box ATPase DDX3 shows a relaxed nucleoside substrate specificity. Proteins 2007, 67, 1128–1137. [Google Scholar] [CrossRef]
- Yedavalli, V.S.; Neuveut, C.; Chi, Y.H.; Kleiman, L.; Jeang, K.T. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 2004, 119, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Ko, C.; Lee, S.; Windisch, M.P.; Ryu, W.-S. DDX3 DEAD-Box rna helicase is a host factor that restricts hepatitis B virus replication at the transcriptional level. J. Virol. 2014, 88, 13689–13698. [Google Scholar] [CrossRef] [PubMed]
- Pek, J.W.; Kai, T. DEAD-box RNA helicase Belle/DDX3 and the RNA interference pathway promote mitotic chromosome segregation. Proc. Natl. Acad. Sci. USA 2011, 108, 12007–12012. [Google Scholar] [CrossRef] [PubMed]
- Topisirovic, I.; Siddiqui, N.; Lapointe, V.L.; Trost, M.; Thibault, P.; Bangeranye, C.; Pinol-Roma, S.; Borden, K.L. Molecular dissection of the eukaryotic initiation factor 4E (eIF4E) export-competent RNP. EMBO J. 2009, 28, 1087–1098. [Google Scholar] [CrossRef]
- Lee, C.-S.; Dias, A.P.; Jedrychowski, M.; Patel, A.H.; Hsu, J.L.; Reed, R. Human DDX3 functions in translation and interacts with the translation initiation factor eIF3. Nucleic Acids Res. 2008, 36, 4708–4718. [Google Scholar] [CrossRef]
- Geissler, R.; Golbik, R.P.; Behrens, S.-E. The DEAD-box helicase DDX3 supports the assembly of functional 80S ribosomes. Nucleic Acids Res. 2012, 40, 4998–5011. [Google Scholar] [CrossRef]
- Shih, J.; Tsai, T.; Chao, C.-H.; Lee, Y.W. Candidate tumor suppressor DDX3 RNA helicase specifically represses cap-dependent translation by acting as an eIF4E inhibitory protein. Oncogene 2008, 27, 700–714. [Google Scholar] [CrossRef]
- Hilliker, A.; Gao, Z.; Jankowsky, E.; Parker, R. The DEAD-Box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Mol. Cell 2011, 43, 962–972. [Google Scholar] [CrossRef]
- Beckham, C.; Hilliker, A.; Cziko, A.-M.; Noueiry, A.; Ramaswami, M.; Parker, R. The DEAD-box RNA helicase Ded1p affects and accumulates in Saccharomyces cerevisiae P-bodies. Mol. Biol. Cell 2008, 19, 984–993. [Google Scholar] [CrossRef]
- Lai, M.-C.; Chang, W.-C.; Shieh, S.-Y.; Tarn, W.-Y. DDX3 regulates cell growth through translational control of cyclin E1. Mol. Cell. Biol. 2010, 30, 5444–5453. [Google Scholar] [CrossRef]
- Soto-Rifo, R.; Rubilar, P.S.; Limousin, T.; De Breyne, S.; Decimo, D.; Ohlmann, T. DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J. 2012, 31, 3745–3756. [Google Scholar] [CrossRef] [PubMed]
- Sen, N.D.; Zhou, F.; Ingolia, N.T.; Hinnebusch, A.G. Genome-wide analysis of translational efficiency reveals distinct but overlapping functions of yeast DEAD-box RNA helicases Ded1 and eIF4A. Genome Res. 2015, 25, 1196–1205. [Google Scholar] [CrossRef]
- Guenther, U.-P.; Weinberg, D.E.; Zubradt, M.M.; Tedeschi, F.A.; Stawicki, B.N.; Zagore, L.L.; Brar, G.A.; Licatalosi, D.D.; Bartel, D.P.; Weissman, J.S. The helicase Ded1p controls use of near-cognate translation initiation codons in 5’ UTRs. Nature 2018, 559, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Iserman, C.; Desroches Altamirano, C.; Jegers, C.; Friedrich, U.; Zarin, T.; Fritsch, A.W.; Mittasch, M.; Domingues, A.; Hersemann, L.; Jahnel, M.; et al. Condensation of Ded1p promotes a translational switch from housekeeping to stress protein production. Cell 2020, 181, 818–831. [Google Scholar] [CrossRef] [PubMed]
- Dhote, V.; Sweeney, T.R.; Kim, N.; Hellen, C.U.; Pestova, T.V. Roles of individual domains in the function of DHX29, an essential factor required for translation of structured mammalian mRNAs. Proc. Natl. Acad. Sci. USA 2012, 109, E3150–E3159. [Google Scholar] [CrossRef] [PubMed]
- Pisareva, V.P.; Pisarev, A.V.; Komar, A.A.; Hellen, C.U.T.; Pestova, T.V. Translation initiation on mammalian mRNAs with structured 5’UTRs requires DExH-Box protein DHX29. Cell 2008, 135, 1237–1250. [Google Scholar] [CrossRef]
- Parsyan, A.; Shahbazian, D.; Martineau, Y.; Petroulakis, E.; Alain, T.; Larsson, O.; Mathonnet, G.; Tettweiler, G.; Hellen, C.U.; Pestova, T.V. The helicase protein DHX29 promotes translation initiation, cell proliferation, and tumorigenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 22217–22222. [Google Scholar] [CrossRef]
- Hashem, Y.; des Georges, A.; Dhote, V.; Langlois, R.; Liao, H.Y.; Grassucci, R.A.; Hellen, C.U.; Pestova, T.V.; Frank, J. Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell 2013, 153, 1108–1119. [Google Scholar] [CrossRef]
- Abaeva, I.S.; Marintchev, A.; Pisareva, V.P.; Hellen, C.U.; Pestova, T.V. Bypassing of stems versus linear base-by-base inspection of mammalian mRNAs during ribosomal scanning. EMBO J. 2011, 30, 115–129. [Google Scholar] [CrossRef]
- Pisareva, V.P.; Pisarev, A.V. DHX29 and eIF3 cooperate in ribosomal scanning on structured mRNAs during translation initiation. RNA 2016, 22, 1859–1870. [Google Scholar] [CrossRef]
- Des Georges, A.; Dhote, V.; Kuhn, L.; Hellen, C.U.; Pestova, T.V.; Frank, J.; Hashem, Y. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature 2015, 525, 491–495. [Google Scholar] [CrossRef]
- Pisareva, V.P.; Pisarev, A.V. DHX29 reduces leaky scanning through an upstream AUG codon regardless of its nucleotide context. Nucleic Acids Res. 2016, 44, 4252–4265. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Tan, P.; Li, Y.; Lin, M.; Li, C.; Mao, J.; Cui, J.; Zhao, W.; Wang, H.Y.; Wang, R.-F. DHX29 functions as a RNA co-sensor for MDA5-mediated EMCV-specific antiviral immunity. PLoS Pathog. 2018, 14, e1006886. [Google Scholar] [CrossRef]
- Giri, B.; Smaldino, P.J.; Thys, R.G.; Creacy, S.D.; Routh, E.D.; Hantgan, R.R.; Lattmann, S.; Nagamine, Y.; Akman, S.A.; Vaughn, J.P. G4 resolvase 1 tightly binds and unwinds unimolecular G4-DNA. Nucleic Acids Res. 2011, 39, 7161–7178. [Google Scholar] [CrossRef] [PubMed]
- Vaughn, J.P.; Creacy, S.D.; Routh, E.D.; Joyner-Butt, C.; Jenkins, G.S.; Pauli, S.; Nagamine, Y.; Akman, S.A. The DEXH protein product of the DHX36 gene is the major source of tetramolecular quadruplex G4-DNA resolving activity in HeLa cell lysates. J. Biol. Chem. 2005, 280, 38117–38120. [Google Scholar] [CrossRef] [PubMed]
- Lattmann, S.; Stadler, M.B.; Vaughn, J.P.; Akman, S.A.; Nagamine, Y. The DEAH-box RNA helicase RHAU binds an intramolecular RNA G-quadruplex in TERC and associates with telomerase holoenzyme. Nucleic Acids Res. 2011, 39, 9390–9404. [Google Scholar] [CrossRef]
- Nie, J.; Jiang, M.; Zhang, X.; Tang, H.; Jin, H.; Huang, X.; Yuan, B.; Zhang, C.; Lai, J.C.; Nagamine, Y. Post-transcriptional regulation of Nkx2–5 by RHAU in heart development. Cell Rep. 2015, 13, 723–732. [Google Scholar] [CrossRef]
- Lai, J.C.; Ponti, S.; Pan, D.; Kohler, H.; Skoda, R.C.; Matthias, P.; Nagamine, Y. The DEAH-box helicase RHAU is an essential gene and critical for mouse hematopoiesis. Blood J. Am. Soc. Hematol. 2012, 119, 4291–4300. [Google Scholar] [CrossRef]
- Sexton, A.N.; Collins, K. The 5’ guanosine tracts of human telomerase RNA are recognized by the G-quadruplex binding domain of the RNA helicase DHX36 and function to increase RNA accumulation. Mol. Cell. Biol. 2011, 31, 736–743. [Google Scholar] [CrossRef]
- Thandapani, P.; Song, J.; Gandin, V.; Cai, Y.; Rouleau, S.G.; Garant, J.-M.; Boisvert, F.-M.; Yu, Z.; Perreault, J.-P.; Topisirovic, I. Aven recognition of RNA G-quadruplexes regulates translation of the mixed lineage leukemia protooncogenes. Elife 2015, 4, e06234. [Google Scholar] [CrossRef]
- Murat, P.; Marsico, G.; Herdy, B.; Ghanbarian, A.; Portella, G.; Balasubramanian, S. RNA G-quadruplexes at upstream open reading frames cause DHX36-and DHX9-dependent translation of human mRNAs. Genome Biol. 2018, 19, 229. [Google Scholar] [CrossRef] [PubMed]
- Vester, K.; Eravci, M.; Serikawa, T.; Schütze, T.; Weise, C.; Kurreck, J. RNAi-mediated knockdown of the Rhau helicase preferentially depletes proteins with a Guanine-quadruplex motif in the 5’-UTR of their mRNA. Biochem. Biophys. Res. Commun. 2019, 508, 756–761. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.C.; Tippana, R.; Demeshkina, N.A.; Murat, P.; Balasubramanian, S.; Myong, S.; Ferré-D’Amaré, A.R. Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36. Nature 2018, 558, 465–469. [Google Scholar] [CrossRef]
- You, H.; Lattmann, S.; Rhodes, D.; Yan, J. RHAU helicase stabilizes G4 in its nucleotide-free state and destabilizes G4 upon ATP hydrolysis. Nucleic Acids Res. 2017, 45, 206–214. [Google Scholar] [CrossRef]
- Booy, E.P.; McRae, E.K.; Howard, R.; Deo, S.R.; Ariyo, E.O.; Dzananovic, E.; Meier, M.; Stetefeld, J.; McKenna, S.A. RNA helicase associated with AU-rich element (RHAU/DHX36) interacts with the 3’-tail of the long non-coding RNA BC200 (BCYRN1). J. Biol. Chem. 2016, 291, 5355–5372. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Pelletier, J. The biology of DHX9 and its potential as a therapeutic target. Oncotarget 2016, 7, 42716. [Google Scholar] [CrossRef]
- Roberts, T.M.; Boris-Lawrie, K. Primary sequence and secondary structure motifs in spleen necrosis virus RU5 confer translational utilization of unspliced human immunodeficiency virus type 1 reporter RNA. J. Virol. 2003, 77, 11973–11984. [Google Scholar] [CrossRef]
- Bolinger, C.; Sharma, A.; Singh, D.; Yu, L.; Boris-Lawrie, K. RNA helicase A modulates translation of HIV-1 and infectivity of progeny virions. Nucleic Acids Res. 2010, 38, 1686–1696. [Google Scholar] [CrossRef]
- Ranji, A.; Shkriabai, N.; Kvaratskhelia, M.; Musier-Forsyth, K.; Boris-Lawrie, K. Features of double-stranded RNA-binding domains of RNA helicase A are necessary for selective recognition and translation of complex mRNAs. J. Biol. Chem. 2011, 286, 5328–5337. [Google Scholar] [CrossRef]
- Short, J.D.; Pfarr, C.M. Translational regulation of the JunD messenger RNA. J. Biol. Chem. 2002, 277, 32697–32705. [Google Scholar] [CrossRef]
- Hartman, T.R.; Qian, S.; Bolinger, C.; Fernandez, S.; Schoenberg, D.R.; Boris-Lawrie, K. RNA helicase A is necessary for translation of selected messenger RNAs. Nat. Struct. Mol. Biol. 2006, 13, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Fritz, D.; Stefanovic, L.; Stefanovic, B. Binding of LARP6 to the conserved 5’ stem–loop regulates translation of mRNAs encoding type I collagen. J. Mol. Biol. 2010, 395, 309–326. [Google Scholar] [CrossRef] [PubMed]
- Manojlovic, Z.; Stefanovic, B. A novel role of RNA helicase A in regulation of translation of type I collagen mRNAs. RNA 2012, 18, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Halaby, M.-J.; Li, Y.; Harris, B.R.; Jiang, S.; Miskimins, W.K.; Cleary, M.P.; Yang, D.-Q. Translational control protein 80 stimulates IRES-mediated translation of p53 mRNA in response to DNA damage. BioMed Res. Int. 2015, 2015, 708158. [Google Scholar] [CrossRef]
- Halaby, M.-J.; Harris, B.R.; Miskimins, W.K.; Cleary, M.P.; Yang, D.-Q. Deregulation of internal ribosome entry site-mediated p53 translation in cancer cells with defective p53 response to DNA damage. Mol. Cell. Biol. 2015, 35, 4006–4017. [Google Scholar] [CrossRef]
- Chakraborty, P.; Grosse, F. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair 2011, 10, 654–665. [Google Scholar] [CrossRef]
- Zhang, Y.; Forys, J.T.; Miceli, A.P.; Gwinn, A.S.; Weber, J.D. Identification of DHX33 as a mediator of rRNA synthesis and cell growth. Mol. Cell. Biol. 2011, 31, 4676–4691. [Google Scholar] [CrossRef]
- Zhang, L.; Yang, S.; Chen, X.; Stauffer, S.; Yu, F.; Lele, S.M.; Fu, K.; Datta, K.; Palermo, N.; Chen, Y. The hippo pathway effector YAP regulates motility, invasion, and castration-resistant growth of prostate cancer cells. Mol. Cell. Biol. 2015, 35, 1350–1362. [Google Scholar] [CrossRef]
- Zhang, Y.; You, J.; Wang, X.; Weber, J. The DHX33 RNA helicase promotes mRNA translation initiation. Mol. Cell. Biol. 2015, 35, 2918–2931. [Google Scholar] [CrossRef][Green Version]
- Wang, H.; Yu, J.; Wang, X.; Zhang, Y. The RNA helicase DHX33 is required for cancer cell proliferation in human glioblastoma and confers resistance to PI3K/mTOR inhibition. Cell. Signal. 2019, 54, 170–178. [Google Scholar] [CrossRef]
- Li, Z.; Zhou, M.; Cai, Z.; Liu, H.; Zhong, W.; Hao, Q.; Cheng, D.; Hu, X.; Hou, J.; Xu, P. RNA-binding protein DDX1 is responsible for fatty acid-mediated repression of insulin translation. Nucleic Acids Res. 2018, 46, 12052–12066. [Google Scholar] [CrossRef] [PubMed]
- Pazo, A.; Pérez-González, A.; Oliveros, J.C.; Huarte, M.; Chavez, J.P.; Nieto, A. hCLE/RTRAF-HSPC117-DDX1-FAM98B: A new cap-binding complex that activates mRNA translation. Front. Physiol. 2019, 10, 92. [Google Scholar] [CrossRef] [PubMed]
- Peters, D.; Radine, C.; Reese, A.; Budach, W.; Sohn, D.; Jänicke, R.U. The DEAD-box RNA helicase DDX41 is a novel repressor of p21WAF1/CIP1 mRNA translation. J. Biol. Chem. 2017, 292, 8331–8341. [Google Scholar] [CrossRef]
- Ostareck, D.H.; Naarmann-de Vries, I.S.; Ostareck-Lederer, A. DDX6 and its orthologs as modulators of cellular and viral RNA expression. Wiley Interdiscip. Rev. RNA 2014, 5, 659–678. [Google Scholar] [CrossRef] [PubMed]
- Mathys, H.; Basquin, J.; Ozgur, S.; Czarnocki-Cieciura, M.; Bonneau, F.; Aartse, A.; Dziembowski, A.; Nowotny, M.; Conti, E.; Filipowicz, W. Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Mol. Cell 2014, 54, 751–765. [Google Scholar] [CrossRef]
- Chen, Y.; Boland, A.; Kuzuoğlu-Öztürk, D.; Bawankar, P.; Loh, B.; Chang, C.-T.; Weichenrieder, O.; Izaurralde, E. A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol. Cell 2014, 54, 737–750. [Google Scholar] [CrossRef] [PubMed]
- Ozgur, S.; Basquin, J.; Kamenska, A.; Filipowicz, W.; Standart, N.; Conti, E. Structure of a human 4E-T/DDX6/CNOT1 complex reveals the different interplay of DDX6-binding proteins with the CCR4-NOT complex. Cell Rep. 2015, 13, 703–711. [Google Scholar] [CrossRef]
- Chapat, C.; Jafarnejad, S.M.; Matta-Camacho, E.; Hesketh, G.G.; Gelbart, I.A.; Attig, J.; Gkogkas, C.G.; Alain, T.; Stern-Ginossar, N.; Fabian, M.R. Cap-binding protein 4EHP effects translation silencing by microRNAs. Proc. Natl. Acad. Sci. USA 2017, 114, 5425–5430. [Google Scholar] [CrossRef]
- Nishimura, T.; Padamsi, Z.; Fakim, H.; Milette, S.; Dunham, W.H.; Gingras, A.-C.; Fabian, M.R. The eIF4E-binding protein 4E-T is a component of the mRNA decay machinery that bridges the 5’ and 3’ termini of target mRNAs. Cell Rep. 2015, 11, 1425–1436. [Google Scholar] [CrossRef]
- Hashimoto, K.; Nakagawa, Y.; Morikawa, H.; Niki, M.; Egashira, Y.; Hirata, I.; Katsu, K.; Akao, Y. Co-overexpression of DEAD box protein rck/p54 and c-myc protein in human colorectal adenomas and the relevance of their expression in cultured cell lines. Carcinogenesis 2001, 22, 1965–1970. [Google Scholar] [CrossRef]
- Lin, C.-J.; Nasr, Z.; Premsrirut, P.K.; Porco, J.A., Jr.; Hippo, Y.; Lowe, S.W.; Pelletier, J. Targeting synthetic lethal interactions between Myc and the eIF4F complex impedes tumorigenesis. Cell Rep. 2012, 1, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Nanbru, C.; Lafon, I.; Audigier, S.; Gensac, M.-C.; Vagner, S.; Huez, G.; Prats, A.-C. Alternative translation of the proto-oncogene c-mycby an internal ribosome entry site. J. Biol. Chem. 1997, 272, 32061–32066. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Hogetsu, K.; Usukura, J.; Sato, T.; Kumasaka, T.; Akao, Y.; Tanaka, N. Structural insight of human DEAD-box protein rck/p54 into its substrate recognition with conformational changes. Genes Cells 2006, 11, 439–452. [Google Scholar] [CrossRef] [PubMed]
- De Vries, S.; Naarmann-de Vries, I.S.; Urlaub, H.; Lue, H.; Bernhagen, J.; Ostareck, D.H.; Ostareck-Lederer, A. Identification of DEAD-box RNA helicase 6 (DDX6) as a cellular modulator of vascular endothelial growth factor expression under hypoxia. J. Biol. Chem. 2013, 288, 5815–5827. [Google Scholar] [CrossRef]
- Bhat, M.; Robichaud, N.; Hulea, L.; Sonenberg, N.; Pelletier, J.; Topisirovic, I. Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 2015, 14, 261–278. [Google Scholar] [CrossRef]
- Pelletier, J.; Graff, J.; Ruggero, D.; Sonenberg, N. Targeting the eIF4F translation initiation complex: A critical nexus for cancer development. Cancer Res. 2015, 75, 250–263. [Google Scholar] [CrossRef]
- Lazaris-Karatzas, A.; Montine, K.S.; Sonenberg, N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5’ cap. Nature 1990, 345, 544–547. [Google Scholar] [CrossRef]
- Ruggero, D.; Montanaro, L.; Ma, L.; Xu, W.; Londei, P.; Cordon-Cardo, C.; Pandolfi, P.P. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat. Med. 2004, 10, 484–486. [Google Scholar] [CrossRef]
- Wendel, H.G.; De Stanchina, E.; Fridman, J.S.; Malina, A.; Ray, S.; Kogan, S.; Cordon-Cardo, C.; Pelletier, J.; Lowe, S.W. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature 2004, 428, 332–337. [Google Scholar] [CrossRef]
- Yángüez, E.; Castello, A.; Welnowska, E.; Carrasco, L.; Goodfellow, I.; Nieto, A. Functional impairment of eIF4A and eIF4G factors correlates with inhibition of influenza virus mRNA translation. Virology 2011, 413, 93–102. [Google Scholar] [CrossRef]
- Surakasi, V.; Nalini, M.; Kim, Y. Host translational control of a polydnavirus, Cotesia plutellae bracovirus, by sequestering host eIF4A to prevent formation of a translation initiation complex. Insect Mol. Biol. 2011, 20, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Hosmillo, M.; Sweeney, T.R.; Chaudhry, Y.; Leen, E.; Curry, S.; Goodfellow, I.; Cho, K.-O. The RNA Helicase eIF4A is required for Sapovirus translation. J. Virol. 2016, 90, 5200–5204. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Pelletier, J. Selective targeting of the DEAD-box RNA helicase eukaryotic initiation factor (eIF) 4A by natural products. Nat. Prod. Rep. 2020. [Google Scholar] [CrossRef]
- Iwasaki, S.; Iwasaki, W.; Takahashi, M.; Sakamoto, A.; Watanabe, C.; Shichino, Y.; Floor, S.N.; Fujiwara, K.; Mito, M.; Dodo, K.; et al. The translation inhibitor rocaglamide targets a bimolecular cavity between eIF4A and polypurine RNA. Mol. Cell 2019, 73, 738–748. [Google Scholar] [CrossRef] [PubMed]
- Bordeleau, M.E.; Matthews, J.; Wojnar, J.M.; Lindqvist, L.; Novac, O.; Jankowsky, E.; Sonenberg, N.; Northcote, P.; Teesdale-Spittle, P.; Pelletier, J. Stimulation of mammalian translation initiation factor eIF4A activity by a small molecule inhibitor of eukaryotic translation. Proc. Natl. Acad. Sci. USA 2005, 102, 10460–10465. [Google Scholar] [CrossRef] [PubMed]
- Chu, J.; Zhang, W.; Cencic, R.; O’Connor, P.B.; Robert, F.; Devine, W.G.; Selznick, A.; Henkel, T.; Merrick, W.C.; Brown, L.E. Rocaglates induce gain-of-function alterations to eIF4A and eIF4F. Cell Rep. 2020, 30, 2481–2488. [Google Scholar] [CrossRef]
- Bordeleau, M.E.; Robert, F.; Gerard, B.; Lindqvist, L.; Chen, S.M.; Wendel, H.G.; Brem, B.; Greger, H.; Lowe, S.W.; Porco, J.A., Jr.; et al. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J. Clin. Investig. 2008, 118, 2651–2660. [Google Scholar] [CrossRef]
- Lindqvist, L.; Oberer, M.; Reibarkh, M.; Cencic, R.; Bordeleau, M.E.; Vogt, E.; Marintchev, A.; Tanaka, J.; Fagotto, F.; Altmann, M.; et al. Selective pharmacological targeting of a DEAD box RNA helicase. PLoS ONE 2008, 3, e1583. [Google Scholar] [CrossRef]
- Iwatani-Yoshihara, M.; Ito, M.; Ishibashi, Y.; Oki, H.; Tanaka, T.; Morishita, D.; Ito, T.; Kimura, H.; Imaeda, Y.; Aparicio, S. Discovery and characterization of a eukaryotic initiation factor 4a-3-selective inhibitor that suppresses nonsense-mediated mrna decay. ACS Chem. Biol. 2017, 12, 1760–1768. [Google Scholar] [CrossRef]
- Cencic, R.; Naineni, S.K.; Pugsley, L.; Senechal, P.; Sahni, A.; Pelletier, J. A CRISPR-based screen links an inhibitor of nonsense-mediated decay to eIF4A3 target engagement. ACS Chem. Biol. 2020. [Google Scholar] [CrossRef]
- Yang, S.N.; Atkinson, S.C.; Audsley, M.D.; Heaton, S.M.; Jans, D.A.; Borg, N.A. RK-33 is a broad-spectrum antiviral agent that targets DEAD-Box RNA helicase DDX3X. Cells 2020, 9, 170. [Google Scholar] [CrossRef] [PubMed]
- Nakao, S.; Nogami, M.; Iwatani, M.; Imaeda, T.; Ito, M.; Tanaka, T.; Tawada, M.; Endo, S.; Cary, D.R.; Ohori, M. Identification of a selective DDX3X inhibitor with newly developed quantitative high-throughput RNA helicase assays. Biochem. Biophys. Res. Commun. 2020, 523, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama-Hirozane, M.; Kondo, M.; Matsumoto, S.-I.; Morikawa-Oki, A.; Morishita, D.; Nakanishi, A.; Kawamoto, T.; Nakayama, M. High-throughput screening to identify inhibitors of DEAD box helicase DDX41. SLAS Discov. Adv. Life Sci. R D 2017, 22, 1084–1092. [Google Scholar] [CrossRef] [PubMed][Green Version]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).