Liver Fibrosis and Inflammation under the Control of ERK2
Abstract
1. Introduction
2. Results
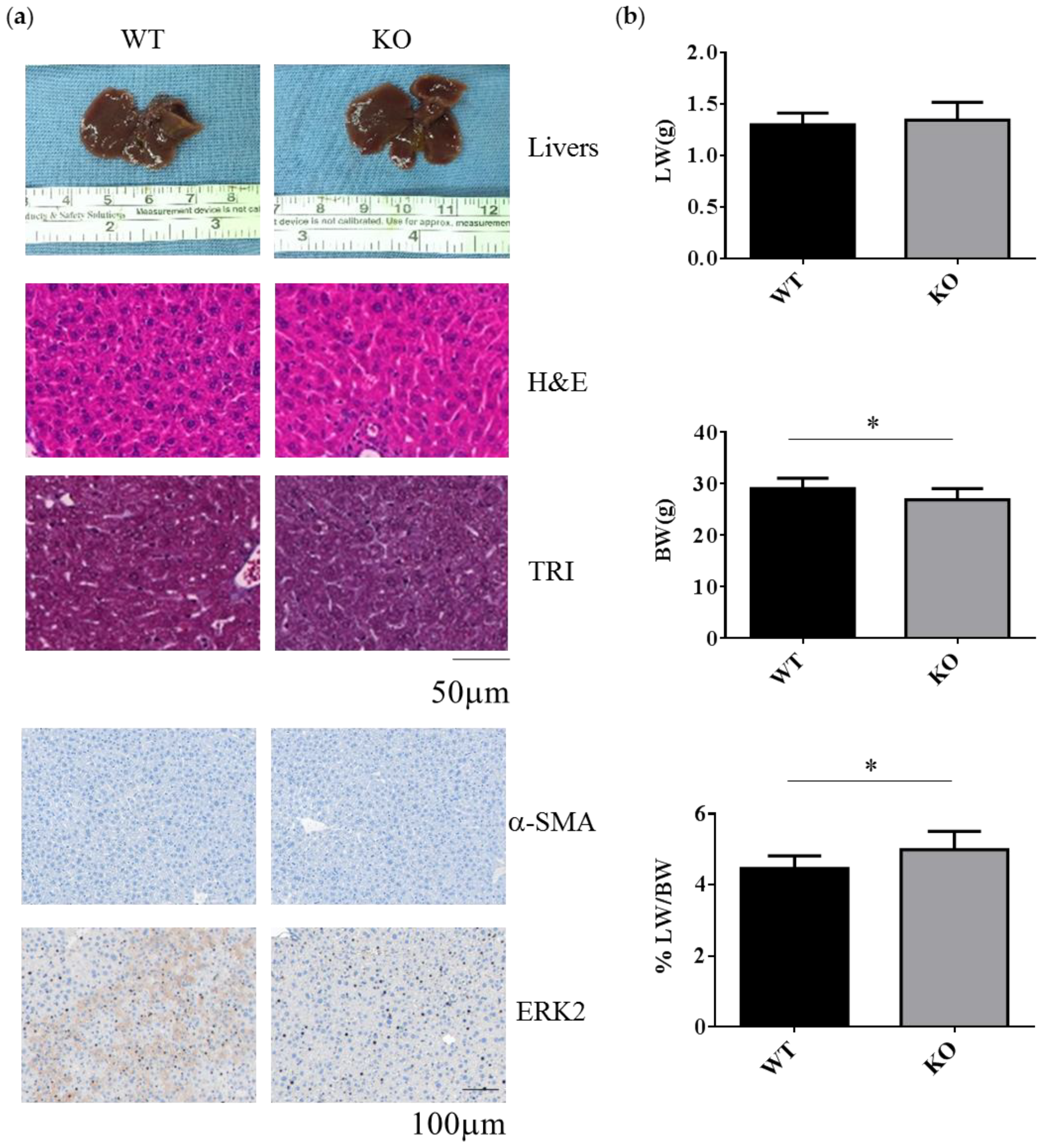
2.1. Erk2-Deficient Mice Displayed Normal Liver but Reduced Body Weight under the Normal Chow Diet
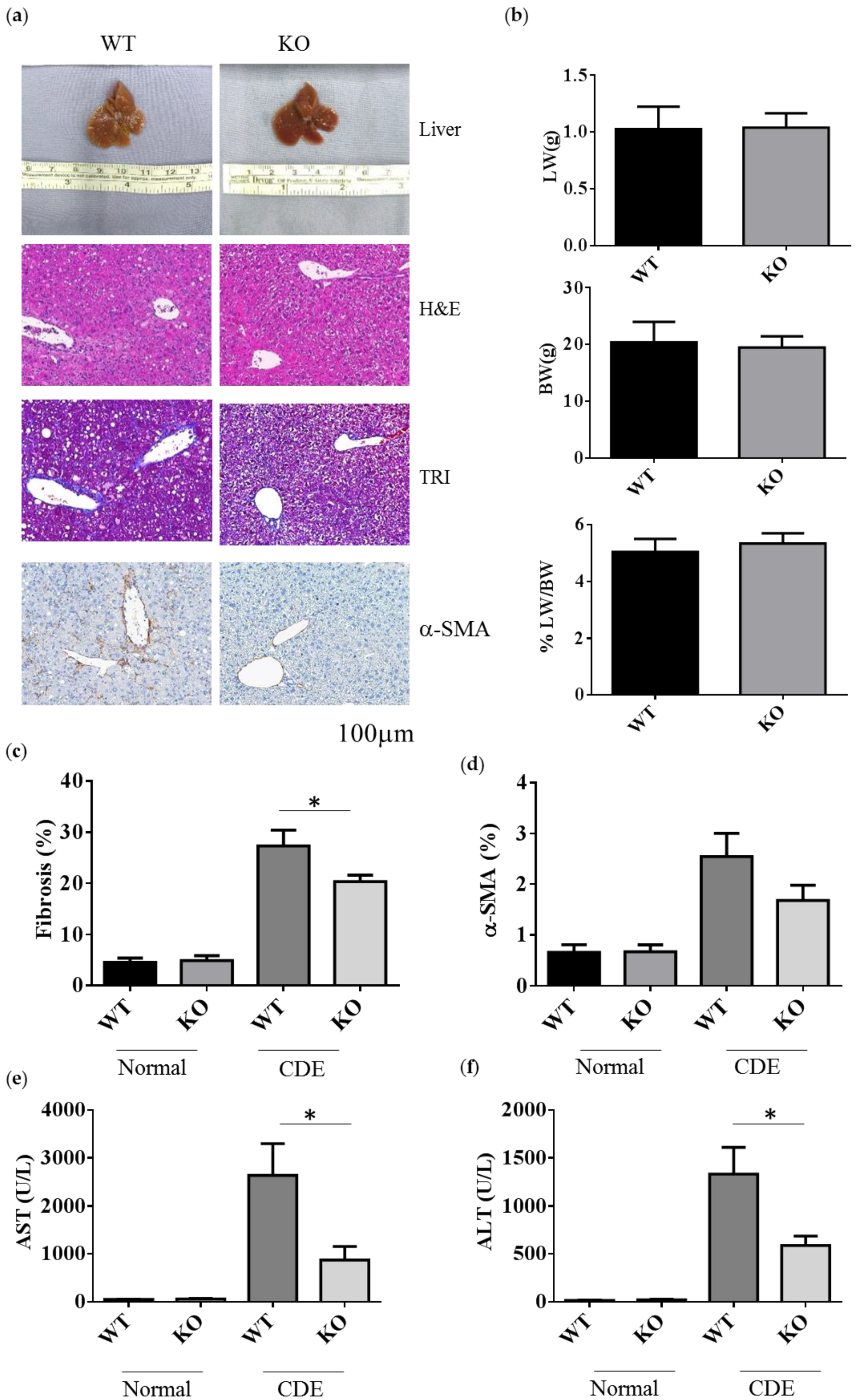
2.2. Erk2-/- Mice Displayed Less Degree of Liver Fibrosis upon Short-Term Liver Injury
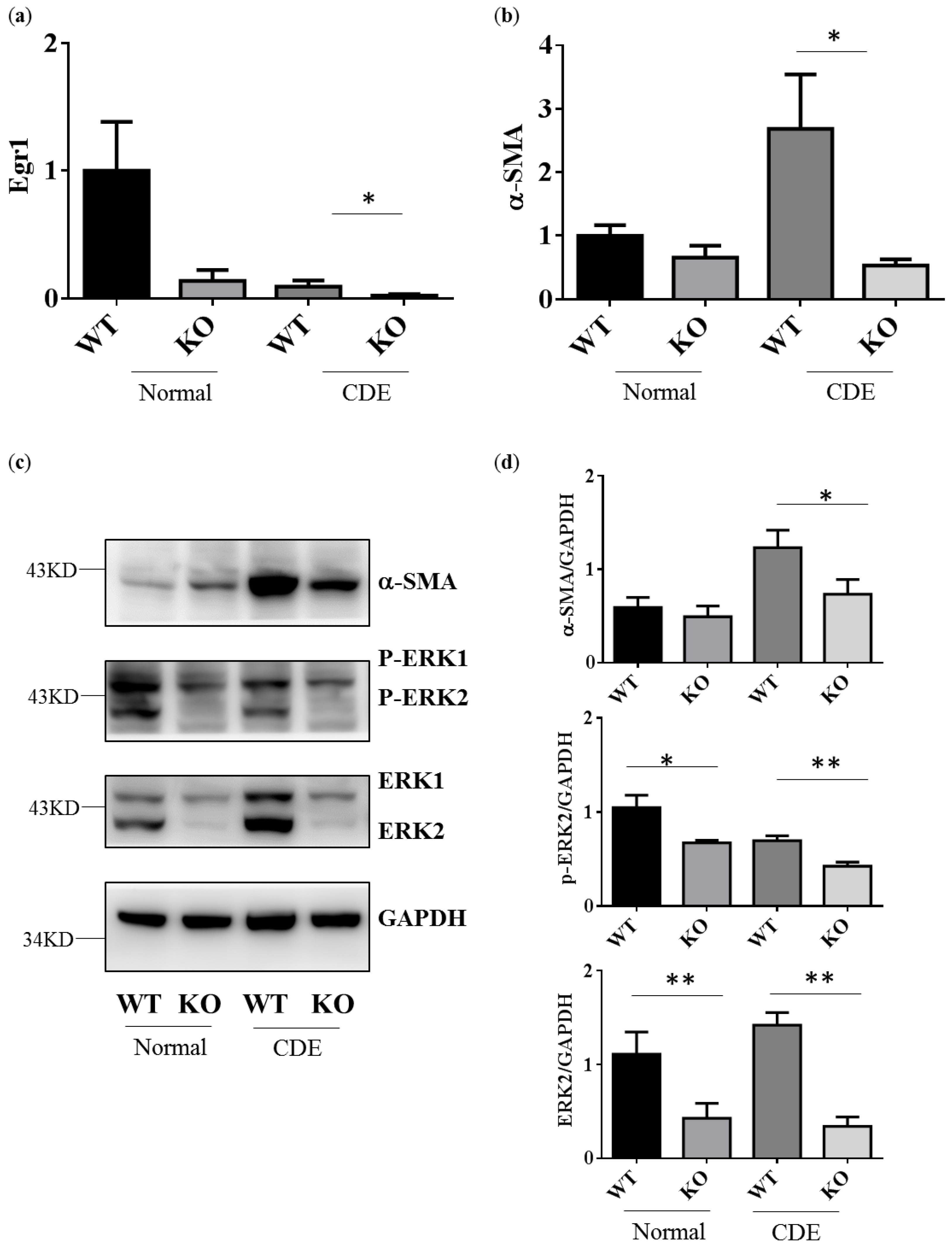
2.3. Reduction of Gene and Protein Expression Level in Fibrosis Genes alpha-SMA in Erk2−/− Livers
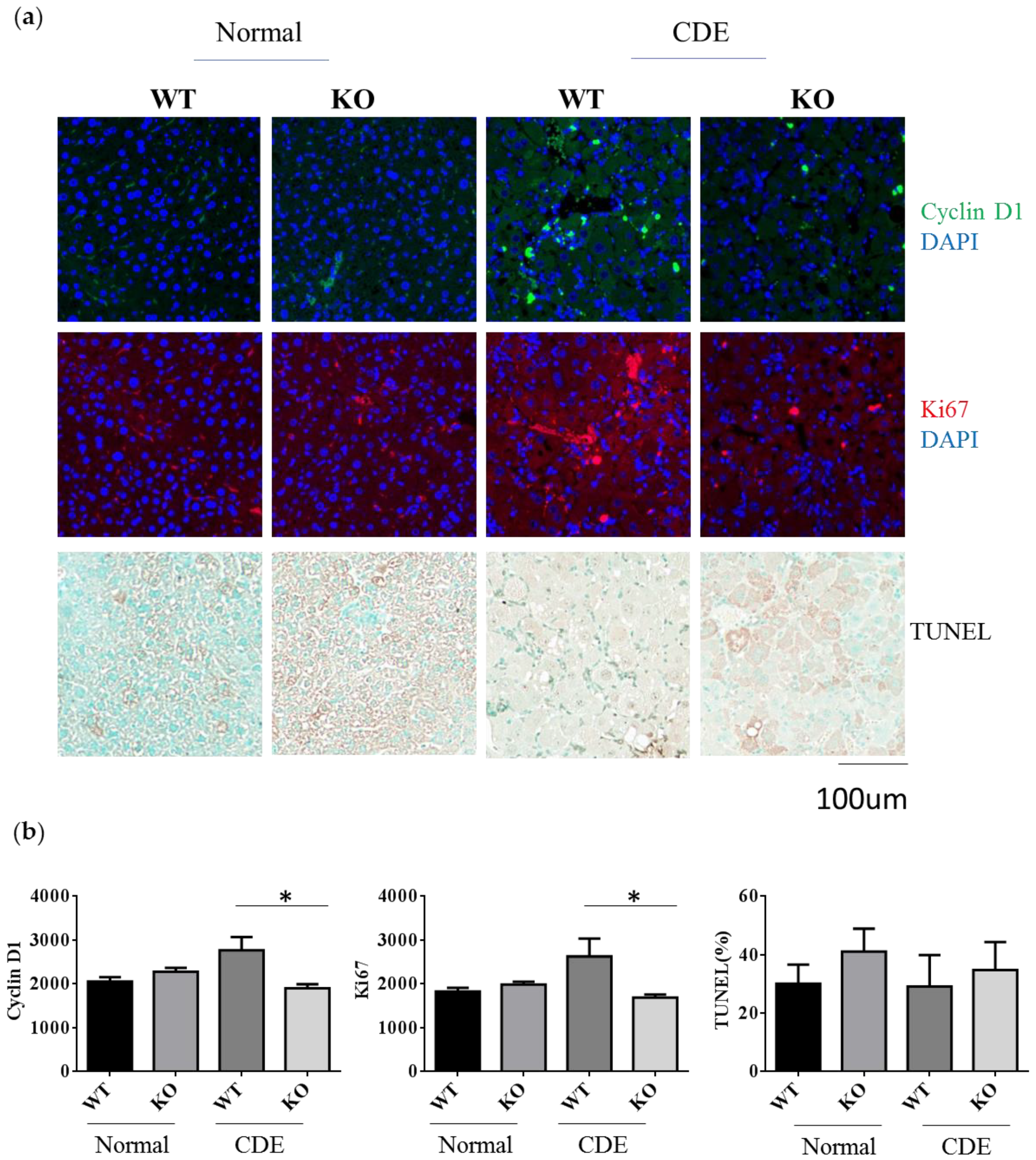
2.4. Reduction of Cell Proliferation in Erk2−/− Livers upon Liver Fibrosis
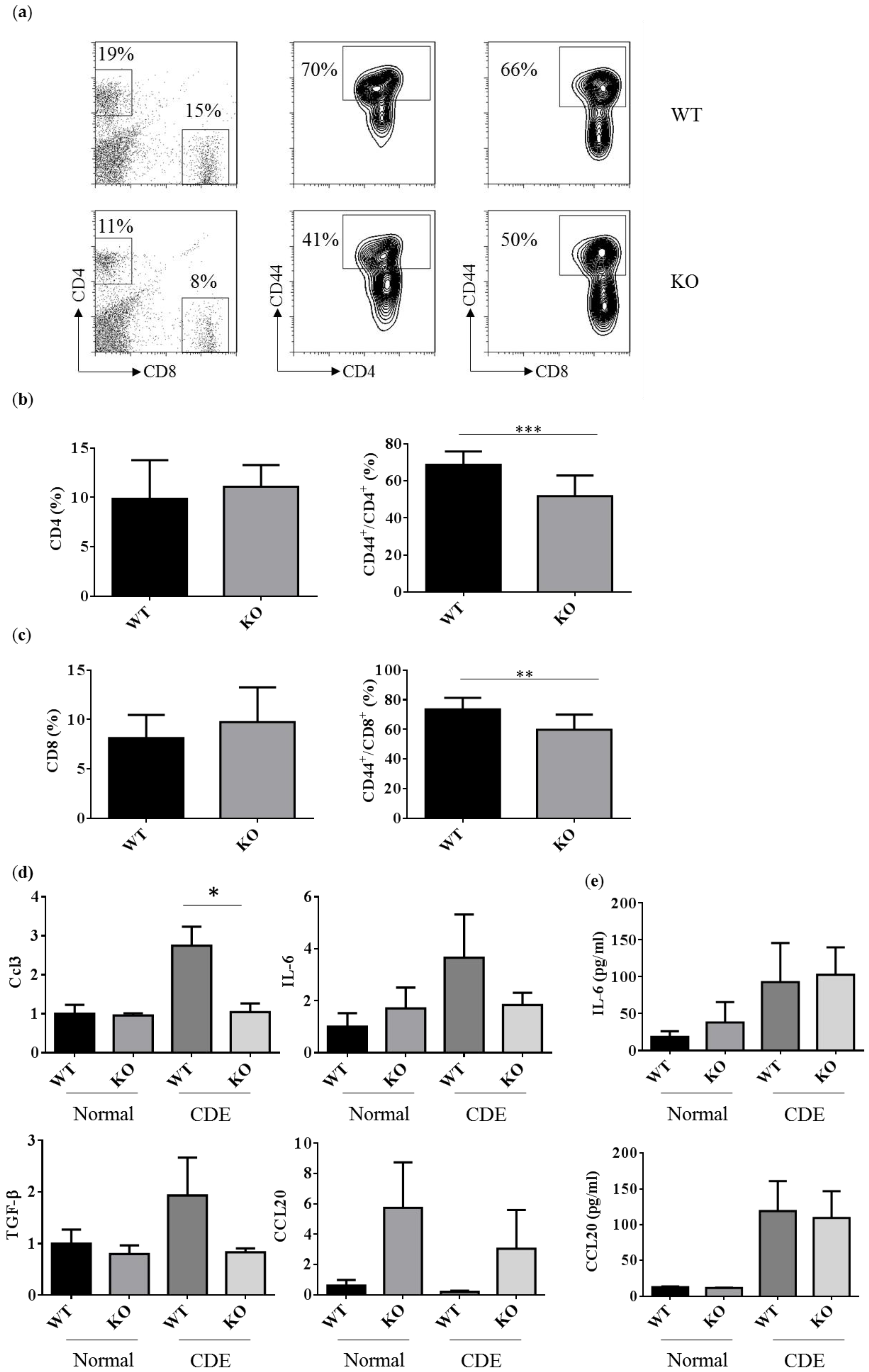
2.5. Reduced Activated T Cells in Erk2−/− (KO) upon the CDE Diet Treatment
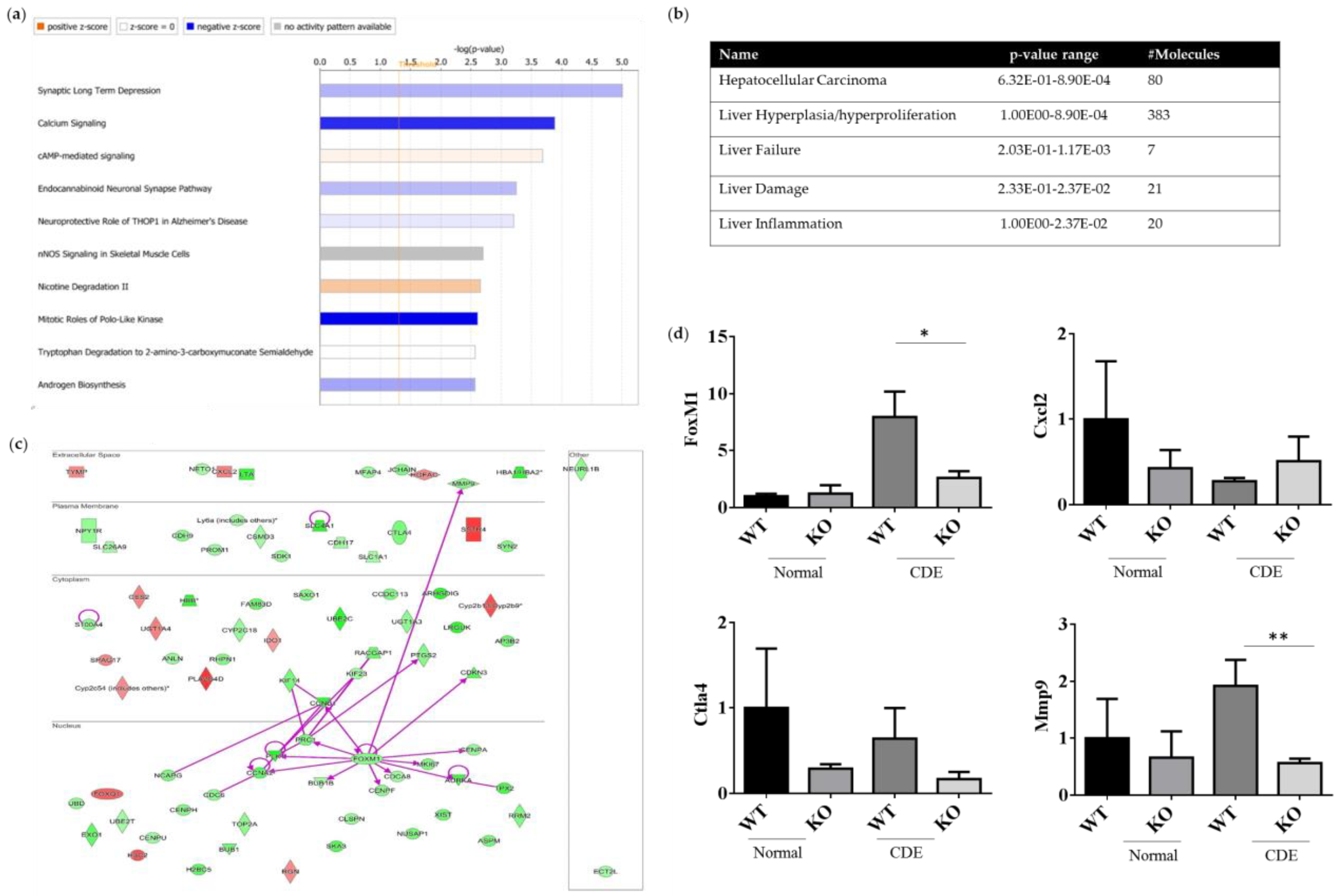
2.6. Differentially Expression Genes (DEGs) of WT and Erk−/− Livers upon Liver Injury
3. Discussion
4. Materials and Methods
4.1. Mice and CDE Diet Treatment
4.2. Histology
4.3. Serum Analysis of ALT and AST
4.4. RNA Isolation and Quantitative Gene Expression
4.5. Western Blotting
4.6. Flow Cytometry
4.7. RNA Sequencing
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| HCC | Hepatocellular carcinoma |
| ERK | Extracellular signal-regulated kinase 2 |
| CDE | Choline-deficient, ethionine-supplemented |
| HSC | Hepatocyte stellate cell |
| alpha-SMA | Alpha smooth muscle |
| MAPK | Mitogen-activated protein kinase |
| RKIP | RAF kinase inhibitory protein |
| HBV | Hepatitis B virus |
| EMT | Epithelial-mesenchymal transition |
| H&E | Hematoxylin and eosin |
| TRI | Masson’s Trichrome |
| LW | Liver weight |
| BW | Body weight |
| CDE | Choline-deficient with supplemented for ethionine |
| AST | Aspartate aminotransferase |
| ALT | Alanine aminotransferase |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| TUNEL | Terminal deoxynucleotidyl transferase dUTP nick end labeling |
| KCs | Kupffer cells |
| DEGs | Differential expression genes |
| FoxM1 | Forkhead Box M1 |
| CNNB1 | Cyclin B1 |
| PRC1 | Protein regulator of cytokinesis |
| PLK1 | Polo-like kinase 1 |
| CCNA2 | Cyclin A2 |
| BUB1B | BUB mitotic checkpoint serine/threonine kinase B |
| CENPF | Centromere protein F |
| CDCA8 | Cell division cycle associated protein 8 |
| AURKA | Aurora kinase A |
| MKI67 | Marker of proliferation Ki-67 |
| CENPA | Centromere protein A |
| SSTR4 | Somatostatin receptor 4 |
| PLA2G4D | Phospholipase A2 group IVD |
| Cyp2b9 | Cytochrome P450 family 2, subfamily b, polypeptide 9 |
| qPCR | quantitative polymerase chain reaction |
| NGS | Next generation sequencing |
| MCD | Methionine-choline deficient |
| NASH | Non-alcoholic steatohepatitis |
| ECM | Extracellular matrix |
| FITC | Fluorescein isothiocyanate |
References
- Tsochatzis, E.A.; Bosch, J.; Burroughs, A.K. Liver cirrhosis. Lancet 2014, 383, 1749–1761. [Google Scholar] [CrossRef]
- Asrani, S.K.; Devarbhavi, H.; Eaton, J.; Kamath, P.S. Burden of liver diseases in the world. J. Hepatol. 2019, 70, 151–171. [Google Scholar] [CrossRef]
- Yoshida, S.; Ikenaga, N.; Liu, S.B.; Peng, Z.W.; Chung, J.; Sverdlov, D.Y.; Miyamoto, M.; Kim, Y.O.; Ogawa, S.; Arch, R.H.; et al. Extrahepatic platelet-derived growth factor-beta, delivered by platelets, promotes activation of hepatic stellate cells and biliary fibrosis in mice. Gastroenterology 2014, 147, 1378–1392. [Google Scholar] [CrossRef]
- Carpino, G.; Morini, S.; Ginanni Corradini, S.; Franchitto, A.; Merli, M.; Siciliano, M.; Gentili, F.; Onetti Muda, A.; Berloco, P.; Rossi, M.; et al. Alpha-SMA expression in hepatic stellate cells and quantitative analysis of hepatic fibrosis in cirrhosis and in recurrent chronic hepatitis after liver transplantation. Dig. Liver Dis. 2005, 37, 349–356. [Google Scholar] [CrossRef]
- Xu, R.; Zhang, Z.; Wang, F.S. Liver fibrosis: Mechanisms of immune-mediated liver injury. Cell. Mol. Immunol. 2012, 9, 296–301. [Google Scholar] [CrossRef]
- Pellicoro, A.; Ramachandran, P.; Iredale, J.P.; Fallowfield, J.A. Liver fibrosis and repair: Immune regulation of wound healing in a solid organ. Nat. Reviews. Immunol. 2014, 14, 181–194. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. Ca Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Llovet, J.M.; Zucman-Rossi, J.; Pikarsky, E.; Sangro, B.; Schwartz, M.; Sherman, M.; Gores, G. Hepatocellular carcinoma. Nat. Rev. Dis Primers 2016, 2, 16018. [Google Scholar] [CrossRef]
- Olsen, S.K.; Brown, R.S.; Siegel, A.B. Hepatocellular carcinoma: Review of current treatment with a focus on targeted molecular therapies. Ther. Adv. Gastroenterol. 2010, 3, 55–66. [Google Scholar] [CrossRef]
- Llovet, J.M.; Ricci, S.; Mazzaferro, V.; Hilgard, P.; Gane, E.; Blanc, J.F.; de Oliveira, A.C.; Santoro, A.; Raoul, J.L.; Forner, A.; et al. Sorafenib in advanced hepatocellular carcinoma. New Engl. J. Med. 2008, 359, 378–390. [Google Scholar] [CrossRef]
- Kim, H.Y.; Park, J.W. Molecularly targeted therapies for hepatocellular carcinoma: Sorafenib as a stepping stone. Dig. Dis. 2011, 29, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, L. Liver regeneration microenvironment of hepatocellular carcinoma for prevention and therapy. Oncotarget 2016. [Google Scholar] [CrossRef] [PubMed]
- Fremin, C.; Ezan, F.; Boisselier, P.; Bessard, A.; Pages, G.; Pouyssegur, J.; Baffet, G. ERK2 but not ERK1 plays a key role in hepatocyte replication: An RNAi-mediated ERK2 knockdown approach in wild-type and ERK1 null hepatocytes. Hepatology 2007, 45, 1035–1045. [Google Scholar] [CrossRef] [PubMed]
- Aroor, A.R.; Jackson, D.E.; Shukla, S.D. Elevated activation of ERK1 and ERK2 accompany enhanced liver injury following alcohol binge in chronically ethanol-fed rats. Alcohol. Clin. Exp. Res. 2011, 35, 2128–2138. [Google Scholar] [CrossRef] [PubMed]
- Jiao, P.; Feng, B.; Li, Y.; He, Q.; Xu, H. Hepatic ERK activity plays a role in energy metabolism. Mol. Cell. Endocrinol. 2013, 375, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Kujiraoka, T.; Satoh, Y.; Ayaori, M.; Shiraishi, Y.; Arai-Nakaya, Y.; Hakuno, D.; Yada, H.; Kuwada, N.; Endo, S.; Isoda, K.; et al. Hepatic extracellular signal-regulated kinase 2 suppresses endoplasmic reticulum stress and protects from oxidative stress and endothelial dysfunction. J. Am. Heart Assoc. 2013, 2, e000361. [Google Scholar] [CrossRef]
- Huang, Q.; Liang, C.; Wei, L.; Nie, J.; Lu, S.; Lu, C.; Zhuo, L.; Lu, Z.; Lin, X. Raf Kinase Inhibitory Protein Down-Expression Exacerbates Hepatic Fibrosis In Vivo and In Vitro. Cell. Physiol. Biochem. 2016, 40, 49–61. [Google Scholar] [CrossRef]
- Min, L.; He, B.; Hui, L. Mitogen-activated protein kinases in hepatocellular carcinoma development. Semin. Cancer Biol. 2011, 21, 10–20. [Google Scholar] [CrossRef]
- Bessard, A.; Fremin, C.; Ezan, F.; Fautrel, A.; Gailhouste, L.; Baffet, G. RNAi-mediated ERK2 knockdown inhibits growth of tumor cells in vitro and in vivo. Oncogene 2008, 27, 5315–5325. [Google Scholar] [CrossRef]
- Gailhouste, L.; Ezan, F.; Bessard, A.; Fremin, C.; Rageul, J.; Langouet, S.; Baffet, G. RNAi-mediated MEK1 knock-down prevents ERK1/2 activation and abolishes human hepatocarcinoma growth in vitro and in vivo. Int. J. Cancer 2010, 126, 1367–1377. [Google Scholar] [CrossRef]
- Son, H.; Moon, A. Epithelial-mesenchymal Transition and Cell Invasion. Toxicol. Res. 2010, 26, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhou, X.; Shen, H.; Wang, D.; Wang, Y. Phosphorylated ERK is a potential predictor of sensitivity to sorafenib when treating hepatocellular carcinoma: Evidence from an in vitro study. BMC Med. 2009, 7, 41. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, K.J.; Wohlschlaeger, J.; Lang, H.; Sotiropoulos, G.C.; Malago, M.; Steveling, K.; Reis, H.; Cicinnati, V.R.; Schmid, K.W.; Baba, H.A. Activation of the ERK and AKT signalling pathway predicts poor prognosis in hepatocellular carcinoma and ERK activation in cancer tissue is associated with hepatitis C virus infection. J. Hepatol. 2008, 48, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Ueda, T.; Honda, M.; Horimoto, K.; Aburatani, S.; Saito, S.; Yamashita, T.; Sakai, Y.; Nakamura, M.; Takatori, H.; Sunagozaka, H.; et al. Gene expression profiling of hepatitis B- and hepatitis C-related hepatocellular carcinoma using graphical Gaussian modeling. Genomics 2013, 101, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ji, T.; Zhao, J.; Li, G.; Zhang, J.; Jin, R.; Liu, J.; Liu, X.; Liang, X.; Huang, D.; et al. Sorafenib-resistant hepatocellular carcinoma stratified by phosphorylated ERK activates PD-1 immune checkpoint. Oncotarget 2016, 7, 41274–41284. [Google Scholar] [CrossRef]
- Yip-Schneider, M.T.; Klein, P.J.; Wentz, S.C.; Zeni, A.; Menze, A.; Schmidt, C.M. Resistance to mitogen-activated protein kinase kinase (MEK) inhibitors correlates with up-regulation of the MEK/extracellular signal-regulated kinase pathway in hepatocellular carcinoma cells. J. Pharmacol. Exp. Ther. 2009, 329, 1063–1070. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, X.; Sun, C.Y.; Wang, J. Delivery of mitogen-activated protein kinase inhibitor for hepatocellular carcinoma stem cell therapy. ACS Appl. Mater. Interfaces 2015, 7, 1012–1020. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Sood, G.K. Hepatocellular carcinoma review: Current treatment, and evidence-based medicine. World J. Gastroenterol. 2014, 20, 4115–4127. [Google Scholar] [CrossRef]
- Xie, Y.; Cui, D.; Sui, L.; Xu, Y.; Zhang, N.; Ma, Y.; Li, Y.; Kong, Y. Induction of forkhead box M1 (FoxM1) by EGF through ERK signaling pathway promotes trophoblast cell invasion. Cell Tissue Res. 2015, 362, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.F.; Zhu, Y.L.; Yuan, H.H.; Wang, J.Y.; Zhao, M.; Gong, X.D.; Liu, F.; Zhang, W.Y.; Wang, C.R.; Jiang, B. FOXM1 regulated by ERK pathway mediates TGF-beta1-induced EMT in NSCLC. Oncol. Res. 2014, 22, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, J.B.; Oakley, F.; Walsh, M.J. Mechanisms and biomarkers of apoptosis in liver disease and fibrosis. Int. J. Hepatol. 2012, 2012, 648915. [Google Scholar] [CrossRef]
- Fabregat, I.; Roncero, C.; Fernandez, M. Survival and apoptosis: A dysregulated balance in liver cancer. Liver Int. 2007, 27, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Foglia, B.; Cannito, S.; Bocca, C.; Parola, M.; Novo, E. ERK Pathway in Activated, Myofibroblast-Like, Hepatic Stellate Cells: A Critical Signaling Crossroad Sustaining Liver Fibrosis. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- Pages, G.; Guerin, S.; Grall, D.; Bonino, F.; Smith, A.; Anjuere, F.; Auberger, P.; Pouyssegur, J. Defective thymocyte maturation in p44 MAP kinase (Erk 1) knockout mice. Science 1999, 286, 1374–1377. [Google Scholar] [CrossRef] [PubMed]
- Hirabaru, M.; Mochizuki, K.; Takatsuki, M.; Soyama, A.; Kosaka, T.; Kuroki, T.; Shimokawa, I.; Eguchi, S. Expression of alpha smooth muscle actin in living donor liver transplant recipients. World J. Gastroenterol. 2014, 20, 7067–7074. [Google Scholar] [CrossRef][Green Version]
- Xie, Y.X.; Liao, R.; Pan, L.; Du, C.Y. ERK pathway activation contributes to the tumor-promoting effects of hepatic stellate cells in hepatocellular carcinoma. Immunol. Lett. 2017, 188, 116–123. [Google Scholar] [CrossRef]
- Wang, Y.; Song, J.; Bian, H.; Bo, J.; Lv, S.; Pan, W.; Lv, X. Apelin promotes hepatic fibrosis through ERK signaling in LX-2 cells. Mol. Cell. Biochem. 2019, 460, 205–215. [Google Scholar] [CrossRef]
- Bhogal, R.K.; Bona, C.A. Regulatory effect of extracellular signal-regulated kinases (ERK) on type I collagen synthesis in human dermal fibroblasts stimulated by IL-4 and IL-13. Int. Rev. Immunol. 2008, 27, 472–496. [Google Scholar] [CrossRef]
- Stankovic, M.N.; Mladenovic, D.R.; Duricic, I.; Sobajic, S.S.; Timic, J.; Jorgacevic, B.; Aleksic, V.; Vucevic, D.B.; Jesic-Vukicevic, R.; Radosavljevic, T.S. Time-dependent changes and association between liver free fatty acids, serum lipid profile and histological features in mice model of nonalcoholic fatty liver disease. Arch. Med. Res. 2014, 45, 116–124. [Google Scholar] [CrossRef]
- D’Souza, W.N.; Chang, C.F.; Fischer, A.M.; Li, M.; Hedrick, S.M. The Erk2 MAPK regulates CD8 T cell proliferation and survival. J. Immunol. 2008, 181, 7617–7629. [Google Scholar] [CrossRef]
- Koyama, Y.; Brenner, D.A. Liver inflammation and fibrosis. J. Clin. Investig. 2017, 127, 55–64. [Google Scholar] [CrossRef]
- Patouraux, S.; Rousseau, D.; Bonnafous, S.; Lebeaupin, C.; Luci, C.; Canivet, C.M.; Schneck, A.S.; Bertola, A.; Saint-Paul, M.C.; Iannelli, A.; et al. CD44 is a key player in non-alcoholic steatohepatitis. J. Hepatol. 2017, 67, 328–338. [Google Scholar] [CrossRef]
- Schulte, L.A.; Lopez-Gil, J.C.; Sainz, B., Jr.; Hermann, P.C. The Cancer Stem Cell in Hepatocellular Carcinoma. Cancers 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Rozeik, M.S.; Hammam, O.A.; Ali, A.I.; Magdy, M.; Khalil, H.; Anas, A.; Abo El Hassan, A.A.; Rahim, A.A.; El-Shabasy, A.I. Evaluation of CD44 and CD133 as markers of liver cancer stem cells in Egyptian patients with HCV-induced chronic liver diseases versus hepatocellular carcinoma. Electron. Physician 2017, 9, 4708–4717. [Google Scholar] [CrossRef] [PubMed]
- Heinrichs, D.; Berres, M.L.; Nellen, A.; Fischer, P.; Scholten, D.; Trautwein, C.; Wasmuth, H.E.; Sahin, H. The chemokine CCL3 promotes experimental liver fibrosis in mice. PLoS ONE 2013, 8, e66106. [Google Scholar] [CrossRef] [PubMed]
- Choi, I.; Kang, H.S.; Yang, Y.; Pyun, K.H. IL-6 induces hepatic inflammation and collagen synthesis in vivo. Clin. Exp. Immunol. 1994, 95, 530–535. [Google Scholar] [CrossRef]
- Schmidt-Arras, D.; Rose-John, S. IL-6 pathway in the liver: From physiopathology to therapy. J. Hepatol. 2016, 64, 1403–1415. [Google Scholar] [CrossRef]
- Chu, X.; Jin, Q.; Chen, H.; Wood, G.C.; Petrick, A.; Strodel, W.; Gabrielsen, J.; Benotti, P.; Mirshahi, T.; Carey, D.J.; et al. CCL20 is up-regulated in non-alcoholic fatty liver disease fibrosis and is produced by hepatic stellate cells in response to fatty acid loading. J. Transl. Med. 2018, 16, 108. [Google Scholar] [CrossRef]
- Jeng, K.S.; Chang, C.F.; Jeng, W.J.; Sheen, I.S.; Jeng, C.J. Heterogeneity of hepatocellular carcinoma contributes to cancer progression. Crit. Rev. Oncol. Hematol. 2015, 94, 337–347. [Google Scholar] [CrossRef]
- Yu, M.; Tang, Z.; Meng, F.; Tai, M.; Zhang, J.; Wang, R.; Liu, C.; Wu, Q. Elevated expression of FoxM1 promotes the tumor cell proliferation in hepatocellular carcinoma. Tumour Biol. 2016, 37, 1289–1297. [Google Scholar] [CrossRef]
- Chai, N.; Xie, H.H.; Yin, J.P.; Sa, K.D.; Guo, Y.; Wang, M.; Liu, J.; Zhang, X.F.; Zhang, X.; Yin, H.; et al. FOXM1 promotes proliferation in human hepatocellular carcinoma cells by transcriptional activation of CCNB1. Biochem. Biophys. Res. Commun. 2018, 500, 924–929. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.D.; Wei, J.C.; Qu, K.; Wang, Z.X.; Wu, Q.F.; Tai, M.H.; Liu, H.C.; Zhang, R.Y.; Liu, C. FoxM1 overexpression promotes epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma. World J. Gastroenterol. 2015, 21, 196–213. [Google Scholar] [CrossRef] [PubMed]
- Kurahashi, T.; Yoshida, Y.; Ogura, S.; Egawa, M.; Furuta, K.; Hikita, H.; Kodama, T.; Sakamori, R.; Kiso, S.; Kamada, Y.; et al. Forkhead Box M1 Transcription Factor Drives Liver Inflammation Linking to Hepatocarcinogenesis in Mice. Cell. Mol. Gastroenterol. Hepatol. 2020, 9, 425–446. [Google Scholar] [CrossRef] [PubMed]
- Duarte, S.; Baber, J.; Fujii, T.; Coito, A.J. Matrix metalloproteinases in liver injury, repair and fibrosis. Matrix Biol. 2015, 44–46, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Chung, T.W.; Lee, Y.C.; Kim, C.H. Hepatitis B viral HBx induces matrix metalloproteinase-9 gene expression through activation of ERK and PI-3K/AKT pathways: Involvement of invasive potential. Faseb J. 2004, 18, 1123–1125. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, S.; Zen, Y.; Fujii, T.; Sato, Y.; Ohta, T.; Aoyagi, Y.; Nakanuma, Y. Characterization of CD133+ parenchymal cells in the liver: Histology and culture. World J. Gastroenterol. 2009, 15, 4896–4906. [Google Scholar] [CrossRef]
- Yin, S.; Li, J.; Hu, C.; Chen, X.; Yao, M.; Yan, M.; Jiang, G.; Ge, C.; Xie, H.; Wan, D.; et al. CD133 positive hepatocellular carcinoma cells possess high capacity for tumorigenicity. Int. J. Cancer 2007, 120, 1444–1450. [Google Scholar] [CrossRef]
- Ding, W.; Mouzaki, M.; You, H.; Laird, J.C.; Mato, J.; Lu, S.C.; Rountree, C.B. CD133+ liver cancer stem cells from methionine adenosyl transferase 1A-deficient mice demonstrate resistance to transforming growth factor (TGF)-beta-induced apoptosis. Hepatology 2009, 49, 1277–1286. [Google Scholar] [CrossRef]
- Fischer, A.M.; Katayama, C.D.; Pages, G.; Pouyssegur, J.; Hedrick, S.M. The role of erk1 and erk2 in multiple stages of T cell development. Immunity 2005, 23, 431–443. [Google Scholar] [CrossRef]
- Chang, C.F.; D’Souza, W.N.; Ch’en, I.L.; Pages, G.; Pouyssegur, J.; Hedrick, S.M. Polar opposites: Erk direction of CD4 T cell subsets. J. Immunol. 2012, 189, 721–731. [Google Scholar] [CrossRef]
- Guo, K.; McMinn, J.E.; Ludwig, T.; Yu, Y.H.; Yang, G.; Chen, L.; Loh, D.; Li, C.; Chua, S., Jr.; Zhang, Y. Disruption of peripheral leptin signaling in mice results in hyperleptinemia without associated metabolic abnormalities. Endocrinology 2007, 148, 3987–3997. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeng, K.-S.; Lu, S.-J.; Wang, C.-H.; Chang, C.-F. Liver Fibrosis and Inflammation under the Control of ERK2. Int. J. Mol. Sci. 2020, 21, 3796. https://doi.org/10.3390/ijms21113796
Jeng K-S, Lu S-J, Wang C-H, Chang C-F. Liver Fibrosis and Inflammation under the Control of ERK2. International Journal of Molecular Sciences. 2020; 21(11):3796. https://doi.org/10.3390/ijms21113796
Chicago/Turabian StyleJeng, Kuo-Shyang, Ssu-Jung Lu, Chih-Hsuan Wang, and Chiung-Fang Chang. 2020. "Liver Fibrosis and Inflammation under the Control of ERK2" International Journal of Molecular Sciences 21, no. 11: 3796. https://doi.org/10.3390/ijms21113796
APA StyleJeng, K.-S., Lu, S.-J., Wang, C.-H., & Chang, C.-F. (2020). Liver Fibrosis and Inflammation under the Control of ERK2. International Journal of Molecular Sciences, 21(11), 3796. https://doi.org/10.3390/ijms21113796





