Abstract
Placental disorders are a major cause of pregnancy loss in humans, and 40–60% of embryos are lost between fertilization and birth. Successful embryo implantation and placental development requires rapid proliferation, invasion, and migration of trophoblast cells. In recent years, microRNAs (miRNAs) have emerged as key regulators of molecular pathways involved in trophoblast function. A miRNA binds its target mRNA in the 3ʹ-untranslated region (3ʹ-UTR), causing its degradation or translational repression. Lethal-7 (let-7) miRNAs induce cell differentiation and reduce cell proliferation by targeting proliferation-associated genes. The oncoprotein LIN28 represses the biogenesis of mature let-7 miRNAs. Proliferating cells have high LIN28 and low let-7 miRNAs, whereas differentiating cells have low LIN28 and high let-7 miRNAs. In placenta, low LIN28 and high let-7 miRNAs can lead to reduced proliferation of trophoblast cells, resulting in abnormal placental development. In trophoblast cells, let-7 miRNAs reduce the expression of proliferation factors either directly by binding their mRNA in 3ʹ-UTR or indirectly by targeting the AT-rich interaction domain (ARID)3B complex, a transcription-activating complex comprised of ARID3A, ARID3B, and histone demethylase 4C (KDM4C). In this review, we discuss regulation of trophoblast function by miRNAs, focusing on the role of LIN28-let-7-ARID3B pathway in placental development.
1. Introduction
Every year, more than 15 million babies are born preterm in the world. A healthy placenta is required for successful establishment of pregnancy and optimal pregnancy outcome [1,2]. Most structural and functional development of the placenta occurs during the first trimester of pregnancy which requires rapid proliferation, invasion, and migration of trophoblast cells [3]. Improper trophoblast function can result in miscarriage, pre-term labor, stillbirth, pre-eclampsia (PE), intrauterine growth restriction (IUGR), and long-term postnatal complication in the mother and fetus [4,5,6,7,8]. The process of rapid trophoblast proliferation and dynamic transformation in placental structure is poorly understood. Previous studies have shown the role of non-coding miRNAs in regulation of trophoblast function. Lethal-7 (Let-7) miRNAs are one of the most studied families of miRNAs and have a well-established role in cell proliferation, invasion, migration, differentiation, and metabolism [9,10]. Let-7 miRNAs reduce cell proliferation by downregulating the proliferation-associated genes [11]. In highly proliferative cells, the RNA binding protein LIN28 represses the production of mature let-7 miRNAs [12,13]. Low LIN28 and increased let-7 miRNAs are thought to be associated with pathogenesis of PE and IUGR [11,14]. During early placental development, dysregulation of miRNAs can lead to reduced proliferation, invasion and migration of trophoblast cells, and contribute to the etiology of placental abnormalities. This review focuses on the role of different miRNAs in trophoblast function, with let-7 miRNAs being the center of discussion.
2. Early Placental Development and Trophoblast Cells
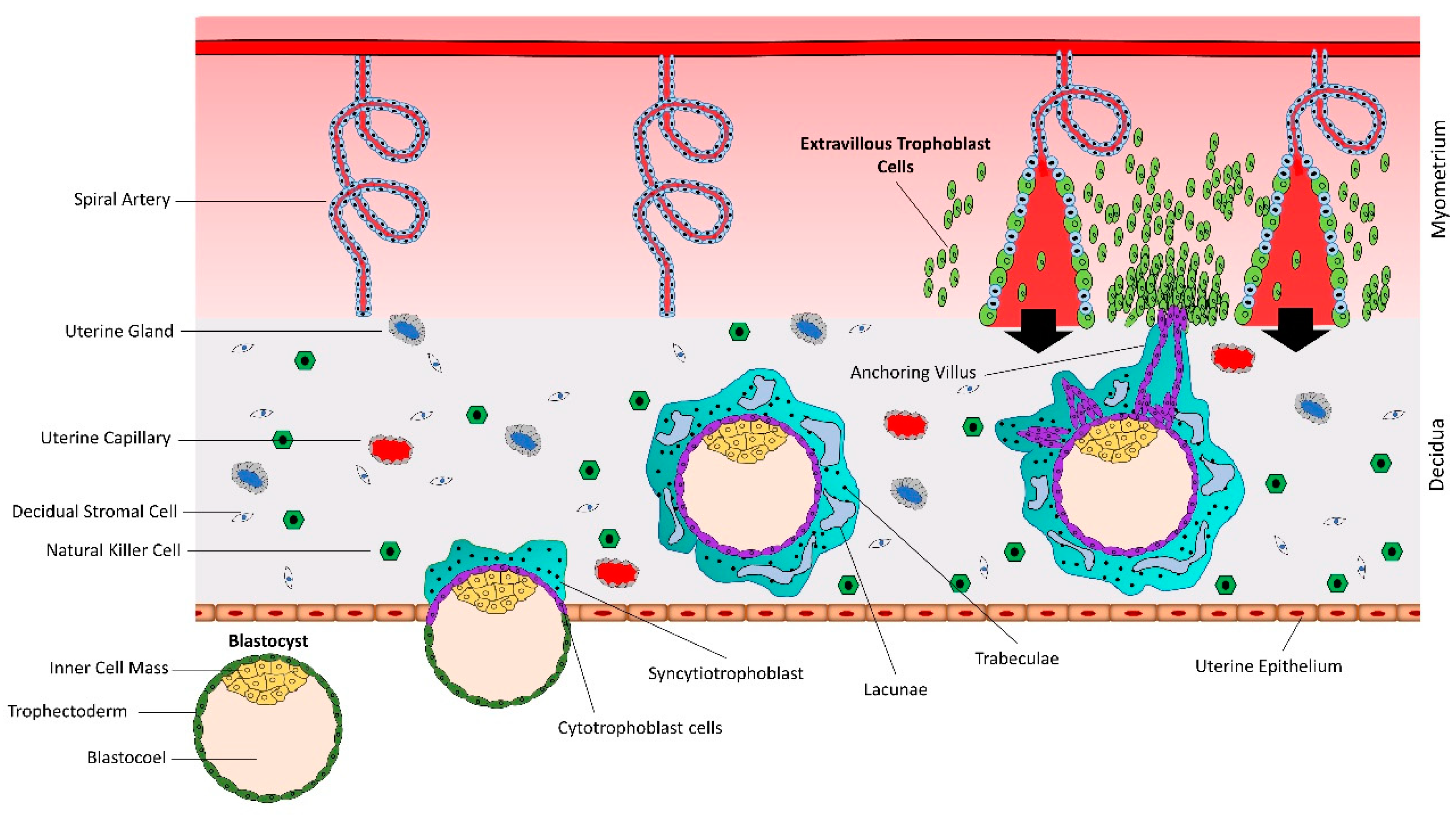
The human blastocyst is formed at day 4–5 after fertilization and contains an outer most layer of zona pellucida, a single layer of mononuclear trophectoderm or trophoblast (TE), a blastocoel cavity, and an inner cell mass (ICM) or embryoblast. The blastocyst sheds the zona pellucida at day 7, exposing the TE [15]. The hatched blastocyst adheres to the endometrial epithelium, subsequently activating the adherent trophoblast cells to proliferate and give rise to different trophoblast lineages [16] (Figure 1). There are two prominent pathways for trophoblast lineages: syncytial pathway and invasive pathway.
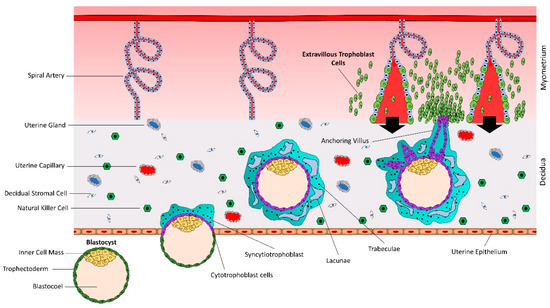
Figure 1.
Early placental development and spiral artery remodeling. Human placental development starts with interaction between hatched blastocyst and uterine epithelium. The trophoblast cells that contact with the uterine epithelium transform into highly proliferative cytotrophoblasts (CTBs). Cytotrophoblasts undergo rapid proliferation and some of them fuse to form a multinucleated syncytiotrophoblast (STB). Within a few hours, STB expands and covers whole blastocyst and helps in blastocyst invasion into the uterine decidua. Continuous proliferation of CTBs results in formation of villi. Some CTBs from the tip of anchoring villi break the STB cover, invade the uterine stroma and myometrium, and transform into extravillous trophoblast cells (EVTs). EVTs remodel the spiral arteries to ensure sufficient flow of blood to the placenta.
2.1. Syncytial Pathway
At the site of attachment, the trophoblast cells transform into rapidly proliferating cytotrophoblasts (CTBs). CTBs undergo rapid proliferation and the newly formed CTBs fuse with each other to form a multinucleated syncytiotrophoblast (STB) [17]. Within a few hours, the STB expands and surrounds the whole blastocyst and mediates the invasion of the blastocyst in the decidualized uterine stroma [18]. The expansion of STB depends upon rapid proliferation and fusion of CTBs [19,20]. By day 21 after conception, the tertiary villi have formed as functional units of the placenta. By week five after conception, the fetoplacental circulation is fully established [21].
2.2. Invasive Pathway
Numerous daughter villi arise from tertiary villi, some of which extend to the maternal tissue and are called anchoring or stem villi [22,23,24]. The anchoring sites can be established as early as the second week of gestation [22]. At proximal ends of anchoring villi, some highly proliferating CTBs break free of the overlying STB layer and invade into maternal endometrium and myometrium [24]. As soon as the detached CTBs make contact with decidual extracellular matrix, they differentiate into interstitial extravillous trophoblast cells (iEVTs) [25]. The iEVTs reach the vascular lumen and differentiate into endovascular extravillous trophoblast cells (enEVTs) [26,27]. The enEVTs remodel the spiral arteries which includes loss of endothelial and smooth muscle cells from arterial walls and their replacement by invasive enEVTs, loss of elasticity, dilation of the arterial lumen, and loss of maternal vasomotor control on the remodeled blood vessels [19,27,28,29].
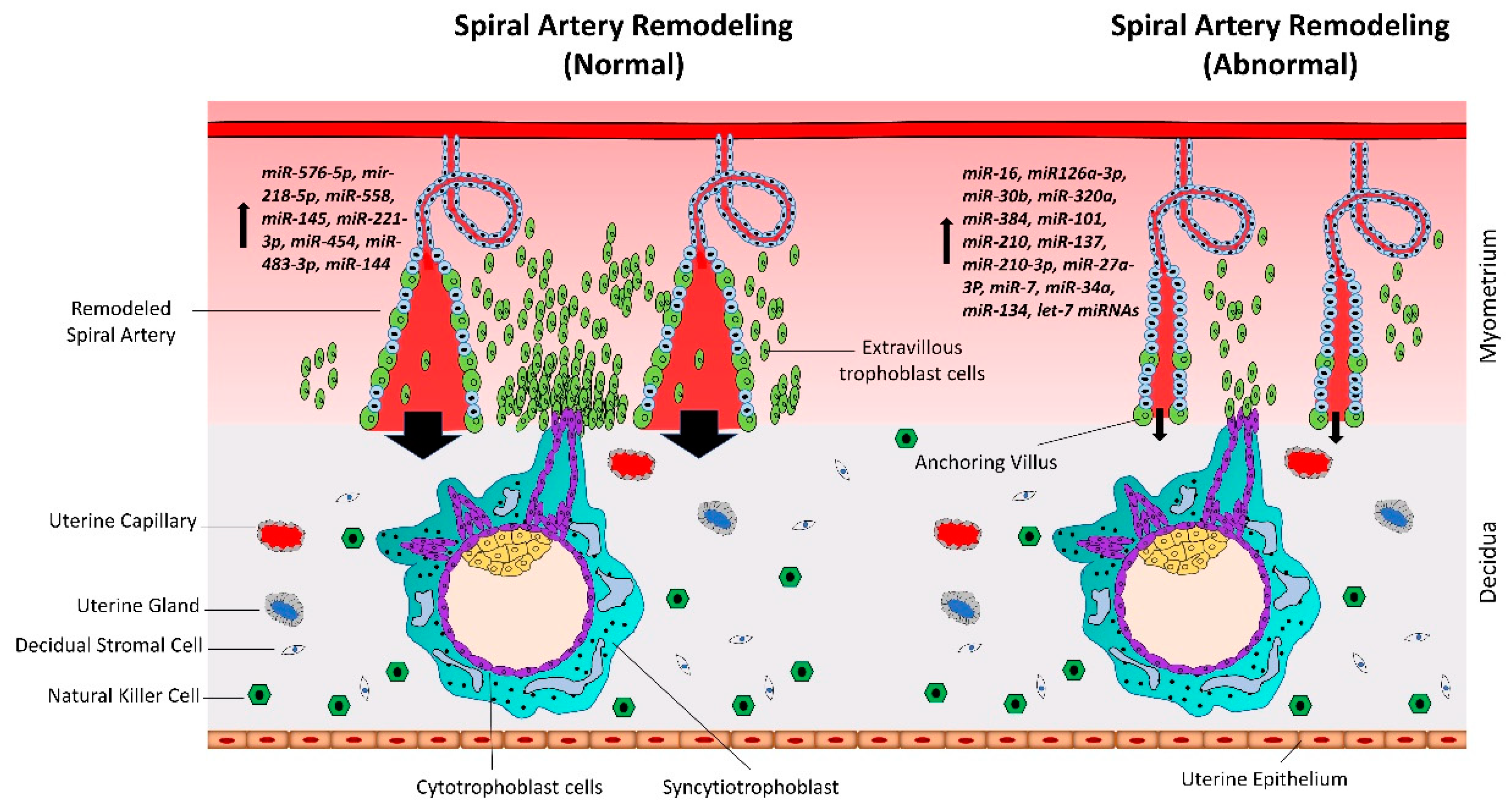
Spiral artery remodeling is crucial for normal placental development and supplying enough nutrients to the fetus (Figure 2). Inadequate remodeling of the spiral arteries is associated with conditions such as preeclampsia (PE), intrauterine growth restriction (IUGR)/fetal growth restriction (FGR), and recurrent miscarriage, that are harmful for both the mother and the fetus. The proliferation and differentiation of trophoblast cells continues throughout gestation. However, unlike cancerous tissues, the proliferation of trophoblast cells is strictly regulated by complex molecular pathways [30,31]. In recent studies, microRNAs (miRNAs) have been shown to play vital roles in trophoblast proliferation and early placental development.
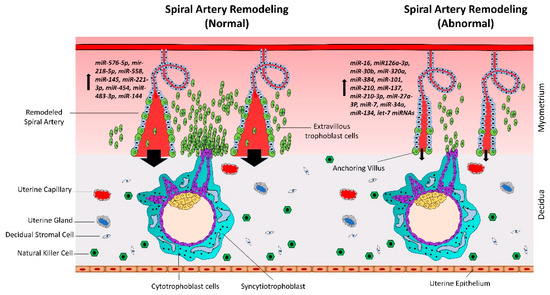
Figure 2.
Normal vs. abnormal spiral artery remodeling. CTBs from anchoring villi break out of the SCT layer and enter the uterine stroma where they differentiate into extravillous trophoblasts (EVTs). Spiral artery remodeling is accomplished by invasion and migration of EVTs. EVTs replace the vascular endothelial cells, remodel the spiral arteries, and ensure sufficient flow of blood to the placenta. In placenta-associated disorders like preeclampsia, reduced proliferation of CTBs results in less availability of EVTs. This leads to insufficient remodeling of spiral arteries and reduced blood flow to the placenta. Based on different studies listed in Table 1, a different set of miRNAs is upregulated in trophoblast cells during normal vs. preeclamptic pregnancies.
3. Functional Analysis of microRNAs in Trophoblast Cells
MicroRNAs are 20–25 nucleotide-long single stranded RNAs which bind 3ʹ-untranslated region (3ʹ-UTR) of the target mRNA causing its degradation or translational repression [32,33,34,35,36]. MicroRNAs have been shown to play important roles in deciding the fate of trophoblast cells [37]. Proliferation, invasion, and migration of trophoblast cells are critical steps during early human placental development. With increasing evidence for the role of miRNAs in regulation of genes associated with cell proliferation, invasion, and migration, several studies have been conducted to investigate the role of miRNAs in placental development and pathogenesis of placenta-associated disorders.
miRNAs impose their effect by regulating the expression of different genes and the effect of a specific miRNA on the phenotype of a cell or tissue depends upon the role of genes targeted by that miRNA. Hence, depending upon the function of their target genes in trophoblast cells, some miRNAs support successful placental development by promoting trophoblast cell proliferation, invasion and migration, and inhibiting the apoptosis of trophoblast cells, whereas some miRNAs can lead to abnormal placental development by reducing cell proliferation, invasion and migration, and increasing apoptosis of trophoblast cells. Table 1 describes the genes regulated by miRNAs and their effect on functionality of trophoblast cells as described in some recent studies. All gene symbols used in Table 1 are according to the Human Genome Organization (HUGO) Gene Nomenclature.

Table 1.
Gene regulation by miRNAs in trophoblast cells.
4. Let-7 miRNAs
The lethal-7 (let-7) family of miRNAs was first discovered in 2002 as a development regulator in Caenorhabditis elegans [91]. The expression of let-7 miRNAs is low in undifferentiated cells and increases gradually as the cells differentiate during development [92]. Therefore, let-7 miRNAs are also referred to as differentiation-inducing miRNAs. The let-7 mutated C. elegans larvae do not mature to the adult stage but keep proliferating and eventually die, earning the name “lethal-7 (let-7)” for this family of miRNAs [91]. Let-7 miRNAs are highly conserved in various animal species [93], suggesting that let-7 miRNAs regulate the same molecular pathways and biological processes in different organisms. In humans, let-7 miRNAs family comprises 12 members including let-7a, let-7b, let-7c, let-7d, let-7e, let-7f, let-7g, let-7i, and miR-98 [94], which originate from eight different genomic loci [95]. Some let-7 miRNAs produced from different genomic loci at different chromosomes have the same sequence. For examples, in humans, let-7a-1, let-7a-2, and let-7a-3 have the same sequence but are encoded by loci on chromosomes 9, 11, and 12, respectively. Similarly, let-7f-1 and let-7f-2 are encoded by different genomic loci but have the same sequence [96]. Let-7 miRNAs have a common seed sequence of seven nucleotides “GAGGUAG” from nucleotide two to eight in all species, which plays an important role in recognizing miRNA response element (MRE) in 3ʹ-UTR of their target mRNA [96]. However, differences in non-seed flanking sequence of let-7 miRNAs affect target specificity [97,98]. Presence of similar seed sequence in all let-7 miRNAs across different species suggests that let-7 miRNAs have the same mechanism for target recognition and might have overlapping targets.
The microarray analysis data from C. elegans show that let-7 miRNAs regulate the expression of thousands of genes, directly and indirectly, indicating their widespread role in biological processes [12]. Let-7 miRNAs play profound roles in embryo development, glucose metabolism, cell pluripotency and differentiation, tumorigenesis, tissue regeneration, age of onset of puberty and menopause in humans, and organ growth [99,100]. Various studies have shown that let-7 miRNAs induce cell differentiation and act as fundamental tumor suppressors by downregulating oncogenes [13,101,102,103]. At early stages of cancer development, let-7 miRNAs are downregulated and let-7 targeted oncofetal genes (LOG) are re-expressed [104]. Comparative bioinformatics analysis shows that let-7 miRNAs target several oncofetal genes including high mobility group AT-hook 2 (HMGA2), insulin like growth factor 2 mRNA binding protein 1 (IMP1), IMP2, IMP3, and malignancy marker nucleosome assembly protein 1 like 1 (NAP1L1) [104]. In hematopoietic stem cells, let-7 miRNAs inhibit transforming growth factor β (TGFβ) pathway and high mobility group AT-hook 2 (HMGA2), decide the fate of these cells, and regulate cell proliferation, self-renewal and differentiation [105,106].
Let-7 miRNAs are synthesized following the same general mechanism for miRNA synthesis. The let-7 loci are transcribed as pri-let-7 miRNA, then processed into 67–80 nucleotide long pre-let-7 miRNA by microprocessor complex [95]. Based on the mechanism of further processing, pre-let-7 miRNAs are divided in two groups: Group I pre-let-7 miRNAs (pre-let-7a-2, 7c, and 7e) are processed in cytoplasm by direct action of Dicer, whereas group II pre-let-7 miRNAs (all remaining let-7s) are mono-uridylated prior to processing by Dicer [107]. Action of Dicer produces 22 nucleotide long mature let-7 miRNAs, called let-7-5p. As a part of miRNA induced silencing complex (miRISC), let-7 miRNAs suppress a wide range of genes involved in development, cell proliferation, metabolism, and other important physiological processes [108]. There is no significant difference in expression of pri-let-7 and pre-let-7 miRNAs between undifferentiated and differentiated cells, however mature let-7 miRNAs are high in differentiated cells compared to undifferentiated cells [109].
Mature let-7 miRNA is a part of hairpin structure in pri- and pre-let-7 miRNA. This hairpin structure contains mature let-7 miRNA (let-7-5p) in the stem and a partially complimentary strand of nucleotides called let-7-3p miRNA, connected by a terminal loop region of different lengths called pre-element (preE) [110]. The process of generation of mature let-7 miRNAs is more precisely regulated compared to the synthesis of other miRNAs. Different proteins regulate posttranscriptional biogenesis of mature let-7 miRNA by binding the preE region of pri- and pre-let-7 miRNAs [111]. One of the most prominent mechanism for regulation of let-7 miRNAs biogenesis is through LIN28 [112].
5. Suppression of let-7 miRNAs by LIN28
LIN28 is a highly conserved RNA binding protein with two paralogues, LIN28A and LIN28B. Both LIN28A and LIN28B have a cold-shock domain (CSD) at the N-terminal and two zinc knuckle domains (ZKDs) at the C-terminal [113]. LIN28 promotes cell proliferation and inhibits cell differentiation [114]. LIN28 is also involved in reprogramming of differentiated somatic cells into tumor or stem cells, hence known as oncoprotein [115,116]. Reduced expression of LIN28 in embryos results in reduced prenatal growth and development and long-term metabolic abnormalities [117]. Knockout of LIN28A in mice leads to perinatal lethality while LIN28B knockout results in postnatal growth abnormalities in males. Knockout of both LIN28A and LIN28B in mice is embryonically lethal at around E13. Conditional knockout of LIN28A and LIN28B in mice at six weeks of age does not produce any evident phenotype [118]. Collectively, these findings show that LIN28 has a more profound role during prenatal development and organogenesis.
LIN28 regulates expression of several genes either directly binding to the mRNA of target genes or by repressing the production of mature let-7 miRNAs, later being a more prevalent mechanism [118,119]. There are conflicting theories about the localization of LIN28A and LIN28B in cells. LIN28A is predominantly localized in the cytoplasm but can be found in the nucleus as well [120,121]; however, according to another study, LIN28A is exclusively localized in the cytoplasm [122]. LIN28B has a nucleolar and nuclear localization signal, while others found it predominantly in cytoplasm with a possibility to shuttle to the nucleus [113,119,123]. LIN28A and LIN28B selectively repress the expression and maturation of let-7 miRNAs by distinct mechanisms, without directly affecting the expression of other miRNAs [124]. LIN28A CSD binds GNGAY motif while ZKDs bind GGAG motif in the stem loop pre-let-7 miRNA in the cytoplasm. After binding to pre-let-7 miRNA, LIN28A recruits terminal uridylyl transferase (TUTase) Zcchc11 (also referred as TUT4). TUT4 causes polyuridylation of pre-let-7 miRNA which blocks the cleavage of pre-let-7 miRNA by Dicer and hence inhibits the production of mature let-7 miRNAs [124,125,126]. Polyuridylated pre-let-7 miRNA is recognized and degraded by exonuclease Dis3L2 [127]. The mechanism of let-7 miRNA suppression by LIN28B remains controversial and there are four different theories. First, LIN28B inhibits maturation of let-7 miRNAs by TUT4 independent mechanism. In the nucleus, LIN28B binds the pri-let-7 miRNA by its CSD and ZKDs and inhibits its processing by a microprocessor [122]. Second, in the cytoplasm, LIN28B binds to pre-let-7 miRNA and inhibits its processing by Dicer [128]. Third, in the cytoplasm, LIN28B binds to pre-let-7 miRNA and leads to its polyuridylation by recruiting an unknown TUTase, leading to its degradation [121]. Fourth, in the nucleolus, LIN28B has the ability to sequester pri-let-7 miRNAs and hence inhibits further processing to mature pre-let-7 miRNAs [122].
In 2018, Ustianenko et al. demonstrated that LIN28 selectively regulates a subclass of let-7 miRNAs [129]. Using single nucleotide resolution, they identified –(U)GAU- as the new binding motif of the CSD. Some pre-let-7 miRNAs with both (U)GAU and GGAG motifs in the stem loop make a stronger and stable interaction with LIN28 and are referred to as CSD+, while the others which do not contain (U)GAU motif are called CSD-. The CSD+ subclass includes pre-let-7b, pre-let-7d, pre-let-7f-1, pre-let-7g, pre-let-7i, and miR-98. The CSD- subclass includes pre-let-7a-1, pre-let-7a-2, pre-let-7-3, pre-let-7c, pre-let-7e, and pre-let-7f-2 [110,129]. Although all let-7 miRNAs express ZKD binding GGAG motif, both LIN28A and LIN28B have shown greater binding affinity for CSD+ pre-let-7 miRNAs, hence leading to their polyuridylation and repression [129].
6. Gene Regulation by LIN28-let-7 miRNA Axis in Trophoblast Cells
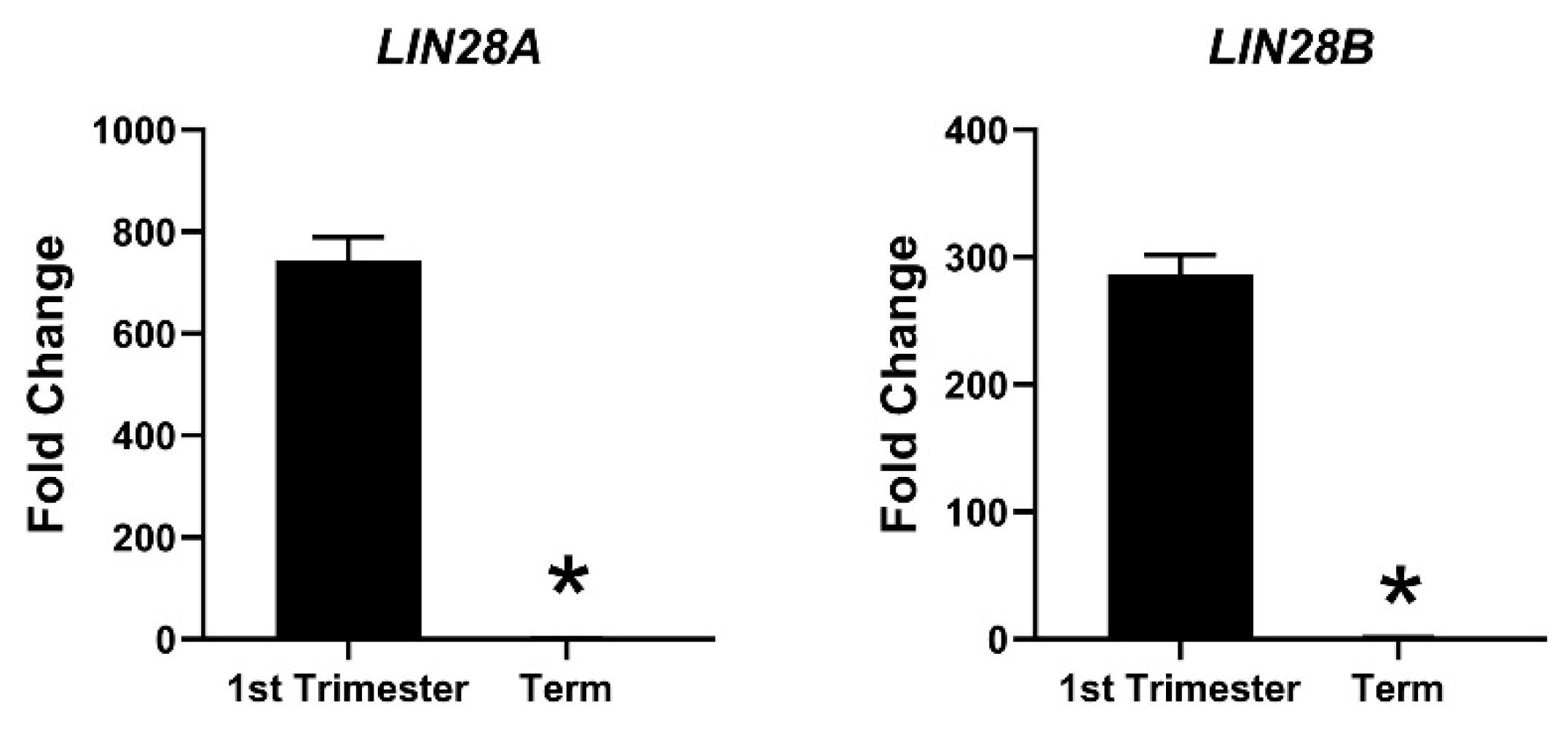
Due to the profound role of let-7 miRNAs as differentiation-inducing miRNAs, the focus of our lab is to investigate the role of LIN28-let-7 miRNA axis in trophoblast cells. Both LIN28A and LIN28B are highly expressed in human placenta and are localized to trophoblast cells [11,130,131,132]. High throughput genotyping array reveals that LIN28B is paternally imprinted in human placenta [133,134]. Using single cell transcriptome profiling, Liu et al. identified 14 different cell types in human placenta and showed that paternally imprinted LIN28B has high expression in CTBs, EVTs and STB, whereas it has no to low expression in mesenchymal cells, macrophages, and blood cells in placenta [135]. They further showed that LIN28B expression in week 24 EVTs was lower compared to week 8 EVTs [135], suggesting that expression of LIN28B in trophoblast cells reduces as the pregnancy progresses. LIN28B is the main paralogue of LIN28 in human placenta and LIN28B mRNA is 1300-fold higher compared to LIN28A mRNA in term human placental tissue [14]. Immunohistochemical analysis of term human placenta shows that LIN28B expression in CTBs and STB is higher compared to placental decidual cells [14]. In 2013, Gu el al. compared the expression of miRNAs between first and third trimester human placentas [136]. They reported that along with many other miRNAs, let-7a, let-7c, let-7d, let-7f, let-7g, and let-7i are upregulated in third trimester compared to first trimester human placenta [136]. We measured LIN28A and LIN28B mRNA in first trimester (11 week) vs. term human placentas and found that LIN28A mRNA was nearly 700-fold higher and LIN28B mRNA was nearly 300-fold higher in first trimester compared to term human placenta (Figure 3). Based on these results, we suggest that increased expression of let-7 miRNAs in term human placentas, reported by Gu et al., is due to reduced expression of LIN28A and LIN28B. Low LIN28 and higher level of let-7 miRNAs in term placenta compared to first trimester placenta suggest that the proliferation rate of trophoblast cells is higher during the first trimester and decreases with advancement in gestational age. As LIN28-let-7 miRNA axis regulates expression of several genes, it would not be surprising to see a difference in gene expression in first trimester vs. third trimester human placenta.
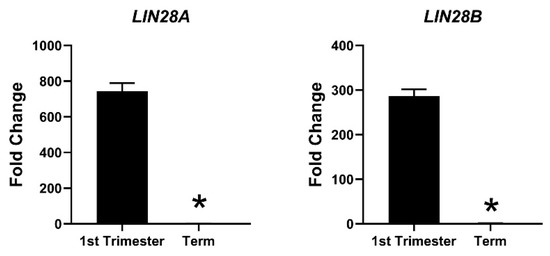
Figure 3.
mRNA was extracted from first trimester (week 11) and term human placentas and LIN28A and LIN28B mRNA levels were measured using real-time RT-PCR, where * p < 0.05.
In IUGR pregnancies, the size of placenta is significantly smaller compared to normal pregnancies [137], which suggests the role of reduced trophoblast proliferation in etiology of IUGR. In a recently published study, we showed that term human placentas from IUGR pregnancies have low LIN28A and LIN28B, and high let-7 miRNAs compared to term human placentas from normal pregnancies [11]. Canfield et al. reported that term human placentas from preeclamptic pregnancies have reduced LIN28B but no change in LIN28A compared to normal term placentas [14]. They further demonstrated that in first trimester human placenta, LIN28B is higher in extravillous cytotrophoblasts compared to villous trophoblast cells, indicating their role in trophoblast cell invasion [14]. Low LIN28 and high let-7 miRNAs during the first trimester of pregnancy can lead to reduced trophoblast proliferation and invasion leading to pregnancy-related disorders.
Due to the limitation that humans cannot be used as experimental models, most studies investigating molecular mechanisms involved in human placental development are conducted using placental cell lines. Commonly used human trophoblast-derived cell lines include BeWo, ACH-3P, JEG3, JAR, Sw.71, and HTR8/SVneo. LIN28A knockdown in immortalized first trimester human trophoblast (ACH-3P) cells drives these cells towards syncytial differentiation and increases the expression of syncytiotrophoblast markers including hCG, LGALS13, and ERVW-1 [132]. Moreover, knockdown of LIN28A increases the expression of let-7 miRNAs including let-7a, let-7c, let-7d, let-7e, let-7g, and let-7i [132], suggesting that differentiation of cells might be due to increased levels of let-7 miRNAs. Overexpression of LIN28B in HTR8 cells increases cell proliferation, invasion, and migration, whereas knockdown of LIN28B in JEG3 cells reduces cell proliferation [14]. In a recently published study, we further investigated the correlation between LIN28 and let-7 miRNAs in trophoblast cells using first trimester human trophoblast-derived ACH-3P and Sw.71 cells. ACH-3P cells were generated by fusing first trimester human trophoblast cells with human choriocarcinoma cells, whereas Sw.71 cells were generated by overexpressing human telomerase reverse transcriptase (h-TERT) in first trimester human trophoblast cells [138,139]. These cell lines have contrasting levels of LIN28 and let-7 miRNAs [11]. ACH-3P cells have high expression of LIN28A and LIN28B whereas these proteins are not detectable in Sw.71 cells [11]. The expression of all let-7 miRNAs is 50–500-fold higher in Sw.71 cells compared to ACH-3P cells, potentially due to depleted LIN28A and LIN28B in Sw.71 cells which are major suppressors of let-7 miRNAs [11]. The contrasting levels of LIN28 and let-7 miRNAs between ACH-3P and Sw.71 cells are potentially due to the difference of methodology used to generate these cell lines. LIN28A knockout in ACH-3P cells increases let-7a, let-7b, let-7c, let-7d, and let-7e, whereas LIN28B knockout in ACH-3P cells increases let-7a, let-7b, let-7c, let-7d, let-7e, and let-7i [11]. According to another study, knockdown of LIN28B in ACH-3P cells increases let-7c, let-7d, let-7e, let-7f, and let-7i [140]. Double knockout of LIN28A and LIN28B in ACH-3P cells results in increased expression of all let-7 miRNAs compared to knockout of either LIN28A or LIN28B [11]. Similarly, LIN28A overexpression in Sw.71 cells decreases let-7d and let-7i, whereas LIN28B overexpression causes reduction in all let-7 miRNAs. However, overexpression of both LIN28A and LIN28B in Sw.71 cells results in decreased expression of all let-7 miRNAs compared to overexpression of either LIN28A or LIN28B [11]. These results suggest that LIN28A and LIN28B work in coordination to suppress let-7 miRNAs and manipulating one paralogue of LIN28 in human trophoblast cells might not induce a similar phenotype compared to if both paralogues are changed.
The majority of let-7-regulated genes are associated with cell proliferation, migration, and invasion—processes which are crucial during early human placental development. We recently demonstrated that double knockout of LIN28A and LIN28B in ACH-3P cells increases in let-7 miRNAs and leads to reduction in expression of proliferation-associated genes including high-mobility group AT-hook 1 (HMGA1), MYC protooncogene (c-MYC), vascular endothelial growth factor A (VEGF-A), and Wnt family member 1 (WNT1). LIN28A/B knockout reduces trophoblast cell proliferation and drives them towards differentiating to a syncytiotrophoblast [11,130]. Similarly, double knockin of LIN28A/B in Sw.71 cells leads to reduction in let-7 miRNAs and increases the expression of HMGA1, c-MYC, VEGF-A, and WNT1 [11]. Other than its role in cell proliferation, VEGF-A is required at all steps of angiogenesis during placental development [21]. Reduced expression of VEGF-A due to high let-7 miRNAs can lead to serious pregnancy complications due to impaired angiogenesis in placenta.
Several studies have demonstrated that let-7 miRNAs bind the 3ʹ-UTR of HMGA2 and reduce its expression in cancer cells [102,141,142]. However, in a recent study, we found a different mechanism of HMGA2 regulation in human trophoblast cells. Double knockout of LIN28A/B in ACH-3P cells increases let-7 miRNAs but does not change HMGA2 expression [130]. Along with increased let-7 miRNAs, LIN28A/B double knockout also increases miR-182. The exact mechanism behind increases in miR-182 in LIN28A/B double knockout ACH-3P cells is not clear. We further showed that HMGA2 expression in trophoblast cells is regulated by a transcription-repressing complex comprised of breast cancer susceptibility gene 1 (BRCA1), CtBP-interacting protein (CtIP), and zinc finger protein 350 (ZNF350). This complex, also called BRCA1 repressor complex, binds the promoter region of HMGA2 and inhibits its transcription [130]. In LIN28A/B double knockout ACH-3P cells, high miR-182 targets BRCA1 leading to inhibition of BRCA1 repressor complex and hence increases HMGA2 expression [130]. Therefore, the expected decrease in HMGA2 due to high let-7 miRNAs is rescued by inhibition of BRCA1 repressor complex. These findings indicate that all genetic pathways demonstrated in the cancer cells might not be applicable in trophoblast cells. It further suggests that rapid proliferation of trophoblast cells during early placental development is more precisely regulated compared to cancer cells.
Although in vitro studies demonstrate the vital role of LIN28-let-7 miRNA axis in trophoblast function, its role in placental development in vivo is not well understood. Using sheep as an experimental model, we investigated the role of LIN28-let-7 miRNA axis in trophoblast proliferation in vivo. In sheep, the hatched blastocyst undergoes a phase of trophectoderm elongation before attachment to the uterine epithelium. The conceptus elongation is accomplished by rapid proliferation of trophoblast cells. Trophoblast proliferation is a critical process in early placental development both in humans and sheep. Trophoblast specific knockdown of LIN28A or LIN28B in sheep leads to reduced conceptus elongation due to reduced proliferation of trophoblast cells [143], suggesting that both LIN28A and LIN28B are equally important in early placental development. Knockdown of LIN28A and LIN28B leads to an increase in let-7 miRNAs and decrease in expression of proliferation-associated genes including insulin like growth factor 2 mRNA binding proteins (IGF2BP1-3), high mobility group AT-hook 1 (HMGA1), AT-rich interaction domain 3B (ARID3B), and MYC protooncogene (c-MYC) [143]. Additionally, overexpression of LIN28A or LIN28B in immortalized ovine trophoblast cells (iOTR) reduces let-7 miRNAs, increases the expression of proliferation associated genes, and increases cell proliferation [143]. These findings further strengthen the data from in vitro studies about the role of LIN28-let-7 miRNA axis in proliferation of human trophoblast cells.
7. LIN28-let-7-ARID3B Pathway in Trophoblast Cells
AT-rich interactive domain (ARID) proteins, first recognized in 1997, are a family of 15 proteins which binds to AT-rich regions of DNA [144,145]. ARID proteins play an important role in cell proliferation, differentiation, and development, and are upregulated in tumorous tissues [146]. The subfamilies of ARID proteins include ARID1, ARID2, ARID3, ARID4, ARID5, JARID1, and JARID2. The ARID3 subfamily has three members including ARID3A, ARID3B, and ARID3C. Although most of the ARID proteins act as tumor suppressors, ARID3A and ARID3B promote tumorigenesis [144]. ARID3A inhibits cell differentiation, promotes cell proliferation, and increases survival potential of cells [146,147], whereas ARID3B promotes proliferation, invasion, and migration of cancer cells [148,149,150]. However, ARID3B is more widely expressed in different tissues compared to ARID3A, suggesting more involvement of ARID3B in biological functions [145]. ARID3A and ARID3B are structurally similar and bind a similar region of DNA. Both ARID3A and ARID3B have an extended central ARID domain and two conserved amino acid domains at the C-terminal, termed REKLES α and REKLES β. Only members of the ARID3 subfamily have REKLES domains [151].
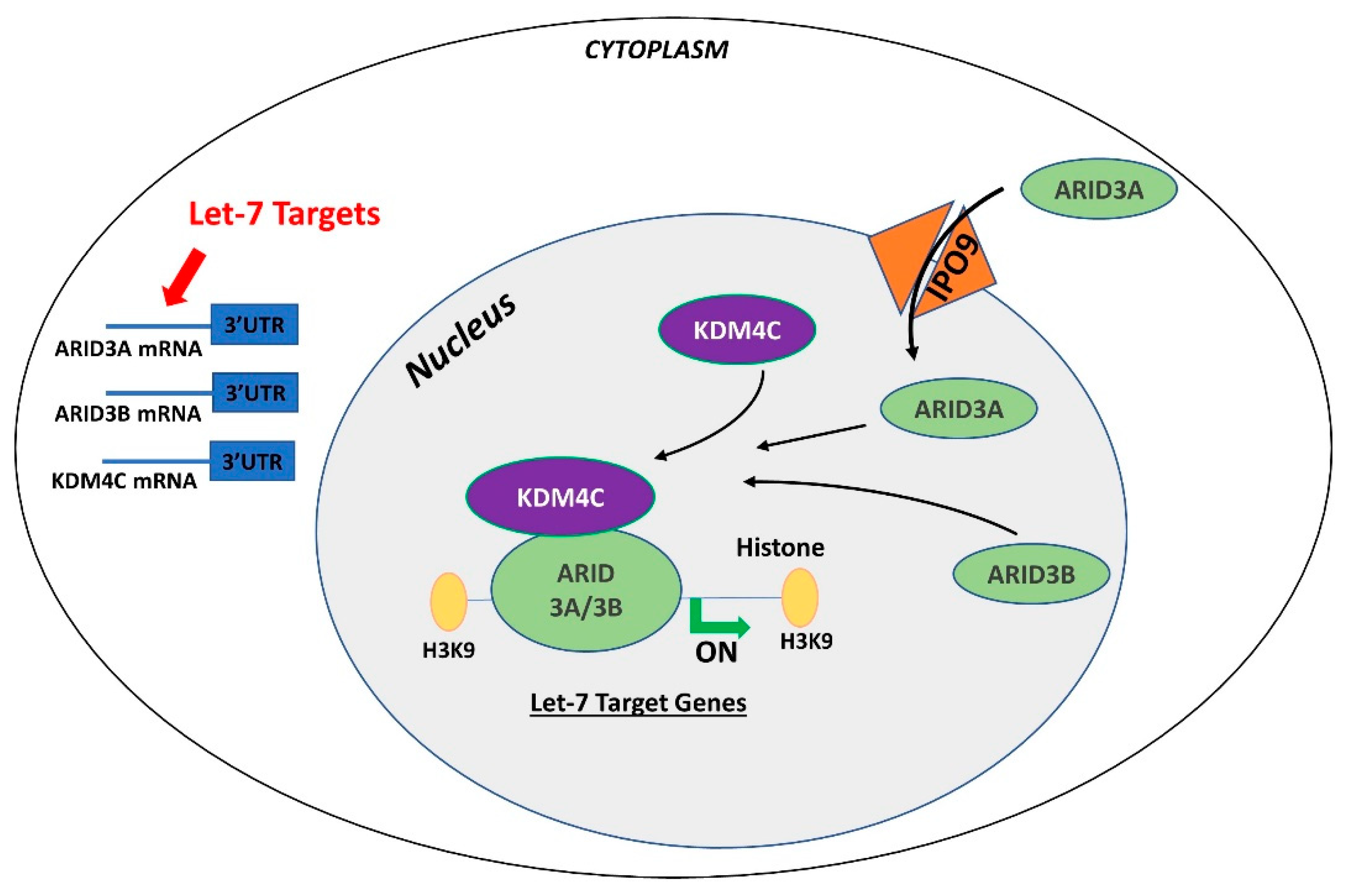
In contrast to ARID3B which is exclusively localized in the nucleus, ARID3A shuttles between the nucleus and cytoplasm. Once in the nucleus, ARID3A interacts with ARID3B through REKLES β domain. Therefore localization of ARID3A in the nucleus is dependent on its interaction with ARID3B in the nucleus, suggesting the dominant role of ARID3B [151,152]. In cancer cells, ARID3A and ARID3B recruit histone demethylase 4C (KDM4C) to make a tri-protein complex, called the ARID3B complex [153]. The ARID3B complex binds in the promoter areas of stemness genes and let-7 target genes, leading to histone demethylation by KDM4C and increased gene expression by initiation of transcription [153]. Therefore, genes regulated by the ARID3B complex also include let-7 miRNA target genes. Liao et al. further demonstrated that both ARID3A and ARID3B are targeted by let-7 miRNAs. Hence, other than directly targeting the mRNAs of target genes, let-7 miRNA can indirectly regulate their target genes by targeting and reducing the expression of ARID3A and ARID3B (Figure 4) [11,153].
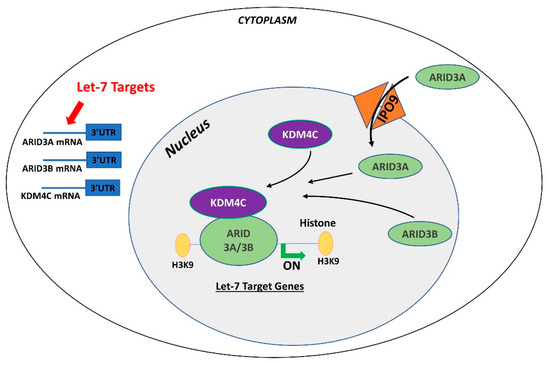
Figure 4.
Gene regulation by the AT-rich interactive domain (ARID)3B complex. ARID3A is imported in the nucleus by importin 9 (IPO9), where it binds ARID3B and histone demethylase 4C (KDM4C) to form the ARID3B complex. The ARID3B complex binds in the promoter regions and activates transcription of let-7 target genes. Other than directly binding the mRNA of their target genes, let-7 miRNAs also target the ARID3B complex and reduce its expression, ultimately leading to reduced expression of let-7 target genes.
Both ARID3A and ARID3B have high expression in human trophoblast cells [154,155]. ARID3A knockout mice have severe structural defects in placenta [156]. In ACH-3P cells, ARID3A, ARID3B, and KDM4C make the tri-protein ARID3B complex [11]. In term human placentas from IUGR pregnancies, LIN28A and LIN28B are low, let-7 miRNAs are high, and ARID3A and ARID3B are low, which suggest a correlation between LIN28-let-7 miRNA axis and the ARID3B complex [11]. Due to the well-established pathway of regulation of let-7 target genes through ARID3B complex and their role in cell proliferation, it is important to understand this phenomenon in early placental development. We recently showed the correlation between LIN28-let-7 miRNA axis and the ARID3B complex using ACH-3P and Sw.71 cells. Double knockout of LIN28A and LIN28B in ACH-3P cells increases let-7 miRNAs and decreases the expression ARID3A, ARID3B, and KDM4C. Similarly, double knockin of LIN28A and LIN28B in Sw.71 cells decreases let-7 miRNAs and increases expression of ARID3A, ARID3B, and KDM4C [11]. In trophoblast cells, the ARID3B complex binds to the promoter areas of proliferation-associated let-7 target genes including HMGA1, c-MYC, VEGF-A, and WNT1, facilitating their transcription via KDM4C mediated histone demethylation. ARID3B knockout ACH-3P cells and KDM4C cannot be recruited in the promoter regions of HMGA1, c-MYC, VEGF-A and WNT1, and expression of these genes is also significantly reduced [73]. Moreover, ARID3B knockout ACH-3P cells have a reduced proliferation rate compared to control cells [11]. Knockdown of LIN28A or LIN28B in sheep trophectoderm increases let-7 miRNAs and reduces the expression of ARID3A and ARID3B [143], showing regulation of the ARID3B complex by LIN28-let-7 miRNA axis in vivo. Collectively these findings show that let-7 miRNAs target the ARID3B complex in trophoblast cells, and the ARID3B complex regulates genes with known importance in placental development.
8. Conclusions
For a long time, transcription activating proteins were thought to be the main regulators of gene expression in cells. However, in recent years microRNAs have emerged as “regulators of the regulators”. miRNAs regulate important processes in trophoblast cells including cell proliferation, differentiation, invasion, and migration. Although identification of widespread let-7 miRNA target genes makes them an important player in placental development, the role of other miRNAs and molecular pathways in placental development cannot be ignored. Although different studies have reported reduced LIN28 and high let-7 miRNAs in term human placentas from preeclamptic and IUGR pregnancies, the exact cause of this dysregulation is not clear. Epigenetic modifications due to adverse uterine environment, random genetic mutations, prenatal insults like hypoxia, oxidative stress, maternal malnutrition, and gestational stress are some of the possible factors which can dysregulate LIN28-let-7 axis in trophoblast cells. LIN28-let-7-ARID3B pathway regulates trophoblast cell proliferation by modulating the expression of proliferation associated genes in vitro and in vivo. The trophoblast cells with low LIN28 will have high let-7 miRNAs and low ARID3B. There are two possible pathways of regulation of proliferation-associated genes by let-7 miRNAs in trophoblast cells (Figure 5). One pathway involves binding of let-7 miRNAs in 3ʹ-UTR of their target mRNA leading to mRNA degradation or translational repression. Secondly, let-7 miRNAs target ARID3A, ARID3B, and KDM4C to inhibit or reduce transcriptional activation of proliferation-associated genes by the ARID3B complex. Therefore, trophoblast cells with high let-7 miRNAs will have reduced expression of proliferation factors and more of a tendency to differentiate. High let-7 miRNAs during early placental development reduce cell proliferation, invasion, and migration of trophoblast cells, leading to placental abnormalities. Let-7 miRNAs might be the major players in pathogenesis of placenta-associated disorders. miRNAs can be readily measured in peripheral blood, tissue biopsies, saliva, cerebrospinal fluid, urine, and other biological samples. High let-7 miRNAs are upregulated in IUGR and preeclamptic placentas, suggesting that let-7 miRNAs can be potential biomarkers for early diagnosis of PE and IUGR. It remains to be explored if early stage placentas from compromised pregnancies will have high let-7 miRNAs and will the increase in let-7 miRNAs in placenta be reflected in maternal blood. Animal models of IUGR can be used to answer these questions.
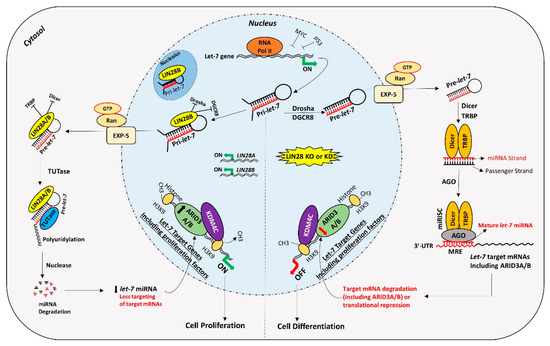
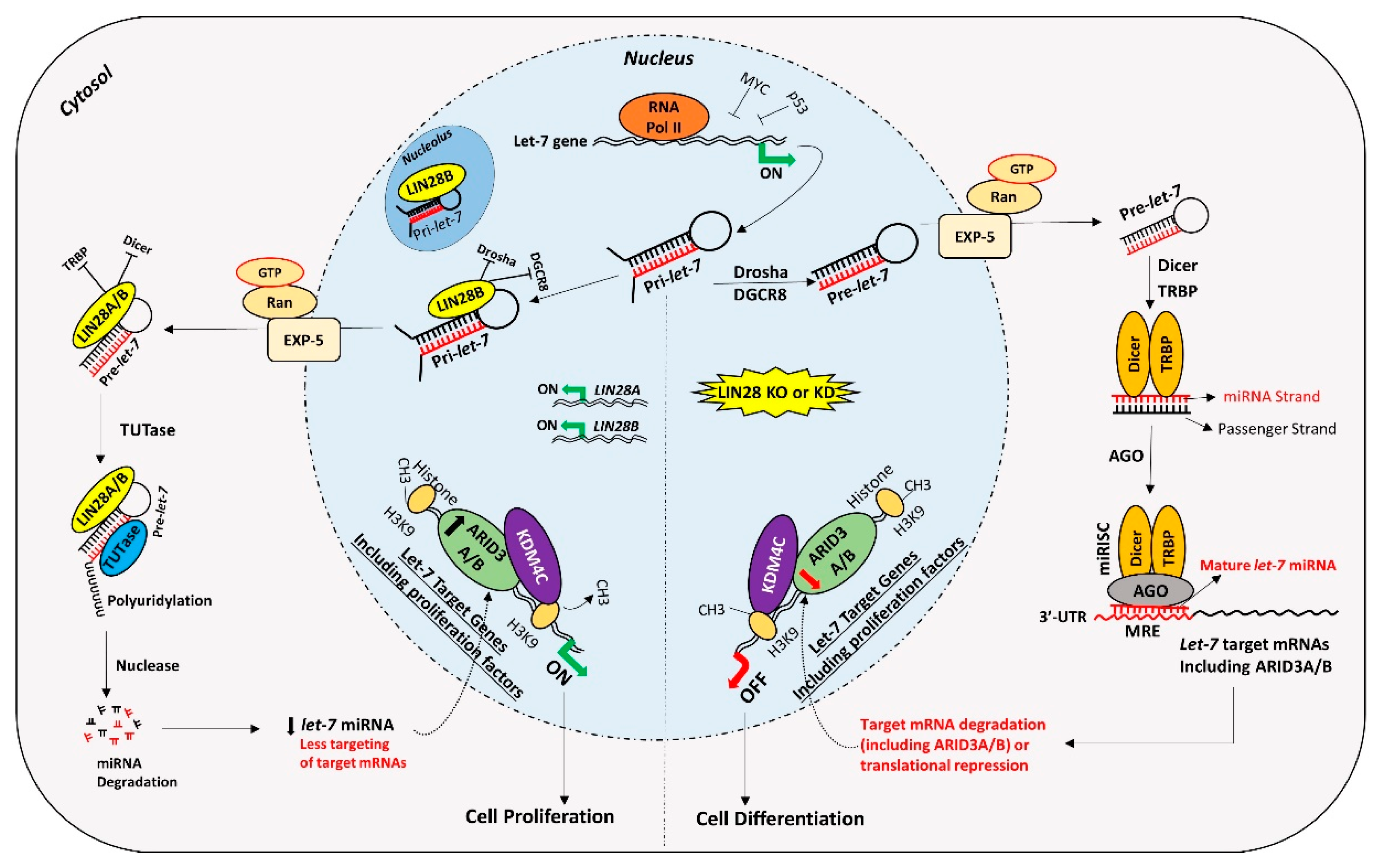
Figure 5.
Proposed mechanism for gene regulation in trophoblast cells. Left panel of figure: LIN28 represses the biogenesis of mature let-7 miRNAs by binding pri-let-7 and pre-let-7 miRNAs and inhibiting their processing. Due to the low level of mature let-7 miRNAs in the cells, there will be less targeting of proliferation-associated genes. Moreover, the ARID3B complex will initiate the transcription of proliferation-associated genes and increase their expression. Increased expression of proliferation-associated genes will lead to increased cell proliferation. Right panel of figure: If LIN28 is knocked-out or knocked-down, there will be no suppression of let-7 miRNA biogenesis. High let-7 miRNAs will target and reduce the expression of proliferation associated genes and the ARDI3B complex, driving the cells towards differentiation.
Author Contributions
A.A. performed research, analyzed the data, and wrote the manuscript; Q.A.W., G.J.B., and R.V.A. edited the manuscript. All authors have read and agree to the published version of the manuscript.
Funding
This project was supported by Agriculture and Food Research Initiative Competitive Grant No. 2017-67015-26460 from the United States Department of Agriculture (USDA) National Institute of Food and Agriculture. This work was supported by the USDA National Institute of Food and Agriculture, Hatch Project COL00293D, accession number 1021217.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Carter, A.M. Evolution of Placental Function in Mammals: The Molecular Basis of Gas and Nutrient Transfer, Hormone Secretion, and Immune Responses. Physiol. Rev. 2012, 92, 1543–1576. [Google Scholar] [CrossRef] [PubMed]
- Gude, N.; Roberts, C.T.; Kalionis, B.; King, R.G. Growth and function of the normal human placenta. Thromb. Res. 2004, 114, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, W.J.; Boyd, J.D. Development of the human placenta in the first three months of gestation. J. Anat. 1960, 94, 297–328. [Google Scholar]
- Crocker, I.P.; Cooper, S.; Ong, S.C.; Baker, P.N. Differences in Apoptotic Susceptibility of Cytotrophoblasts and Syncytiotrophoblasts in Normal Pregnancy to Those Complicated with Preeclampsia and Intrauterine Growth Restriction. Am. J. Pathol. 2003, 162, 637–643. [Google Scholar] [CrossRef]
- Longtine, M.S.; Chen, B.; Odibo, A.; Zhong, Y.; Nelson, D. Villous trophoblast apoptosis is elevated and restricted to cytotrophoblasts in pregnancies complicated by preeclampsia, IUGR, or preeclampsia with IUGR. Placenta 2012, 33, 352–359. [Google Scholar] [CrossRef]
- Burton, G.J.; Fowden, A.L.; Thornburg, K.L. Placental Origins of Chronic Disease. Physiol. Rev. 2016, 96, 1509–1565. [Google Scholar] [CrossRef]
- Barker, D.J.P. The developmental origins of well-being. Philos. Trans. R. Soc. B Biol. Sci. 2004, 359, 1359–1366. [Google Scholar] [CrossRef] [PubMed]
- Barker, D.J.P. The developmental origins of chronic adult disease. Acta Paediatr. 2004, 93, 26–33. [Google Scholar] [CrossRef]
- Jiang, S. A Regulator of Metabolic Reprogramming: MicroRNA Let-7. Transl. Oncol. 2019, 12, 1005–1013. [Google Scholar] [CrossRef]
- Boyerinas, B.; Park, S.-M.; Hau, A.; Murmann, A.E.; Peter, M.E. The role of let-7 in cell differentiation and cancer. Endocr.-Relat. Cancer 2010, 17, F19–F36. [Google Scholar] [CrossRef]
- Ali, A.; Anthony, R.V.; Bouma, G.J.; Winger, Q.A. LIN28-let-7 axis regulates genes in immortalized human trophoblast cells by targeting the ARID3B-complex. FASEB J. 2019, 33, 12348–12363. [Google Scholar] [CrossRef] [PubMed]
- Hunter, S.E.; Finnegan, E.F.; Zisoulis, D.G.; Lovci, M.T.; Melnik-Martinez, K.V.; Yeo, G.W.; Pasquinelli, A.E. Functional Genomic Analysis of the let-7 Regulatory Network in Caenorhabditis elegans. PLoS Genet. 2013, 9, e1003353. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cao, L.; Wang, Y.; Wang, X.; Liu, N.; You, Y. Regulation of let-7 and its target oncogenes (Review). Oncol. Lett. 2012, 3, 955–960. [Google Scholar] [CrossRef] [PubMed]
- Canfield, J.; Arlıer, S.; Mong, E.F.; Lockhart, J.; Van Wye, J.; Guzeloglu-Kayisli, O.; Schatz, F.; Magness, R.R.; Lockwood, C.J.; Tsibris, J.C.M.; et al. Decreased LIN28B in preeclampsia impairs human trophoblast differentiation and migration. FASEB J. 2018, 33, 2759–2769. [Google Scholar] [CrossRef]
- Bazer, F.W.; Spencer, T.E.; Johnson, G.A.; Burghardt, R.C.; Wu, G. Comparative aspects of implantation. Reproduction 2009, 138, 195–209. [Google Scholar] [CrossRef]
- Herzog, M. A contribution to our knowledge of the earliest known stages of placentation and embryonic development in man. Am. J. Anat. 2005, 9, 361–400. [Google Scholar] [CrossRef]
- Pötgens, A.; Schmitz, U.; Bose, P.; Versmold, A.; Kaufmann, P.; Frank, H.-G. Mechanisms of Syncytial Fusion: A Review. Placenta 2002, 23, S107–S113. [Google Scholar] [CrossRef]
- Kim, S.-M.; Kim, J.-S. A Review of Mechanisms of Implantation. Dev. Reprod. 2017, 21, 351–359. [Google Scholar] [CrossRef]
- Benirschke, K.; Burton, G.J.; Baergen, R.N. The Pathology of the Human Placenta; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2012; pp. 97–571. [Google Scholar]
- Frank, H.-G. 10-Placental Development. In Fetal and Neonatal Physiology, 5th ed.; Polin, R.A., Abman, S.H., Rowitch, D.H., Benitz, W.E., Fox, W.W., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 101–113. [Google Scholar]
- Chen, D.-B.; Zheng, J. Regulation of placental angiogenesis. Microcirculation 2014, 21, 15–25. [Google Scholar] [CrossRef]
- Vicovac, L. Trophoblast differentiation during formation of anchoring villi in a model of the early human placenta in vitro. Placenta 1995, 16, 41–56. [Google Scholar] [CrossRef]
- Damsky, C.H.; Fitzgerald, M.L.; Fisher, S.J. Distribution patterns of extracellular matrix components and adhesion receptors are intricately modulated during first trimester cytotrophoblast differentiation along the invasive pathway, in vivo. J. Clin. Investig. 1992, 89, 210–222. [Google Scholar] [CrossRef]
- Prakobphol, A.; Genbacev, O.; Gormley, M.; Kapidzic, M.; Fisher, S.J. A role for the L-selectin adhesion system in mediating cytotrophoblast emigration from the placenta. Dev. Biol. 2006, 298, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Kemp, B.; Kertschanska, S.; Kadyrov, M.; Rath, W.; Kaufmann, P.; Huppertz, B. Invasive depth of extravillous trophoblast correlates with cellular phenotype:A comparison of intra- and extrauterine implantation sites. Histochem. Cell Biol. 2002, 117, 401–414. [Google Scholar] [CrossRef]
- Espinoza, J.; Romero, R.; Kim, Y.M.; Kusanovic, J.P.; Hassan, S.; Erez, O.; Gotsch, F.; Than, N.G.; Papp, Z.; Kim, C.J. Normal and abnormal transformation of the spiral arteries during pregnancy. J. Périnat. Med. 2006, 34, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Anin, S.; Vince, G.; Quenby, S. Trophoblast invasion. Hum. Fertil. 2004, 7, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Pijnenborg, R.; Vercruysse, L.; Hanssens, M. The Uterine Spiral Arteries in Human Pregnancy: Facts and Controversies. Placenta 2006, 27, 939–958. [Google Scholar] [CrossRef]
- Lyall, F.; Bulmer, J.N.; Duffie, E.; Cousins, F.; Thériault, A.; Robson, S.C. Human Trophoblast Invasion and Spiral Artery Transformation. Am. J. Pathol. 2001, 158, 1713–1721. [Google Scholar] [CrossRef]
- Red-Horse, K.; Zhou, Y.; Genbacev, O.; Prakobphol, A.; Foulk, R.; McMaster, M.; Fisher, S.J. Trophoblast differentiation during embryo implantation and formation of the maternal-fetal interface. J. Clin. Investig. 2004, 114, 744–754. [Google Scholar] [CrossRef]
- Reynolds, L.; Redmer, D.A. Angiogenesis in the Placenta1. Biol. Reprod. 2001, 64, 1033–1040. [Google Scholar] [CrossRef]
- Kobayashi, H.; Tomari, Y. RISC assembly: Coordination between small RNAs and Argonaute proteins. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2016, 1859, 71–81. [Google Scholar] [CrossRef]
- Braun, J.E.; Truffault, V.; Boland, A.; Huntzinger, E.; Chang, C.-T.; Haas, G.; Weichenrieder, O.; Coles, M.; Izaurralde, E. A direct interaction between DCP1 and XRN1 couples mRNA decapping to 5’ exonucleolytic degradation. Nat. Struct. Mol. Biol. 2012, 19, 1324–1331. [Google Scholar] [CrossRef]
- Christie, M.; Boland, A.; Huntzinger, E.; Weichenrieder, O.; Izaurralde, E. Structure of the PAN3 Pseudokinase Reveals the Basis for Interactions with the PAN2 Deadenylase and the GW182 Proteins. Mol. Cell 2013, 51, 360–373. [Google Scholar] [CrossRef] [PubMed]
- Behm-Ansmant, I.; Rehwinkel, J.; Doerks, T.; Stark, A.; Bork, P.; Izaurralde, E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genome Res. 2006, 20, 1885–1898. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs:Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Doridot, L.; Miralles, F.; Barbaux, S.; Vaiman, D. Trophoblasts, invasion, and microRNA. Front. Genet. 2013, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Wang, H.; Yang, J.; Long, W.; Zhang, B.; Liu, J.; Yu, B. Down-regulated circPAPPA suppresses the proliferation and invasion of trophoblast cells via the miR-384/STAT3 pathway. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Li, J.; Wang, J.M.; Liu, Y.H.; Zhang, Z.; Han, N.; Xue, S.H.; Wang, P. Effect of microRNA-106b on the invasion and proliferation of trophoblasts through targeting MMP-2. Zhonghua Fu Chan Ke Za Zhi 2017, 52, 327–332. [Google Scholar]
- Liu, F.; Wu, K.; Wu, W.; Chen, Y.; Wu, H.; Wang, H.; Zhang, W. miR-203 contributes to pre-eclampsia via inhibition of VEGFA expression. Mol. Med. Rep. 2018, 17, 5627–5634. [Google Scholar] [CrossRef]
- Jiang, L.; Long, A.; Tan, L.; Hong, M.; Wu, J.; Cai, L.; Li, Q. Elevated microRNA-520g in pre-eclampsia inhibits migration and invasion of trophoblasts. Placenta 2017, 51, 70–75. [Google Scholar] [CrossRef]
- Wang, R.; Liu, W.; Liu, X.; Liu, X.; Tao, H.; Wu, D.; Zhao, Y.; Zou, L. MicroRNA-210 regulates human trophoblast cell line HTR-8/SVneo function by attenuating Notch1 expression: Implications for the role of microRNA-210 in pre-eclampsia. Mol. Reprod. Dev. 2019, 86, 896–907. [Google Scholar] [CrossRef]
- Yuan, Y.; Wang, X.; Sun, Q.; Dai, X.; Cai, Y. MicroRNA-16 is involved in the pathogenesis of pre-eclampsia via regulation of Notch2. J. Cell. Physiol. 2019, 235, 4530–4544. [Google Scholar] [CrossRef] [PubMed]
- Xie, N.; Jia, Z.; Li, L. miR-320a upregulation contributes to the development of preeclampsia by inhibiting the growth and invasion of trophoblast cells by targeting interleukin 4. Mol. Med. Rep. 2019, 20, 3256–3264. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Meng, Q.; Shi, Y.-P.; Xu, H.-S. Regulatory role of microRNA-320a in the proliferation, migration, invasion, and apoptosis of trophoblasts and endothelial cells by targeting estrogen-related receptor γ. J. Cell. Physiol. 2018, 234, 682–691. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Huang, X.; He, Z.; Xiong, Y.; Fang, Q. miRNA-210-3p regulates trophoblast proliferation and invasiveness through fibroblast growth factor 1 in selective intrauterine growth restriction. J. Cell. Mol. Med. 2019, 23, 4422–4433. [Google Scholar] [CrossRef]
- Shih, J.-C.; Lin, H.-H.; Hsiao, A.-C.; Su, Y.-T.; Tsai, S.; Chien, C.-L.; Kung, H.-N. Unveiling the role of microRNA-7 in linking TGF-β-Smad-mediated epithelial-mesenchymal transition with negative regulation of trophoblast invasion. FASEB J. 2019, 33, 6281–6295. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.J.; Wu, P.Y.; Gao, R.Q. MiR-218 inhibits HTR-8 cells migration and invasion by targeting SOX4. Zhongguo Ying Yong Sheng Li Xue Za Zhi 2017, 33, 169–173. [Google Scholar] [PubMed]
- Xue, F.; Yang, J.; Li, Q.; Zhou, H. Down-regulation of microRNA-34a-5p promotes trophoblast cell migration and invasion via targetting Smad4. Biosci. Rep. 2019, 39, 39. [Google Scholar] [CrossRef]
- Zhou, X.; Li, Q.; Xu, J.; Zhang, X.; Zhang, H.; Xiang, Y.; Fang, C.; Wang, T.; Xia, S.; Zhang, Q.; et al. The aberrantly expressed miR-193b-3p contributes to preeclampsia through regulating transforming growth factor-β signaling. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Sun, M.; Chen, H.; Liu, J.; Tong, C.; Meng, T. MicroRNA-34a inhibits human trophoblast cell invasion by targeting MYC. BMC Cell Biol. 2015, 16. [Google Scholar] [CrossRef]
- Ding, J.; Huang, F.; Wu, G.; Han, T.; Xu, F.; Weng, D.; Wu, C.; Zhang, X.; Yao, Y.; Zhu, X. MiR-519d-3p Suppresses Invasion and Migration of Trophoblast Cells via Targeting MMP-2. PLoS ONE 2015, 10, e0120321. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, Y.; Li, J.; Zhong, Q.; Li, Y. MiR-101 inhibits migration and invasion of trophoblast HTR-8/SVneo cells by targeting CXCL6 in preeclampsia. Minerva Med. 2019. [Google Scholar] [CrossRef]
- Liu, J.-J.; Zhang, L.; Zhang, F.-F.; Luan, T.; Yin, Z.-M.; Rui, C.; Ding, H.-J. Influence of miR-34a on preeclampsia through the Notch signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 923–931. [Google Scholar] [PubMed]
- Yang, X.; Meng, T. MicroRNA-431 affects trophoblast migration and invasion by targeting ZEB1 in preeclampsia. Gene 2018, 683, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Chen, Y.; Long, Y.; Yu, J.; Li, M. Tumor necrosis factor-alpha suppresses the invasion of HTR-8/SVneo trophoblast cells through microRNA-145-5p-mediated downregulation of Cyr61. Life Sci. 2018, 209, 132–139. [Google Scholar] [CrossRef]
- Zou, A.-X.; Chen, B.; Li, Q.-X.; Liang, Y.-C. MiR-134 inhibits infiltration of trophoblast cells in placenta of patients with preeclampsia by decreasing ITGB1 expression. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2199–2206. [Google Scholar]
- Ding, J.; Cheng, Y.; Zhang, Y.; Liao, S.; Yin, T.; Yang, J. The miR-27a-3p/USP25 axis participates in the pathogenesis of recurrent miscarriage by inhibiting trophoblast migration and invasion. J. Cell. Physiol. 2019, 234, 19951–19963. [Google Scholar] [CrossRef]
- Wang, N.; Feng, Y.; Xu, J.; Zou, J.; Chen, M.; He, Y.; Liu, H.; Xue, M.; Feng, Y.-L. miR-362-3p regulates cell proliferation, migration and invasion of trophoblastic cells under hypoxia through targeting Pax3. Biomed. Pharmacother. 2018, 99, 462–468. [Google Scholar] [CrossRef]
- Wu, L.; Song, W.-Y.; Xie, Y.; Hu, L.-L.; Hou, X.-M.; Wang, R.; Gao, Y.; Zhang, J.-N.; Zhang, L.; Li, W.-W.; et al. miR-181a-5p suppresses invasion and migration of HTR-8/SVneo cells by directly targeting IGF2BP2. Cell Death Dis. 2018, 9, 16. [Google Scholar] [CrossRef]
- Peng, H.-Y.; Li, M.-Q.; Li, H.-P. MiR-137 Restricts the Viability and Migration of HTR-8/SVneo Cells by Downregulating FNDC5 in Gestational Diabetes Mellitus. Curr. Mol. Med. 2019, 19, 494–505. [Google Scholar] [CrossRef]
- Qian, S.; Liu, R. miR-30b facilitates preeclampsia through targeting MXRA5 to inhibit the viability, invasion and apoptosis of placental trophoblast cells. Int. J. Clin. Exp. Pathol. 2019, 12, 4057–4065. [Google Scholar]
- Niu, Z.-R.; Han, T.; Sun, X.-L.; Luan, L.-X.; Gou, W.; Zhu, X. MicroRNA-30a-3p is overexpressed in the placentas of patients with preeclampsia and affects trophoblast invasion and apoptosis by its effects on IGF-1. Am. J. Obstet. Gynecol. 2018, 218, 249. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, X.; Weng, Z.; Zhang, S.; Ning, H.; Li, B. Expression and role of microRNA 18b and hypoxia inducible factor-1α in placental tissues of preeclampsia patients. Exp. Ther. Med. 2017, 14, 4554–4560. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gao, Y.; She, R.; Wang, Q.; Li, Y.; Zhang, H. Up-regulation of miR-299 suppressed the invasion and migration of HTR-8/SVneo trophoblast cells partly via targeting HDAC2 in pre-eclampsia. Biomed. Pharmacother. 2018, 97, 1222–1228. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; She, K.; Li, H.; Yuan, X.; Han, X.; Wang, Y. MicroRNA-454 contributes to sustaining the proliferation and invasion of trophoblast cells through inhibiting Nodal/ALK7 signaling in pre-eclampsia. Chem. Interact. 2019, 298, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Chi, Z.; Zhang, M. Exploration of the regulation and control mechanisms of miR-145 in trophoblast cell proliferation and invasion. Exp. Ther. Med. 2018, 16, 5298–5304. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, H.; Ma, Y.; Zhu, X.; Zhang, S.; Li, J. MiR-221-3p is down-regulated in preeclampsia and affects trophoblast growth, invasion and migration partly via targeting thrombospondin 2. Biomed. Pharmacother. 2019, 109, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Wang, J.; Cao, Z.; Gao, L.; Zheng, Y.; Wang, J.; Liu, X. miR-126a-3p induces proliferation, migration and invasion of trophoblast cells in pre-eclampsia-like rats by inhibiting A Disintegrin and Metalloprotease 9. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Li, J.; Fu, Z.; Jiang, H.; Chen, L.; Wu, X.; Ding, H.; Xia, Y.; Wang, X.; Tang, Q.; Wu, W. IGF2-derived miR-483-3p contributes to macrosomia through regulating trophoblast proliferation by targeting RB1CC1. Mol. Hum. Reprod. 2018, 24, 444–452. [Google Scholar] [CrossRef]
- Xiao, J.; Tao, T.; Yin, Y.; Zhao, L.; Yang, L.; Hu, L. miR-144 may regulate the proliferation, migration and invasion of trophoblastic cells through targeting PTEN in preeclampsia. Biomed. Pharmacother. 2017, 94, 341–353. [Google Scholar] [CrossRef]
- Liu, M.; Wang, Y.; Lu, H.; Wang, H.; Shi, X.; Shao, X.; Li, Y.-X.; Zhao, Y.; Wang, Y.-L. miR-518b Enhances Human Trophoblast Cell Proliferation Through Targeting Rap1b and Activating Ras-MAPK Signal. Front. Endocrinol. 2018, 9, 100. [Google Scholar] [CrossRef]
- Brkić, J.; Dunk, C.; O’Brien, J.; Fu, G.; Nadeem, L.; Wang, Y.-L.; Rosman, D.; Salem, M.; Shynlova, O.; Yougbaré, I.; et al. MicroRNA-218-5p Promotes Endovascular Trophoblast Differentiation and Spiral Artery Remodeling. Mol. Ther. 2018, 26, 2189–2205. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhao, Y.; Luo, R.; Bian, X.; Wang, Y.; Shao, X.; Li, Y.-X.; Liu, M.; Wang, Y.-L. A positive feedback self-regulatory loop between miR-210 and HIF-1α mediated by CPEB2 is involved in trophoblast syncytiolization:Implication of trophoblast malfunction in preeclampsia. Biol. Reprod. 2019. [Google Scholar] [CrossRef]
- Kumar, P.; Luo, Y.; Tudela, C.; Alexander, J.M.; Mendelson, C.R. The c-Myc-Regulated MicroRNA-17∼92 (miR-17∼92) and miR-106a∼363 Clusters Target hCYP19A1 and hGCM1 To Inhibit Human Trophoblast Differentiation. Mol. Cell. Biol. 2013, 33, 1782–1796. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.; Jiang, S.; Chen, J.; Li, J.; Ao, L.; Zhang, Y. Upregulated long noncoding RNA Linc00261 in pre-eclampsia and its effect on trophoblast invasion and migration via regulating miR-558/TIMP4 signaling pathway. J. Cell. Biochem. 2019, 120, 13243–13253. [Google Scholar] [CrossRef]
- Wang, X.; Peng, S.; Cui, K.; Hou, F.; Ding, J.; Li, A.; Wang, M.; Geng, L. MicroRNA-576-5p enhances the invasion ability of trophoblast cells in preeclampsia by targeting TFAP2A. Mol. Genet. Genom. Med. 2019, 8, e1025. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, J.; Li, M.-Q.; Xu, J.; Zhang, J.-P.; Jin, L.-P. MicroRNA-184 promotes apoptosis of trophoblast cells via targeting WIG1 and induces early spontaneous abortion. Cell Death Dis. 2019, 10, 223. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.-M.; Cao, P.; Xin, L.; Zhang, Y.; Liu, Z.; Yao, N.; Ma, Y.-Y. Effect of miR-133 on apoptosis of trophoblasts in human placenta tissues via Rho/ROCK signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 10600–10608. [Google Scholar] [PubMed]
- Zhang, L.; Yuan, J.-M.; Zhao, R.-H.; Wang, L.-M.; Tu, Z.-B. Correlation of MiR-152 expression with VEGF expression in placental tissue of preeclampsia rat and its influence on apoptosis of trophoblast cells. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3553–3560. [Google Scholar] [PubMed]
- Li, X.; Lu, J.; Dong, L.; Lv, F.; Liu, W.; Liu, G.; Zhu, W.; Diao, X. Effects of MiR-155 on trophoblast apoptosis in placental tissues of preeclampsia rats through HIF-1α signaling pathway. Panminerva Med. 2019. [Google Scholar] [CrossRef]
- Du, E.; Cao, Y.; Feng, C.; Lu, J.; Yang, H.; Zhang, Y. The Possible Involvement of miR-371a-5p Regulating XIAP in the Pathogenesis of Recurrent Pregnancy Loss. Reprod. Sci. 2019, 26, 1468–1475. [Google Scholar] [CrossRef]
- Dong, X.; Yang, L.; Wang, H. miR-520 promotes DNA-damage-induced trophoblast cell apoptosis by targeting PARP1 in recurrent spontaneous abortion (RSA). Gynecol. Endocrinol. 2016, 33, 274–278. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Zhao, X.; Yuan, X.; Li, P. Elevated microRNA-34a contributes to trophoblast cell apoptosis in preeclampsia by targeting BCL-2. J. Hum. Hypertens. 2017, 31, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, S.; Li, Y.; Han, B.; Ma, Y. Inhibition of miR-18a increases expression of estrogen receptor 1 and promotes apoptosis in human HTR8 trophoblasts. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2017, 33, 1102–1107. [Google Scholar] [PubMed]
- West, R.; Russ, J.E.; Bouma, G.J.; Winger, Q.A. BRCA1 regulates HMGA2 levels in the Swan71 trophoblast cell line. Mol. Reprod. Dev. 2019, 86, 1663–1670. [Google Scholar] [CrossRef]
- Li, L.; Hou, A.; Gao, X.; Zhang, J.; Zhang, L.; Wang, J.; Li, H.; Song, Y. Lentivirus-mediated miR-23a overexpression induces trophoblast cell apoptosis through inhibiting X-linked inhibitor of apoptosis. Biomed. Pharmacother. 2017, 94, 412–417. [Google Scholar] [CrossRef]
- Mao, Z.; Yao, M.; Li, Y.; Fu, Z.; Li, S.; Zhang, L.; Zhou, Z.; Tang, Q.; Han, X.; Xia, Y. miR-96-5p and miR-101-3p as potential intervention targets to rescue TiO2 NP-induced autophagy and migration impairment of human trophoblastic cells. Biomater. Sci. 2018, 6, 3273–3283. [Google Scholar] [CrossRef]
- Zhang, X.; Ge, Y.-W.; Wang, Z.-X.; Xu, Q.-L.; Guo, R.; Xu, H.-Y. MiR-200c regulates apoptosis of placental trophoblasts in preeclampsia rats through Wnt/β-catenin signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7209–7216. [Google Scholar]
- Gu, Y.; Meng, J.; Zuo, C.; Wang, S.; Li, H.; Zhao, S.; Huang, T.; Wang, X.-T.; Yan, J. Downregulation of MicroRNA-125a in Placenta Accreta Spectrum Disorders Contributes Antiapoptosis of Implantation Site Intermediate Trophoblasts by Targeting MCL1. Reprod. Sci. 2019, 26, 1582–1589. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef]
- Liu, S.; Xia, Q.; Zhao, P.; Cheng, T.; Hong, K.; Xiang, Z. Characterization and expression patterns of let-7 microRNA in the silkworm (Bombyx mori). BMC Dev. Biol. 2007, 7, 88. [Google Scholar] [CrossRef]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.J.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.; Degnan, B.M.; Müller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A Mammalian microRNA Expression Atlas Based on Small RNA Library Sequencing. Cell 2007, 129, 1401–1414. [Google Scholar] [CrossRef] [PubMed]
- Roush, S.; Slack, F.J. The let-7 family of microRNAs. Trends Cell Biol. 2008, 18, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Han, S.; Kwon, C.S.; Lee, D. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell 2015, 7, 100–113. [Google Scholar] [CrossRef]
- Zhang, H.; Artiles, K.L.; Fire, A.Z. Functional relevance of “seed” and “non-seed” sequences in microRNA-mediated promotion of C. elegans developmental progression. RNA 2015, 21, 1980–1992. [Google Scholar] [CrossRef]
- Kehl, T.; Backes, C.; Kern, F.; Fehlmann, T.; Ludwig, N.; Meese, E.; Lenhof, H.-P.; Keller, A. About miRNAs, miRNA seeds, target genes and target pathways. Oncotarget 2017, 8, 107167–107175. [Google Scholar] [CrossRef]
- Shyh-Chang, N.; Zhu, H.; De Soysa, T.Y.; Shinoda, G.; Seligson, M.T.; Tsanov, K.M.; Nguyen, L.; Asara, J.M.; Cantley, L.C.; Daley, G.Q. Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell 2013, 155, 778–792. [Google Scholar] [CrossRef]
- Shyh-Chang, N.; Daley, G.Q. Lin28:Primal regulator of growth and metabolism in stem cells. Cell Stem Cell 2013, 12, 395–406. [Google Scholar] [CrossRef]
- Caygill, E.E.; Johnston, L.A. Temporal Regulation of Metamorphic Processes in Drosophila by the let-7 and miR-125 Heterochronic MicroRNAs. Curr. Biol. 2008, 18, 943–950. [Google Scholar] [CrossRef]
- Wagner, S.; Ngezahayo, A.; Murua Escobar, H.; Nolte, I. Role of miRNA let-7 and its major targets in prostate cancer. Biomed Res. Int. 2014, 2014, 14. [Google Scholar] [CrossRef]
- Takamizawa, J.; Chamoto, K.; Tsuji, T.; Funamoto, H.; Kosaka, A.; Matsuzaki, J.; Sato, T.; Konishi, H.; Fujio, K.; Yamamoto, K.; et al. Reduced Expression of the let-7 MicroRNAs in Human Lung Cancers in Association with Shortened Postoperative Survival. Cancer Res. 2004, 64, 3753–3756. [Google Scholar] [CrossRef] [PubMed]
- Boyerinas, B.; Park, S.-M.; Shomron, N.; Hedegaard, M.M.; Vinther, J.; Andersen, J.S.; Feig, C.; Xu, J.; Burge, C.B.; Peter, M.E. Identification of Let-7-Regulated Oncofetal Genes. Cancer Res. 2008, 68, 2587–2591. [Google Scholar] [CrossRef]
- Copley, M.R.; Babovic, S.; Benz, C.; Knapp, D.J.; Beer, P.A.; Kent, D.; Wohrer, S.; Treloar, D.Q.; Day, C.; Rowe, K.; et al. The Lin28b–let-7–Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat. Cell Biol. 2013, 15, 916–925. [Google Scholar] [CrossRef] [PubMed]
- Emmrich, S.; Rasche, M.; Schöning, J.; Reimer, C.; Keihani, S.; Maroz, A.; Xie, Y.; Li, Z.; Schambach, A.; Reinhardt, D.; et al. miR-99a/100∼125b tricistrons regulate hematopoietic stem and progenitor cell homeostasis by shifting the balance between TGFβ and Wnt signaling. Genes Dev. 2014, 28, 858–874. [Google Scholar] [CrossRef] [PubMed]
- Heo, I.; Ha, M.; Lim, J.; Yoon, M.-J.; Park, J.-E.; Kwon, S.C.; Chang, H.; Kim, V.N. Mono-Uridylation of Pre-MicroRNA as a Key Step in the Biogenesis of Group II let-7 MicroRNAs. Cell 2012, 151, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Büssing, I.; Slack, F.J.; Großhans, H. let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 2008, 14, 400–409. [Google Scholar] [CrossRef]
- Thomson, J.M.; Newman, M.; Parker, J.S.; Morin-Kensicki, E.M.; Wright, T.; Hammond, S.M. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genome Res. 2006, 20, 2202–2207. [Google Scholar] [CrossRef]
- Nam, Y.; Chen, C.; Gregory, R.I.; Chou, J.J.; Sliz, P. Molecular Basis for Interaction of let-7 MicroRNAs with Lin28. Cell 2011, 147, 1080–1091. [Google Scholar] [CrossRef]
- Zhang, P.; Elabd, S.; Hammer, S.; Solozobova, V.; Yan, H.; Bartel, F.; Inoue, S.; Henrich, T.; Wittbrodt, J.; Loosli, F.; et al. TRIM25 has a dual function in the p53/Mdm2 circuit. Oncogene 2015, 34, 5729–5738. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Meister, G. Publisher Correction: Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 321. [Google Scholar] [CrossRef]
- Guo, Y.; Chen, Y.; Ito, H.; Watanabe, A.; Ge, S.X.; Kodama, T.; Aburatani, H. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene 2006, 384, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Tsialikas, J.; Romer-Seibert, J. LIN28: Roles and regulation in development and beyond. Development 2015, 142, 2397–2404. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Baltimore, D. RNA-binding protein Lin28 in cancer and immunity. Cancer Lett. 2016, 375, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Shinoda, G.; Shyh-Chang, N.; De Soysa, T.Y.; Zhu, H.; Seligson, M.T.; Shah, S.P.; Abo-Sido, N.; Yabuuchi, A.; Hagan, J.P.; Gregory, R.I.; et al. Fetal deficiency of lin28 programs life-long aberrations in growth and glucose metabolism. Stem Cells 2013, 31, 1563–1573. [Google Scholar] [CrossRef]
- Mayr, F.; Heinemann, U. Mechanisms of Lin28-Mediated miRNA and mRNA Regulation—A Structural and Functional Perspective. Int. J. Mol. Sci. 2013, 14, 16532–16553. [Google Scholar] [CrossRef]
- Zhang, J.; Ratanasirintrawoot, S.; Chandrasekaran, S.; Wu, Z.; Ficarro, S.B.; Yu, C.; Ross, C.A.; Cacchiarelli, D.; Xia, Q.; Seligson, M.; et al. LIN28 Regulates Stem Cell Metabolism and Conversion to Primed Pluripotency. Cell Stem Cell 2016, 19, 66–80. [Google Scholar] [CrossRef]
- Viswanathan, S.; Daley, G.Q.; Gregory, R. Selective Blockade of MicroRNA Processing by Lin28. Science 2008, 320, 97–100. [Google Scholar] [CrossRef]
- Heo, I.; Joo, C.; Cho, J.; Ha, M.; Han, J.; Kim, V.N. Lin28 Mediates the Terminal Uridylation of let-7 Precursor MicroRNA. Mol. Cell 2008, 32, 276–284. [Google Scholar] [CrossRef]
- Piskounova, E.; Polytarchou, C.; Thornton, J.E.; Lapierre, R.J.; Pothoulakis, C.; Hagan, J.P.; Iliopoulos, D.; Gregory, R. Lin28A and Lin28B Inhibit let-7 MicroRNA Biogenesis by Distinct Mechanisms. Cell 2011, 147, 1066–1079. [Google Scholar] [CrossRef]
- Molenaar, J.J.; Domingo-Fernández, R.; Ebus, M.E.; Lindner, S.; Koster, J.; Drabek, K.; Mestdagh, P.; van Sluis, P.; Valentijn, L.J.; van Nes, J.; et al. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat. Genet. 2012, 44, 1199–1206. [Google Scholar] [CrossRef] [PubMed]
- Thornton, J.E.; Chang, H.-M.; Piskounova, E.; Gregory, R. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7). RNA 2012, 18, 1875–1885. [Google Scholar] [CrossRef] [PubMed]
- Heo, I.; Joo, C.; Kim, Y.-K.; Ha, M.; Yoon, M.-J.; Cho, J.; Yeom, K.-H.; Han, J.; Kim, V.N. TUT4 in Concert with Lin28 Suppresses MicroRNA Biogenesis through Pre-MicroRNA Uridylation. Cell 2009, 138, 696–708. [Google Scholar] [CrossRef] [PubMed]
- Hagan, J.P.; Piskounova, E.; Gregory, R. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2009, 16, 1021–1025. [Google Scholar] [CrossRef]
- Faehnle, C.R.; Walleshauser, J.; Joshua-Tor, L. Mechanism of Dis3l2 substrate recognition in the Lin28–let-7 pathway. Nature 2014, 514, 252–256. [Google Scholar] [CrossRef]
- Rybak, A.; Fuchs, H.; Smirnova, L.; Brandt, C.; Pohl, E.E.; Nitsch, R.; Wulczyn, F.G. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nature 2008, 10, 987–993. [Google Scholar] [CrossRef]
- Ustianenko, D.; Chiu, H.-S.; Treiber, T.; Weyn-Vanhentenryck, S.M.; Treiber, N.; Meister, G.; Sumazin, P.; Zhang, C. LIN28 Selectively Modulates a Subclass of Let-7 MicroRNAs. Mol. Cell 2018, 71, 271–283.e5. [Google Scholar] [CrossRef]
- West, R.; McWhorter, E.S.; Ali, A.; Goetzman, L.N.; Russ, J.; Gonzalez-Berrios, C.L.; Anthony, R.V.; Bouma, G.J.; A Winger, Q. HMGA2 is regulated by LIN28 and BRCA1 in human placental cells. Biol. Reprod. 2018, 100, 227–238. [Google Scholar] [CrossRef]
- Seabrook, J.L.; Cantlon, J.D.; Cooney, A.J.; McWhorter, E.E.; Fromme, B.A.; Bouma, G.J.; Anthony, R.V.; Winger, Q.A. Role of LIN28A in mouse and human trophoblast cell differentiation. Biol. Reprod. 2013, 89, 95. [Google Scholar] [CrossRef]
- Chan, H.-W.; Lappas, M.; Yee, S.Y.; Vaswani, K.; Mitchell, M.; Rice, G.E. The expression of the let-7 miRNAs and Lin28 signalling pathway in human term gestational tissues. Placenta 2013, 34, 443–448. [Google Scholar] [CrossRef]
- Barbaux, S.; Gascoin-Lachambre, G.; Buffat, C.; Monnier, P.; Mondon, F.; Tonanny, M.-B.; Pinard, A.; Auer, J.; Bessières, B.; Barlier, A.; et al. A genome-wide approach reveals novel imprinted genes expressed in the human placenta. Epigenetics 2012, 7, 1079–1090. [Google Scholar] [CrossRef] [PubMed]
- Monk, D. Genomic imprinting in the human placenta. Am. J. Obstet. Gynecol. 2015, 213, S152–S162. [Google Scholar] [CrossRef]
- Liu, Y.; Fan, X.; Wang, R.; Lu, X.; Dang, Y.-L.; Wang, H.; Lin, H.-Y.; Zhu, C.; Ge, H.; Cross, J.C.; et al. Single-cell RNA-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell Res. 2018, 28, 819–832. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Sun, J.; Groome, L.J.; Wang, Y. Differential miRNA expression profiles between the first and third trimester human placentas. Am. J. Physiol. Metab. 2013, 304, E836–E843. [Google Scholar] [CrossRef] [PubMed]
- Souza, M.A.; Brizot, M.L.; Biancolin, S.E.; Schultz, R.; De Carvalho, M.H.B.; Francisco, R.P.V.; Zugaib, M. Placental weight and birth weight to placental weight ratio in monochorionic and dichorionic growth-restricted and non-growth-restricted twins. Clinics 2017, 72, 265–271. [Google Scholar] [CrossRef]
- Hiden, U.; Wadsack, C.; Prutsch, N.; Gauster, M.; Weiss, U.; Frank, H.-G.; Schmitz, U.; Fast-Hirsch, C.; Hengstschläger, M.; Pötgens, A.; et al. The first trimester human trophoblast cell line ACH-3P: A novel tool to study autocrine/paracrine regulatory loops of human trophoblast subpopulations – TNF-α stimulates MMP15 expression. BMC Dev. Biol. 2007, 7, 137. [Google Scholar] [CrossRef] [PubMed]
- Chavez, S.L.; Abrahams, V.M.; Alvero, A.B.; Aldo, P.B.; Ma, Y.; Guller, S.; Romero, R.; Mor, G. The Isolation and Characterization of a Novel Telomerase Immortalized First Trimester Trophoblast Cell Line, Swan 71. Placenta 2009, 30, 939–948. [Google Scholar] [CrossRef]
- McWhorter, E.S.; West, R.; Russ, J.E.; Ali, A.; Winger, Q.A.; Bouma, G.J. LIN28B regulates androgen receptor in human trophoblast cells through Let-7c. Mol. Reprod. Dev. 2019, 86, 1086–1093. [Google Scholar] [CrossRef]
- Madison, B.B.; Jeganathan, A.N.; Mizuno, R.; Winslow, M.M.; Castells, A.; Cuatrecasas, M.; Rustgi, A.K. Let-7 Represses Carcinogenesis and a Stem Cell Phenotype in the Intestine via Regulation of Hmga2. PLoS Genet. 2015, 11. [Google Scholar] [CrossRef]
- Lee, Y.S.; Dutta, A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genome Res. 2007, 21, 1025–1030. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Stenglein, M.D.; Spencer, T.E.; Bouma, G.J.; Anthony, R.V.; Winger, Q.A. Trophectoderm-Specific Knockdown of LIN28 Decreases Expression of Genes Necessary for Cell Proliferation and Reduces Elongation of Sheep Conceptus. Int. J. Mol. Sci. 2020, 21, 2549. [Google Scholar] [CrossRef] [PubMed]
- Wilsker, D.; Patsialou, A.; Dallas, P.; Moran, E. ARID proteins:A diverse family of DNA binding proteins implicated in the control of cell growth, differentiation, and development. Cell Growth Differ. Mol. Biol. J. Am. Assoc. Cancer Res. 2002, 13, 95–106. [Google Scholar]
- Hong, Z.; Wei, S.; Bi, X.; Zhao, J.; Huang, Z.; Li, Z.; Zhou, J.; Cai, J.; Chen, L.; Lin, C.; et al. Recent advances in the ARID family:Focusing on roles in human cancer. OncoTargets Ther. 2014, 7, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Fukuyo, Y.; Takahashi, A.; Hara, E.; Horikoshi, N.; Pandita, T.K.; Nakajima, T. E2FBP1 antagonizes the p16INK4A-Rb tumor suppressor machinery for growth suppression and cellular senescence by regulating promyelocytic leukemia protein stability. Int. J. Oral Sci. 2011, 3, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Fukuyo, Y.; Mogi, K.; Tsunematsu, Y.; Nakajima, T. E2FBP1/hDril1 modulates cell growth through downregulation of promyelocytic leukemia bodies. Cell Death Differ. 2004, 11, 747–759. [Google Scholar] [CrossRef]
- Kobayashi, K.; Jakt, L.M.; Nishikawa, S.-I. Epigenetic regulation of the neuroblastoma genes, Arid3b and Mycn. Oncogene 2012, 32, 2640–2648. [Google Scholar] [CrossRef][Green Version]
- Bobbs, A.; Gellerman, K.; Hallas, W.M.; Joseph, S.; Yang, C.; Kurkewich, J.; Dahl, K.C. ARID3B Directly Regulates Ovarian Cancer Promoting Genes. PLoS ONE 2015, 10, e0131961. [Google Scholar] [CrossRef]
- Nakahara, S.; Fukushima, S.; Yamashita, J.; Kubo, Y.; Tokuzumi, A.; Miyashita, A.; Harada, M.; Nakamura, K.; Jinnin, M.; Ihn, H. AT-rich Interaction Domain-containing Protein 3B is a New Tumour Marker for Melanoma. Acta Derm. Venereol. 2017, 97, 112–114. [Google Scholar] [CrossRef]
- Kim, D.; Probst, L.; Das, C.; Tucker, P.W. REKLES Is an ARID3-restricted Multifunctional Domain. J. Biol. Chem. 2007, 282, 15768–15777. [Google Scholar] [CrossRef]
- Webb, C.F.; Bryant, J.; Popowski, M.; Allred, L.; Kim, D.; Harriss, J.; Schmidt, C.; Miner, C.A.; Rose, K.; Cheng, H.-L.; et al. The ARID Family Transcription Factor Bright Is Required for both Hematopoietic Stem Cell and B Lineage Development. Mol. Cell. Biol. 2011, 31, 1041–1053. [Google Scholar] [CrossRef]
- Liao, T.-T.; Hsu, W.-H.; Ho, C.-H.; Hwang, W.-L.; Lan, H.-Y.; Lo, T.; Chang, C.-C.; Tai, S.; Yang, M.-H. Let-7 Modulates Chromatin Configuration and Target Gene Repression through Regulation of the ARID3B Complex. Cell Rep. 2016, 14, 520–533. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Oksvold, P.; Fagerberg, L.; Lundberg, E.; Jonasson, K.; Forsberg, M.; Zwahlen, M.; Kampf, C.; Wester, K.; Hober, S.; et al. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 2010, 28, 1248–1250. [Google Scholar] [CrossRef] [PubMed]
- Rhee, C.; Lee, B.-K.; Beck, S.; Anjum, A.; Cook, K.R.; Popowski, M.; Tucker, H.O.; Kim, J. Arid3a is essential to execution of the first cell fate decision via direct embryonic and extraembryonic transcriptional regulation. Genome Res. 2014, 28, 2219–2232. [Google Scholar] [CrossRef] [PubMed]
- Rhee, C.; Edwards, M.; Dang, C.; Harris, J.; Brown, M.; Kim, J.; Tucker, H.O. ARID3A is required for mammalian placenta development. Dev. Biol. 2016, 422, 83–91. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).