Abstract
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease with no known cure. Approximately 90% of ALS cases are sporadic, although multiple genetic risk factors have been recently revealed also in sporadic ALS (SALS). The pathological expansion of a hexanucleotide repeat in chromosome 9 open reading frame 72 (C9orf72) is the most common genetic mutation identified in familial ALS, detected also in 5–10% of SALS patients. C9orf72-related ALS phenotype appears to be dependent on several modifiers, including demographic factors. Sex has been reported as an independent factor influencing ALS development, with men found to be more susceptible than women. Exposure to both female and male sex hormones have been shown to influence disease risk or progression. Moreover, interplay between genetics and sex has been widely investigated in ALS preclinical models and in large populations of ALS patients carrying C9orf72 repeat expansion. In light of the current need for reclassifying ALS patients into pathologically homogenous subgroups potentially responsive to targeted personalized therapies, we aimed to review the recent literature on the role of genetics and sex as both independent and synergic factors, in the pathophysiology, clinical presentation, and prognosis of ALS. Sex-dependent outcomes may lead to optimizing clinical trials for developing patient-specific therapies for ALS.
1. Amyotrophic Lateral Sclerosis (ALS): A Multifactorial Neurodegenerative Disease
Amyotrophic Lateral Sclerosis (ALS), also known as Lou Gehrig’s disease, is a fatal neurodegenerative disease characterized by varying phenotypic manifestations due to degeneration of both upper and lower motor neurons [1]. Clinical symptoms include muscle weakness, spasticity, dysphagia, dysarthria, and respiratory failure; up to 50% of the patients develop cognitive and/or behavioral impairment during the disease course, and about 13% present with concomitant behavioral variant frontotemporal dementia (bv-FTD) [1].
Although our understanding of ALS is improving, the pathological pathways that lead to the disease and affect the overall integrity of brain networks are still incompletely understood. Extensive literature provides convincing evidence for a number of cellular and molecular processes found to be implicated in ALS pathogenesis, including, among the most largely studied, excitotoxicity, oxidative stress, impaired protein degradation (proteasome and/or autophagy), toxic protein aggregation, mitochondrial dysfunction, axonal transport, prion-like spreading, dysfunction of non-neuronal cells, neuroinflammation, defects in RNA metabolism, and RNA toxicity [2]. However, defects in some of these pathways may represent a secondary phenomenon, and taking into account that 5–15% of ALS patients have a familial disease (FALS) and that multiple genetic risk factors have been recently revealed for sporadic ALS (SALS) [3], genetics has provided relevant insights regarding pathophysiology underlying ALS. In particular, it is considered a multifactorial disease, influenced by genetic and environmental factors.
Two twin studies, one population- and one clinic-based, have suggested that heritability contributes about 60% to the risk of developing ALS and the unshared environmental component approximately 40% [4], although an analysis applying the genome-wide complex trait analysis algorithm to three genome-wide association studies (GWAS) datasets estimated the overall heritability of ALS to be approximately 21% [5]. More recently, a population-based study by Chiò et al. [6] demonstrated that the spatial–temporal combination of motor and cognitive events leading to the onset and progression of ALS is influenced by demographic (mainly age and sex) and genetic factors. Remarkably, data from ALS population registers in Italy and the Republic of Ireland confirmed that deficits in multiple pathways, reflecting a “multistep” model of disease consistent with a six-step process, are required to develop ALS, although they revealed that a reduced number of steps are detected in patients with ALS with genetic mutations compared to those without mutations [7]. Therefore, these findings from the interrogation of population-based registers further emphasize the need to study the interaction between genetic, demographic, environmental, and lifestyle risk factors.
Among demographic factors, sex has been reported as an independent factor influencing the development of ALS, with men reported to be 2-3 times more susceptible than women [8], especially in case of flail limbs and respiratory phenotypes, with a trend toward a higher frequency in older age [6]. Moreover, the role of sex in clinical presentation and prognosis of ALS patients carrying superoxide dismutase-1 (SOD1) mutations and chromosome 9 open reading frame 72 (C9orf72) repeat expansion has also been explored [6,7,8,9,10].
With this background, in the light of the current need to reclassify ALS patients into pathologically homogenous subgroups potentially responsive to targeted personalized therapies, we aimed to review the recent literature on the role of genetics and sex, as both independent and synergic factors, on the pathophysiology, clinical presentation, and prognosis of ALS. With regard to the search strategy and the selection criteria adopted, we searched Pubmed (2010 to April 2020) using the search terms “amyotrophic lateral sclerosis” or “motor neuron disease” in combination with “epidemiology”, “genetics”, “sex”, and “gender”. Further articles were included from reference lists and review articles. The final reference list was generated on the basis of originality and relevance to the topics covered in this manuscript. Emphasis was placed on publications from the last 5 years but did not exclude highly regarded older publications. Table 1 summarizes the most relevant articles among the reviewed and referenced ones, which addressed the interplay between sex and genetics in the ALS pathogenesis.

Table 1.
Research studies analyzing the role of sex–genetics interplay in the pathogenesis and clinical phenotype of ALS.
2. The Role of Genetics in ALS Pathogenesis
An increasing number of genetic variants are recognized as associated with ALS [3,11,12]. In 60–80% of patients with FALS, gene mutations of large effect, such as repeat expansions in C9orf72 (40%), mutations of SOD1 (20%), fused in sarcoma (FUS) (1–5%), and transactive response DNA binding protein (TARBDP) (1–5%), can be most commonly identified [10,11]. In recent years, advances in massive parallel sequencing approaches such as whole-genome sequencing (WGS) and whole-exome sequencing (WES) [12,13,14], derived from large-scale collaborations which have previously led to GWAS in ALS [15,16,17] and to the international WGS project MINE (www.ProjectMine.com) [18], shed more light on the genetic architecture of ALS. In particular, the latest WGS and WES studies underlined that in ALS, one or a few large-effect, relatively rare variants in each patient seem to contribute most to the disease risk, which is different from conditions such as Alzheimer disease or schizophrenia, in which multiple small-effect variants add up to create risk [12,19,20], suggesting an oligogenic model in ALS [21]. Moreover, this evidence is consistent with the observation of multiple ALS-associated genes co-segregating in some kindreds [21]. Another relevant aspect is the incomplete penetrance in many ALS pedigrees, causing unreliable heritability, especially in the case of reduction of family sizes (i.e., “apparently” SALS) [22], and shown to be age- and sex-dependent in patients carrying C9orf72 repeat expansion [23]. Of note, ALS-associated genes may be pleiotropic (e.g., linked to several phenotypes), such as for C9orf72 repeat expansions [24] or other genetic variants [25,26].
The pathological expansion of a hexanucleotide repeat in C9orf72 is the most common genetic mutation identified in European and Caucasian patients with ALS, reported in about 40% of patients with FALS, 25% of patients with familial FTD and 5–10% of patients with SALS [16,17]. C9orf72 expansion influences clinical presentation, with patients carrying the expansion exhibiting higher prevalence of bulbar onset, earlier age at onset, and reduced survival, with higher incidence of comorbid FTD and/or family history of dementia or ALS [27,28,29,30,31,32,33]. Moreover, the detection of repeat expansion in C9orf72 has been also associated with Parkinsonism, Huntington phenocopies, Alzheimer’s disease, corticobasal degeneration, and psychosis [24]. Although the function of the protein encoded by C9orf72 is still unknown, there are several theories regarding how the C9orf72 repeat expansion leads to ALS and FTD [34] (Figure 1): i) loss of C9orf72 protein expression could inhibit autophagy and promote neuroinflammation (Figure 1A) [35,36]; ii) expanded sense and antisense RNA, derived from the expanded gene, form toxic RNA foci that sequester RNA binding proteins, impairing RNA metabolism (Figure 1B) [15,36]; iii) non-canonical repeat-associated non-ATG (RAN) translation of toxic dipeptide repeat proteins aggregates (Figure 1C) [37]. Disease severity and phenotype appear to be dependent on the size of the repeat expansion (which may vary between cell types within an individual (mosaicism)), methylation level at CpG sites [38] and the co-occurrence of genetic variation in other genes (e.g., transmembrane protein 106B -TMEM106b, ataxin-2- ATXN2, C6orf10/LOC101929163/HLA-DRB1 pathway, and others) [24,39,40].
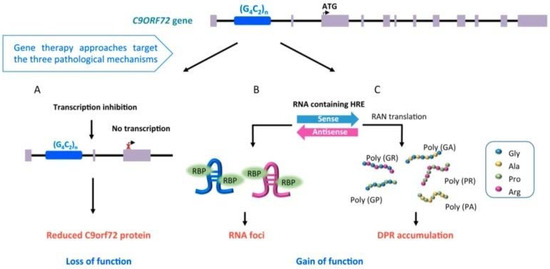
Figure 1.
Pathological mechanisms involved in C9orf72-ALS: (A) transcription inhibition (loss of function); (B) detection of RNA foci, involved in impairment of RNA metabolism (gain of function); (C) accumulation of toxic dipeptide repeat proteins (DRP) aggregates (gain of function) (derived from Cappella et al. [34], open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/)).
Mutations in copper/zinc SOD1 gene were identified as causative of ALS in 1993 [41] and account for 12–23.5% of FALS and about 7.3% of apparent SALS in Caucasian and European populations [42]. Among Asian populations, in Chinese ALS patients, in which C9orf72 repeat expansion account for 0–1.5% of SALS and 5.98% of FALS, SOD1 gene has been revealed as the most common causative gene, accounting for 1.45% of SALS and 25.33% of FALS [43]. Currently, the best-studied mechanism for ALS involves SOD1 [44], and more than 180 missense mutations have been reported in SOD1 (http://alsod.iop.kcl.ac.uk) to date. The reported heterogeneity in the age of onset and phenotype in SOD1-mutated patients [45] is due to different types of mutations that result in a gain of toxicity rather than a loss of enzymatic activity [46] and may be influenced by epigenetic factors such as sex, modifier genes, or environmental factors [47]. With regard to the contribution of rare genetic variants in regulating phenotypic variability, in a WGS analysis on SOD1-mutated patients and in a WES analysis on SALS patients, Pang et al. [48] observed that SOD1-mutated patients with additional rare variants had shorter survival and that the cumulative effect of rare genetic variants was significant on reducing survival probability in both FALS and SALS.
Mutations in the TARDBP gene, encoding the 43 kDa transactive response DNA-binding protein (TDP-43), are described in 1–5% of FALS [49]. Interestingly, phosphorylated TDP-43 (pTDP-43)-positive inclusion bodies have been detected in ubiquitin-positive, tau-negative ALS and FTD cytoplasmatic aggregates, representing the pathological hallmark of the ALS–FTD continuum of neurodegeneration [50,51]. pTDP-43 is, therefore, thought to have an essential role in ALS pathogenesis, suggesting its involvement in both loss-of-function (i.e., perturbation of nuclear TDP-43 function) [52] and gain-of-function (i.e., splicing of selected RNA targets; proteasome inhibition) [53] pathological mechanisms. Moreover, progression of ALS pathology can be divided into four stages on the basis of changes in the brain and spine distribution of intraneuronal pTDP-43 aggregates [51,54]. Mutations in the gene FUS, encoding a second RNA/DNA-binding protein, have been associated with 1–5% of FALS. Similarly to the mechanisms associated with pTDP-43 pathology, FUS toxicity in ALS has been related to dysfunction in RNA processing and in maintaining the DNA damage response [49,55].
More recently, identification of several other genes carrying rare causal variants, including valosin-containing protein (VCP), sequestosome 1 (SQTSM1), ubiquilin 2 (UBQLN2), TANK-binding kinase 1 (TBK1), optineurin (OPTN), coiled-coil-helix-coiled-coil-helix domain-containing 10 (CHCHD10), tubulin alpha 4A (TUBA4A), matrin 3 (MATR3), cyclin F (CCNF), never in mitosis, gene A -related kinase 1 (NEK1), chromosome 21 open reading frame 2 (C21orf2), Annexin A11 (ANXA11), T cell-restricted intracellular antigen-1 (TIA1), and glycosyltransferase 8 domain containing 1 (GLT8D1), has contributed to shedding more light on the genetic architecture of ALS and FTD and the related molecular pathways [12,56,57]. In particular, among the most recently described mutations in the ALS–FTD spectrum of neurodegeneration, TBK1 loss-of-function mutations, producing premature termination codons that cause nonsense-mediated mRNA decay and resulting in the loss of mutant transcript and subsequent loss or reduction of TBK1 protein, has been associated with both ALS and FTD [58,59]. From a phenotypic point of view, a genotype–phenotype analysis of a Belgian population shows that TBK1 mutated carriers may present ALS and/or FTD with a relatively late age at onset or extrapyramidal symptoms, and associated memory deficits [60]. Of note, a WGS analysis performed on pathologically confirmed Frontotemporal Lobar Degeneration-TDP-43 patients, negative for C9orf72 and GRN mutations, revealed a heterozygous missense mutation in TBK1 in three patients and a loss-of-function mutation in TBK1 associated with deletion of exons 13–15 of OPTN [61], thereby corroborating the idea of an oligogenic model of disease in the ALS–FTD spectrum caused by multiple rare variants with additive or synergistic effects on disease presentation (i.e., both TBK1 and OPTN are involved in the autophagy pathways) [12]. Rare variants in NEK1 were identified in 3–5% of ALS populations [13,58,62,63,64]. With regard to the pathogenic pathways involving NEK proteins, which belong to the protein family of NIMA-related serine/threonine kinases, they participate in maintaining the cytoskeleton network [65] together with TUBA4A [66] and profilin 1 [67]. Moreover, NEK1 interacts with the chromosome 21 open reading frame 2 (C21orf2) in DNA damage repair, together with vesicle-associated membrane-protein-associated protein B/C (VAPB) and alsin (ALS2) [13,68]. Remarkably, C21orf2 was also recently identified as an ALS-associated gene [14], and autosomal recessive mutations in both NEK1 and C21orf2 are linked to a skeletal disorder, axial spondylometaphyseal dysplasia [69,70], further highlighting the genetic and functional link between these genes and proteins. Of note, in ALS and ALS–FTD patients, additional mutations in ALS genes may be revealed in NEK1 carriers, as identified in two Belgian siblings with FALS and cognitive impairment in which NEK1 p.Ser1036* loss-of-function mutation was associated with C9orf72 repeat expansion and TUBA4A p.Thr381Met variant, presenting with early disease onsets [63,71]. More recently, mutations in GLT8D1, encoding a widely expressed glycosyltransferase enzyme, has been associated with FALS [57], resulting in higher substrate affinity of the mutated enzyme and in a dominant-negative effect with a competitive antagonism of wild-type enzyme function [57].
With this multifaceted genetic background and bearing in mind also the recently proposed “multistep” model of disease in ALS consistent with a six-step process, in the patients carrying genetic mutations associated with ALS, the number of steps necessary to start the neurodegenerative process has been shown to be reduced compared to cases without mutation (i.e., lower for SOD1, intermediate for C9orf72, and higher for TARDBP) [7]. These findings provide evidence for the investigation of demographic, environmental, and lifestyle risk factors that could interact with genetics, especially in individuals with identified mutations rather than in those apparently with SALS, because such additional factors will be fewer in number per person in the former ones and likely of larger effect size [7]. Among non-genetic triggers of pathological steps, the role of sex has been also investigated and debated [72].
3. The Role of Sex in ALS Pathogenesis
ALS is characterized by a sexual dimorphism, the most obvious being the higher risk of developing the disease in men [8]. However, a recent population-based study on prospective data from the Piemonte and Valle d’Aosta (in Northern Italy) ALS register has revealed that the age-adjusted incidence rate of ALS increased from 1994 to 2014 and the adjusted rate ratio of men to women decreased from 1.27:1 (1995–2004) to 1.17:1 (2005–2014) [73], probably due to the effect of the modified lifestyle in women across time, which may have caused a larger exposure of women to exogenous risk factors (e.g., cigarette smoking, engagement in manual work) [73]. Importantly, an epidemiologic analysis on demographic, clinical, and lifestyle data from 2008 to 2012 derived from European population-based ALS registries (i.e., from France, Ireland, Italy, United Kingdom, and Serbia) revealed an inverse correlation between the odds of having ALS and sport-related physical activity in women but not in men [74]. However, this different association between ALS and sport-related physical activity in the two sexes has been shown to be less evident after menopause [75], thereby suggesting the potential influence of female hormones, in particular estrogen and progesterone, as protective factors against ALS triggers. In a population-based, case-control study in the Netherlands between 2006 and 2009, de Jong et al. [76] demonstrated that longer reproductive time-span and lifetime endogenous estrogen exposure were associated with a longer survival in ALS patients. Conversely, contrasting results are available on the role of exposure to exogenous estrogens and progestogens as risk factors for ALS, with some studies reporting a decreased risk in women treated with oral contraceptives [77] and others reporting no association between postmenopausal hormone use and development of ALS [78]. Moreover, among sex hormone drugs, in a population-based case-control study of 10,450 U.S. Medicare participants, tamoxifen was related to lower ALS risk and testosterone to a higher risk in women [79].
Evidence from ALS preclinical models have been useful in demonstrating the protective effects of female hormones, mainly attributed to their direct influence on neural and muscular cells [80], preventing cell death, and to a reduction of neuroinflammation, among the main pathogenic mechanisms related to ALS [81]. In particular, a study performed on SOD1 G93A mice revealed that progesterone slowed down the progression of the disease and extended the life span of the affected male mice, without delaying the symptom onset [80]. Progesterone might delay the neurodegeneration process by activating autophagy degradation of mutant SOD1. On the other hand, Heitzer et al. [81] revealed that in male SOD1 G93A ALS mice, the treatment with 17β-Estradiol increases the survival of motoneurons, suggesting that this could be related to the downregulation of several components of the inflammatory response (e.g., NLRP3, IL1beta, and activated caspase 1) abnormally elevated in the spinal cord of the affected mice [81].
With regard to the effects of male sex hormones on the ALS risk, it has been hypothesized that androgens might affect motoneurons organization and that prenatal testosterone, irrespective of gender, might play a role in the development of ALS later in life [82]. Considering that the ratio between the lengths of the index finger and the ring finger (2D:4D ratio) may help to estimate the exposure to testosterone in utero for both males and female, with a reduced ratio associating with increased testosterone, Vivekananda et al. [82] demonstrated that, independently of sex, in ALS patients, the 2D:4D ratio was lower than in controls. This finding suggested that increased prenatal testosterone levels could be an independent risk factor for ALS [82]. Moreover, Herron et al. [83] revealed that, in the SOD1 G93A mice model, only males presented enlarged C-boutons, a specific class of synaptic input that originates from a small cluster of spinal interneurons: perturbations in their inputs to motoneurons may contribute to altered excitability and degeneration of motoneurons. In particular, testosterone-mediated effects on cholinergic C-boutons might lead to their selective perturbation in male SOD1 G93A mice [83], since testosterone has been shown to influence the number and size of synapses on motoneurons [84]. Furthermore, the potential role played by circulating androgens in the pathogenesis of ALS may also be suggested by the absence of androgen receptors in neurons spared by ALS (cranial nerves III, IV, and VI) [85]. With regard to the investigations on the correlation between androgens exposure and ALS risk in humans, Gargiulo-Monachelli et al. [86] showed that female ALS patients had an increased level of circulating testosterone compared to healthy females and that the circulating levels of testosterone, dehydroepiandrosterone (and its sulfate), and progesterone were elevated with age in the patient group, differently from healthy females, in whom the three hormones significantly declined across time. In addition, in ALS patients who showed higher testosterone levels and a lower progesterone/free testosterone ratio, a faster worsening of respiratory parameters was observed [86]. In line with these findings, Pfeiffer et al. [79] have described in a cohort of U.S. Medicare beneficiaries an association between testosterone use and elevated ALS risk, particularly in women.
Large population-based studies on data from the Piemonte and Valle d’Aosta register [6,87] revealed an association between sex and motor and cognitive phenotypes in ALS. Male sex was found to be associated with flail arm and respiratory phenotypes, and female sex with bulbar phenotype, but only through an interaction with age [6,87]. With regard to cognitive phenotypes, in the ALS–FTD subgroup, women were found to be more affected than men with increasing age [6]. In agreement with this evidence of an increased vulnerability in cognitive functions in ALS females, Palmieri et al. [88] revealed a greater executive impairment in ALS females than in ALS males. Conversely, Flaherty et al. [89] reported a protective role of estrogens against Frontotemporal Lobar Degeneration, resulting in a preservation of executive abilities in ALS females.
Interestingly, with regard to the investigations on the potential correlation between sex and disease duration/survival in ALS, Pupillo et al. [90] performed a population-based study on data from the Lombardia ALS register, collected from 1998 to 2002, addressing the role of sex, among other demographic and clinical variables, as a predictor of survival: longer survival was predicted by the male gender, considering that standardized mortality ratios at 10 years, used to assess the 10-year excess mortality of ALS patients, were found to be higher until age 75 year in comparison to the general population, predominately in women, and became non-significant for males thereafter. Of note, the Dutch case-control study by de Jong et al. [76] revealed that, among the ALS females, a longer survival was described only in those with more protracted reproductive time-span, exposed longer to endogenous estrogens. However, although largely explored, data about the potential association between sex and risk of developing ALS in humans are still scattered, needing the assessment of co-occurrent risk factors, such as the potential interplay with genetics.
Key Points
Question: Are the Female and/or the Male Hormones Associated with ALS Development and Prognosis?
Findings in favor of the neuroprotective role of the female hormones, differently from the male hormones [74,75,76,77,79,80,81,82,83,84,85,86,89,90]
There is an inverse correlation between the odds of having ALS and sport-related physical activity in women but not in men, and this correlation becomes less evident after menopause. Longer reproductive time-span and lifetime endogenous estrogen exposure are associated with a longer survival in ALS patients. A decreased risk of developing ALS is reported also in women treated with oral contraceptives. Furthermore, tamoxifen use is related to lower ALS risk, and testosterone use to a higher risk in women. Higher testosterone levels and lower progesterone/free testosterone ratio are associated with a faster worsening of respiratory parameters.
With regard to cognitive phenotypes, estrogens are protective against Frontotemporal Lobar Degeneration.
Preclinical models (i.e., SOD1 G93A mice or rats) support the protective role of progesterone and 17β-estradiol, mainly attributed to their direct effect on neural and muscular cells, preventing cell death, and to a reduction of neuroinflammation. Conversely, androgens seem to affect motoneurons organization, and prenatal exposure to testosterone, irrespective of gender, might play a role in the development of ALS later in life. Finally, neurons spared by ALS (cranial nerves III, IV, and VI) do not show androgen receptors.
Findings contrasting with the neuroprotective role of the female hormones [6,73,78,88].
The crude incidence rate of ALS increased by 9% during the 1995 to 2014 period, after age and sex adjustment and mostly limited to women. This increase can be partly ascribed to the effect of the modified lifestyle in women across time, which may have caused a larger exposure of women to exogenous risk factors. No association is described between postmenopausal hormone use and the development of ALS.
With regard to cognitive phenotypes, among ALS–FTD patients, women may become more affected than men at increasing age. Moreover, ALS females may exhibit a greater executive impairment than males.
4. Interplay between Sex and Genetics in ALS Pathogenesis
Several studies have addressed the hypothesis that sex may influence ALS triggers due to genetic mutations in ALS preclinical models and in humans. In particular, SOD1 mutations seem to have a sex-specific effect on the proliferation and differentiation rate of motoneuron cells, according to the study performed in neural progenitor cells (rNPCs) from SOD1 G93A transgenic rats [91]. Li et al. [91] showed that in male rNPCs, but not in female rNPCs, the overexpression of mutant SOD1 decreased significantly their proliferative and differentiating potential, and sensitivity to oxidative stress was increased, thereby suggesting that sexual dimorphism in ALS can be attributed to intrinsic cellular differences of sex and SOD1 G93A transgene, more than to gonadal steroids. Accordingly, Hayes-Punzo et al. [92] revealed that in SOD1 G93A transgenic, gonadectomy had no significant effect on disease onset or progression, corroborating the idea that gonadal steroids are not key modulators in ALS. On the other hand, a more recent study reported that ovariectomy resulted in earlier disease onset and attenuated anti-inflammatory and anti-apoptotic actions of the estrogens in SOD1 G39A transgenic mice, as a consequence of downregulation of aromatase and estrogen receptor α expression and inhibition of anti-inflammatory and anti-apoptotic factors (e.g., arginase 1, transforming growth factor beta -TGFβ, B-cell lymphoma 2 -Bcl 2) [93].
Another hypothesis regards the role of allelic variants of Chromogranin B (CHGB), a component of secretory vesicles that acts as a chaperone-like protein, in exacerbating sexual differences in the presence of SOD1 mutations [94]. Ohta et al. [94] found that overexpression of the allelic variant CHGBL413 transgene in SOD1 G37R mice precipitated disease onset and increased its duration specifically in female mice, likely by restraining the secretion of misfolded SOD1 and therefore attenuating microgliosis. Gender differences were hypothesized to be due to a sex-determining region Y element in the CHGB promoter that might act as a suppressor of transcription in males [94]. Of note, overexpression of the more common allelic variant CHGBP413 in SOD1 G37R mice significantly accelerated disease progression and pathological changes, similarly to the ALS course described in women of Japanese and French/Canadian origins carrying CHGBP413L allele, who presented an earlier ALS disease onset [94]. Moreover, peroxisome proliferator-activated receptor-γ Coactivator (PGC) -1α, a transcriptional co-activator that regulates the cellular response to metabolic demands, has been described as a disease modifier of human and experimental ALS, with a sex-dependent effect [95]. In ALS-transgenic mice, deficiency of PGC-1α accelerated disease onset and had a borderline survival effect only in the male group [95]. This modulating effect of PGC-1α on age of onset and survival was also revealed in two large ALS populations, confirming that it acts in a male-specific manner [95]. In contrast to evidence of these disease-modifying effects of PGC-1α in males, Kaneb et al. [96] reported an unexpected dose-dependent negative effect of metformin, an inductor of the production of PGC-1α, on the onset and disease progression in female SOD1 G93A transgenic mice, and this harmful effect of metformin only in female mice has been hypothesized to be due to inhibition of 17β-estradiol production caused by metformin itself [97].
Sex differences in disease phenotype have also been explored in light of the effects on mitochondrial function of mutant SOD1 in male and female ALS transgenic mice and in cellular models overexpressing SOD1 with the G93A mutation [98]. In particular, in SOD1 G93A transgenic mice, mutant SOD1 has been shown to be accumulated in the mitochondrial intermembrane space (IMS), thus determining mitochondrial dysfunction. A distinct mitochondrial unfolded protein response (UPRmt), induced by misfolded proteins in the IMS and mediated by the estrogen receptor alpha (ERα), was found to upregulate the proteasome and the IMS serine protease Omi (also known as high-temperature requirement factor A2 -HtrA2) activity [99]. A higher activity of these two mitochondrial pathways was detected in female SOD1G39A mice, but not in males, highlighting a significant sex difference in the IMS-UPRmt [99]. Furthermore, this increased response was not observed in the absence of ERα, suggesting that sex differences in the disease phenotype could be linked to differential activation of the ERα axis of the IMS-UPRmt [99].
More recently, studies on animal models have also been helpful in exploring, together with the motor phenotype, behavior differences between the two sexes: SOD1 G93A males have been shown to display reduced locomotion and exploration and increased anxiety-like behaviors, as well as an increased conditioned cue-freezing response, while impaired intermediate-term spatial memory may be reported in SOD1 G93A females [100].
Crucially, taking into account the sexual dimorphism hypothesized to be involved in pathogenic mechanisms underlying ALS, gender has also emerged as a possible factor influencing response to treatment. As an example, Torres et al. [101] studied the effect of a dietary supplementation of Docosahexaenoic acid (DHA), an essential fatty acid modulating key nervous system functions, including neuroinflammation, in SOD1 G93A mice. The Kaplan–Meyer survival analysis revealed a longer survival of G93A male mice under DHA-supplemented diet, while female G93A mice survival did not exhibit any differences. In the same way, weight loss, indicating motor neuron impairment in this model, was significantly influenced by diet in G93A male mice, but not in females [101]. On the contrary, anti-inflammatory indexes were equally affected in males and females, suggesting that only males, in addition to changes in fatty acid profiles, are benefited by yet-unknown mechanisms dependent on dietary DHA content [101]. Moreover, a previous study by Cacabelos et al. [102] has shown that high content of unsaturated fatty acids in the diet may have a negative effect on survival of SOD1 G93A female mice, causing an increased production of mitochondrial free radical species. In addition, Bame et al. [103] demonstrated that the beneficial effect of a dietary supplementation of methionine sulfoximine on the survival of SOD1 G39A mice, mostly observed in females, was completely abolished by ovariectomization or castration, suggesting that the therapeutic effect of this supplementation may involve enzymes and/or pathways influenced by sex hormones [103].
In humans, most population-based or cohorts studies mainly focused on the effects of sex–genetics interplay in patients carrying C9orf72 repeat expansion. Remarkably, Rooney et al. [9] performed an analysis of the prognostic characteristics of the C9orf72 repeat expansion in 4925 ALS cases from Dutch, Irish, and Italian population-based national registers and from two (Belgian and UK) clinical research center cohorts. In this study, the authors revealed a reduced survival rate in male spinal onset patients carrying C9orf72 repeat expansion when compared to females with the same type of onset. Moreover, this group of patients showed a shorter diagnostic delay, suggesting a rapidly progressing disease. These results were in line with those from Trojsi et al. [10], who performed a multicenter case-control study on data collected from 1417 Italian patients, stratified on the basis of the genetic data. In particular, the patients carrying C9orf72 repeat expansion were younger at onset, at first clinical observation, and at diagnosis and exhibited higher odds of bulbar onset, FTD diagnosis, and family history of ALS, FTD, and Alzheimer’s disease. When stratifying the ALS population by sex, only men carrying C9orf72 repeat expansion showed a shorter survival compared to apparently SALS men (Figure 2).
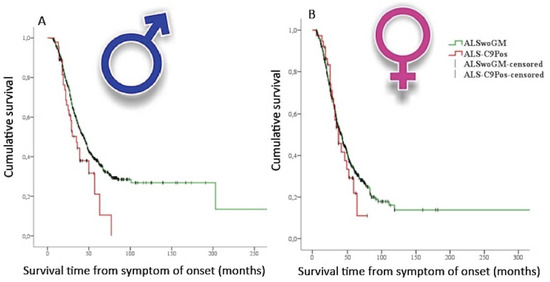
Figure 2.
Kaplan–Meier plots of survival probabilities, stratifying the studied sample by sex [10]: shorter survival time is displayed in amyotrophic lateral sclerosis (ALS) patients carrying C9orf72 repeat expansion (ALS-C9Pos) (red line) compared to ALS patients without documented genetic mutations (ALSwoGM) (green line) only for males. (A): (male): Log-rank χ2 = 4.33, p = 0.037; median survival was 35 months (95% CI. 26–44) for ALS-C9Pos (n = 47) and 44 months (95% CI. 40–48) for ALSwoGM (n = 681). (B): (female): Log-rank χ2 = 0.43, p = 0.510; median survival was 37 months (95% CI. 26–47) for ALS-C9Pos (n = 37) and 42 months (95% CI. 37–46) for ALSwoGM (n = 559). +: censored cases. (Derived and modified from Trojsi et al. [10], open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/)).
With regard to evidence of potential sex–genetics interplay in C9orf72 repeat expansion (C9+) carriers, a recent meta-analysis on sex differences in genetic mutations in ALS and FTD showed that in women there has been a higher prevalence of expanded C9orf72-related ALS [104]. This finding may be explained on the basis of potential sex-related factors, including environmental, lifestyle, or hormonal factors, that may moderate pathogenic mechanisms and influence an older age of onset and longer survival in women, allowing for reaching the age of complete penetrance and expression of the C9orf72-related ALS phenotype [23,104], considering that penetrance of C9orf72 expansions has been shown to increase with age [23,27]. Conversely, men carrying C9orf72 expansion have been recognized to be more prone to develop ALS at a younger age [105]. Current explanations for higher male prevalence of SALS include their greater exposure to environmental risk factors (e.g., pesticides, greater amounts of physical activity, and occupations in the armed forces) [106,107], and there is no reason to assume that men with C9orf72-related ALS may have less exposure to these risk factors [104]. Compounding this sex difference in life expectancy, men with C9orf72 mutations probably may have a shorter disease course [108].
Other studies did not confirm the potential interaction between sex and occurrence of C9orf72 repeat expansion. In a population-based study by Chiò et al. [6], genetic mutations did not modify the effect of sex on clinical phenotype, neither motor nor cognitive. Similarly, Glasmacher et al. [109] did not find an association between sex and survival in C9orf72 repeat expansion carriers.
5. Conclusions and Future Perspectives
ALS is a complex disease, and this complexity is mostly due to the heterogeneity of its phenotype, influenced by genetics through oligogenic mechanisms, that is, through gene–gene (or protein–protein) interactions [12,19] and by demographic factors, including age and sex, and by environment, which have longstanding negative effects, probably interacting with genetics through epigenetic mechanisms (i.e., interactomics) [38,47,110]. It is important that this complex scenario is taken into account when exploring the multiple disease pathways involved in ALS and when approaching therapy development. As personalized healthcare is taking hold in the common medical procedures, sex-specific differences, together with omics approaches (i.e., genomics, proteomics, metabolomics, epigenomics, and interactomics) have to be taken into account to shed further light on unresolved genetic causes, postgenomic effects, and molecular pathways involved in ALS and associated therapeutic strategies in the perspective of optimizing a precision medicine approach.
Most differences between the two sexes in the pathogenic mechanisms triggering ALS have been reported in preclinical models, mainly investigating disease pathways in SOD1G93A transgenic mice. Future research should explore potential sex differences in the ALS-related pathogenic mechanisms, such as, with regard to C9orf72-related ALS, gain of neurotoxic protein (RNA or dipeptide repeat) aggregates. Moreover, longitudinal incidence studies could be a relevant manner through which sex differences in survival and disease course may be accounted for when examining genetically well-characterized ALS populations, especially for C9orf72-related ALS.
Author Contributions
Conceptualization, F.T. and G.D.; resources, F.T. and G.D.; data curation, F.T., G.D., S.B., and G.T.; writing—original draft preparation, F.T., G.D., S.B., and G.T.; writing—review and editing, F.T., G.D., S.B., and G.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Takeda, T.; Kitagawa, K.; Arai, K. Phenotypic variability and its pathological basis in amyotrophic lateral sclerosis. Neuropathology 2020, 40, 40–56. [Google Scholar] [CrossRef] [PubMed]
- Robberecht, W.; Philips, T. The changing scene of amyotrophic lateral sclerosis. Nat. Rev. Neurosci. 2013, 14, 248–264. [Google Scholar] [CrossRef] [PubMed]
- Chia, R.; Chiò, A.; Traynor, B.J. Novel genes associated with amyotrophic lateral sclerosis: Diagnostic and clinical implications. Lancet Neurol. 2018, 17, 94–102. [Google Scholar] [CrossRef]
- Al-Chalabi, A.; Fang, F.; Hanby, M.F.; Leigh, P.N.; Shaw, C.E.; Ye, W.; Rijsdijk, F. An estimate of amyotrophic lateral sclerosis heritability using twin data. J. Neurol. Neurosurg. Psychiatry 2010, 81, 1324–1326. [Google Scholar] [CrossRef]
- Keller, M.F.; Ferrucci, L.; Singleton, A.B.; Tienari, P.J.; Laaksovirta, H.; Restagno, G.; Chiò, A.; Traynor, B.J.; Nalls, M.A. Genome-wide analysis of the heritability of amyotrophic lateral sclerosis. JAMA Neurol. 2014, 71, 1123–1134. [Google Scholar] [CrossRef]
- Chiò, A.; Moglia, C.; Canosa, A.; Manera, U.; D’Ovidio, F.; Vasta, R.; Grassano, M.; Brunetti, M.; Barberis, M.; Corrado, L.; et al. ALS phenotype is influenced by age, sex, and genetics: A population-based study. Neurology 2020, 94, e802–e810. [Google Scholar] [CrossRef]
- Chiò, A.; Mazzini, L.; D’Alfonso, S.; Corrado, L.; Canosa, A.; Moglia, C.; Manera, U.; Bersano, E.; Brunetti, M.; Barberis, M.; et al. The multistep hypothesis of ALS revisited: The role of genetic mutations. Neurology 2018, 91, 635–642. [Google Scholar] [CrossRef]
- Pape, J.A.; Grose, J.H. The effects of diet and sex in amyotrophic lateral sclerosis. Rev. Neurol. (Paris) 2020, 176, 301–315, pii:S0035-3787(20)30397-0. [Google Scholar] [CrossRef]
- Rooney, J.; Fogh, I.; Westeneng, H.J.; Vajda, A.; McLaughlin, R.; Heverin, M.; Jones, A.; van Eijk, R.; Calvo, A.; Mazzini, L.; et al. C9orf72 expansion differentially affects males with spinal onset amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry 2017, 88, 295–300. [Google Scholar] [CrossRef]
- Trojsi, F.; Siciliano, M.; Femiano, C.; Santangelo, G.; Lunetta, C.; Calvo, A.; Moglia, C.; Marinou, K.; Ticozzi, N.; Ferro, C.; et al. Comparative analysis of C9orf72 and sporadic disease in a large multicenter ALS population: The effect of male sex on survival of C9orf72 positive patients. Front. Neurosci. 2019, 13, 485. [Google Scholar] [CrossRef]
- Su, X.W.; Broach, J.R.; Connor, J.R.; Gerhard, G.S.; Simmons, Z. Genetic heterogeneity of amyotrophic lateral sclerosis: Implications for clinical practice and research. Muscle Nerve 2014, 49, 786–803. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.P.; Van Broeckhoven, C.; van der Zee, J. ALS Genes in the genomic era and their implications for FTD. Trends Genet. 2018, 34, 404–423. [Google Scholar] [CrossRef] [PubMed]
- Kenna, K.P.; van Doormaal, P.T.C.; Dekker, A.M.; Ticozzi, N.; Kenna, B.J.; Diekstra, F.P.; van Rheenen, W.; van Eijk, K.R.; Jones, A.R.; Keagle, P.; et al. NEK1 variants confer susceptibility to amyotrophic lateral sclerosis. Nat. Genet. 2016, 48, 1037–1042. [Google Scholar] [CrossRef] [PubMed]
- Van Rheenen, W.; Shatunov, A.; Dekker, A.M.; McLaughlin, R.L.; Diekstra, F.P.; Pulit, S.L.; van der Spek, A.A.; Võsa, U.; de Jong, S.; Robinson, M.R.; et al. Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat. Genet. 2016, 48, 1043–1048. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, A.; Kenna, K.P.; Renton, A.E.; Ticozzi, N.; Faghri, F.; Chia, R.; Dominov, J.A.; Kenna, B.J.; Nalls, M.A.; Keagle, O.; et al. Genome-wide Analyses Identify KIF5A as a Novel ALS Gene. Neuron 2018, 97, 1268–1283. [Google Scholar] [CrossRef] [PubMed]
- DeJesus-Hernandez, M.; Mackenzie, I.R.; Boeve, B.F.; Boxer, A.L.; Baker, M.; Rutherford, N.J.; Nicholson, A.M.; Finch, N.A.; Flynn, H.; Adamson, J.; et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 2011, 72, 245–256. [Google Scholar] [CrossRef]
- Renton, A.E.; Majounie, E.; Waite, A.; Simón-Sánchez, J.; Rollinson, S.; Gibbs, J.R.; Schymick, J.C.; Laaksovirta, H.; van Swieten, J.C.; Myllykangas, L.; et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 2011, 72, 257–268. [Google Scholar] [CrossRef]
- Project MinE ALS Sequencing Consortium. Project MinE: Study design and pilot analyses of a large-scale whole-genome sequencing study in amyotrophic lateral sclerosis. Eur. J. Hum. Genet. 2018, 26, 1537–1546. [Google Scholar] [CrossRef]
- Malkki, H. Motor Neuron Disease: New insights into genetic risk factors for amyotrophic lateral sclerosis. Nat. Rev. Neurol. 2016, 12, 491. [Google Scholar] [CrossRef]
- Loh, P.R.; Bhatia, G.; Gusev, A.; Finucane, H.K.; Bulik-Sullivan, B.K.; Pollack, S.J.; Schizophrenia Working Group of Psychiatric Genomics Consortium; de Candia, T.R.; Hong Lee, S.; Wray, N.R.; et al. Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat. Genet. 2015, 47, 1385–1392. [Google Scholar] [CrossRef]
- Van Blitterswijk, M.; van Es, M.A.; Hennekam, E.A.M.; Dooijes, D.; van Rheenen, W.; Medic, J.; Bourque, P.R.; Schelhaas, H.J.; van der Kooi, A.J.; de Visser, M.; et al. Evidence for an oligogenic basis of amyotrophic lateral sclerosis. Hum. Mol. Genet. 2012, 21, 3776–3784. [Google Scholar] [CrossRef] [PubMed]
- Al-Chalabi, A.; Lewis, C.M. Modelling the effects of penetrance and family size on rates of sporadic and familial disease. Hum. Hered. 2011, 71, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Murphy, N.A.; Arthur, K.C.; Tienari, P.J.; Houlden, H.; Chiò, A.; Traynor, B.J. Age-related penetrance of the C9orf72 repeat expansion. Sci. Rep. 2017, 7, 2116. [Google Scholar] [CrossRef] [PubMed]
- Cooper-Knock, J.; Shaw, P.J.; Kirby, J. The widening spectrum of C9ORF72-related disease; genotype/phenotype correlations and potential modifiers of clinical phenotype. Acta Neuropathol. 2014, 127, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Tripolszki, K.; Gampawar, P.; Schmidt, H.; Nagy, Z.F.; Nagy, D.; Klivényi, P.; Engelhardt, J.I.; Széll, M. Comprehensive Genetic Analysis of a Hungarian Amyotrophic Lateral Sclerosis Cohort. Front. Genet. 2019, 10, 732. [Google Scholar] [CrossRef] [PubMed]
- Karch, C.M.; Wen, N.; Fan, C.C.; Yokoyama, J.S.; Kouri, N.; Ross, O.A.; Höglinger, G.; Müller, U.; Ferrari, R.; Hardy, J.; et al. International Frontotemporal Dementia (FTD)–Genomics Consortium, International Collaboration for Frontotemporal Dementia, Progressive Supranuclear Palsy (PSP) Genetics Consortium, and International Parkinson’s Disease Genomics Consortium. Selective Genetic overlap between Amyotrophic Lateral Sclerosis and diseases of the Frontotemporal Dementia spectrum. JAMA Neurol. 2018, 75, 860–875. [Google Scholar]
- Majounie, E.; Renton, A.E.; Mok, K.; Dopper, E.G.; Waite, A.; Rollinson, S.; Chiò, A.; Restagno, C.; Nicolaou, N.; Simon-Sanchez, J.; et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: A cross-sectional study. Lancet Neurol. 2012, 11, 323–330. [Google Scholar] [CrossRef]
- Chiò, A.; Borghero, G.; Restagno, G.; Mora, G.; Drepper, C.; Traynor, B.J.; Sendtner, M.; Brunetti, M.; Ossola, I.; Calvo, A.; et al. Clinical characteristics of patients with familial amyotrophic lateral sclerosis carrying the pathogenic GGGGCC hexanucleotide repeat expansion of C9ORF72. Brain 2012, 135, 784–793. [Google Scholar] [CrossRef]
- Byrne, S.; Elamin, M.; Bede, P.; Shatunov, A.; Walsh, C.; Corr, B.; Heverin, M.; Jordan, N.; Kenna, K.; Lynch, C.; et al. Cognitive and clinical characteristics of patients with amyotrophic lateral sclerosis carrying a C9orf72 repeat expansion: A population-based cohort study. Lancet Neurol. 2012, 11, 232–240. [Google Scholar] [CrossRef]
- Byrne, S.; Heverin, M.; Elamin, M.; Bede, P.; Lynch, C.; Kenna, K.; MacLaughlin, R.; Walsh, C.; Al Chalabi, A.; Hardimanet, O. Aggregation of neurologic and neuropsychiatric disease in amyotrophic lateral sclerosis kindreds: A population-based case-control cohort study of familial and sporadic amyotrophic lateral sclerosis. Ann. Neurol. 2013, 74, 699–708. [Google Scholar] [CrossRef]
- Sabatelli, M.; Conforti, F.L.; Zollino, M.; Mora, G.; Monsurrò, M.R.; Volanti, P.; Marinou, K.; Salvi, F.; Corbo, M.; Giannini, F.; et al. C9ORF72 hexanucleotide repeat expansions in the Italian sporadic ALS population. Neurobiol. Aging 2012, 33, 1848.e15–1848.e20. [Google Scholar] [CrossRef] [PubMed]
- Irwin, D.J.; McMillan, C.T.; Brettschneider, J.; Libon, D.J.; Powers, J.; Rascovsky, K.; Toledo, J.B.; Boller, A.; Bekisz, J.; Chandrasekaran, K.; et al. Cognitive decline and reduced survival in C9orf72 expansion frontotemporal degeneration and amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry 2013, 84, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Umoh, M.E.; Fournier, C.; Li, Y.; Polak, M.; Shaw, L.; Landers, J.E.; Hu, W.; Gearing, M.; Glass, J.D. Comparative analysis of C9orf72 and sporadic disease in an ALS clinic population. Neurology 2016, 87, 1024–1030. [Google Scholar] [CrossRef] [PubMed]
- Cappella, M.; Ciotti, C.; Cohen-Tannoudji, M.; Biferi, M.G. Gene Therapy for ALS-A Perspective. Int. J. Mol. Sci. 2019, 20, 4388. [Google Scholar] [CrossRef] [PubMed]
- Webster, C.P.; Smith, E.F.; Bauer, C.S.; Moller, A.; Hautbergue, G.M.; Ferraiuolo, L.; Myszczynska, M.A.; Higginbottom, A.; Walsh, M.J.; Whitworth, A.J.; et al. The C9orf72 protein interacts with Rab 1 a and the ULK 1 complex to regulate initiation of autophagy. EMBO J. 2016, 35, 1656–1676. [Google Scholar] [CrossRef]
- Zhu, Q.; Jiang, J.; Gendron, T.F.; McAlonis-Downes, M.; Jiang, L.; Taylor, A.; Diaz Garcia, S.; Ghosh Dastidar, S.; Rodriguez, M.J.; King, P.; et al. Reduced C9ORF72 function exacerbates gain of toxicity from ALS/FTD-causing repeat expansion in C9orf72. Nat. Neurosci. 2020, 23, 615–624. [Google Scholar] [CrossRef]
- Gitler, A.D.; Tsuiji, H. There has been an awakening: Emerging mechanisms of C9orf72 mutations in FTD/ALS. Brain Res. 2016, 1647, 19–29. [Google Scholar] [CrossRef]
- Zhang, M.; Tartaglia, M.C.; Moreno, D.; Sato, C.; McKeever, P.; Weichert, A.; Keith, J.; Robertson, J.; Zinman, L.; Rogaeva, E. DNA methylation age-acceleration is associated with disease duration and age at onset in C9orf72 patients. Acta Neuropathol. 2017, 134, 271–279. [Google Scholar] [CrossRef]
- Van Blitterswijk, M.; Mullen, B.; Heckman, M.G.; Baker, M.C.; DeJesus-Hernandez, M.; Brown, P.H.; Murray, M.E.; Hsiung, G.-Y.R.; Stewart, H.; Karydas, A.M.; et al. Ataxin-2 as potential disease modifier in C9ORF72 expansion carriers. Neurobiol. Aging 2014, 35, 2421.e13–2421.e17. [Google Scholar] [CrossRef]
- Nicholson, A.M.; Rademakers, R. What we know about TMEM106B in neurodegeneration. Acta Neuropathol. 2016, 132, 639–651. [Google Scholar] [CrossRef]
- Rosen, D.R. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 1993, 364, 362. [Google Scholar] [CrossRef]
- Andersen, P.M. Amyotrophic lateral sclerosis associated with mutations in the CuZn superoxide dismutase gene. Curr. Neurol. Neurosci. Rep. 2006, 6, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Zhou, Q.; Chen, Y.; Ou, R.W.; Cao, B.; Xu, Y.Q.; Yang, J.; Shang, H.F. Analysis of SOD1 mutations in a Chinese population with amyotrophic lateral sclerosis: A case-control study and literature review. Sci. Rep. 2017, 7, 44606. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, D.W.; Rothstein, J.D. From Charcot to Lou Gehrig: Deciphering selective motor neuron death in ALS. Nat. Rev. Neurosci. 2001, 2, 806–819. [Google Scholar] [CrossRef] [PubMed]
- Cudkowicz, M.E.; McKenna-Yasek, D.; Sapp, P.E.; Chin, W.; Geller, B.; Hayden, D.L.; Schoenfeld, D.A.; Hosler, B.A.; Horvitz, H.R.; Brown, R.H. Epidemiology of mutations in superoxide dismutase in amyotrophic lateral sclerosis. Ann. Neurol. 1997, 41, 210–221. [Google Scholar] [CrossRef]
- Bruijn, L.I.; Houseweart, M.K.; Kato, S.; Anderson, K.L.; Anderson, S.D.; Ohama, E.; Reaume, A.G.; Scott, R.W.; Cleveland, D.W. Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science 1998, 281, 1851–1854. [Google Scholar] [CrossRef]
- Radunovic, A.; Leigh, P.N. Cu/Zn superoxide dismutase gene mutations in amyotrophic lateral sclerosis: Correlation between genotype and clinical features. J. Neurol. Neurosurg. Psychiatry 1996, 61, 565–572. [Google Scholar] [CrossRef]
- Pang, S.Y.; Hsu, J.S.; Teo, K.C.; Li, Y.; Kung, M.H.W.; Cheah, K.S.E.; Chan, D.; Cheung, K.M.C.; Li, M.; Sham, P.C.; et al. Burden of rare variants in ALS genes influences survival in familial and sporadic ALS. Neurobiol. Aging 2017, 58, 238.e9–238.e15. [Google Scholar] [CrossRef]
- Lattante, S.; Rouleau, G.A.; Kabashi, E. TARDBP and FUS mutations associated with amyotrophic lateral sclerosis: Summary and update. Hum. Mutat. 2013, 34, 812–826. [Google Scholar] [CrossRef]
- Neumann, M.; Sampathu, D.M.; Kwong, L.K.; Truax, A.C.; Micsenyi, M.C.; Chou, T.T.; Bruce, J.; Schuck, T.; Grossman, M.; Clark, C.M.; et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006, 314, 130–133. [Google Scholar] [CrossRef]
- Brettschneider, J.; Del Tredici, K.; Toledo, J.B.; Robinson, J.L.; Irwin, D.J.; Grossman, M.; Suh, E.R.; Van Deerlin, V.M.; Wood, E.M.; Baek, Y.; et al. Stages of pTDP-43 pathology in amyotrophic lateral sclerosis. Ann. Neurol. 2013, 74, 20–38. [Google Scholar] [CrossRef]
- Igaz, L.M.; Kwong, L.K.; Lee, E.B.; Chen-Plotkin, A.; Swanson, E.; Unger, T.; Malunda, J.; Xu, Y.; Winton, M.J.; Trojanowski, J.Q.; et al. Dysregulation of the ALS associated gene TDP-43 leads to neuronal death and degeneration in mice. J. Clin. Invest. 2011, 121, 726–738. [Google Scholar] [CrossRef]
- Arnold, E.S.; Ling, S.C.; Huelga, S.C.; Lagier-Tourenne, C.; Polymenidou, M.; Ditsworth, D.; Kordasiewicz, H.B.; McAlonis-Downes, M.; Platoshyn, O.; Parone, P.A.; et al. ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc. Natl. Acad. Sci. USA 2013, 110, 736–745. [Google Scholar] [CrossRef] [PubMed]
- Braak, H.; Brettschneider, J.; Ludolph, A.C.; Lee, V.M.; Trojanowski, J.Q.; Del Tredici, K. Amyotrophic lateral sclerosis--a model of corticofugal axonal spread. Nat. Rev. Neurol. 2013, 9, 708–714. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Gao, K.; Jankovic, J. The role of FUS gene variants in neurodegenerative diseases. Nat. Rev. Neurol. 2014, 10, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Sabatelli, M.; Marangi, G.; Conte, A.; Tasca, G.; Zollino, M.; Lattante, S. New ALS-related genes expand the spectrum paradigm of Amyotrophic Lateral Sclerosis. Brain. Pathol. 2016, 26, 266–275. [Google Scholar] [CrossRef]
- Cooper-Knock, J.; Moll, T.; Ramesh, T.; Castelli, L.; Beer, A.; Robins, H.; Fox, I.; Niedermoser, I.; Van Damme, P.; Moisse, M.; et al. Mutations in the glycosyltransferase domain of GLT8D1 are associated with familial Amyotrophic Lateral Sclerosis. Cell Rep. 2019, 26, 2298–2306.e5. [Google Scholar] [CrossRef]
- Cirulli, E.T.; Lasseigne, B.N.; Petrovski, S.; Sapp, P.C.; Dion, P.A.; Leblond, C.S.; Couthous, J.; Lu, Y.-F.; Wang, Q.; Krueger, B.J.; et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science 2015, 347, 1436–1441. [Google Scholar] [CrossRef]
- Freischmidt, A.; Wieland, T.; Richter, B.; Ruf, W.; Schaeffer, V.; Müller, K.; Marroquin, N.; Nordin, F.; Hubers, A.; Weydt, P.; et al. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat. Neurosci. 2015, 18, 631–636. [Google Scholar] [CrossRef]
- Van Mossevelde, S.; van der Zee, J.; Gijselinck, I.; Engelborghs, S.; Sieben, A.; Van Langenhove, T.; De Bleecker, J.; Baets, J.; Vandenbulcke, M.; Van Laere, K.; et al. Belgian Neurology consortium. Clinical features of TBK1 carriers compared with C9orf72, GRN and non-mutation carriers in a Belgian cohort. Brain 2016, 139, 452–467. [Google Scholar] [CrossRef]
- Pottier, C.; Bieniek, K.F.; Finch, N.; van de Vorst, M.; Baker, M.; Perkersen, R.; Brown, P.; Ravenscroft, T.; van Blitterswijk, M.; Nicholson, A.M.; et al. Whole-genome sequencing reveals important role for TBK1 and OPTN mutations in frontotemporal lobar degeneration without motor neuron disease. Acta Neuropathol. 2015, 130, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Brenner, D.; Müller, K.; Wieland, T.; Weydt, P.; Böhm, S.; Lulé, D.; Hübers, A.; Neuwirth, C.; Weber, M.; Borck, G.; et al. NEK1 mutations in familial amyotrophic lateral sclerosis. Brain 2016, 139, e28. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.P.; Van Mossevelde, S.; Dillen, L.; De Bleecker, J.L.; Moisse, M.; Van Damme, P.; Van Broeckhoven, C.; van der Zee, J.; BELNEU Consortium. NEK1 genetic variability in a Belgian cohort of ALS and ALS-FTD patients. Neurobiol. Aging 2017, 61, 255.e1–255.e7. [Google Scholar] [CrossRef] [PubMed]
- Black, H.A.; Leighton, D.J.; Cleary, E.M.; Rose, E.; Stephenson, L.; Colville, S.; Ross, D.; Warner, J.; Porteous, M.; Gorrie, G.H.; et al. Genetic epidemiology of motor neuron disease-associated variants in the Scottish population. Neurobiol. Aging 2017, 51, 178.e11–178.e20. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Baloh, R.H.; Milbrandt, J. The NIMA-family kinase Nek3 regulates microtubule acetylation in neurons. J. Cell Sci. 2009, 122, 2274–2282. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.N.; Ticozzi, N.; Fallini, C.; Soragia Gkazi, A.; Topp, S.; Kenna, K.P.; Scotter, E.L.; Kost, J.; Keagle, P.; Miller, J.W.; et al. Exome-wide rare variant analysis identifies TUBA4A mutations associated with familial ALS. Neuron 2014, 84, 324–331. [Google Scholar] [CrossRef]
- Wu, C.H.; Fallini, C.; Ticozzi, N.; Keagle, P.J.; Sapp, P.C.; Piotrowska, K.; Lowe, P.; Koppers, M.; McKenna-Yasek, D.; Baron, D.M.; et al. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature 2012, 488, 499–503. [Google Scholar] [CrossRef]
- Fang, X.; Lin, H.; Wang, X.; Zuo, Q.; Qin, J.; Zhang, P. The NEK1 interactor, C21ORF2, is required for efficient DNA damage repair. Acta Biochim. Biophys. Sin. (Shanghai) 2015, 47, 834–841. [Google Scholar] [CrossRef]
- Wang, Z.; Iida, A.; Miyake, N.; Nishiguchi, K.M.; Fujita, K.; Nakazawa, T.; Alswaid, A.; Albalwi, M.A.; Kim, O.H.; Cho, T.J.; et al. Axial spondylometaphyseal dysplasia is caused by C21orf2 mutations. PLoS ONE 2016, 11, e0150555. [Google Scholar] [CrossRef]
- Wang, Z.; Horemuzova, E.; Iida, A.; Guo, L.; Liu, Y.; Matsumoto, N.; Nishimura, G.; Nordgren, A.; Miyake, N.; Tham, E.; et al. Axial spondylometaphyseal dysplasia is also caused by NEK1 mutations. J. Hum. Genet. 2017, 62, 503–506. [Google Scholar] [CrossRef]
- Perrone, F.; Nguyen, H.P.; Van Mossevelde, S.; Moisse, M.; Sieben, A.; Santens, P.; De Bleecker, J.; Vandenbulcke, M.; Engelborghs, S.; Baets, J.; et al. Investigating the role of ALS genes CHCHD10 and TUBA4A in Belgian FTD-ALS spectrum patients. Neurobiol. Aging 2017, 51, 177.e9–177.e16. [Google Scholar] [CrossRef] [PubMed]
- Siu, J.; Perkins, E.; Cashman, N.R. Effects of sex and family history on the amyotrophic lateral sclerosis (ALS) multistep model. Amyotroph. Lateral Scler. 2016, 17, 94. [Google Scholar]
- Chiò, A.; Mora, G.; Moglia, C.; Manera, U.; Canosa, A.; Cammarosano, S.; Ilardi, A.; Bertuzzo, D.; Bersano, E.; Cugnasco, P.; et al. Secular trends of amyotrophic lateral sclerosis: The Piemonte and Valle d’Aosta Register. JAMA Neurol. 2017, 74, 1097–1104. [Google Scholar] [CrossRef] [PubMed]
- Pupillo, E.; Messina, P.; Giussani, G.; Logroscino, G.; Zoccolella, S.; Chiò, A.; Calvo, A.; Corbo, M.; Lunetta, C.; Marin, B.; et al. Physical Activity and Amyotrophic Lateral Sclerosis: A European Population-Based Case–Control Study. Ann. Neurol. 2014, 75, 708–716. [Google Scholar] [CrossRef] [PubMed]
- Manjaly, Z.R.; Scott, K.M.; Abhinav, K.; Wijesekera, L.; Ganesalingam, J.; Goldstein, L.H.; Janssen, A.; Dougherty, A.; Willey, E.; Stanton, B.R.; et al. The sex ratio in amyotrophic lateral sclerosis: A population based study. Amyotroph. Lateral Scler. 2010, 11, 439–442. [Google Scholar] [CrossRef]
- De Jong, S.; Huisman, M.; Sutedja, N.; van der Kooi, A.; de Visser, M.; Schelhaas, J.; van der Schouw, Y.; Veldink, J.; van den Berget, L. Endogenous female reproductive hormones and the risk of amyotrophic lateral sclerosis. J. Neurol. 2013, 260, 507–512. [Google Scholar] [CrossRef]
- Rooney, J.P.K.; Visser, A.E.; D’Ovidio, F.; Vermeulen, R.; Beghi, E.; Chio, A.; Veldink, J.H.; Logroscino, G.; van den Berg, L.H.; Hardiman, O.; et al. A case-control study of hormonal exposures as etiologic factors for ALS in women Euro-MOTOR. Neurology 2017, 89, 1283–1290. [Google Scholar] [CrossRef]
- Popat, R.A.; Van Den Eeden, S.K.; Tanner, C.M.; Bernstein, A.L.; Bloch, D.A.; Leimpeter, A.; McGuire, V.; Nelson, L.M. Effect of reproductive factors and postmenopausal hormone use on the risk of amyotrophic lateral sclerosis. Neuroepidemiology 2006, 27, 117–121. [Google Scholar] [CrossRef]
- Pfeiffer, R.M.; Mayer, B.; Kuncl, R.W.; Check, D.P.; Cahoon, E.K.; Rivera, D.R.; Freedman, D.M. Identifying potential targets for prevention and treatment of amyotrophic lateral sclerosis based on a screen of medicare prescription drugs. Amyotroph. Lateral Sclero. Frontotemporal Degenerat. 2019, 1–11. [Google Scholar] [CrossRef]
- Kim, J.; Kim, T.Y.; Cho, K.S.; Kim, H.N.; Koh, J.Y. Autophagy activation and neuroprotection by progesterone in the G93A-SOD1 transgenic mouse model of amyotrophic lateral sclerosis. Neurobiol. Dis. 2013, 59, 80–85. [Google Scholar] [CrossRef]
- Heitzer, M.; Kaiser, S.; Kanagaratnam, M.; Zendedel, A.; Hartmann, P.; Beyer, C.; Johann, S. Administration of 17beta-Estradiol Improves Motoneuron Survival and Down-regulates Inflammasome Activation in Male SOD1(G93A) ALS Mice. Mol. Neurobiol. 2017, 54, 8429–8443. [Google Scholar] [CrossRef]
- Vivekananda, U.; Manjalay, Z.R.; Ganesalingam, J.; Simms, J.; Shaw, C.E.; Leigh, P.N.; Turner, M.R.; Al-Chalabi, A. Low index-to-ring finger length ratio in sporadic ALS supports prenatally defined motor neuronal vulnerability. J. Neurol. Neurosurg. Psychiatry 2011, 82, 635–637. [Google Scholar] [CrossRef]
- Herron, L.R.; Miles, G.B. Gender-specific perturbations in modulatory inputs to motoneurons in a mouse model of amyotrophic lateral sclerosis. Neuroscience 2012, 226, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, A. Androgen stimulates neuronal plasticity in the perineal motoneurons of aged male rats. J. Comp. Neurol. 2011, 430, 389–395. [Google Scholar] [CrossRef]
- Weiner, L.P. Possible role of androgen receptors in amyotrophic lateral sclerosis. A hypothesis. Arch. Neurol. 1980, 37, 129–131. [Google Scholar] [CrossRef]
- Gargiulo-Monachelli, G.M.; Sivori, M.; Meyer, M.; Sica, R.E.; De Nicola, A.F.; Gonzalez-Deniselle, M.C. Circulating gonadal and adrenal steroids in amyotrophic lateral sclerosis: Possible markers of susceptibility and outcome. Horm. Metab. Res. 2014, 46, 433–439. [Google Scholar] [CrossRef]
- Chiò, A.; Calvo, A.; Moglia, C.; Mazzini, L.; Mora, G.; PARALS Study Group. Phenotypic heterogeneity of amyotrophic lateral sclerosis: A population based study. J. Neurol. Neurosurg. Psychiatry 2011, 82, 740e–746e. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, A.; Mento, G.; Calvo, V.; Querin, G.; D’Ascenzo, C.; Volpato, C.; Kleinbub, J.R.; Bisiacchi, P.S.; Soraru, G. Female gender doubles executive dysfunction risk in ALS: A case-control study in 165 patients. J. Neurol. Neurosurg. Psychiatry 2015, 86, 574–579. [Google Scholar] [CrossRef] [PubMed]
- Flaherty, C.; Kraft, J.; Brothers, A.; Harrison, M.; Legro, R.S.; Manni, A.; Yang, C.; Simmons, Z. for the VALUES Consortium. The relationship between oestrogen and executive functioning in ALS females with emerging Frontotemporal Lobar Degeneration (FTLD) supports a neuroendocrine model of FTLD attenuation. Amyotroph. Lateral Scler. Frontotemporal Degener. 2017, 18, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Pupillo, E.; Messina, P.; Logroscino, G.; Beghi, E.; The SLALOM Group. Long-Term Survival in Amyotrophic Lateral Sclerosis: A Population-Based Study. Ann. Neurol. 2014, 75, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Strykowski, R.; Meyer, M.; Mulcrone, P.; Krakora, D.; Suzuki, M. Male-specific differences in proliferation, neurogenesis, and sensitivity to oxidative stress in neural progenitor cells derived from a rat model of ALS. PLoS ONE 2012, 7, e48581. [Google Scholar] [CrossRef] [PubMed]
- Hayes-Punzo, A.; Mulcrone, P.; Meyer, M.; McHugh, J.; Svendsen, C.N.; Suzuki, M. Gonadectomy and dehydroepiandrosterone (DHEA) do not modulate disease progression in the G93A mutant SOD1 rat model of amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. 2012, 13, 311–314. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Liu, Y.; Sun, C.; Zheng, Q.; Hao, P.; Zhai, J.; Liu, Y. Effects of Ovariectomy in an hSOD1-G93A Transgenic Mouse Model of Amyotrophic Lateral Sclerosis (ALS). Med. Sci. Monit. 2018, 24, 678–686. [Google Scholar] [CrossRef] [PubMed]
- Ohta, Y.; Soucy, G.; Phaneuf, D.; Audet, J.N.; Gros-Louis, F.; Rouleau, G.A.; Blasco, H.; Corcia, P.; Andersen, P.M.; Nordin, F.; et al. Sex-dependent effects of chromogranin B P413L allelic variant as disease modifier in amyotrophic lateral sclerosis. Hum. Mol. Genet. 2016, 25, 4771–4786. [Google Scholar] [CrossRef][Green Version]
- Eschbach, J.; Schwalenstocker, B.; Soyal, S.M.; Bayer, H.; Wiesner, D.; Akimoto, C.; Nilsson, A.C.; Birve, A.; Meyer, T.; Dupuis, L.; et al. PGC-1a is a male-specific disease modifier of human and experimental amyotrophic lateral sclerosis. Hum. Mol. Genet. 2013, 22, 3477–3484. [Google Scholar] [CrossRef]
- Kaneb, H.M.; Sharp, P.S.; Rahmani-Kondori, N.; Wells, D.J. Metformin treatment has no beneficial effect in a dose-response survival study in the SOD1G93A mouse model of ALS and is harmful in female mice. PLoS ONE 2011, 6, e24189. [Google Scholar] [CrossRef]
- Mansfield, R.; Galea, R.; Brincat, M.; Hole, D.; Mason, H. Metformin has direct effects on human ovarian steroidogenesis. Fertil. Steril. 2003, 79, 956–962. [Google Scholar] [CrossRef]
- Cacabelos, S.; Ramírez-Núñez, O.; Granado-Serrano, A.B.; Torres, P.; Ayala, V.; Moiseeva, V.; Povedano, M.; Ferrer, I.; Pamplona, R.; Portero-Otin, M.; et al. Early and gender-specific differences in spinal cord mitochondrial function and oxidative stress markers in a mouse model of ALS. Acta Neuropathol. Commun. 2016, 4, 3. [Google Scholar] [CrossRef]
- Riar, A.K.; Burstein, S.R.; Palomo, G.M.; Arreguin, A.; Manfredi, G.; Germain, D. Sex specific activation of the ERalpha axis of the mitochondrial UPR (UPRmt) in the G93A-SOD1 mouse model of familial ALS. Hum. Mol. Genet. 2017, 26, 1318–1327. [Google Scholar] [CrossRef]
- Kreilaus, F.; Guerra, S.; Masanetz, R.; Menne, V.; Yerbury, J.; Karl, T. Novel behavioural characteristics of the superoxide dismutase1 G93A (SOD1G93A) mouse model of amyotrophic lateral sclerosis include sex-dependent phenotypes. Genes Brain Behav. 2020, 19, e12604. [Google Scholar] [CrossRef]
- Torres, P.; Cacabelos, D.; Pairada, J.; Bauer, K.C.; Boada, J.; Fontdevila, L.; Rossi, C.; Povedano, M.; Ferrer, I.; Pamplona, R.; et al. Gender-specific beneficial effects of docosahexaenoic acid dietary supplementation in G93A-SOD1 Amyotrophic Lateral Sclerosis mice. Neurotherapeutics 2020, 17, 269–281. [Google Scholar] [CrossRef]
- Cacabelos, B.; Ayala, V.; Ramırez-Nunez, O.; Granado-Serrano, A.B.; Boada, J.; Serrano, J.C.E.; Cabre’, R.; Nadal-Rey, G.; Bellmunt, M.G.; Ferrer, I.; et al. Dietary lipid unsaturation influences survival and oxidative modifications of an Amyotrophic Lateral Sclerosis model in a gender-specific manner. Neuromol. Med. 2014, 16, 669–685. [Google Scholar] [CrossRef]
- Bame, M.; Pentiak, P.A.; Needleman, R.; Brusilow, W.S.A. Effect of sex on lifespan, disease progression, and the response to Methionine Sulfoximine in the SOD1 G93A mouse model for ALS. Gend. Med. 2012, 9, 524–535. [Google Scholar] [CrossRef]
- Curtis, A.F.; Masellis, M.; Hsiung, G.R.; Moineddin, R.; Zhang, K.; Au, B.; Millett, G.; Mackenzie, I.; Rogaeva, E.; Tierney, M.C. Sex differences in the prevalence of genetic mutations in FTD and ALS. A meta-analysis. Neurology 2017, 89, 1633–1642. [Google Scholar] [CrossRef]
- Williams, K.L.; Fifita, J.A.; Vucic, S.; Durnall, J.C.; Kiernan, M.C.; Blair, I.P.; Nicholson, G.A. Pathophysiological insights into ALS with C9ORF72 expansions. J. Neurol. Neurosurg. Psychiatry 2013, 84, 931–935. [Google Scholar] [CrossRef]
- Al-Chalabi, A.; Hardiman, O. The epidemiology of ALS: A conspiracy of genes, environment and time. Nat. Rev. Neurol. 2013, 9, 617–628. [Google Scholar] [CrossRef]
- Gallo, V.; Bueno-De-Mesquita, H.B.; Vermeulen, R.; Andersen, P.M.; Kyrozis, A.; Linseisen, J.; Kaaks, R.; Allen, N.E.; Roddam, A.W.; Boshuizen, H.C.; et al. Smoking and risk for amyotrophic lateral sclerosis: Analysis of the EPIC cohort. Ann. Neurol. 2009, 65, 378–385. [Google Scholar] [CrossRef]
- Rooney, J.; McLaughlin, R.; Vajda, A.; Fogh, I.; Jones, A.; Westeneng, H.J.; Heverin, M.; van Eijk, R.; Robberecht, W.; Van Damme, P.; et al. Novel gender selective survival effect of C9orf72 in European ALS cohorts (P5. 093). Neurology 2016, 86, P5.093. [Google Scholar]
- Glasmacher, S.A.; Wong, C.; Pearson, I.E.; Pal, S. Survival and Prognostic Factors in C9orf72 Repeat Expansion Carriers. A Systematic Review and Meta-analysis. JAMA Neurol. 2020, 77, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Bradley, W.G.; Andrew, A.S.; Traynor, B.J.; Chiò, A.; Butt, T.H.; Stommel, E.W. Gene environment-time interactions in neurodegenerative diseases: Hypotheses and research approaches. Ann. Neurosci. 2018, 25, 261–267. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).