Histone Deacetylase (HDAC) Gene Family in Allotetraploid Cotton and Its Diploid Progenitors: In Silico Identification, Molecular Characterization, and Gene Expression Analysis under Multiple Abiotic Stresses, DNA Damage and Phytohormone Treatments
Abstract
1. Introduction
2. Results
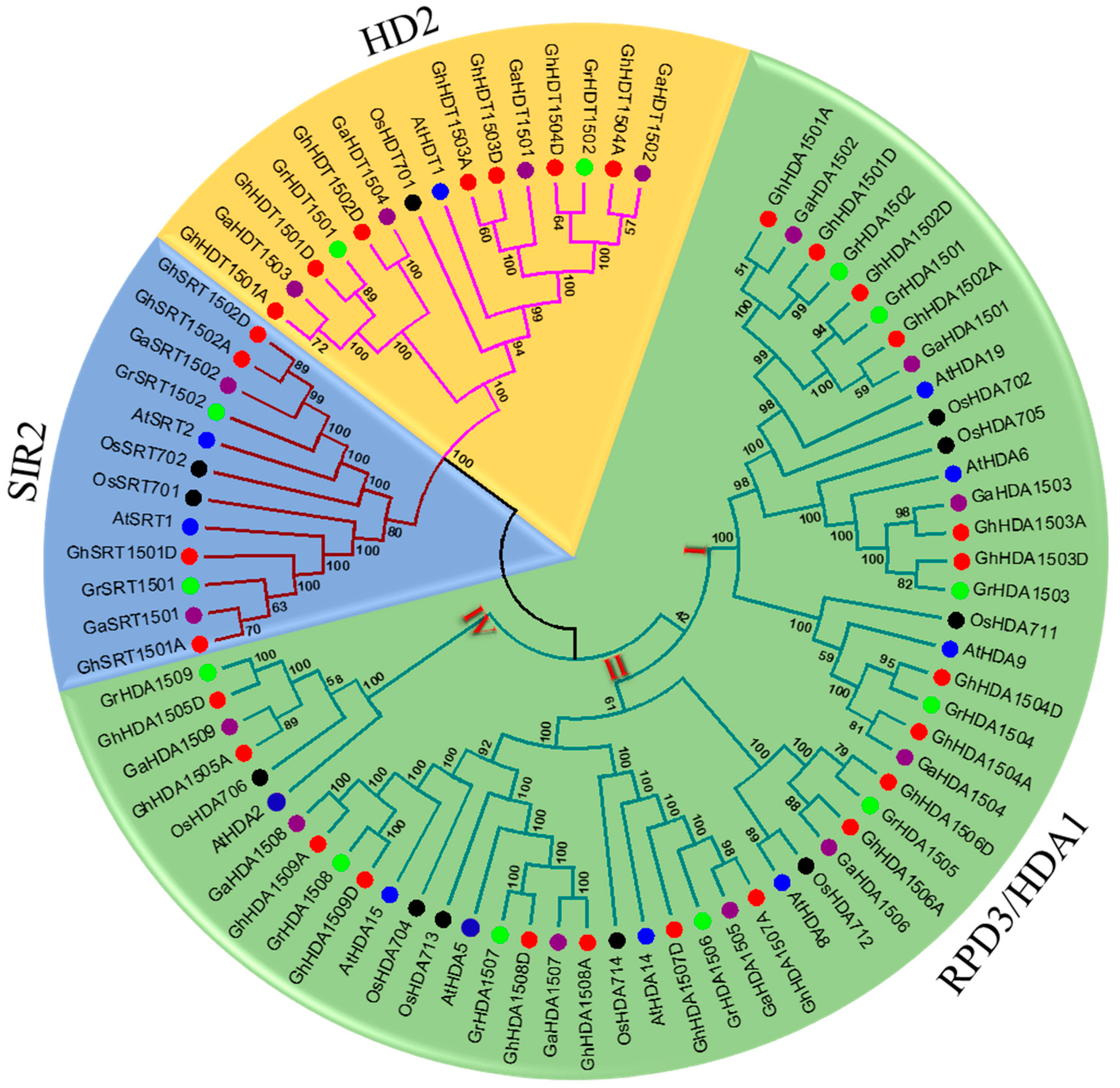
2.1. Identification and Phylogenetic Analysis of Gossypium HDAC
2.2. Phylogenetic Classification, Gene Structure, and Conserved Domain Analysis of Gossypium HDAC Genes
2.3. Genomic Localization and Gene Duplication Analysis of Gossypium HDACs
2.4. Cis-Elements in the Promoter of G. hirsutum HDAC Genes
2.5. Gene Expression Analysis
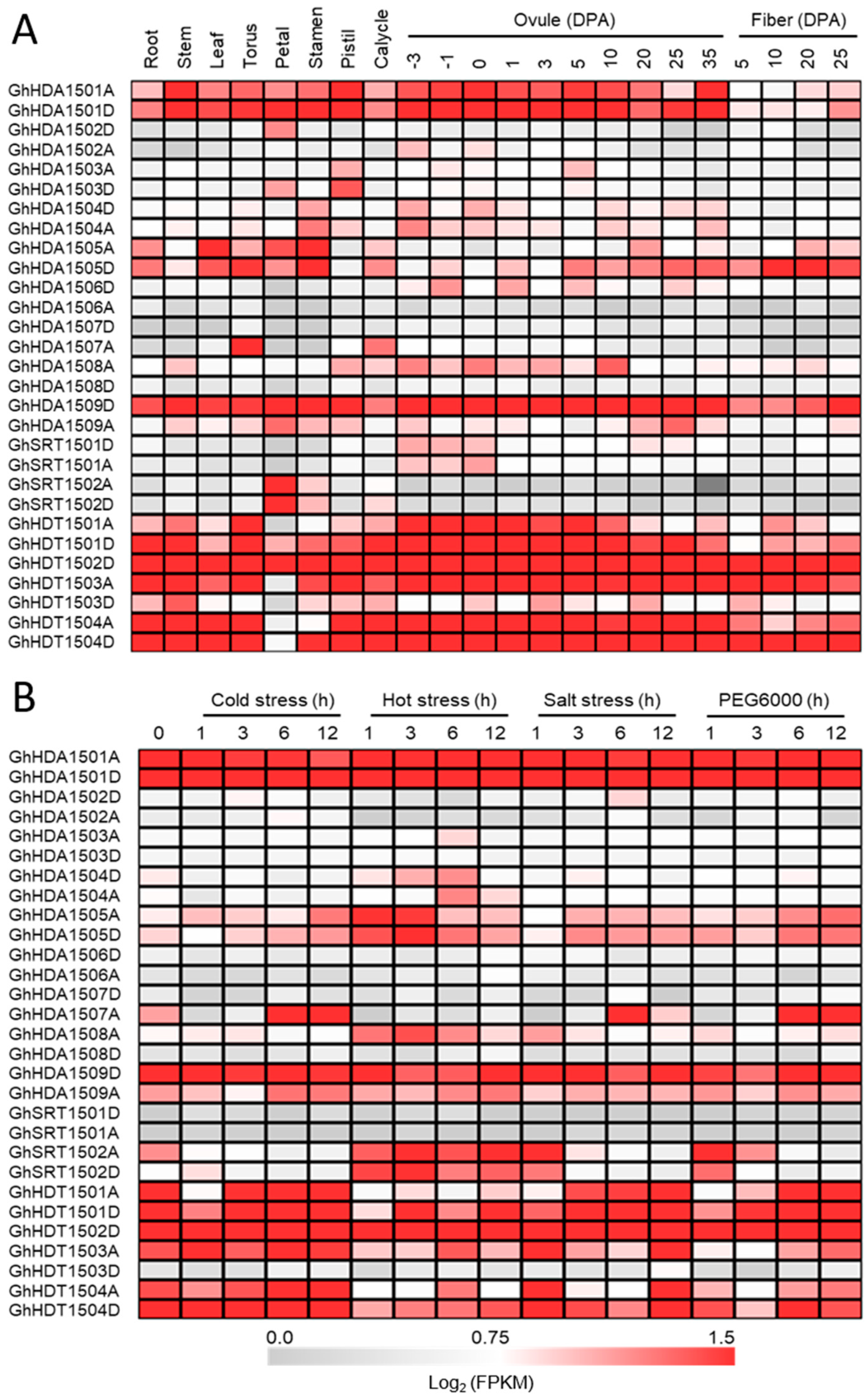
2.5.1. Expression Patterns of G. hirsutum HDACs in Different Tissues and Under Multiple Stresses
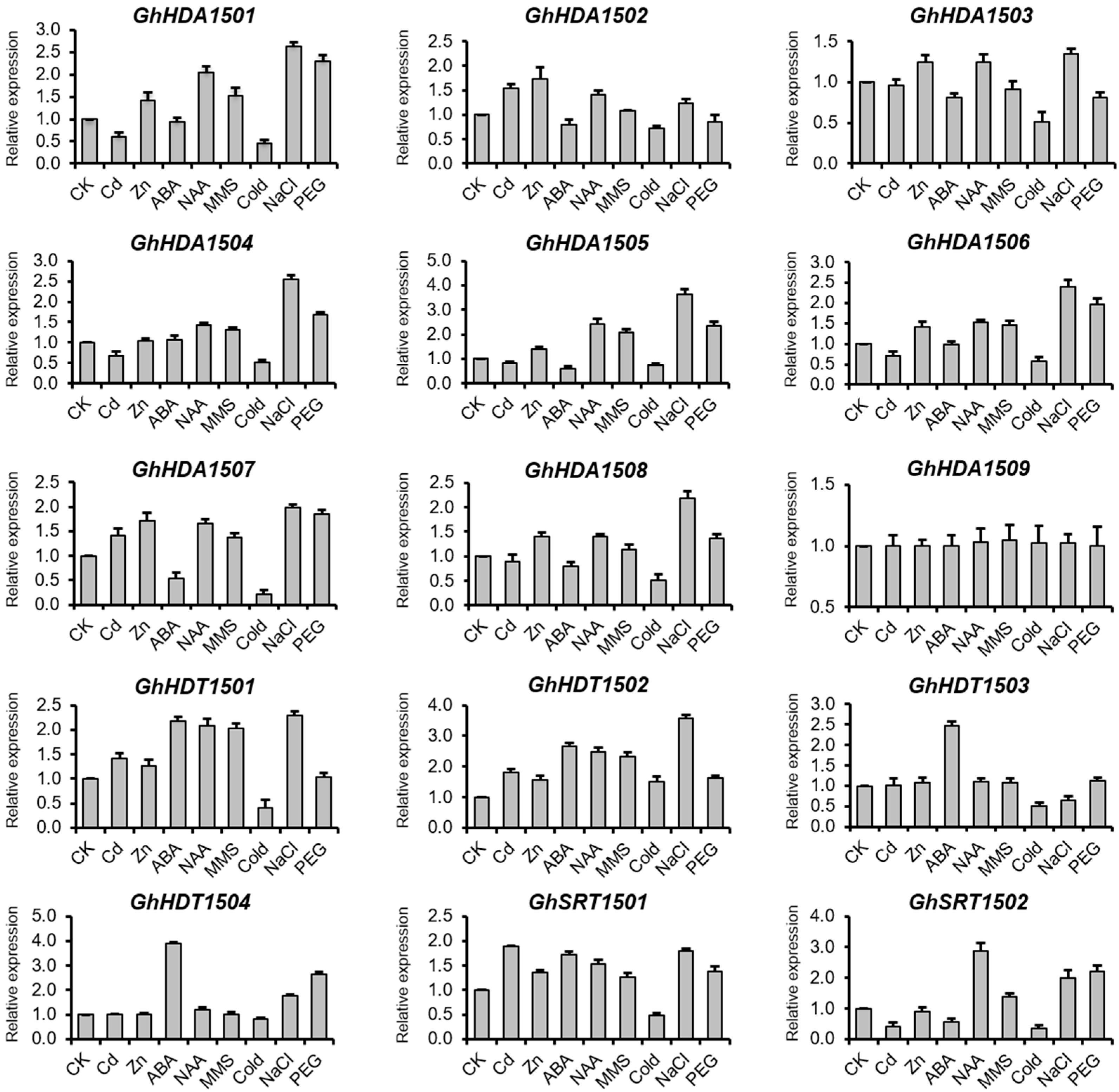
2.5.2. Validation of the Expression of G. hirsutum HDACs Genes Using qRT-PCR
Tissue-Specific Expression of GhHDAC Genes
GhHDACs Expression Pattern at Different Fiber Developmental Stages
Expression of GhHDACs in Response to Phytohormones
Expression of GhHDACs in Response to DNA Damage
Expression of GhHDACs in Response to Abiotic Stresses
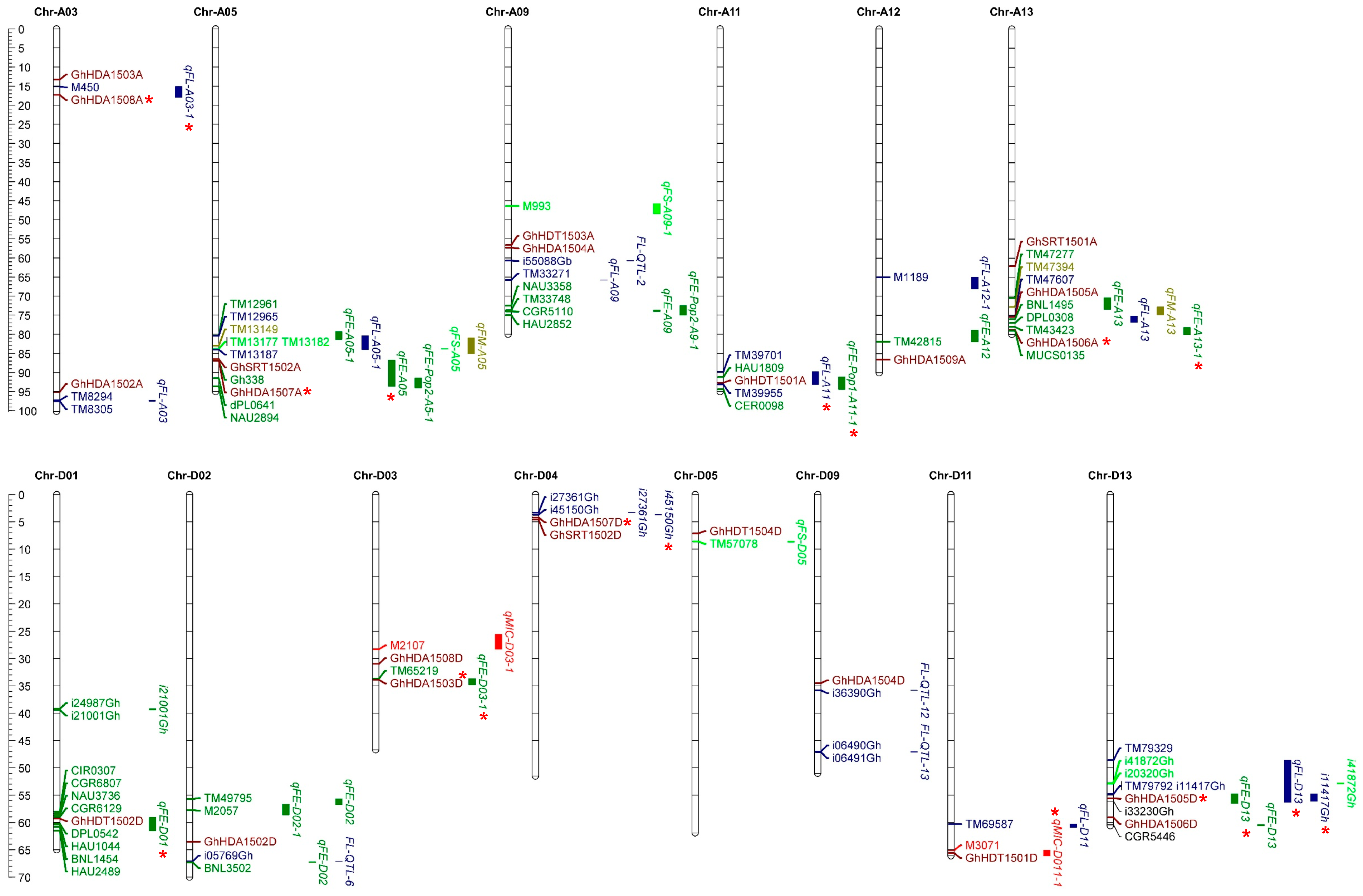
2.6. Co-Localization of GhHDACs with Quantitative Trait Loci and Single-Nucleotide Polymorphisms of Fiber-Related Traits
3. Discussion
4. Materials and Methods
4.1. Identification of the HDACs Gene Family
4.2. Chromosome Location and Duplication of HDACs
4.3. Phylogenetic Analyses
4.4. Gene Structure, Domain Composition and Cis-Element Analysis
4.5. Transcriptome Data Analysis and Gene Expression Heatmap
4.6. Plant Materials, Stress Treatments and qRT-PCR
4.7. Co-Localization of HDACs with Fiber-Related QTLs
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Cao, X. Whole genome sequencing of cotton—A new chapter in cotton genomics. Sci. China Life Sci. 2015, 58, 515–516. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J.; Scheffler, B.E.; Dennis, E.; Triplett, B.A.; Zhang, T.; Guo, W.; Chen, X.; Stelly, D.M.; Rabinowicz, P.D.; Town, C.D.; et al. Toward sequencing cotton (Gossypium) genomes. Plant Physiol. 2007, 145, 1303–1310. [Google Scholar] [CrossRef] [PubMed]
- Wendel, J.F. New World tetraploid cottons contain Old World cytoplasm. Proc. Natl. Acad. Sci. USA 1989, 86, 4132–4136. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.L.; Llewellyn, D.J.; Furbank, R.T. Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation, and seed development. Plant Cell 2003, 15, 952–964. [Google Scholar] [CrossRef]
- Shi, Y.H.; Zhu, S.W.; Mao, X.Z.; Feng, J.X.; Qin, Y.M.; Zhang, L.; Cheng, J.; Wei, L.P.; Wang, Z.Y.; Zhu, Y.X. Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 2006, 18, 651–664. [Google Scholar] [CrossRef]
- Qin, Y.M.; Zhu, Y.X. How cotton fibers elongate: A tale of linear cell-growth mode. Curr. Opin. Plant Biol. 2011, 14, 106–111. [Google Scholar] [CrossRef]
- Imran, M.; Shakeel, A.; Farooq, J.; Saeed, A.; Farooq, A.; Riaz, M. Genetic studies of fiber quality parameter and earliness related traits in upland cotton (Gossypium hirsutum L.). Adv. Agric. Bot. 2011, 3, 151–159. [Google Scholar]
- Imran, M.; Shakeel, A.; Azhar, F.; Farooq, J.; Saleem, M.; Saeed, A.; Nazeer, W.; Riaz, M.; Naeem, M.; Javaid, A. Combining ability analysis for within-boll yield components in upland cotton (Gossypium hirsutum L.). Genet. Mol. Res. 2012, 11, 2790–2800. [Google Scholar] [CrossRef]
- Li, F.; Fan, G.; Lu, C.; Xiao, G.; Zou, C.; Kohel, R.J.; Ma, Z.; Shang, H.; Ma, X.; Wu, J.; et al. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 2015, 33, 524–530. [Google Scholar] [CrossRef]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef]
- Li, F.; Fan, G.; Wang, K.; Sun, F.; Yuan, Y.; Song, G.; Li, Q.; Ma, Z.; Lu, C.; Zou, C.; et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 2014, 46, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Wang, Z.; Li, F.; Ye, W.; Wang, J.; Song, G.; Yue, Z.; Cong, L.; Shang, H.; Zhu, S.; et al. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 2012, 44, 1098–1103. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.J.; Wang, Z. Readout of epigenetic modifications. Annu. Rev. Biochem. 2013, 82, 81–118. [Google Scholar] [CrossRef] [PubMed]
- Berr, A.; Shafiq, S.; Shen, W.H. Histone modifications in transcriptional activation during plant development. Biochim. Biophys. Acta 2011, 1809, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Ho, L.; Crabtree, G.R. Chromatin remodelling during development. Nature 2010, 463, 474–484. [Google Scholar] [CrossRef] [PubMed]
- Peterson, C.L.; Laniel, M.A. Histones and histone modifications. Curr. Biol. CB 2004, 14, R546–R551. [Google Scholar] [CrossRef] [PubMed]
- Ekwall, K. Genome-wide analysis of HDAC function. Trends Genet. TIG 2005, 21, 608–615. [Google Scholar] [CrossRef]
- Hollender, C.; Liu, Z. Histone deacetylase genes in Arabidopsis development. J. Integr. Plant Biol. 2008, 50, 875–885. [Google Scholar] [CrossRef]
- Hu, Y.; Qin, F.; Huang, L.; Sun, Q.; Li, C.; Zhao, Y.; Zhou, D.X. Rice histone deacetylase genes display specific expression patterns and developmental functions. Biochem. Biophys. Res. Commun. 2009, 388, 266–271. [Google Scholar] [CrossRef]
- Liew, L.C.; Singh, M.B.; Bhalla, P.L. An RNA-seq transcriptome analysis of histone modifiers and RNA silencing genes in soybean during floral initiation process. PLoS ONE 2013, 8, e77502. [Google Scholar] [CrossRef]
- Aquea, F.; Timmermann, T.; Arce-Johnson, P. Analysis of histone acetyltransferase and deacetylase families of Vitis vinifera. Plant Physiol. Biochem. 2010, 48, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Ying, P.; Liu, X.; Li, C.; Xia, R.; Li, J.; Zhao, M. Genome-Wide Identification of Histone Modifiers and Their Expression Patterns during Fruit Abscission in Litchi. Front. Plant Sci. 2017, 8, 639. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Lu, J.; Zhang, J.; Wu, P.Y.; Yang, S.; Wu, K. Identification and characterization of histone deacetylases in tomato (Solanum lycopersicum). Front. Plant Sci. 2014, 5, 760. [Google Scholar] [CrossRef] [PubMed]
- Lusser, A.; Brosch, G.; Loidl, A.; Haas, H.; Loidl, P. Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science 1997, 277, 88–91. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Liu, X.; Singh, P.; Cui, Y.; Zimmerli, L.; Wu, K. Chromatin modifications and remodeling in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Yu, C.W.; Chen, F.F.; Zhao, L.; Tian, G.; Liu, X.; Cui, Y.; Yang, J.Y.; Wu, K. Histone deacetylase HDA6 is functionally associated with AS1 in repression of KNOX genes in Arabidopsis. PLoS Genet. 2012, 8, e1003114. [Google Scholar] [CrossRef]
- Yu, C.W.; Tai, R.; Wang, S.C.; Yang, P.; Luo, M.; Yang, S.; Cheng, K.; Wang, W.C.; Cheng, Y.S.; Wu, K. HISTONE DEACETYLASE6 Acts in Concert with Histone Methyltransferases SUVH4, SUVH5, and SUVH6 to Regulate Transposon Silencing. Plant. Cell 2017, 29, 1970–1983. [Google Scholar] [CrossRef]
- Kim, J.M.; To, T.K.; Matsui, A.; Tanoi, K.; Kobayashi, N.I.; Matsuda, F.; Habu, Y.; Ogawa, D.; Sakamoto, T.; Matsunaga, S.; et al. Erratum: Acetate-mediated novel survival strategy against drought in plants. Nat. Plants 2017, 3, 17119. [Google Scholar] [CrossRef]
- Devoto, A.; Nieto-Rostro, M.; Xie, D.; Ellis, C.; Harmston, R.; Patrick, E.; Davis, J.; Sherratt, L.; Coleman, M.; Turner, J.G. COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 2002, 32, 457–466. [Google Scholar] [CrossRef]
- Zhu, Z.; An, F.; Feng, Y.; Li, P.; Xue, L.; A, M.; Jiang, Z.; Kim, J.M.; To, T.K.; Li, W.; et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 12539–12544. [Google Scholar] [CrossRef]
- Chen, X.; Lu, L.; Mayer, K.S.; Scalf, M.; Qian, S.; Lomax, A.; Smith, L.M.; Zhong, X. POWERDRESS interacts with HISTONE DEACETYLASE 9 to promote aging in Arabidopsis. Elife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Wang, R.; Gao, L.; Li, D.; Xu, C.; Mang, H.; Jeon, J.; Chen, X.; Zhong, X.; Kwak, J.M.; et al. POWERDRESS and HDA9 interact and promote histone H3 deacetylation at specific genomic sites in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 14858–14863. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Ding, Y.; Sun, X.; Xie, S.; Wang, D.; Liu, X.; Su, L.; Wei, W.; Pan, L.; Zhou, D.X. Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in Arabidopsis. J. Exp. Bot. 2016, 67, 1703–1713. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Yu, H.; Zhao, Z.; Hunter, D.; Luo, X.; Duan, J.; Tian, L. AtHD2D Gene Plays a Role in Plant Growth, Development, and Response to Abiotic Stresses in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 310. [Google Scholar] [CrossRef] [PubMed]
- Sridha, S.; Wu, K. Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J. 2006, 46, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Bellizzi Mdel, R.; Ning, Y.; Meyers, B.C.; Wang, G.L. HDT701, a histone H4 deacetylase, negatively regulates plant innate immunity by modulating histone H4 acetylation of defense-related genes in rice. Plant Cell 2012, 24, 3783–3794. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, J.; Zhang, W.; Wu, K.; Zheng, F.; Tian, L.; Liu, X.; Duan, J. Expression and functional analysis of the plant-specific histone deacetylase HDT701 in rice. Front. Plant Sci. 2014, 5, 764. [Google Scholar] [CrossRef]
- Zhao, J.; Li, M.; Gu, D.; Liu, X.; Zhang, J.; Wu, K.; Zhang, X.; Teixeira da Silva, J.A.; Duan, J. Involvement of rice histone deacetylase HDA705 in seed germination and in response to ABA and abiotic stresses. Biochem. Biophys. Res. Commun. 2016, 470, 439–444. [Google Scholar] [CrossRef]
- Luo, M.; Wang, Y.Y.; Liu, X.; Yang, S.; Wu, K. HD2 proteins interact with RPD3-type histone deacetylases. Plant Signal. Behav. 2012, 7, 608–610. [Google Scholar] [CrossRef]
- Liu, X.; Yu, C.W.; Duan, J.; Luo, M.; Wang, K.; Tian, G.; Cui, Y.; Wu, K. HDA6 directly interacts with DNA methyltransferase MET1 and maintains transposable element silencing in Arabidopsis. Plant Physiol. 2012, 158, 119–129. [Google Scholar] [CrossRef]
- Yu, C.W.; Liu, X.; Luo, M.; Chen, C.; Lin, X.; Tian, G.; Lu, Q.; Cui, Y.; Wu, K. HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis. Plant Physiol. 2011, 156, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.H.; Li, D.M.; Yin, M.H.; Li, X.B.; Zhang, M.; Wang, Y.J.; Dong, J.; Zhao, J.; Luo, M.; Luo, X.Y.; et al. Gibberellin 20-oxidase promotes initiation and elongation of cotton fibers by regulating gibberellin synthesis. J. Plant Physiol. 2010, 167, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhu, Y.; Hu, W.; Zhang, X.; Cai, C.; Guo, W. Comparative Transcriptomics Reveals Jasmonic Acid-Associated Metabolism Related to Cotton Fiber Initiation. PLoS ONE 2015, 10, e0129854. [Google Scholar] [CrossRef] [PubMed]
- Samuel Yang, S.; Cheung, F.; Lee, J.J.; Ha, M.; Wei, N.E.; Sze, S.H.; Stelly, D.M.; Thaxton, P.; Triplett, B.; Town, C.D.; et al. Accumulation of genome-specific transcripts, transcription factors and phytohormonal regulators during early stages of fiber cell development in allotetraploid cotton. Plant J. 2006, 47, 761–775. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zheng, X.; Song, S.; Zeng, Q.; Hou, L.; Li, D.; Zhao, J.; Wei, Y.; Li, X.; Luo, M.; et al. Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nat. Biotechnol. 2011, 29, 453–458. [Google Scholar] [CrossRef]
- Maury, S.; Sow, M.D.; Le Gac, A.L.; Genitoni, J.; Lafon-Placette, C.; Mozgova, I. Phytohormone and Chromatin Crosstalk: The Missing Link For Developmental Plasticity? Front. Plant Sci. 2019, 10, 395. [Google Scholar] [CrossRef]
- Anzola, J.M.; Sieberer, T.; Ortbauer, M.; Butt, H.; Korbei, B.; Weinhofer, I.; Mullner, A.E.; Luschnig, C. Putative Arabidopsis transcriptional adaptor protein (PROPORZ1) is required to modulate histone acetylation in response to auxin. Proc. Natl. Acad. Sci. USA 2010, 107, 10308–10313. [Google Scholar] [CrossRef]
- Manzano, C.; Ramirez-Parra, E.; Casimiro, I.; Otero, S.; Desvoyes, B.; De Rybel, B.; Beeckman, T.; Casero, P.; Gutierrez, C.; J, C.D.P. Auxin and epigenetic regulation of SKP2B, an F-box that represses lateral root formation. Plant Physiol. 2012, 160, 749–762. [Google Scholar] [CrossRef]
- Petricka, J.J.; Winter, C.M.; Benfey, P.N. Control of Arabidopsis root development. Annu. Rev. Plant Biol. 2012, 63, 563–590. [Google Scholar] [CrossRef]
- Nguyen, H.N.; Kim, J.H.; Jeong, C.Y.; Hong, S.W.; Lee, H. Inhibition of histone deacetylation alters Arabidopsis root growth in response to auxin via PIN1 degradation. Plant Cell Rep. 2013, 32, 1625–1636. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, Y. Cross-talk between the H3K36me3 and H4K16ac histone epigenetic marks in DNA double-strand break repair. J. Biol. Chem. 2017, 292, 11951–11959. [Google Scholar] [CrossRef] [PubMed]
- Toiber, D.; Erdel, F.; Bouazoune, K.; Silberman, D.M.; Zhong, L.; Mulligan, P.; Sebastian, C.; Cosentino, C.; Martinez-Pastor, B.; Giacosa, S.; et al. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol. Cell 2013, 51, 454–468. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Cheng, K.; Xu, Y.; Yang, S.; Wu, K. Plant Responses to Abiotic Stress Regulated by Histone Deacetylases. Front. Plant Sci. 2017, 8, 2147. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhang, L.; Zhao, L.; Li, J.; He, S.; Zhou, K.; Yang, F.; Huang, M.; Jiang, L.; Li, L. Trichostatin A selectively suppresses the cold-induced transcription of the ZmDREB1 gene in maize. PLoS ONE 2011, 6, e22132. [Google Scholar] [CrossRef]
- Imran, M.; Shafiq, S.; Farooq, M.A.; Naeem, M.K.; Widemann, E.; Bakhsh, A.; Jensen, K.B.; Wang, R.-C. Comparative Genome-wide Analysis and Expression Profiling of Histone Acetyltransferase (HAT) Gene Family in Response to Hormonal Applications, Metal and Abiotic Stresses in Cotton. Int. J. Mol. Sci. 2019, 20, 5311. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Imran, M.; Tang, K.; Liu, J.-Y. Comparative Genome-Wide Analysis of the Malate Dehydrogenase Gene Families in Cotton. PLoS ONE 2016, 11, e0166341. [Google Scholar] [CrossRef]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Imran, M.; Liu, J.-Y. Genome-wide identification and expression analysis of the malate dehydrogenase gene family in Gossypium arboreum. Pak. J. Bot. 2016, 48, 1081–1090. [Google Scholar]
- Blanc, G.; Wolfe, K.H. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 2004, 16, 1667–1678. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Guo, A.; Zhu, Q.; Chen, X.; Luo, J. GSDS: A gene structure display server. Yi Chuan Hered. 2007, 29, 1023–1026. [Google Scholar] [CrossRef]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Saeed, A.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M. TM4: A free, open-source system for microarray data management and analysis. Biotechniques 2003, 34, 374–378. [Google Scholar] [CrossRef] [PubMed]
- Shafiq, S.; Chen, C.; Yang, J.; Cheng, L.; Ma, F.; Widemann, E.; Sun, Q. DNA Topoisomerase 1 prevents R-loop accumulation to modulate auxin-regulated root development in rice. Mol. Plant 2017, 10, 821–833. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Wang, X.; Liu, Z.; Gu, Q.; Zhang, Y.; Li, Z.; Ke, H.; Yang, J.; Wu, J.; Wu, L.; et al. Genome-wide association study discovered genetic variation and candidate genes of fibre quality traits in Gossypium hirsutum L. Plant Biotechnol. J. 2017, 15, 982–996. [Google Scholar] [CrossRef] [PubMed]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Imran, M.; Shafiq, S.; Naeem, M.K.; Widemann, E.; Munir, M.Z.; Jensen, K.B.; Wang, R.R.-C. Histone Deacetylase (HDAC) Gene Family in Allotetraploid Cotton and Its Diploid Progenitors: In Silico Identification, Molecular Characterization, and Gene Expression Analysis under Multiple Abiotic Stresses, DNA Damage and Phytohormone Treatments. Int. J. Mol. Sci. 2020, 21, 321. https://doi.org/10.3390/ijms21010321
Imran M, Shafiq S, Naeem MK, Widemann E, Munir MZ, Jensen KB, Wang RR-C. Histone Deacetylase (HDAC) Gene Family in Allotetraploid Cotton and Its Diploid Progenitors: In Silico Identification, Molecular Characterization, and Gene Expression Analysis under Multiple Abiotic Stresses, DNA Damage and Phytohormone Treatments. International Journal of Molecular Sciences. 2020; 21(1):321. https://doi.org/10.3390/ijms21010321
Chicago/Turabian StyleImran, Muhammad, Sarfraz Shafiq, Muhammad Kashif Naeem, Emilie Widemann, Muhammad Zeeshan Munir, Kevin B. Jensen, and Richard R.-C. Wang. 2020. "Histone Deacetylase (HDAC) Gene Family in Allotetraploid Cotton and Its Diploid Progenitors: In Silico Identification, Molecular Characterization, and Gene Expression Analysis under Multiple Abiotic Stresses, DNA Damage and Phytohormone Treatments" International Journal of Molecular Sciences 21, no. 1: 321. https://doi.org/10.3390/ijms21010321
APA StyleImran, M., Shafiq, S., Naeem, M. K., Widemann, E., Munir, M. Z., Jensen, K. B., & Wang, R. R.-C. (2020). Histone Deacetylase (HDAC) Gene Family in Allotetraploid Cotton and Its Diploid Progenitors: In Silico Identification, Molecular Characterization, and Gene Expression Analysis under Multiple Abiotic Stresses, DNA Damage and Phytohormone Treatments. International Journal of Molecular Sciences, 21(1), 321. https://doi.org/10.3390/ijms21010321








