Rice Senescence-Induced Receptor-Like Kinase (OsSRLK) Is Involved in Phytohormone-Mediated Chlorophyll Degradation
Abstract
1. Introduction
2. Results
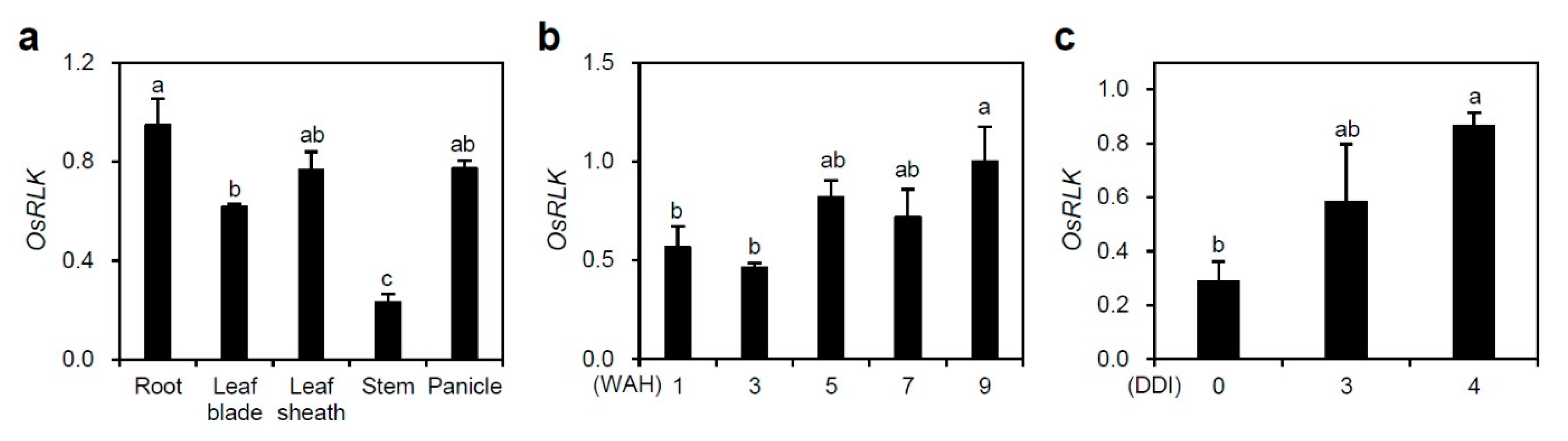
2.1. OsSRLK Is Upregulated during Leaf Senescence
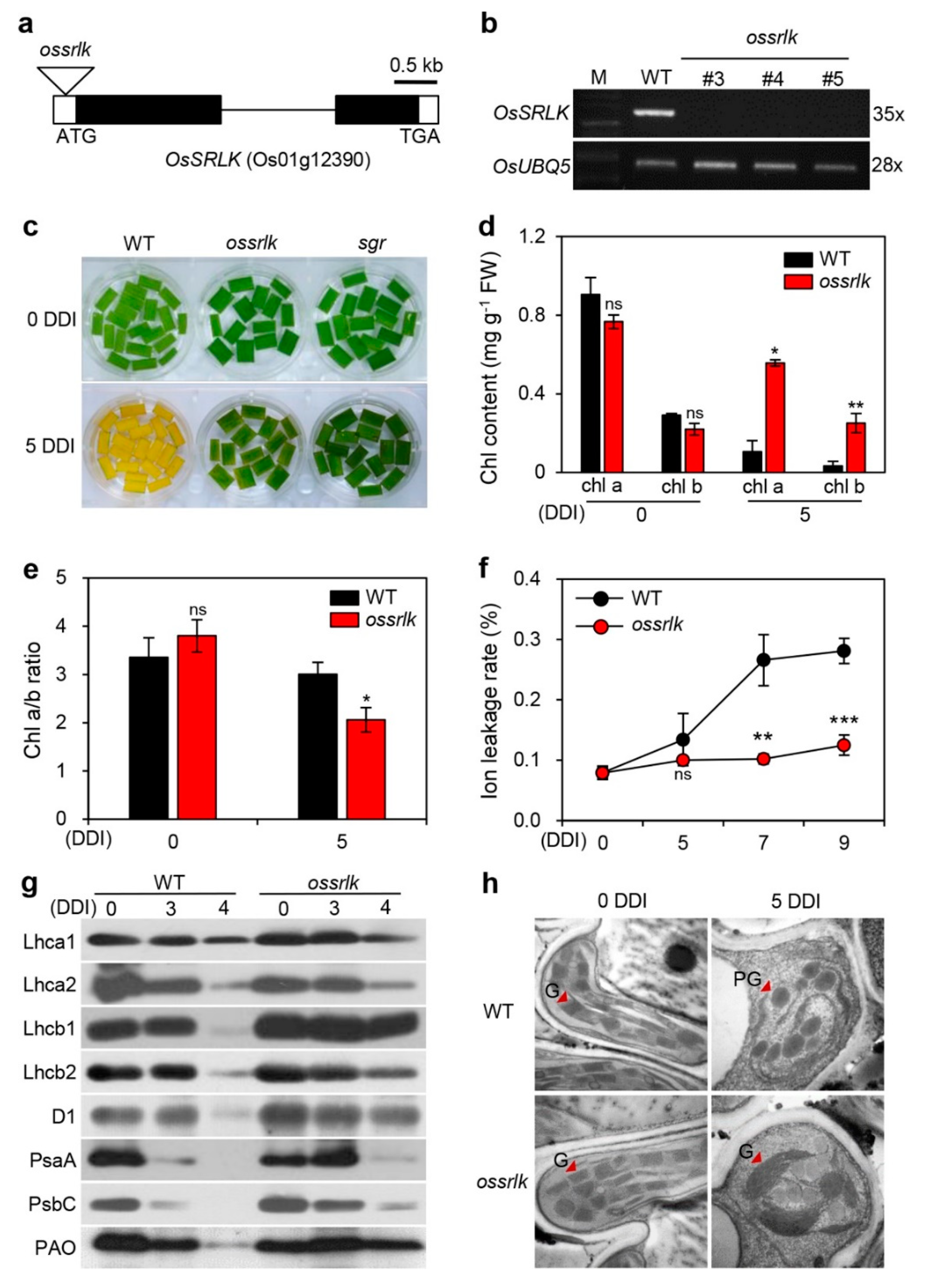
2.2. ossrlk Mutant Delays Leaf Yellowing during DIS
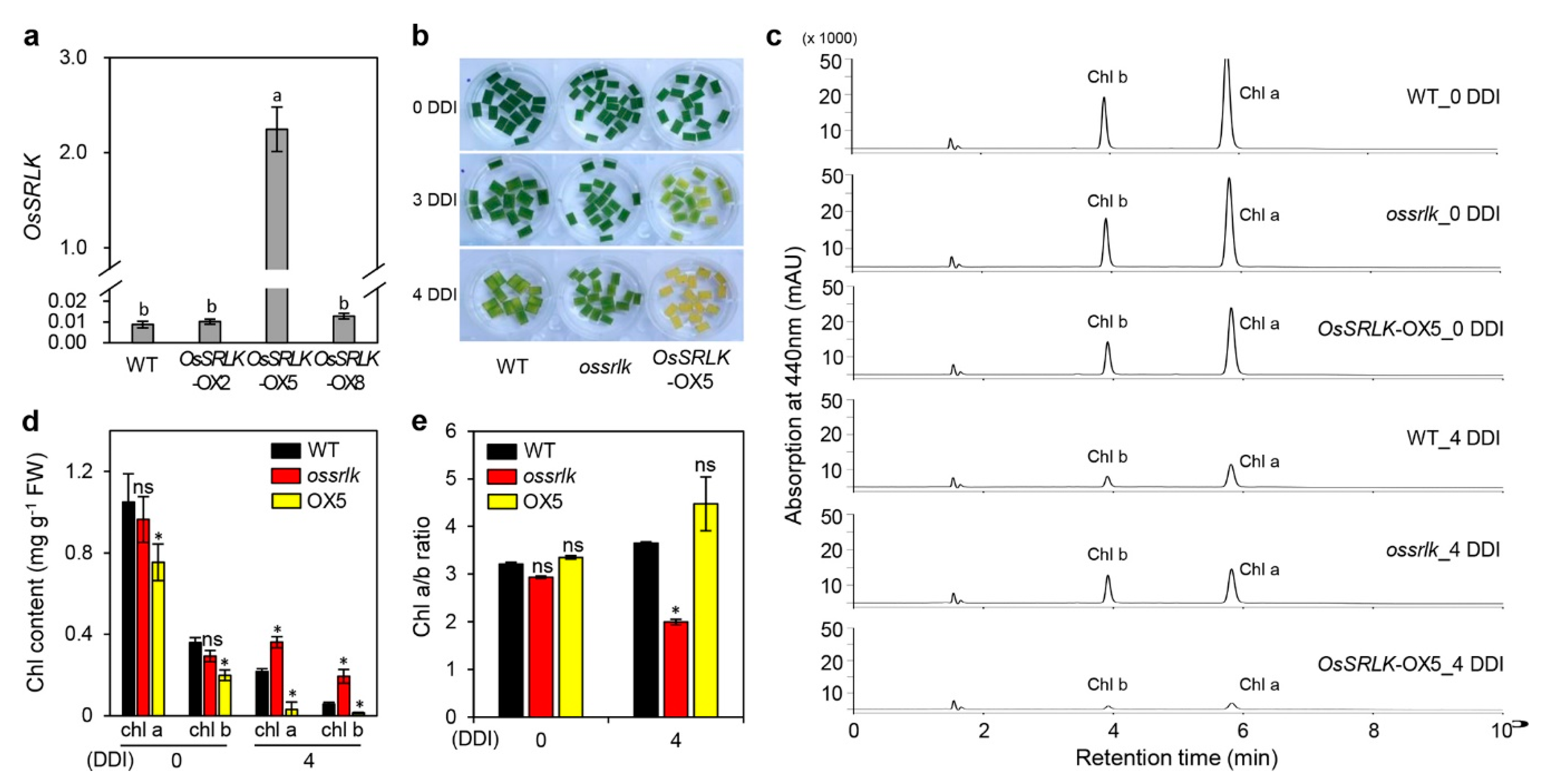
2.3. Overexpression of OsSRLK Accelerates Leaf Yellowing under DIS Conditions
2.4. OsSRLK Regulates the Expression of Phytohormone-Related Genes during DIS
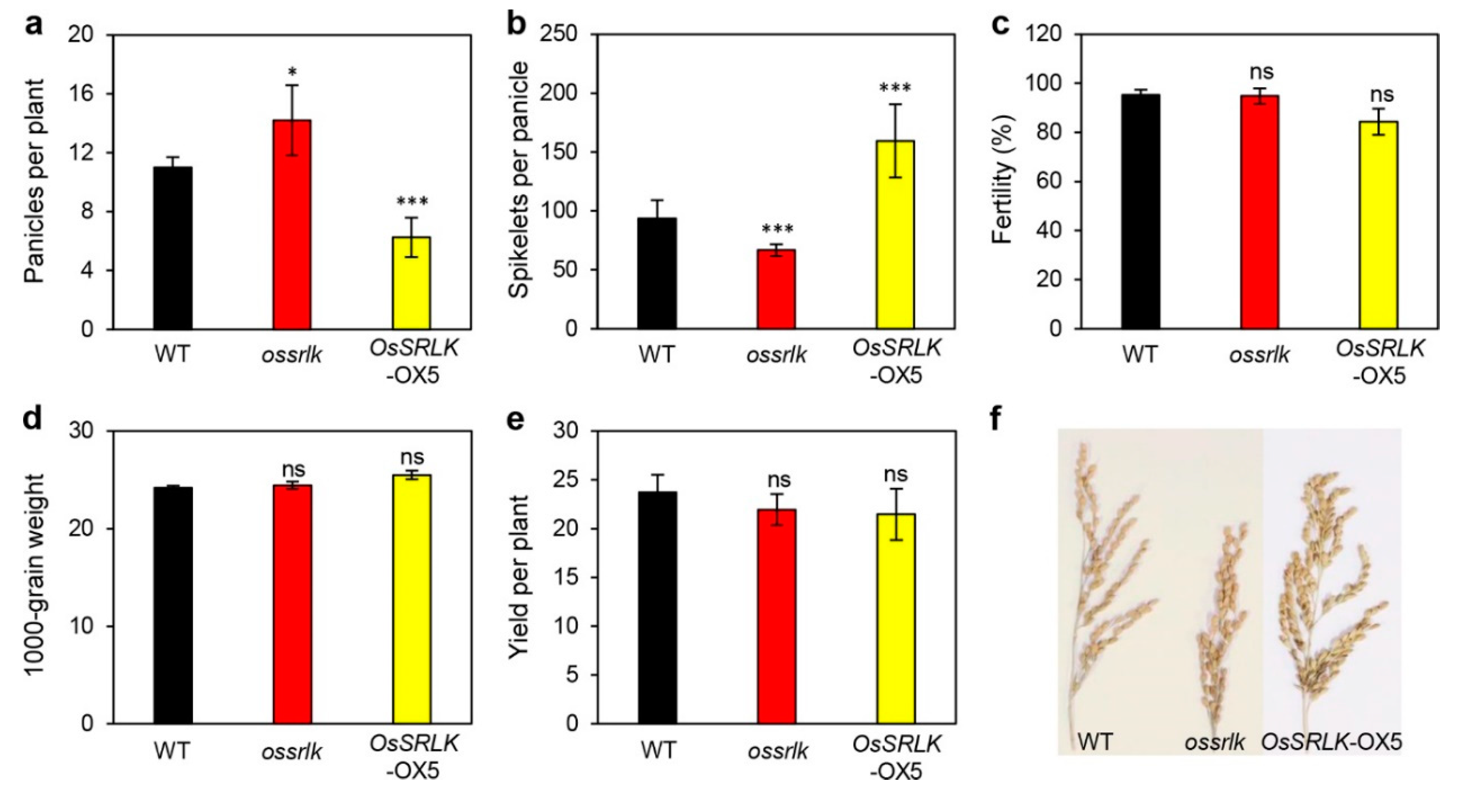
2.5. Effects of OsSRLK on the Panicle Count Per Plant and Spikelets Per Panicle
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Experimental Treatments
4.2. Determination of Chlorophyll Content, Photosynthetic Activity, SPAD Value, and Ion Leakage Rate
4.3. RNA Isolation and RT-qPCR Analysis
4.4. Transmission Electron Microscopy Analysis
4.5. HPLC Analysis
4.6. RNA-Seq and Analysis
4.7. Plasmid Construction and Transformation into Rice
4.8. SDS-PAGE and Immunoblot Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| WT | wild type |
| Chl | chlorophyll |
| NLD | natural long day |
| LD | long day |
| RT | reverse transcriptase |
| DIS | dark-induced senescence |
| DDI | day(s) of dark incubation |
| CDGs | chlorophyll degradation genes |
| SAGs | senescence-associated genes |
| DEGs | differential expressed genes |
References
- Tanaka, R.; Tanaka, A. Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 2007, 58, 321–346. [Google Scholar] [CrossRef]
- Kusaba, M.; Ito, H.; Morita, R.; Iida, S.; Sato, Y.; Fujimoto, M.; Kawasaki, S.; Tanaka, R.; Hirochika, H.; Nishimura, M.; et al. Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 2007, 19, 1362–1375. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.F.; Forsberg, J. Molecular recognition in thylakoid structure and function. Trends Plant Sci. 2001, 6, 317–326. [Google Scholar] [CrossRef]
- Standfuss, J.; Terwisscha van Scheltinga, A.C.; Lamborghini, M.; Kühlbrandt, W. Mechanisms of photoprotection and nonphotochemical quenching in pea light-harvesting complex at 2.5 A resolution. EMBO J. 2005, 24, 919–928. [Google Scholar] [CrossRef] [PubMed]
- Morita, R.; Sato, Y.; Masuda, Y.; Nishimura, M.; Kusaba, M. Defect in non-yellow coloring 3, an α/β hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J. 2009, 59, 940–952. [Google Scholar] [CrossRef]
- Hörtensteiner, S.; Kräutler, B. Chlorophyll breakdown in higher plants. Biochimica et Biophysica Acta (BBA)-Bioenergetics 2011, 1807, 977–988. [Google Scholar] [CrossRef]
- Park, S.Y.; Yu, J.W.; Park, J.S.; Li, J.; Yoo, S.C.; Lee, N.Y.; Lee, S.K.; Jeong, S.W.; Seo, H.S.; Koh, H.J.; et al. The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 2007, 19, 1649–1664. [Google Scholar] [CrossRef]
- Horie, Y.; Ito, H.; Kusaba, M.; Tanaka, R.; Tanaka, A. Participation of chlorophyll b reductase in the initial step of the degradation of light-harvesting chlorophyll a/b-protein complexes in Arabidopsis. J. Biol. Chem. 2009, 284, 17449–17456. [Google Scholar] [CrossRef]
- Sato, Y.; Morita, R.; Katsuma, S.; Nishimura, M.; Tanaka, A.; Kusaba, M. Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 2009, 57, 120–131. [Google Scholar] [CrossRef]
- Meguro, M.; Ito, H.; Takabayashi, A.; Tanaka, R.; Tanaka, A. Identification of the 7-hydroxymethyl chlorophyll a reductase of the chlorophyll cycle in Arabidopsis. Plant Cell 2011, 23, 3442–3453. [Google Scholar] [CrossRef]
- Armstead, I.; Donnison, I.; Aubry, S.; Harper, J.; Hörtensteiner, S.; James, C.; Mani, J.; Moffet, M.; Ougham, H.; Roberts, L.; et al. Cross-species identification of Mendel’s I locus. Science 2007, 315, 73. [Google Scholar] [CrossRef] [PubMed]
- Shimoda, Y.; Ito, H.; Tanaka, A. Arabidopsis STAY-GREEN, mendel’s green cotyledon gene, encodes magnesium-dechelatase. Plant Cell 2016, 28, 2147–2160. [Google Scholar] [CrossRef]
- Schelbert, S.; Aubry, S.; Burla, B.; Agne, B.; Kessler, F.; Krupinska, K.; Hörtensteiner, S. Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 2009, 21, 767–785. [Google Scholar] [CrossRef] [PubMed]
- Pruzinská, A.; Tanner, G.; Anders, I.; Roca, M.; Hörtensteiner, S. Chlorophyll breakdown: Pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc. Natl. Acad. Sci. USA 2003, 100, 15259–15264. [Google Scholar] [CrossRef] [PubMed]
- Takeda, T.; Suwa, Y.; Suzuki, M.; Kitano, H.; Ueguchi-Tanaka, M.; Ashikari, M.; Matsuoka, M.; Ueguchi, C. The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 2003, 33, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Pružinská, A.; Anders, I.; Aubry, S.; Schenk, N.; Tapernoux-Lüthi, E.; Müller, T.; Kräutler, B.; Hörtensteiner, S. In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell 2007, 19, 369–387. [Google Scholar] [CrossRef]
- Hörtensteiner, S. Chlorophyll degradation during senescence. Annu. Rev. Plant Biol. 2006, 57, 55–77. [Google Scholar] [CrossRef]
- Lim, P.O.; Kim, H.J.; Gil Nam, H. Leaf senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef]
- Guo, Y.; Gan, S.-S. Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence-promoting hormonal, pathological and environmental stress treatments. Plant Cell Environ. 2012, 35, 644–655. [Google Scholar] [CrossRef]
- Li, Z.; Peng, J.; Wen, X.; Guo, H. Gene network analysis and functional studies of senescence-associated genes reveal novel regulators of Arabidopsis leaf senescence. J. Integr. Plant Biol. 2012, 54, 526–539. [Google Scholar] [CrossRef]
- Kim, T.; Kang, K.; Kim, S.-H.; An, G.; Paek, N.-C. OsWRKY5 promotes rice leaf senescence via senescence-associated NAC and abscisic acid biosynthesis pathway. Int. J. Mol. Sci 2019, 20, 4437. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.H.; Lim, J.H.; Kim, S.S.; Cho, S.H.; Yoo, S.C.; Koh, H.J.; Sakuraba, Y.; Paek, N.C. Mutation of SPOTTED LEAF3 (SPL3) impairs abscisic acid-responsive signalling and delays leaf senescence in rice. J. Exp. Bot. 2015, 66, 7045–7059. [Google Scholar] [CrossRef] [PubMed]
- Shim, Y.; Kang, K.; An, G.; Paek, N.C. Rice DNA-binding one zinc finger 24 (OsDOF24) delays leaf Senescence in a jasmonate-mediated pathway. Plant Cell Physiol. 2019, 60, 2065–2076. [Google Scholar] [CrossRef] [PubMed]
- Ke, S.; Liu, S.; Luan, X.; Xie, X.M.; Hsieh, T.F.; Zhang, X.Q. Mutation in a putative glycosyltransferase-like gene causes programmed cell death and early leaf senescence in rice. Rice 2019, 12, 7. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.-H.; Karlowski, W.M.; Pan, R.; Tzeng, Y.-H.; Mayer, K.F.X.; Li, W.H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 2004, 16, 1220–1234. [Google Scholar] [CrossRef] [PubMed]
- Becraft, P.W.; Stinard, P.S.; McCarty, D.R. CRINKLY4: A TNFR-like receptor kinase involved in maize epidermal differentiation. Science 1996, 273, 1406–1409. [Google Scholar] [CrossRef]
- Torii, K.U.; Mitsukawa, N.; Oosumi, T.; Matsuura, Y.; Yokoyama, R.; Whittier, R.F.; Komeda, Y. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 1996, 8, 735–746. [Google Scholar]
- Clark, S.E.; Williams, R.W.; Meyerowitz, E.M. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 1997, 89, 575–585. [Google Scholar] [CrossRef]
- Jinn, T.L.; Stone, J.M.; Walker, J.C. HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 2000, 14, 108–117. [Google Scholar]
- Li, J.; Wen, J.; Lease, K.A.; Doke, J.T.; Tax, F.E.; Walker, J.C. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 2002, 110, 213–222. [Google Scholar] [CrossRef]
- Matsubayashi, Y.; Ogawa, M.; Morita, A.; Sakagami, Y. An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science 2002, 296, 1470–1472. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.L.; Song, W.Y.; Ruan, D.L.; Sideris, S.; Ronald, P.C. The cloned gene, Xa21, confers resistance to multiple Xanthomonas oryzae pv. oryzae isolates in transgenic plants. Mol. Plant Microbe Interact. 1996, 9, 850–855. [Google Scholar] [CrossRef] [PubMed]
- Feuillet, C.; Schachermayr, G.; Keller, B. Molecular cloning of a new receptor-like kinase gene encoded at the Lr10 disease resistance locus of wheat. Plant J. 1997, 11, 45–52. [Google Scholar] [CrossRef]
- Park, D.-Y.; Shim, Y.; Gi, E.; Lee, B.-D.; An, G.; Kang, K.; Paek, N.C. The MYB-related transcription factor RADIALIS-LIKE3 (OsRL3) functions in ABA-induced leaf senescence and salt sensitivity in rice. Environ. Exp. Bot. 2018, 156, 86–95. [Google Scholar] [CrossRef]
- Yoo, Y.H.; Choi, H.K.; Jung, K.H. Genome-wide identification and analysis of genes associated with lysigenous aerenchyma formation in rice roots. J. Plant Biol. 2015, 58, 117–127. [Google Scholar] [CrossRef]
- Yan, Y.S.; Chen, X.Y.; Yang, K.; Sun, Z.X.; Fu, Y.P.; Zhang, Y.M.; Fang, R.X. Overexpression of an F-box protein gene reduces abiotic stress tolerance and promotes root growth in rice. Mol. Plant 2011, 4, 190–197. [Google Scholar] [CrossRef]
- Sharma, G.; Giri, J.; Tyagi, A.K. Rice OsiSAP7 negatively regulates ABA stress signalling and imparts sensitivity to water-deficit stress in Arabidopsis. Plant Sci. 2015, 237, 80–92. [Google Scholar] [CrossRef]
- Imai, R.; Ali, A.; Pramanik, M.H.R.; Nakaminami, K.; Sentoku, N.; Kato, H. A distinctive class of spermidine synthase is involved in chilling response in rice. J. Plant Physiol. 2004, 161, 883–886. [Google Scholar] [CrossRef]
- Sperotto, R.A.; Ricachenevsky, F.K.; Duarte, G.L.; Boff, T.; Lopes, K.L.; Sperb, E.R.; Grusak, M.A.; Fett, J.P. Identification of up-regulated genes in flag leaves during rice grain filling and characterization of OsNAC5, a new ABA-dependent transcription factor. Planta 2009, 230, 985–1002. [Google Scholar] [CrossRef]
- Jisha, V.; Dampanaboina, L.; Vadassery, J.; Mithöfer, A.; Kappara, S.; Ramanan, R. Overexpression of an AP2/ERF type transcription factor OsEREBP1 confers biotic and abiotic stress tolerance in rice. PLoS ONE 2015, 10, e0127831. [Google Scholar] [CrossRef]
- Yu, S.; Ligang, C.; Liping, Z.; Diqiu, Y. Overexpression of OsWRKY72 gene interferes in the abscisic acid signal and auxin transport pathway of Arabidopsis. J. Biosci. 2010, 35, 459–471. [Google Scholar] [PubMed]
- Zang, A.; Xu, X.; Neill, S.; Cai, W. Overexpression of OsRAN2 in rice and Arabidopsis renders transgenic plants hypersensitive to salinity and osmotic stress. J. Exp. Bot. 2010, 61, 777–789. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Nguyen, B.H.; Xie, Y.; Xiao, B.; Tang, N.; Zhu, W.; Mou, T.; Xiong, L. Co-overexpression of the constitutively active form of OsbZIP46 and ABA-activated protein kinase SAPK6 improves drought and temperature stress resistance in rice. Front. Plant Sci. 2017, 8, 1102. [Google Scholar] [CrossRef] [PubMed]
- Akhter, D.; Qin, R.; Nath, U.; Alamin, M.; Jin, X.; Shi, C. The brown midrib leaf (bml) mutation in rice (Oryza sativa L.) causes premature leaf senescence and the induction of defense responses. Genes 2018, 9, 203. [Google Scholar] [CrossRef]
- Shi, B.; Ni, L.; Liu, Y.; Zhang, A.; Tan, M.; Jiang, M. OsDMI3-mediated activation of OsMPK1 regulates the activities of antioxidant enzymes in abscisic acid signalling in rice. Plant Cell Environ. 2014, 37, 341–352. [Google Scholar] [CrossRef]
- Nakagawa, H.; Ohmiya, K.; Hattori, T. A rice bZIP protein, designated OSBZ8, is rapidly induced by abscisic acid. Plant J. 1996, 9, 217–227. [Google Scholar] [CrossRef]
- Huang, L.; Hong, Y.; Zhang, H.; Li, D.; Song, F. Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance. BMC Plant Biol. 2016, 16, 203. [Google Scholar] [CrossRef]
- Bhatnagar, N.; Min, M.K.; Choi, E.H.; Kim, N.; Moon, S.J.; Yoon, I.; Kwon, T.; Jung, K.H.; Kim, B.G. The protein phosphatase 2C clade a protein OsPP2C51 positively regulates seed germination by directly inactivating OsbZIP10. Plant Mol. Biol. 2017, 93, 389–401. [Google Scholar] [CrossRef]
- Du, H.; Wang, N.; Cui, F.; Li, X.; Xiao, J.; Xiong, L. Characterization of the beta-carotene hydroxylase gene DSM2 conferring drought and oxidative stress resistance by increasing xanthophylls and abscisic acid synthesis in rice. Plant Physiol. 2010, 154, 1304–1318. [Google Scholar] [CrossRef]
- Wang, Y.J.; Zhang, Z.G.; He, X.J.; Zhou, H.L.; Wen, Y.X.; Dai, J.X.; Zhang, J.S.; Chen, S.Y. A rice transcription factor OsbHLH1 is involved in cold stress response. Theor. Appl. Genet. 2003, 107, 1402–1409. [Google Scholar] [CrossRef]
- Kim, S.G.; Kim, S.T.; Wang, Y.; Kim, S.K.; Lee, C.H.; Kim, K.K.; Kim, J.K.; Lee, S.Y.; Kang, K.Y. Overexpression of rice isoflavone reductase-like gene (OsIRL) confers tolerance to reactive oxygen species. Physiol. Plant. 2010, 138, 1–9. [Google Scholar] [CrossRef]
- Miyamoto, K.; Shimizu, T.; Lin, F.; Sainsbury, F.; Thuenemann, E.; Lomonossoff, G.; Nojiri, H.; Yamane, H.; Okada, K. Identification of an E-box motif responsible for the expression of jasmonic acid-induced chitinase gene OsChia4a in rice. J. Plant Physiol. 2012, 169, 621–627. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Hu, Y.; Tang, X.; Zhou, P.; Deng, X.; Wang, H.; Guo, Z. Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta 2012, 236, 1485–1498. [Google Scholar] [CrossRef] [PubMed]
- He, S.L.; Jiang, J.Z.; Chen, B.-H.; Kuo, C.H.; Ho, S.L. Overexpression of a constitutively active truncated form of OsCDPK1 confers disease resistance by affecting OsPR10a expression in rice. Sci. Rep. 2018, 8, 403. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.M.; Shin, S.H.; Kim, K.S.; Kim, Y.C.; Eun, M.Y.; Cho, B.H. Enhanced expression of a gene encoding a nucleoside diphosphate kinase 1 (OsNDPK1) in rice plants upon infection with bacterial pathogens. Mol. Cells 2004, 18, 390–395. [Google Scholar] [PubMed]
- Jung, Y.H.; Agrawal, G.K.; Rakwal, R.; Kim, J.A.; Lee, M.O.; Choi, P.G.; Kim, Y.J.; Kim, M.J.; Shibato, J.; Kim, S.H.; et al. Functional characterization of OsRacB GTPase—A potentially negative regulator of basal disease resistance in rice. Plant Physiol. Biochem. 2006, 44, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Yokotani, N.; Ichikawa, T.; Kondou, Y.; Iwabuchi, M.; Matsui, M.; Hirochika, H.; Oda, K. Role of the rice transcription factor JAmyb in abiotic stress response. J. Plant Res. 2013, 126, 131–139. [Google Scholar] [CrossRef]
- Zhang, X.; Bao, Y.; Shan, D.; Wang, Z.; Song, X.; Wang, Z.; Wang, J.; He, L.; Wu, L.; Zhang, Z.; et al. Magnaporthe oryzae induces the expression of a microRNA to suppress the immune response in rice. Plant Physiol. 2018, 177, 352–368. [Google Scholar] [CrossRef]
- Wang, R.; Shen, W.; Liu, L.; Jiang, L.; Liu, Y.; Su, N.; Wan, J. A novel lipoxygenase gene from developing rice seeds confers dual position specificity and responds to wounding and insect attack. Plant Mol. Biol. 2008, 66, 401–414. [Google Scholar] [CrossRef]
- Wong, H.L.; Sakamoto, T.; Kawasaki, T.; Umemura, K.; Shimamoto, K. Down-regulation of metallothionein, a reactive oxygen scavenger, by the small GTPase OsRac1 in rice. Plant Physiol. 2004, 135, 1447–1456. [Google Scholar] [CrossRef]
- Zhang, W.; Zhou, X.; Wen, C.-K. Modulation of ethylene responses by OsRTH1 overexpression reveals the biological significance of ethylene in rice seedling growth and development. J. Exp. Bot. 2012, 63, 4151–4164. [Google Scholar] [CrossRef] [PubMed]
- Yau, C.P.; Wang, L.; Yu, M.; Zee, S.Y.; Yip, W.K. Differential expression of three genes encoding an ethylene receptor in rice during development, and in response to indole-3-acetic acid and silver ions. J. Exp. Bot. 2004, 55, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.; Saigusa, M.; Hase, S.; Hayakawa, T.; Satoh, S. Cloning of a cDNA encoding an ETR2-like protein (Os-ERL1) from deep water rice (Oryza sativa L.) and increase in its mRNA level by submergence, ethylene, and gibberellin treatments. J. Exp. Bot. 2004, 55, 1145–1148. [Google Scholar] [CrossRef] [PubMed]
- Lian, H.L.; Yu, X.; Ye, Q.; Ding, X.; Kitagawa, Y.; Kwak, S.S.; Su, W.A.; Tang, Z.C.; Ding, X.S. The role of aquaporin RWC3 in drought avoidance in rice. Plant Cell Physiol. 2004, 45, 481–489. [Google Scholar] [CrossRef]
- Sauter, M.; Lorbiecke, R.; Ouyang, B.; Pochapsky, T.C.; Rzewuski, G. The immediate-early ethylene response gene OsARD1 encodes an acireductone dioxygenase involved in recycling of the ethylene precursor S-adenosylmethionine. Plant J. 2005, 44, 718–729. [Google Scholar] [CrossRef]
- Sauter, M.; Rzewuski, G.; Marwedel, T.; Lorbiecke, R. The novel ethylene-regulated gene OsUsp1 from rice encodes a member of a plant protein family related to prokaryotic universal stress proteins. J. Exp. Bot. 2002, 53, 2325–2331. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, S.; Guo, H.; Wang, S.; Xu, L.; Li, C.; Qian, Q.; Chen, F.; Geisler, M.; Qi, Y.; et al. OsABCB14 functions in auxin transport and iron homeostasis in rice (Oryza sativa L.). Plant J. 2014, 79, 106–117. [Google Scholar] [CrossRef]
- Jung, H.; Lee, D.K.; Choi, Y.D.; Kim, J.K. OsIAA6, a member of the rice Aux/IAA gene family, is involved in drought tolerance and tiller outgrowth. Plant Sci. 2015, 236, 304–312. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, S.; Sun, C.; Xu, Y.; Chen, Y.; Yu, C.; Qian, Q.; Jiang, D.A.; Qi, Y. Auxin response factor (OsARF12), a novel regulator for phosphate homeostasis in rice (Oryza sativa). New Phytol. 2014, 201, 91–103. [Google Scholar] [CrossRef]
- Yoshikawa, T.; Ito, M.; Sumikura, T.; Nakayama, A.; Nishimura, T.; Kitano, H.; Yamaguchi, I.; Koshiba, T.; Hibara, K.-I.; Nagato, Y.; et al. The rice FISH BONE gene encodes a tryptophan aminotransferase, which affects pleiotropic auxin-related processes. Plant J. 2014, 78, 927–936. [Google Scholar] [CrossRef]
- Xu, M.; Zhu, L.; Shou, H.; Wu, P. A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 2005, 46, 1674–1681. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, S.; Xu, Y.; Yu, C.; Shen, C.; Qian, Q.; Geisler, M.; Jiang, D.A.; Qi, Y. The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ. 2015, 38, 638–654. [Google Scholar] [CrossRef] [PubMed]
- Moons, A. Ospdr9, which encodes a PDR-type ABC transporter, is induced by heavy metals, hypoxic stress and redox perturbations in rice roots. FEBS Lett. 2003, 553, 370–376. [Google Scholar] [CrossRef]
- Chen, Q.; Westfall, C.S.; Hicks, L.M.; Wang, S.; Jez, J.M. Kinetic basis for the conjugation of auxin by a GH3 family indole-acetic acid-amido synthetase. J. Biol. Chem. 2010, 285, 29780–29786. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zong, W.; Hou, X.; Yao, J.; Liu, H.; Li, X.; Zhao, Y.; Xiong, L. OsARID3, an AT-rich Interaction Domain-containing protein, is required for shoot meristem development in rice. Plant J. 2015, 83, 806–817. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Piao, W.; Lim, J.H.; Han, S.H.; Kim, Y.S.; An, G.; Paek, N.C. Rice ONAC106 inhibits leaf senescence and increases salt tolerance and tiller angle. Plant Cell Physiol. 2015, 56, 2325–2339. [Google Scholar] [CrossRef]
- Chattopadhyay, T.; Bhattacharyya, S.; Das, A.K.; Maiti, M.K. A structurally novel hemopexin fold protein of rice plays role in chlorophyll degradation. Biochem. Biophys. Res. Commun. 2012, 420, 862–868. [Google Scholar] [CrossRef]
- Qin, R.; Zeng, D.; Liang, R.; Yang, C.; Akhter, D.; Alamin, M.; Jin, X.; Shi, C. Rice gene SDL/RNRS1, encoding the small subunit of ribonucleotide reductase, is required for chlorophyll synthesis and plant growth development. Gene 2017, 627, 351–362. [Google Scholar] [CrossRef]
- Feng, Z.; Zhang, L.; Yang, C.; Wu, T.; Lv, J.; Chen, Y.; Liu, X.; Liu, S.; Jiang, L.; Wan, J. EF8 is involved in photoperiodic flowering pathway and chlorophyll biogenesis in rice. Plant Cell Rep. 2014, 33, 2003–2014. [Google Scholar] [CrossRef]
- He, Y.; Fukushige, H.; Hildebrand, D.F.; Gan, S. Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 2002, 128, 876–884. [Google Scholar] [CrossRef] [PubMed]
- Nick, S.; Meurer, J.; Soll, J.; Ankele, E. Nucleus-encoded light-harvesting chlorophyll a/b proteins are imported normally into chlorophyll b-free chloroplasts of Arabidopsis. Mol Plant 2013, 6, 860–871. [Google Scholar] [CrossRef] [PubMed]
- Horn, R.; Grundmann, G.; Paulsen, H. Consecutive binding of chlorophylls a and b during the assembly in vitro of light-harvesting chlorophyll-a/b protein (LHCIIb). J. Mol. Biol. 2007, 366, 1045–1054. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Guo, Y.; Liu, Y.; Zhang, F.; Wang, Z.; Wang, H.; Wang, F.; Li, D.; Mao, D.; Luan, S.; et al. 9-cis-epoxycarotenoid dioxygenase 3 regulates plant growth and enhances multi-abiotic stress tolerance in rice. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Jeong, J.; Kang, M.Y.; Kim, J.; Paek, N.C.; Choi, G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat Commun. 2014, 5, 4636. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.; Shim, Y.; Gi, E.; An, G.; Paek, N.C. Mutation of ONAC096 enhances grain yield by increasing panicle number and delaying leaf senescence during grain filling in rice. Int. J. Mol. Sci. 2019, 20, 5241. [Google Scholar] [CrossRef]
- Lee, S.-H.; Sakuraba, Y.; Lee, T.; Kim, K.-W.; An, G.; Lee, H.Y.; Paek, N.-C. Mutation of Oryza sativa CORONATINE INSENSITIVE 1b (OsCOI1b) delays leaf senescence. J. Integr. Plant Biol. 2015, 57, 562–576. [Google Scholar] [CrossRef]
- Koyama, T. The roles of ethylene and transcription factors in the regulation of onset of leaf senescence. Front. Plant Sci. 2014, 5, 650. [Google Scholar] [CrossRef]
- Nam, K.H.; Li, J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 2002, 110, 203–212. [Google Scholar] [CrossRef]
- Badshah, M.A.; Naimei, T.; Zou, Y.; Ibrahim, M.; Wang, K. Yield and tillering response of super hybrid rice Liangyoupeijiu to tillage and establishment methods. Crop J. 2014, 2, 79–86. [Google Scholar] [CrossRef]
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X.; et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef]
- Jeon, J.-S.; Lee, S.; Jung, K.-H.; Jun, S.-H.; Jeong, D.-H.; Lee, J.; Kim, C.; Jang, S.; Lee, S.; Yang, K.; et al. T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 2000, 22, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Jeong, D.H.; An, S.; Park, S.; Kang, H.G.; Park, G.G.; Kim, S.R.; Sim, J.; Kim, Y.O.; Kim, M.K.; Kim, S.R.; et al. Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice: Activation tagging in rice. Plant J. 2006, 45, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H.K. Chlorophylls and Carotenoids: Pigments of Photosynthetic Biomembranes. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1987; Volume 148, pp. 350–382. ISBN 978-0-12-182048-0. [Google Scholar]
- Porra, R.J.; Thompson, W.A.; Kriedemann, P.E. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochimica et Biophysica Acta (BBA) Bioenergetics 1989, 975, 384–394. [Google Scholar] [CrossRef]
- Fan, L.; Zheng, S.; Wang, X. Antisense suppression of phospholipase D alpha retards abscisic acid- and ethylene-promoted senescence of postharvest Arabidopsis leaves. Plant Cell 1997, 9, 2183–2196. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Sharabi-Schwager, M.; Samach, A.; Porat, R. Overexpression of the CBF2 transcriptional activator in Arabidopsis counteracts hormone activation of leaf senescence. Plant Signal Behav. 2010, 5, 296–299. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Cao, P.; Jung, K.-H.; Choi, D.; Hwang, D.; Zhu, J.; Ronald, P.C. The Rice Oligonucleotide Array Database: An atlas of rice gene expression. Rice 2012, 5, 17. [Google Scholar] [CrossRef]
- Jung, K.-H.; Dardick, C.; Bartley, L.E.; Cao, P.; Phetsom, J.; Canlas, P.; Seo, Y.S.; Shultz, M.; Ouyang, S.; Yuan, Q.; et al. Refinement of light-responsive transcript lists using rice oligonucleotide arrays: Evaluation of gene-redundancy. PLoS ONE 2008, 3, e3337. [Google Scholar] [CrossRef]
- Yoo, Y.H.; Hong, W.J.; Jung, K.H. A Systematic view exploring the role of chloroplasts in plant abiotic stress responses. BioMed Res. Int. 2019, 2019, 6534745. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.R.; Lee, D.Y.; Yang, J.I.; Moon, S.; An, G. Cloning vectors for rice. J. Plant Biol. 2009, 52, 73–78. [Google Scholar] [CrossRef]

| Locus_ID (LOC_) | Gene symbol | Fold Change (Log2) | p-Value | References |
|---|---|---|---|---|
| ABA | ||||
| Os02g44990 | MAIF1 | 2.374286 | 0.00005 | [36] |
| Os03g57900 | OsiSAP7 | 1.427566 | 0.02315 | [37] |
| Os06g33710 | OsSPDS2 | 1.394406 | 0.01064 | [38] |
| Os11g08210 | OsNAC5 | 1.354165 | 0.03263 | [39] |
| Os02g54160 | OsEREBP1 | −1.019618 | 0.00204 | [40] |
| Os11g29870 | OsWRKY72 | −1.040532 | 0.02098 | [41] |
| Os05g49890 | OsRAN2 | −1.061454 | 0.00769 | [42] |
| Os02g34600 | SAPK6 | −1.221792 | 0.00969 | [43] |
| Os05g02020 | OsRLCK176 | −1.391996 | 0.00094 | [44] |
| Os05g41090 | OsCCaMK | −1.436117 | 0.00770 | [45] |
| Os01g46970 | OSBZ8 | −1.510641 | 0.04456 | [46] |
| Os06g51070 | ONAC095 | −1.621916 | 0.03699 | [47] |
| Os05g49730 | OsPP2C51 | −2.253431 | 0.02230 | [48] |
| Os03g03370 | DSM2 | −2.439699 | 0.00472 | [49] |
| Os07g43530 | OsbHLH1 | −2.468823 | 0.00065 | [50] |
| Os01g01660 | OsIRL | −4.027866 | 0.00066 | [51] |
| JA | ||||
| Os04g41680 | OsChia4a | 4.944696 | 0.00223 | [52] |
| Os08g38990 | OsWRKY30 | 1.860935 | 0.02102 | [53] |
| Os03g03660 | OsCDPK1 | −1.027442 | 0.02197 | [54] |
| Os07g30970 | OsNDPK1 | −1.148937 | 0.00489 | [55] |
| Os02g02840 | OsRacB | −1.201440 | 0.01898 | [56] |
| Os11g45740 | JAmyb | −1.409207 | 0.00710 | [57] |
| Os03g08220 | OsLOX2 | −1.575594 | 0.00335 | [58] |
| Os03g49380 | OsLOX1 | −2.426267 | 0.01822 | [59] |
| Ethylene | ||||
| Os01g12900 | OsRac1 | 2.952107 | 0.01944 | [60] |
| Os01g51430 | OsRTH1 | −1.087656 | 0.00187 | [61] |
| Os02g57530 | ETR3 | −1.181857 | 0.03265 | [62] |
| Os04g08740 | ETR2 | −1.220128 | 0.04523 | [63] |
| Os02g57720 | RWC3 | −2.322009 | 0.00321 | [64] |
| Os10g28350 | OsARD1 | −3.650923 | 0.00420 | [65] |
| Os07g47620 | OsUsp1 | −3.842188 | 0.00163 | [66] |
| Auxin | ||||
| Os04g38570 | OsABCB14 | 2.802053 | 0.03772 | [67] |
| Os01g53880 | IAA6 | 1.486924 | 0.01248 | [68] |
| Os04g57610 | OsARF12 | −1.274296 | 0.03383 | [69] |
| Os01g07500 | FIB | −1.395635 | 0.01820 | [70] |
| Os02g50960 | OsPIN1 | −1.538501 | 0.00160 | [71] |
| Os06g48950 | OsARF19 | −1.720215 | 0.00083 | [72] |
| Os01g42380 | Ospdr9 | −2.046885 | 0.00035 | [73] |
| Os07g40290 | OsGH3.8 | −2.265828 | 0.00652 | [74] |
| Os05g47840 | OsIPT7 | −2.424619 | 0.04842 | [75] |
| Locus_ID (LOC_) | Gene Symbol | Fold Change (Log2) | p-Value | References |
|---|---|---|---|---|
| Senescence | ||||
| Os08g33670 | ONAC106 | 0.611718 | 0.01580 | [76] |
| Os11g29870 | OsWRKY72 | −1.040532 | 0.02098 | [41] |
| Os05g02020 | OsRLCK176 | −1.391996 | 0.00094 | [44] |
| Chlorophyll biogenesis | ||||
| Os04g13540 | OsHFP | 4.025662 | 0.00903 | [77] |
| Os06g14620 | RNRS1 | −1.282421 | 0.03238 | [78] |
| Os08g07740 | EF8 | −2.647314 | 0.00042 | [79] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, N.-H.; Trang, D.T.; Hong, W.-J.; Kang, K.; Chuluuntsetseg, J.; Moon, J.-K.; Yoo, Y.-H.; Jung, K.-H.; Yoo, S.-C. Rice Senescence-Induced Receptor-Like Kinase (OsSRLK) Is Involved in Phytohormone-Mediated Chlorophyll Degradation. Int. J. Mol. Sci. 2020, 21, 260. https://doi.org/10.3390/ijms21010260
Shin N-H, Trang DT, Hong W-J, Kang K, Chuluuntsetseg J, Moon J-K, Yoo Y-H, Jung K-H, Yoo S-C. Rice Senescence-Induced Receptor-Like Kinase (OsSRLK) Is Involved in Phytohormone-Mediated Chlorophyll Degradation. International Journal of Molecular Sciences. 2020; 21(1):260. https://doi.org/10.3390/ijms21010260
Chicago/Turabian StyleShin, Na-Hyun, Do Thi Trang, Woo-Jong Hong, Kiyoon Kang, Jadamba Chuluuntsetseg, Joon-Kwan Moon, Yo-Han Yoo, Ki-Hong Jung, and Soo-Cheul Yoo. 2020. "Rice Senescence-Induced Receptor-Like Kinase (OsSRLK) Is Involved in Phytohormone-Mediated Chlorophyll Degradation" International Journal of Molecular Sciences 21, no. 1: 260. https://doi.org/10.3390/ijms21010260
APA StyleShin, N.-H., Trang, D. T., Hong, W.-J., Kang, K., Chuluuntsetseg, J., Moon, J.-K., Yoo, Y.-H., Jung, K.-H., & Yoo, S.-C. (2020). Rice Senescence-Induced Receptor-Like Kinase (OsSRLK) Is Involved in Phytohormone-Mediated Chlorophyll Degradation. International Journal of Molecular Sciences, 21(1), 260. https://doi.org/10.3390/ijms21010260





