Function of miR825 and miR825* as Negative Regulators in Bacillus cereus AR156-elicited Systemic Resistance to Botrytis cinerea in Arabidopsis thaliana
Abstract
:1. Introduction
2. Results
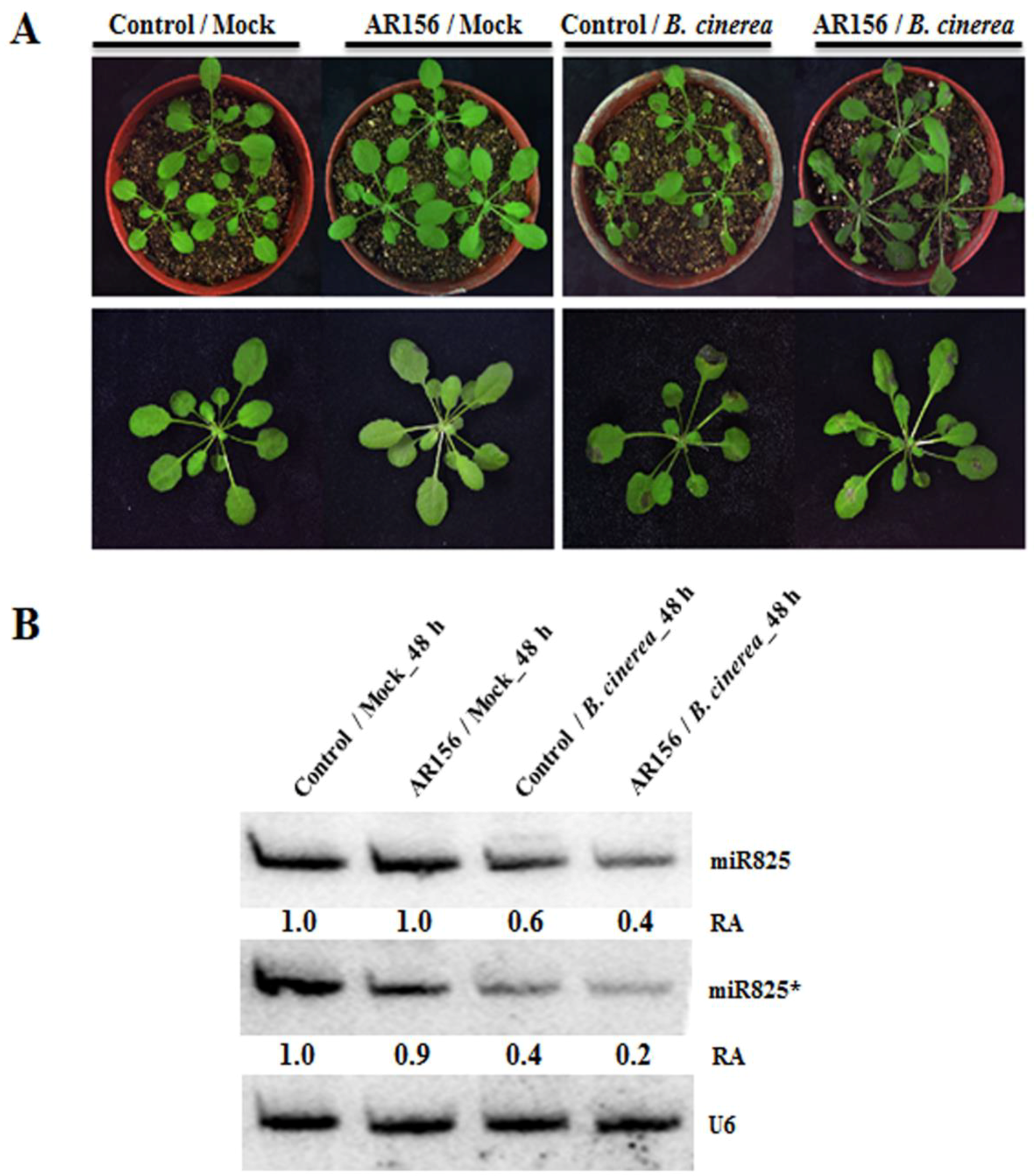
2.1. miR825 and miR825* Expression was Suppressed in AR156-induced ISR to Prevent B. Cinerea in Arabidopsis
2.2. miR825 and miR825* Participate in AR156-induced ISR
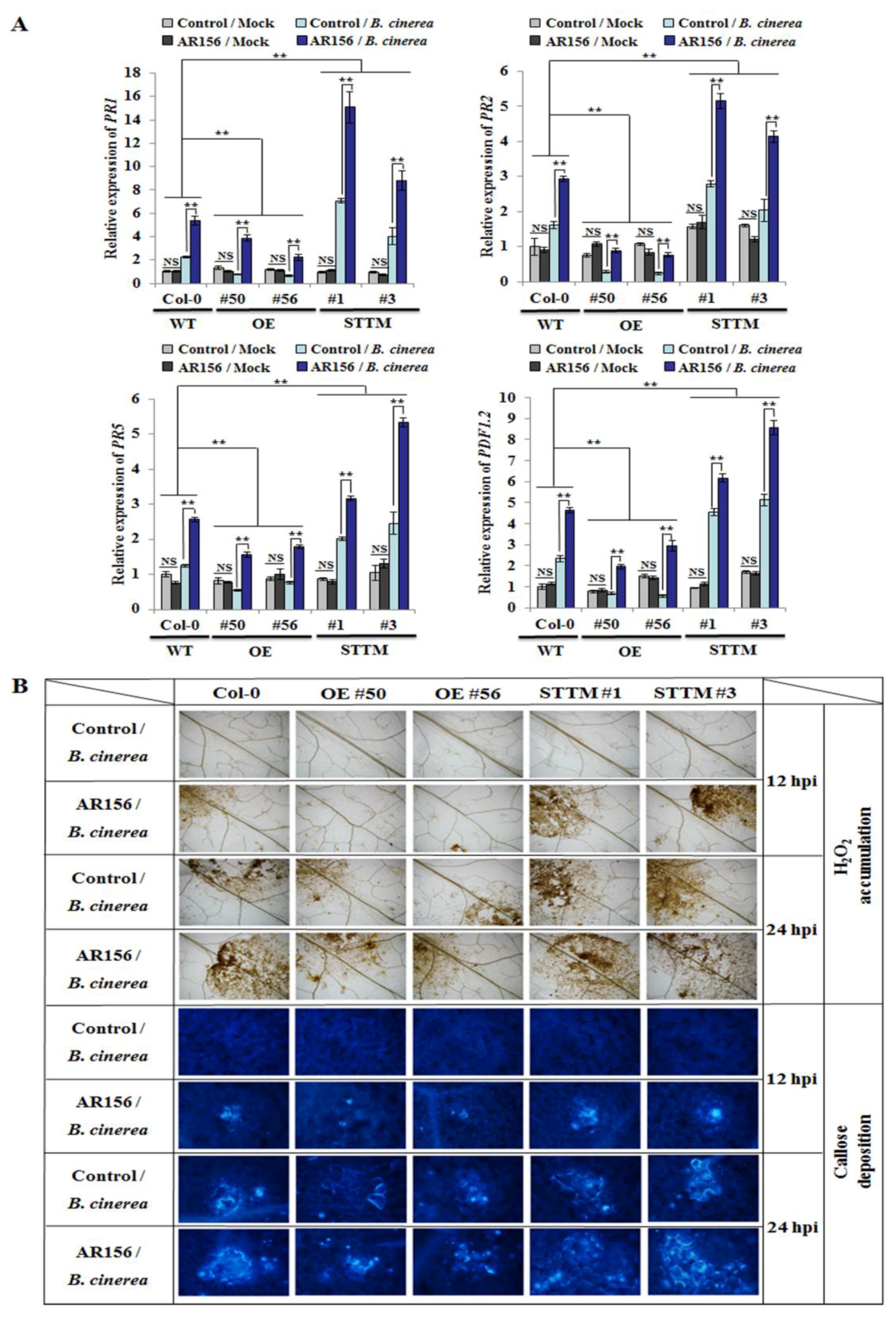
2.3. miR825 and miR825* Affect Defense-related Gene Expression and Cellular Defense Responses in AR156-primed Plants upon Pathogen Attack
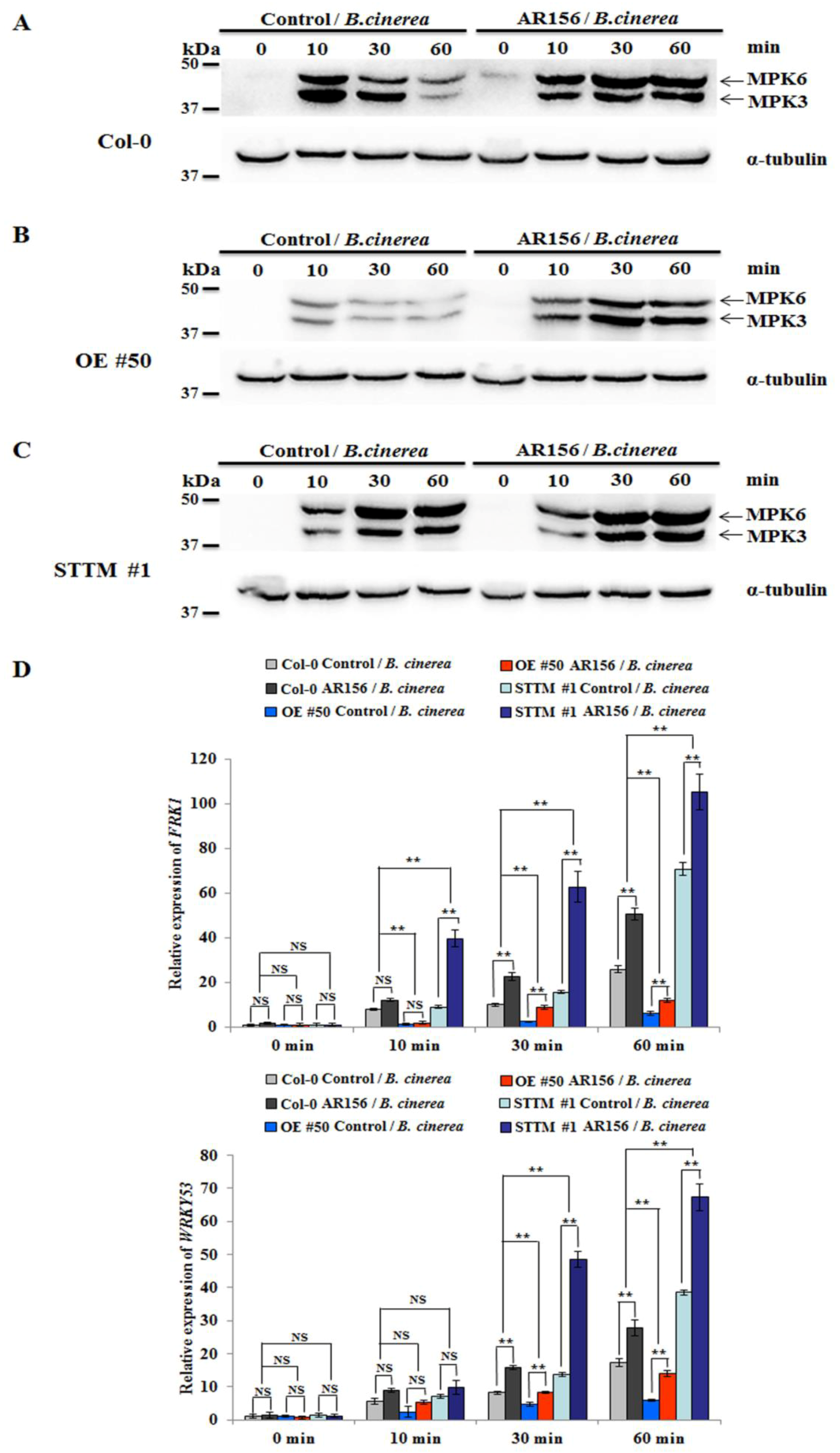
2.4. miR825 and miR825* Participate in Regulation of PTI Components in AR156-elicited ISR
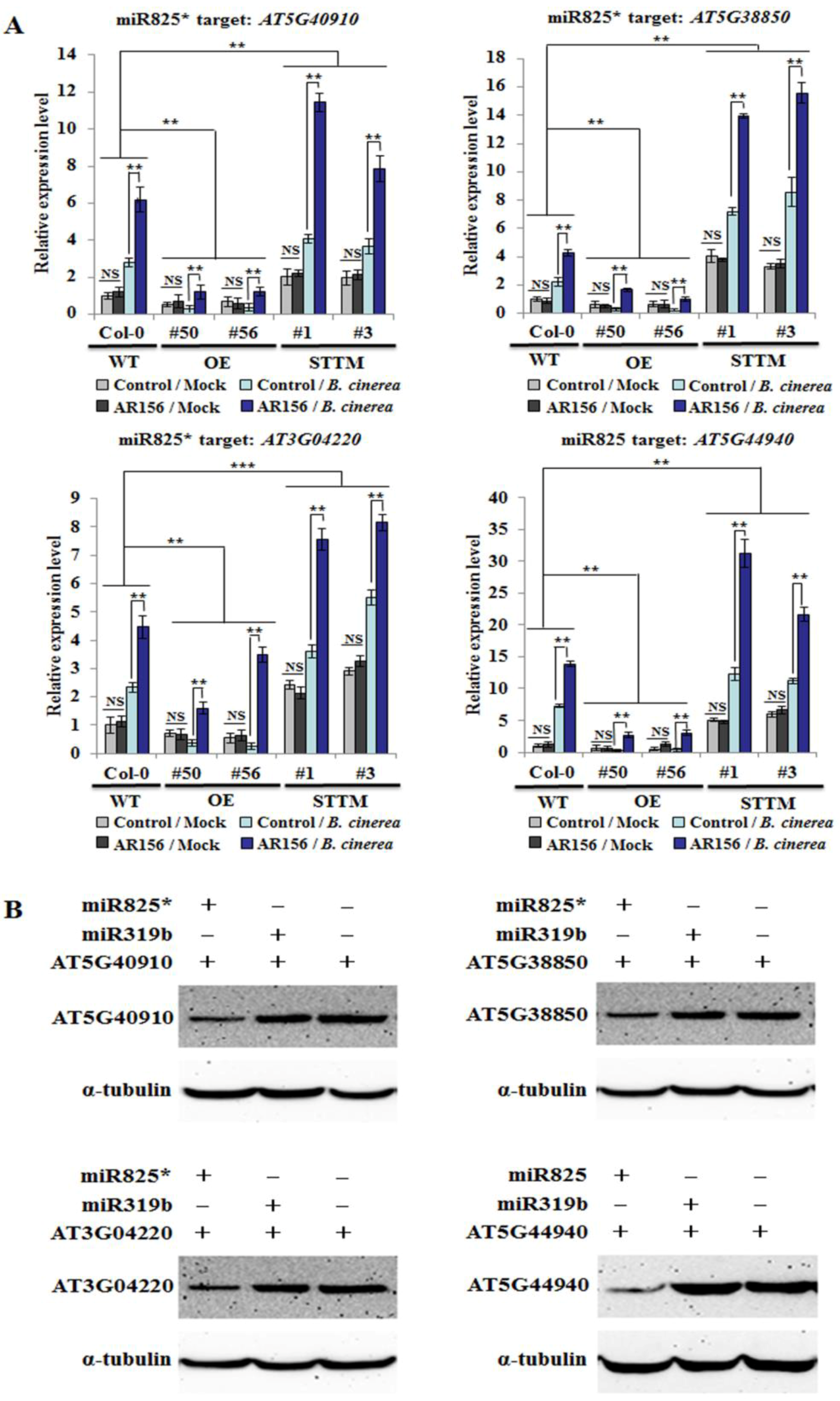
2.5. Target Genes of miR825 and miR825* are Expressed in a Similar Manner in AR156-induced ISR
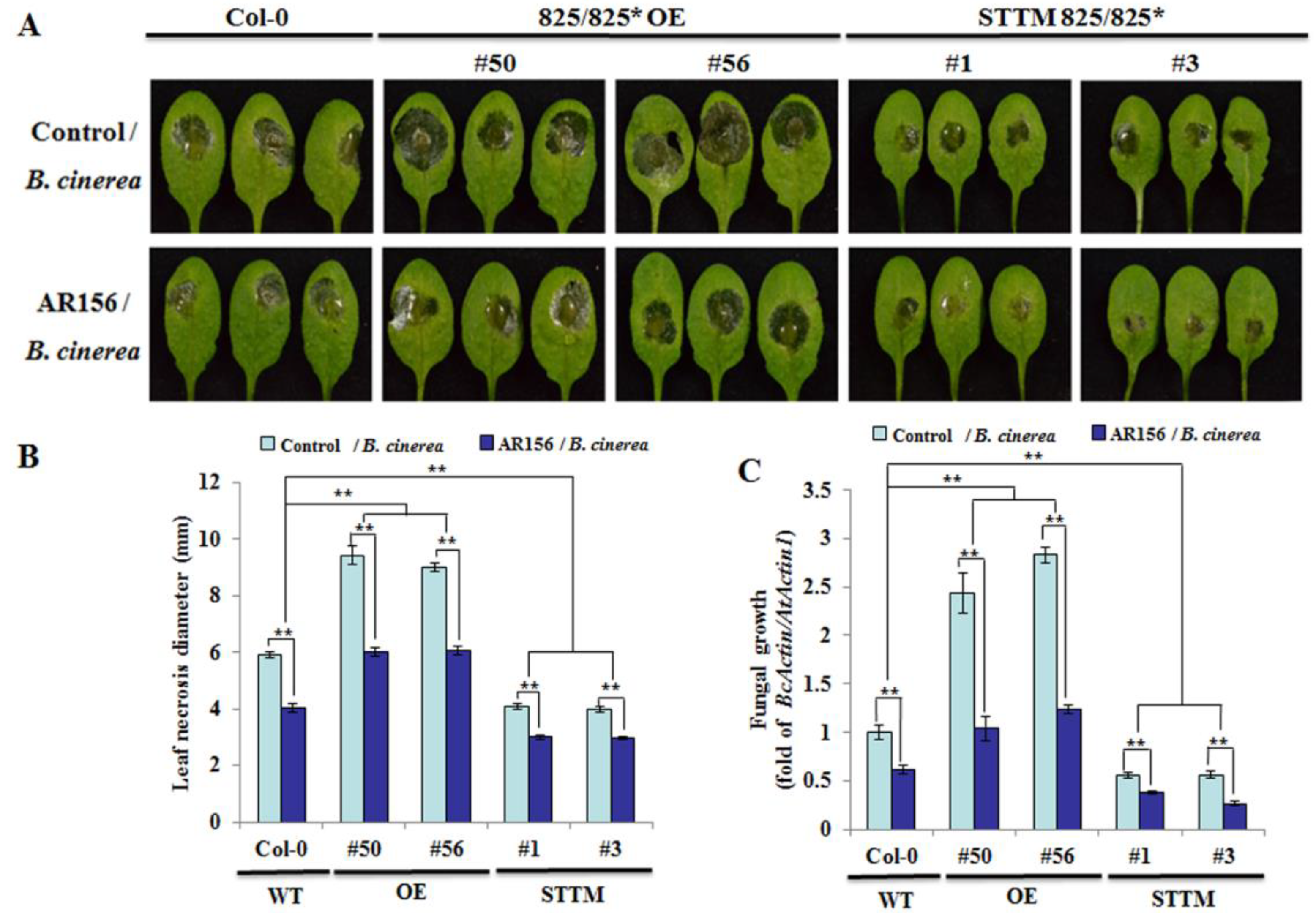
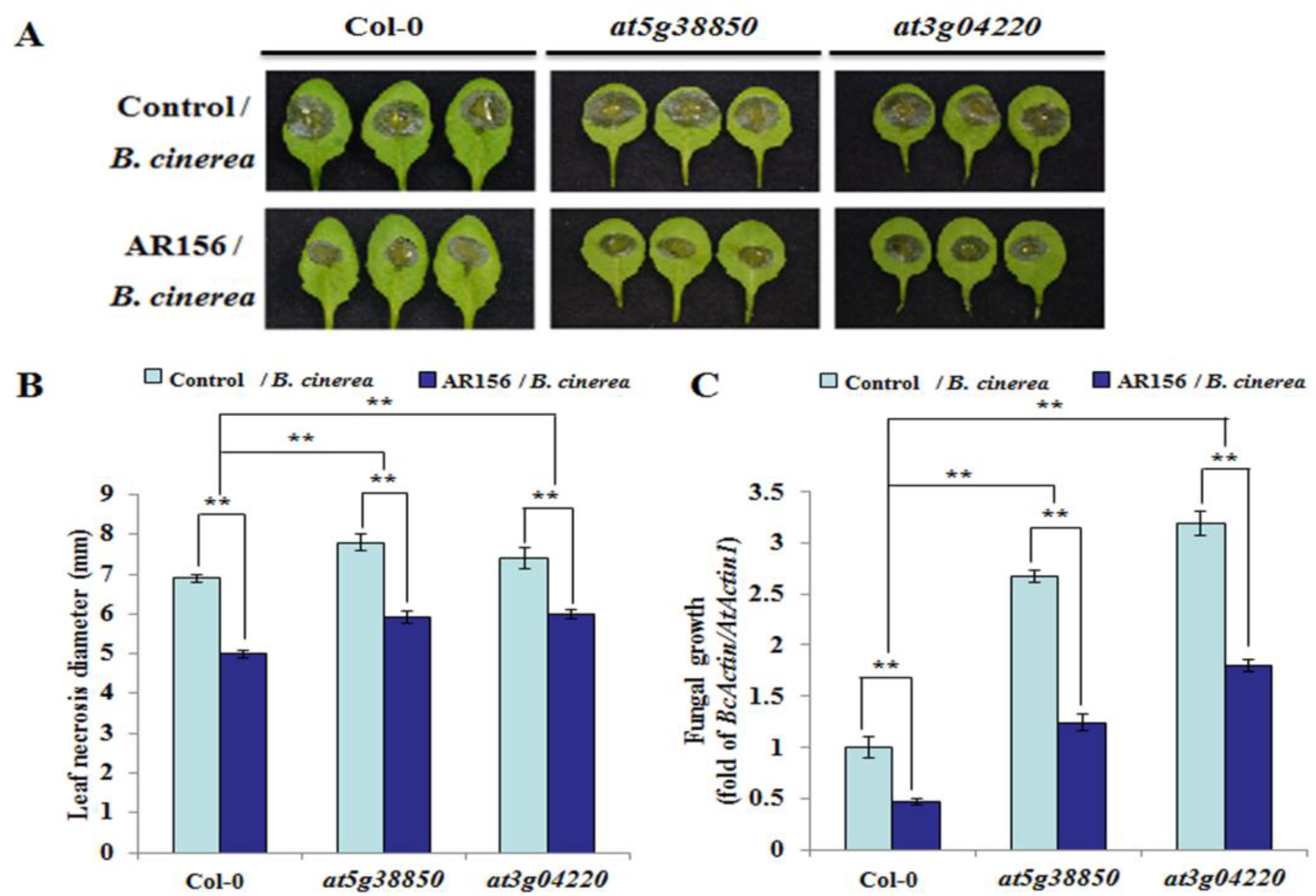
2.6. Plants Silencing miR825/miR825*-targeted Genes are More Susceptible to B. cinerea
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Induction Treatments and Pathogen Infection
4.3. RNA Extraction, Quantitative RT-PCR, and RNA Gel Blot Analysis
4.4. Detection of Hydrogen Peroxide Accumulation and Callose Deposition
4.5. Protein Extraction and Western Blotting Analysis
4.6. Transient Expression Analysis in Nicotiana benthamiana
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| PRRs | pattern recognition receptors |
| PAMPs | pathogen-associated molecular patterns |
| PTI | PAMP-triggered immunity |
| MAPK | mitogen-activated protein kinase |
| ETI | effector-triggered immunity |
| HR | hypersensitive response |
| SAR | systemic acquired resistance |
| ISR | induced systemic resistance |
| PGPR | plant-growth-promoting rhizobacteria |
| SA | salicylic acid |
| JA | jasmonic acid |
| ET | ethylene |
| PR | pathogenesis-related |
| PDF1.2 | plant defensin 1.2 |
| B. cinerea | Botrytis cinerea |
| sRNAs | small RNAs |
| siRNAs | small interfering RNAs |
| miRNAs | microRNAs |
| AGO | argonaute |
| TuMV | Turnip mosaic virus |
| TMV | tobacco mosaic virus |
| P. capsici | Phytophthora capsici |
| CMV | cucumber mosaic virus |
| RPS5 | resistance Pseudomonas syringae 5 |
| CC | coiled-coil |
| Pst | Pseudomonas syringae pv. tomato |
| OE | overexpressing |
| STTM | short tandem target mimic |
| dpt | days post-treatment |
| dpi | days post-inoculation |
| hpi | hours post-inoculation |
| mpi | min post-inoculation |
| qRT-PCR | quantitative real time PCR |
| FRK1 | flg22-induced receptor-like kinase 1 |
| MAPKs | mitogen-activated protein kinases |
| TIR1 | transport inhibitor response 1 |
| Aux/IAA | auxin/indole-3-acetic acid |
References
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ichimura, K.; Shinozaki, K.; Tena, G.; Sheen, J.; Henry, Y.; Champion, A.; Kreis, M.; Zhang, S.Q.; Hirt, H.; Wilson, C.; et al. Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 2002, 7, 301–308. [Google Scholar]
- Altenbach, D.; Robatzek, S. Pattern recognition receptors: From the cell surface to intracellular dynamics. Mol. Plant-Microbe Interact. 2007, 20, 1031–1039. [Google Scholar] [CrossRef] [PubMed]
- Schwessinger, B.; Zipfel, C. News from the frontline: Recent insights into PAMP-triggered immunity in plants. Curr. Opin. Plant Biol. 2008, 11, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, C.; Zhang, X.M.; Jin, H.L. Host small RNAs are big contributors to plant innate immunity. Curr. Opin. Plant Biol. 2009, 12, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Weiberg, A.; Wan, M.; Lin, F.M.; Zhao, H.W.; Zhang, Z.H.; Kaloshian, I.; Huang, H.D.; Jin, H.L. Fungal small RNAs suppress plant immunity by Hijacking host RNA interference pathways. Science 2013, 342, 118–123. [Google Scholar] [CrossRef]
- Katiyar, A.S.; Jin, H.L. Role of small RNAs in host-microbe interactions. Annu. Rev. Phytopathol. 2010, 48, 225–246. [Google Scholar] [CrossRef]
- Goodman, T.P.; Behn, R.; Camenen, Y.; Coda, S.; Fable, E.; Henderson, M.A.; Nikkola, P.; Rosse, l.J.; Sauter, O.; Scarabosio, A.; et al. Safety factor profile requirements for electron ITB formation in TCV. Plasma Phys. Control. Fusion 2005, 47, B107–B120. [Google Scholar] [CrossRef]
- Niu, D.D.; Liu, H.X.; Jiang, C.H.; Wang, Y.P.; Wang, Q.Y.; Jin, H.L.; Guo, J.H. The plant growth-promoting rhizobacterium Bacillus cereus AR156 induces systemic resistance in Arabidopsis thaliana by simultaneously activating salicylate- and jasmonate/ethylene-dependent signaling pathways. Mol. Plant.-Microbe Interact. 2011, 24, 533–542. [Google Scholar] [CrossRef]
- Walters, D.R.; Ratsep, J.; Havis, N.D. controlling crop diseases using induced resistance: Challenges for the future. J. Exp. Bot. 2013, 64, 1263–1280. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.; Bakke, P.A. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef] [PubMed]
- Mishra, M.K.; Chaturvedi, P.; Singh, R.; Singh, G.; Sharma, L.K.; Pandey, V.; Kumari, N.; Misra, P. Overexpression of WsSGTL1 gene of Withania somnifera enhances salt tolerance, heat tolerance and cold acclimation ability in transgenic Arabidopsis plants. PLoS ONE 2013, 8, e63064. [Google Scholar] [CrossRef] [PubMed]
- Conrath, U.; Pieterse, C.M.; Mauch, M.B. Priming in plant-pathogen interactions. Trends Plant Sci. 2002, 7, 210–216. [Google Scholar] [CrossRef]
- Durrant, W.E.; Dong, X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, D.K.; Johri, B.N. Interactions of Bacillus spp. and plants with special reference to induced systemic resistance (ISR). Microbiol. Res. 2009, 164, 493–513. [Google Scholar] [CrossRef] [PubMed]
- Van der Ent, S.; Verhagen, B.W.; Van Doorn, R.; Bakker, D.; Verlaan, M.G.; Pel, M.J.; Joosten, R.G.; Proveniers, M.C.; Van Loon, L.C.; Ton, J.; et al. MYB72 is required in early signaling steps of rhizobacteria-induced systemic resistance in Arabidopsis. Plant. Physiol. 2008, 146, 1293–1304. [Google Scholar] [CrossRef]
- Van Loon, L.C.; Bakker, P.A.; Pieters, C.M. Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 1998, 36, 453–483. [Google Scholar] [CrossRef] [PubMed]
- Heil, M.; Bostock, R.M. Induced systemic resistance (ISR) against pathogens in the context of induced plant defences. Ann. Bot. 2002, 89, 503–512. [Google Scholar] [CrossRef]
- Ahn, I.P.; Lee, S.W.; Suh, S.C. Rhizobacteria-induced priming in Arabidopsis is dependent on ethylene, jasmonic acid, and NPR1. Mol. Plant-Microbe Interact. 2007, 20, 759–768. [Google Scholar] [CrossRef]
- Rahman, A.; Uddin, W.; Wenner, N.G. Induced systemic resistance responses in perennial ryegrass against Magnaporthe oryzae elicited by semi-purified surfactin lipopeptides and live cells of Bacillus amyloliquefaciens. Mol. Plant. Pathol. 2014, 16, 546–558. [Google Scholar] [CrossRef] [PubMed]
- Yedidia, I.; Shoresh, M.; Kerem, Z.; Benhamou, N.; Kapulnik, Y.; Chet, I. Concomitant induction of systemic resistance to Pseudomonas syringae pv. lachrymans in cucumber by Trichoderma asperellum (T-203) and accumulation of phytoalexins. Appl. Env. Microbiol. 2003, 69, 7343–7353. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J. Genes controlling expression of defense responses in Arabidopsis - 2001 status. Curr. Opin. Plant. Biol. 2001, 4, 301–308. [Google Scholar] [CrossRef]
- Ward, E.R.; Uknes, S.J.; Williams, S.C.; Dincher, S.S.; Wiederhold, D.L.; Alexande, D.C.; Ahlgoy, P.; Metraux, J.P.; Ryals, J.A. Coordinate gene activity in response to agents that Induce systemic acquired-resistance. Plant Cell 1991, 3, 1085–1094. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, L.C. Induced resistance in plants and the role of pathogenesis-related proteins. Eur. J. Plant Pathol. 1997, 103, 753–765. [Google Scholar] [CrossRef]
- Van Oosten, V.R.; Bodenhausen, N.; Reymond, P.; Van Pelt, J.A.; Van Loon, L.C.; Dicke, M.; Pieterse, C.M. Differential effectiveness of microbially induced resistance against herbivorous insects in Arabidopsis. Mol. Plant-Microbe Interact. 2008, 21, 919–930. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.; Van Wees, S.C.; Ton, J.; Van Pelt, J.A.; Van Loon, L.C. Signalling in rhizobacteria-induced systemic resistance in Arabidopsis thaliana. Plant Biol. 2002, 4, 535–544. [Google Scholar] [CrossRef]
- Nie, P.P.; Li, X.; Wang, S.E.; Guo, J.H.; Zhao, H.W.; Niu, D.D. Induced systemic resistance against Botrytis cinerea by Bacillus cereus AR156 through a JA/ET- and NPR1-dependent signaling pathway and activates PAMP-triggered immunity in Arabidopsis. Front. Plant. Sci. 2017, 8, 238. [Google Scholar] [CrossRef]
- Salas-Marina, M.A.; Silva-Flores, M.A.; Uresti-Rivera, E.E.; Castro-Longoria, E.; Herrera-Estrella, A.; Casas-Flores, S. Colonization of Arabidopsis roots by Trichoderma atroviride promotes growth and enhances systemic disease resistance through jasmonic acid/ethylene and salicylic acid pathways. Eur. J. Plant. Pathol. 2011, 131, 15–26. [Google Scholar] [CrossRef]
- Jin, H.L. Endogenous small RNAs and antibacterial immunity in plants. FEBS Lett. 2008, 582, 2679–2684. [Google Scholar] [CrossRef] [Green Version]
- Pumplin, N.; Voinnet, O. RNA silencing suppression by plant pathogens: Defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 2013, 11, 745–760. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.K.; Wu, J.G.; Lii, Y.F.; Li, Y.; Jin, H.L. Contribution of small RNA pathway components in pant immunity. Mol. Plant-Microbe Interact. 2013, 26, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Niu, D.D.; Lii, Y.E.; Chellappan, P.; Lei, L.; Peralta, K.; Jiang, C.H.; Guo, J.H.; Coaker, G.; Jin, H.L. miRNA863-3p sequentially targets negative immune regulator ARLPKs and positive regulator SERRATE upon bacterial infection. Nat. Commun. 2016, 7, 11324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navarro, L.; Dunoyer, P.; Jay, F.; Arnold, B.; Dharmasiri, N.; Estelle, M.; Voinnet, O.; Jones, J.D. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 2006, 312, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Q.Q.; Zhang, J.G.; Wu, L.; Qi, Y.J.; Zhou, J.M. Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant. Physiol. 2010, 152, 2222–2231. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, H.; Gao, S.; Wang, W.C.; Katiyar Agarwal, S.; Huang, H.D.; Raikhel, N.; Jin, H. Arabidopsis Argonaute 2 regulates innate immunity via miRNA393(*)-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol. Cell 2011, 42, 356–366. [Google Scholar] [CrossRef] [PubMed]
- Zhai, J.X.; Jeong, D.H.; De Paoli, E.; Park, S.; Rose, B.D.; Li, Y.P.; Gonzalez, A.J.; Yan, Z.; Kitto, S.L.; Grusak, M.A.; et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011, 25, 2540–2553. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Pignatta, D.; Bendix, C.; Brunkard, J.O.; Cohn, M.M.; Tung, J.; Sun, H.Y.; Kumar, P.; Baker, B. MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA 2012, 109, 1790–1795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, X.F.; Fang, Y.Y.; Feng, L.; Guo, H.S. Characterization of conserved and novel microRNAs and their targets, including a TuMV-induced TIR-NBS-LRR class R gene-derived novel miRNA in Brassica. FEBS Lett. 2008, 582, 2445–2452. [Google Scholar] [CrossRef]
- Shivaprasad, P.V.; Chen, H.M.; Patel, K.; Bond, D.M.; Santos, B.A.; Baulcombe, D.C. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 2012, 24, 859–874. [Google Scholar] [CrossRef]
- Boccara, M.; Sarazin, A.; Thiebeauld, O.; Jay, F.; Voinnet, O.; Navarro, L.; Colot, V. The Arabidopsis miR472-RDR6 silencing pathway modulates PAMP- and effector-triggered immunity through the post-transcriptional control of disease resistance genes. PLoS Pathog. 2014, 10, e1004814. [Google Scholar] [CrossRef] [PubMed]
- Niu, D.D.; Xia, J.; Jiang, C.H.; Qi, B.B.; Ling, X.Y.; Lin, S.Y.; Zhang, W.X.; Guo, J.H.; Jin, H.L.; Zhao, H.W. Bacillus cereus AR156 primes induced systemic resistance by suppressing miR825/825*and activating defense-related genes in Arabidopsis. J. Integr. Plant. Biol. 2016, 58, 426–439. [Google Scholar] [CrossRef] [PubMed]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.L.; Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 2002, 415, 977–983. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Kuo, Y.C.; Mishra, S.; Tsa, C.H.; Chien, C.C.; Chen, C.W.; Desclos-Theveniaun, M.; Chu, P.W.; Schulze, B.; Chinchilla, D.; et al. The lectin receptor kinase-VI.2 is required for priming and positively regulates Arabidopsis pattern-triggered immunity. Plant. Cell 2012, 24, 1256–1270. [Google Scholar] [CrossRef] [PubMed]
- Niu, D.D.; Wang, C.J.; Guo, Y.H.; Jiang, C.H.; Zhang, W.Z.; Wang, Y.P.; Guo, J.H. The plant growth-promoting rhizobacterium Bacillus cereus AR156 induces resistance in tomato with induction and priming of defence response. Biocontrol Sci. Technol. 2012, 22, 991–1004. [Google Scholar] [CrossRef]
- Ruiz-Ferrer, V.; Voinnet, O. Roles of plant small RNAs in biotic stress responses. Annu. Rev. Plant Biol. 2009, 60, 485–510. [Google Scholar] [CrossRef]
- Su, Z.Z.; Mao, L.J.; Li, N.; Feng, X.X.; Yuan, Z.L.; Wang, L.W.; Lin, F.C.; Zhang, C.L. Evidence for biotrophic lifestyle and biocontrol potential of dark septate endophyte Harpophora oryzae to Rice Blast Disease. PLoS ONE 2013, 8, e61332. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, M.; Chun, S.C. Expression of PR-protein genes and induction of defense-related enzymes by Bacillus subtilis CBR05 in tomato (Solanum lycopersicum) plants challenged with Erwinia carotovora subsp carotovora. Biosci. Biotechnol. Biochem. 2016, 80, 2277–2283. [Google Scholar] [CrossRef] [PubMed]
- Mengiste, T. Plant immunity to necrotrophs. Annu. Rev. Phytopathol. 2012, 50, 267–294. [Google Scholar] [CrossRef]
- Schwessinger, B.; Ronald, P.C. Plant innate immunity: Perception of conserved microbial signatures. Annu. Rev. Plant. Biol. 2012, 63, 451–482. [Google Scholar] [CrossRef]
- Eckardt, N.A. BIK1 function in plant growth and defense signaling. Plant Cell 2011, 23, 2806. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.Z.; Zhang, S.Q. MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 2013, 51, 245–266. [Google Scholar] [CrossRef] [PubMed]
- Schweighofer, A.; Meskiene, I. Regulation of stress hormones jasmonates and ethylene by MAPK pathways in plants. Mol. Biosyst. 2008, 4, 799–803. [Google Scholar] [CrossRef] [PubMed]
- Tena, G.; Boudsocq, M.; Sheen, J. Protein kinase signaling networks in plant innate immunity. Curr. Opin. Plant Biol. 2011, 14, 519–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, D.T.; Liu, Y.D.; Yang, K.Y.; Han, L.; Mao, G.H.; Glazebrook, J.; Zhang, S.Q. A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2008, 105, 5638–5643. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Li, G.J.; Yang, K.Y.; Mao, G.H.; Wang, R.Q.; Liu, Y.D.; Zhang, S.Q. Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. Plant J. 2010, 64, 114–127. [Google Scholar] [CrossRef] [PubMed]
- Mendez, B.A.; Calderon, V.C.; Ibarra, L.E.; Raya, G.J.; Ramirez, C.E.; Molina, T.J.; Guevara, G.A.; Lopez, B.J.; Herrera, E.L. Alkamides activate jasmonic acid biosynthesis and signaling pathways and confer resistance to Botrytis cinerea in Arabidopsis thaliana. PLoS ONE 2011, 6, e27251. [Google Scholar]
- Yang, X.Y.; Wang, L.C.; Yuan, D.J.; Lindsey, K.; Zhang, X.L. Small RNA and degradome sequencing reveal complex miRNA regulation during cotton somatic embryogenesis. J. Exp. Bot. 2013, 64, 1521–1536. [Google Scholar] [CrossRef]
- Kundu, A.; Paul, S.; Dey, A.; Pal, A. High throughput sequencing reveals modulation of microRNAs in Vigna mungo upon Mungbean Yellow Mosaic India Virus inoculation highlighting stress regulation. Plant. Sci. 2017, 257, 96–105. [Google Scholar] [CrossRef]
- Ouyang, S.; Park, G.; Atamian, H.S.; Han, C.S.; Stajich, J.E.; Kaloshian, I.; Borkovich, K.A. microRNAs suppress NB domain genes in tomato that confer resistance to Fusarium oxysporum. PLoS Pathog. 2014, 10, e1004464. [Google Scholar] [CrossRef]
- Yang, L.; Mu, X.; Liu, C.; Cai, J.; Shi, K.; Zhu, W.; Yang, Q. Overexpression of potato miR482e enhanced plant sensitivity to Verticillium dahliae infection. J. Intergative Plant. Biol. 2015, 57, 1078–1088. [Google Scholar] [CrossRef]
- Lu, S.; Sun, Y.H.; Shi, R.; Clark, C.; Li, L.; Chiang, V.L. Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 2005, 17, 2186–2203. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Liu, M.; Li, X.; Li, F. microRNA-mediated R gene regulation: Molecular scabbards for double-edged swords. Sci. China Life Sci. 2018, 61, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Chen, L.L.; Tao, K.; Sun, N.N.; Wu, Y.R.; Lu, X.X.; Wang, Y.C.; Dou, D.L. Intracelluar and extracelluar phosphatidylinositol 3-phosphate produced by Phytophthora species is important for infection. Mol. Plant 2013, 6, 1592–1604. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Zhu, J.H.; Chen, T.; Zhang, L. MicroRNA profiling identifies miR-146a and miR-29a and their target genes CD40L and LPL in the regulation of oxLDL-treated dendritic cell maturation, inflammatory response and lipid uptake. Eur. Heart J. 2011, 32, 27–28. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nie, P.; Chen, C.; Yin, Q.; Jiang, C.; Guo, J.; Zhao, H.; Niu, D. Function of miR825 and miR825* as Negative Regulators in Bacillus cereus AR156-elicited Systemic Resistance to Botrytis cinerea in Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 5032. https://doi.org/10.3390/ijms20205032
Nie P, Chen C, Yin Q, Jiang C, Guo J, Zhao H, Niu D. Function of miR825 and miR825* as Negative Regulators in Bacillus cereus AR156-elicited Systemic Resistance to Botrytis cinerea in Arabidopsis thaliana. International Journal of Molecular Sciences. 2019; 20(20):5032. https://doi.org/10.3390/ijms20205032
Chicago/Turabian StyleNie, Pingping, Chen Chen, Qian Yin, Chunhao Jiang, Jianhua Guo, Hongwei Zhao, and Dongdong Niu. 2019. "Function of miR825 and miR825* as Negative Regulators in Bacillus cereus AR156-elicited Systemic Resistance to Botrytis cinerea in Arabidopsis thaliana" International Journal of Molecular Sciences 20, no. 20: 5032. https://doi.org/10.3390/ijms20205032
APA StyleNie, P., Chen, C., Yin, Q., Jiang, C., Guo, J., Zhao, H., & Niu, D. (2019). Function of miR825 and miR825* as Negative Regulators in Bacillus cereus AR156-elicited Systemic Resistance to Botrytis cinerea in Arabidopsis thaliana. International Journal of Molecular Sciences, 20(20), 5032. https://doi.org/10.3390/ijms20205032




