ORESARA15 Acts Synergistically with ANGUSTIFOLIA3 and Separately from AINTEGUMENTA to Promote Cell Proliferation during Leaf Growth
Abstract
1. Introduction
2. Results
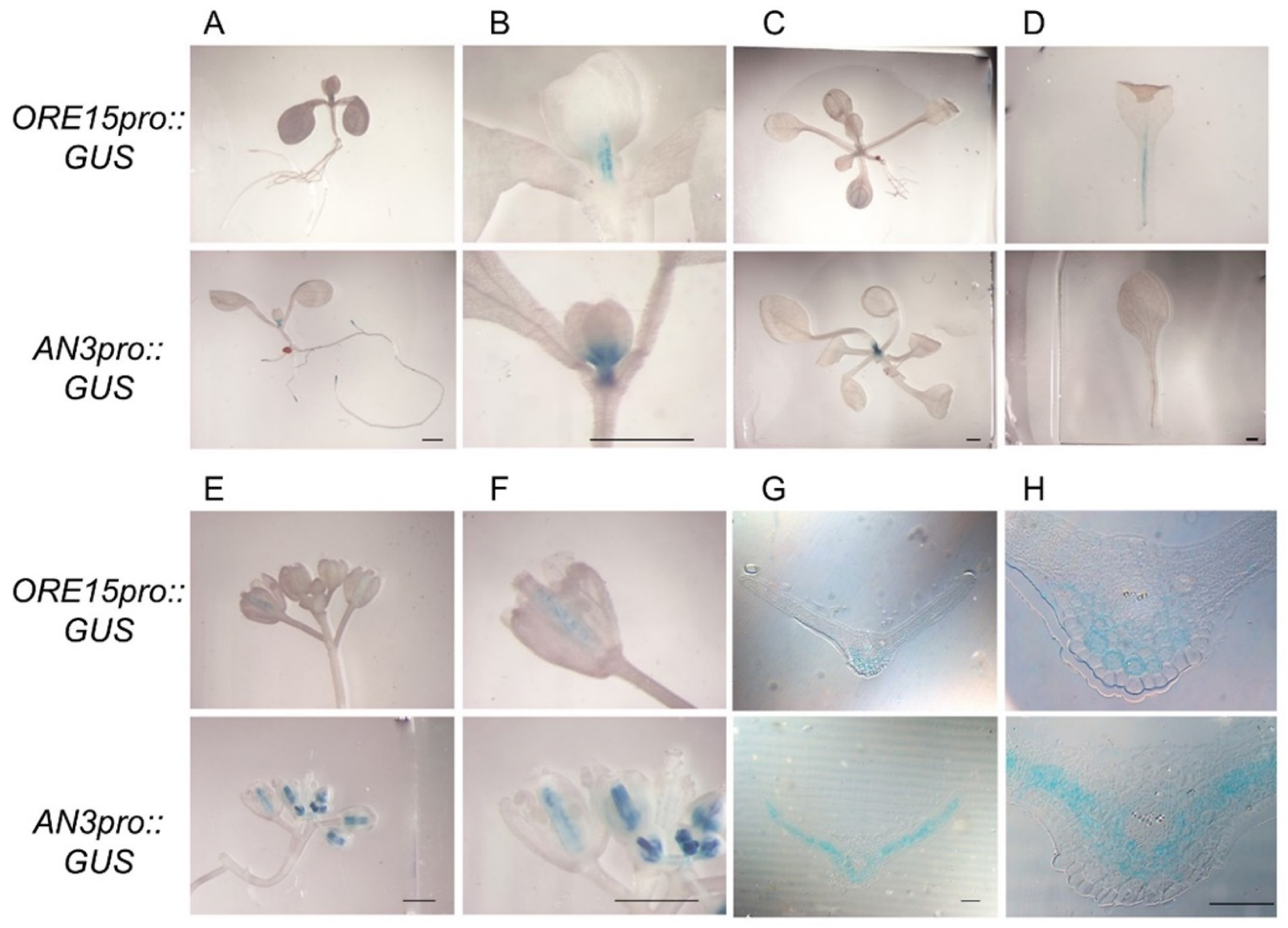
2.1. ORE15 Is Expressed in the Proximal Region of the Leaf Blade and in Petioles of Young Leaves
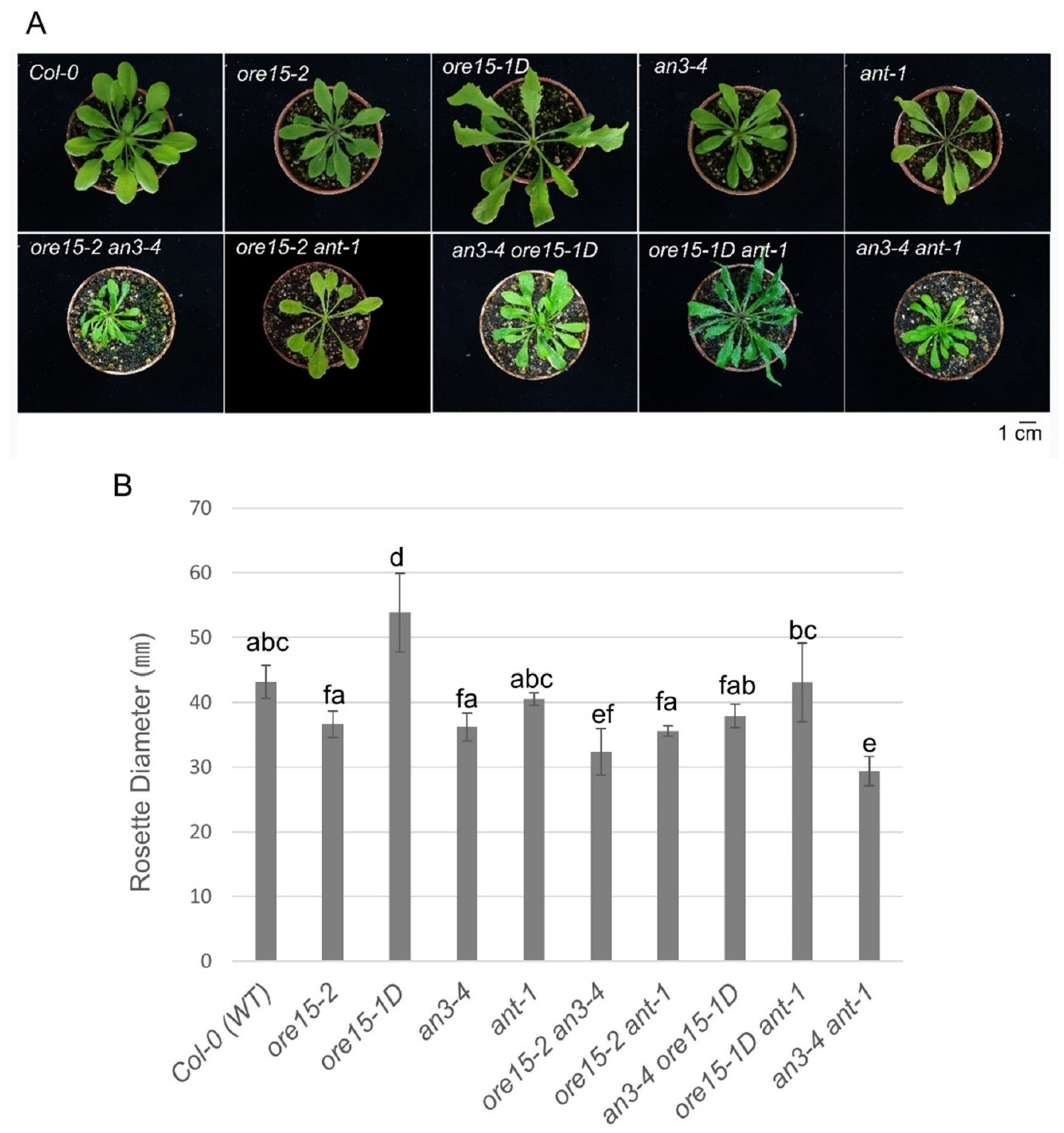
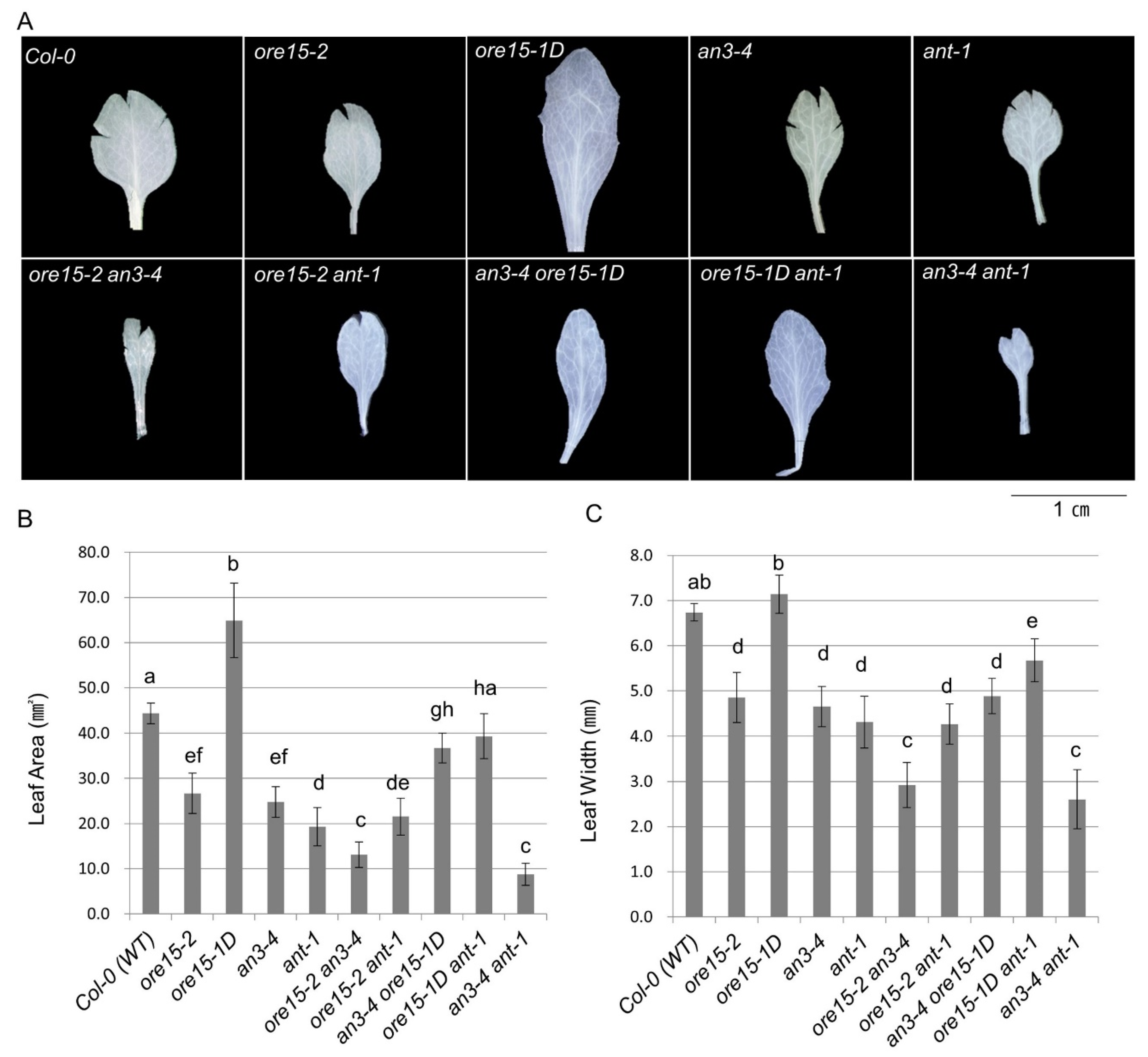
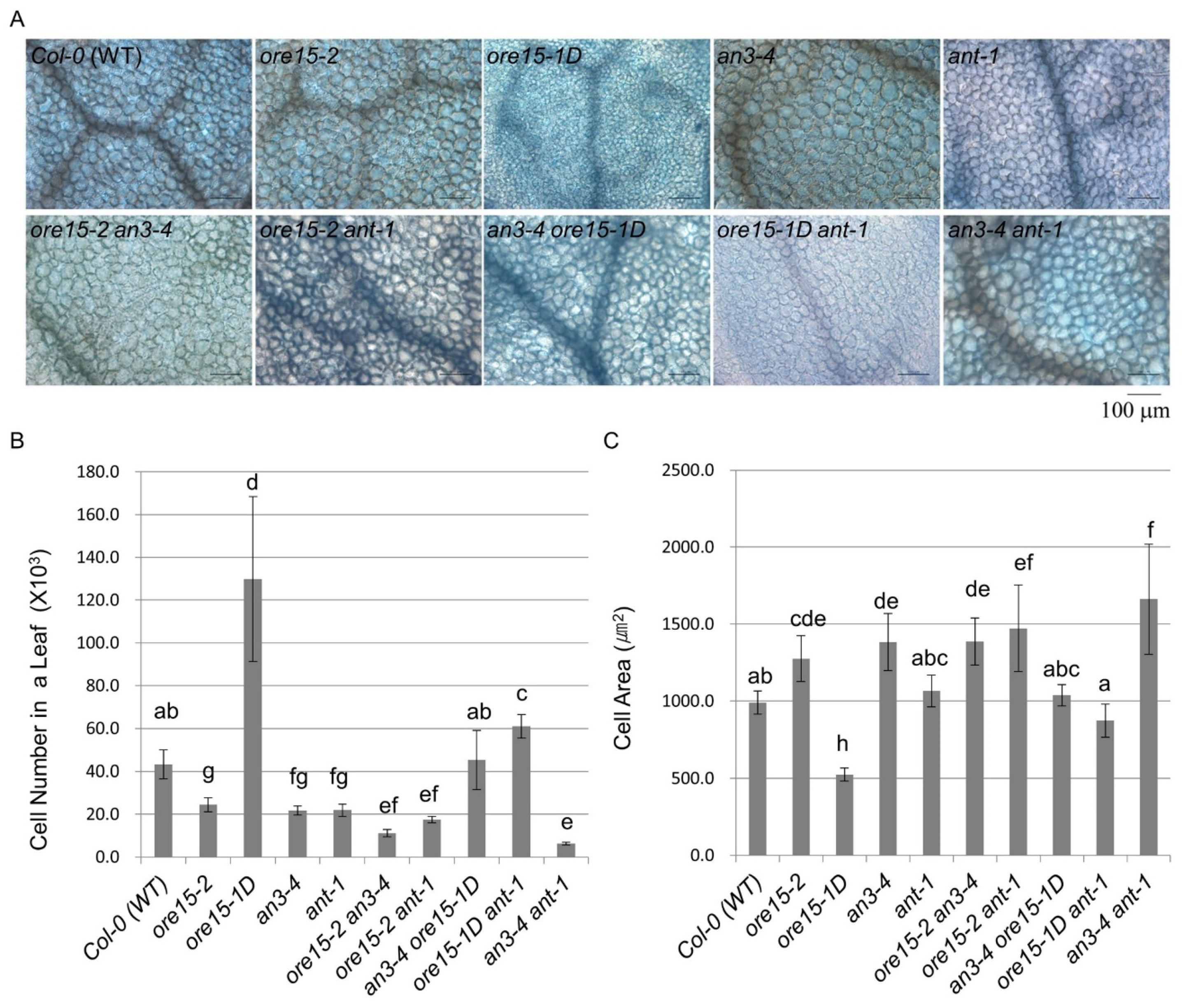
2.2. ORE15 Acts Synergically with AN3 and Separately from ANT to Promote Leaf Growth
2.3. ORE15 Regulates Cell Proliferation during Leaf Growth Independently to ANT
2.4. AN3 Genetically Interacts with ANT to Promote Cell Proliferation during Leaf Growth
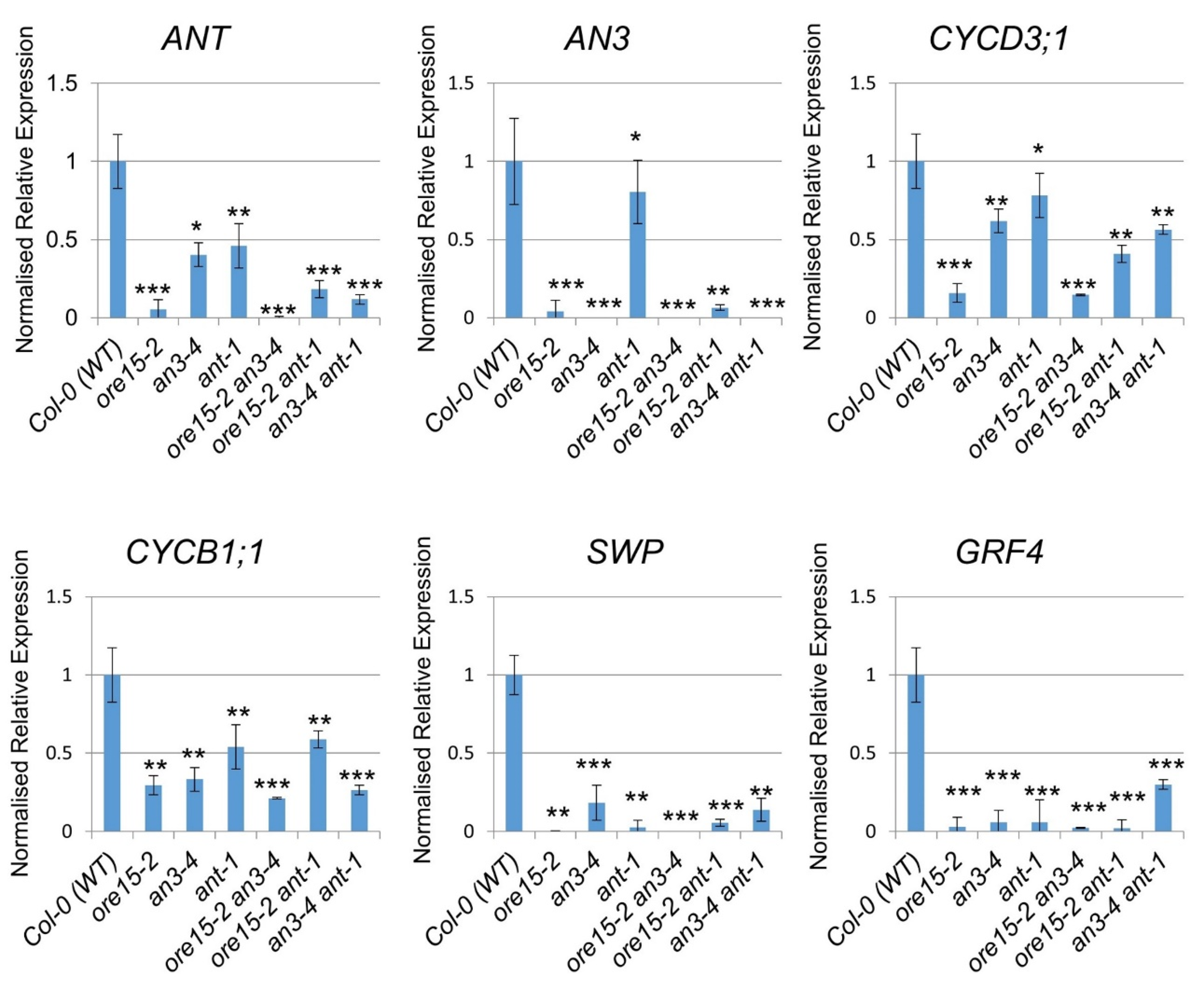
2.5. ORE15 Influences Cell Proliferation by Affecting Genes That Regulate Cell Division
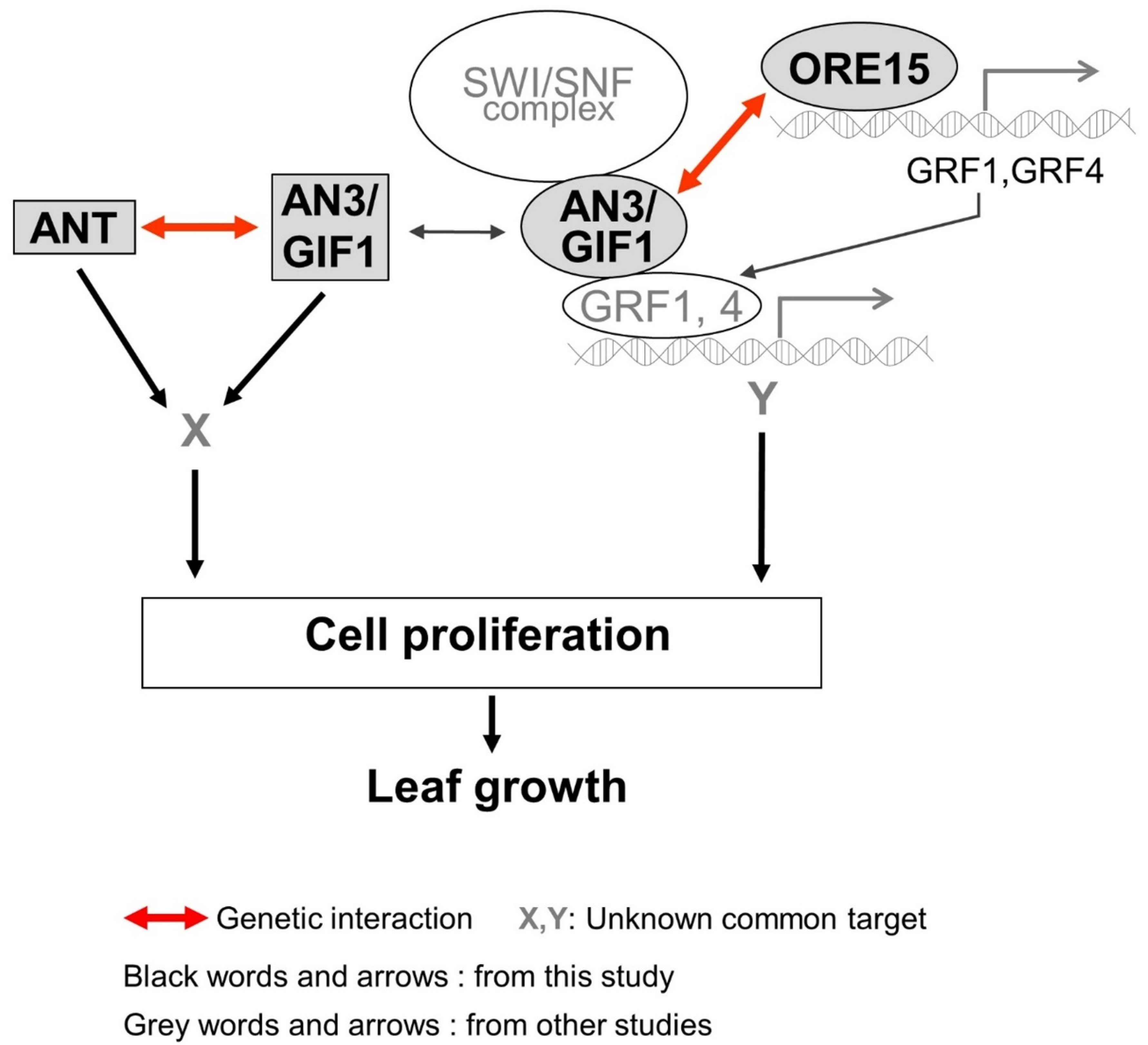
3. Discussion
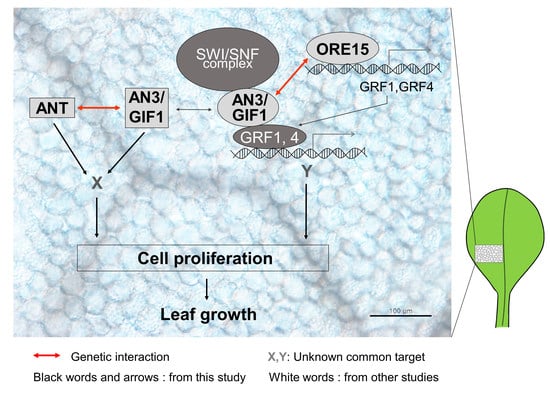
3.1. ORE15 May Act to Maintain AN3 and ANT Expression in the Later Stage of Cell Proliferation during Leaf Growth
3.2. Functional Redendancy between ORE15 and AN3, and between AN3 and ANT in Cell Proliferation Regulatory Pathways during Leaf Growth
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Histochemical Staining for GUS Activity and Anatomy of Leaves
4.3. RNA Isolation and Analysis of Gene Expression
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Col-0 | Columbia-0 |
| ORE15 | ORESARA15 |
| AN3 | ANGUSTIFOLIA3 |
| ANT | AINTEGUMENTA |
| GIF | GRFINTERACTING FACTOR |
| GRF | GROWTH-REGULATING FACTOR |
| TCP | TEOSINTE BRANCHED 1, CYCLOIDEA, and PROLIFERATING CELL FACTORS 1 and 2 |
| KRP1 | Kip-related protein 1 |
| WT | Wild type |
| LOF | Loss of function |
| GOF | Gain of function |
| SWP | STRUWWELPETER |
References
- Breuninger, H.; Lenhard, M. Control of Tissue and Organ Growth in Plants. In Current Topics in Developmental Biology; Timmermans, M., Ed.; Elsevier: Amsterdam, The Netherlands, 2010; Volume 91, pp. 185–220. [Google Scholar] [CrossRef]
- Sablowski, R.; Carnier Dornelas, M. Interplay between Cell Growth and Cell Cycle in Plants. J. Exp. Bot. 2014, 65, 2703–2714. [Google Scholar] [CrossRef] [PubMed]
- Schnittger, A.; Schobinger, U.; Bouyer, D.; Weinl, C.; Stierhof, Y.D.; Hulskamp, M. Ectopic D-Type Cyclin Expression Induces Not Only DNA Replication but Also Cell Division in Arabidopsis Trichomes. Proc. Natl. Acad. Sci. USA 2002, 99, 6410–6415. [Google Scholar] [CrossRef] [PubMed]
- Dewitte, W.; Riou-Khamlichi, C.; Scofield, S.; Healy, J.M.S.; Jacqmard, A.; Kilby, N.J.; Murray, J.A.H. Altered Cell Cycle Distribution, Hyperplasia, and Inhibited Differentiation in Arabidopsis Caused by the D-Type Cyclin CYCD3. Plant Cell 2003, 15, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Mizukami, Y.; Fischer, R.L. Plant Organ Size Control: AINTEGUMENTA Regulates Growth and Cell Numbers during Organogenesis. Proc. Natl. Acad. Sci. USA 2000, 97, 942–947. [Google Scholar] [CrossRef] [PubMed]
- Dewitte, W.; Scofield, S.; Alcasabas, A.A.; Maughan, S.C.; Menges, M.; Braun, N.; Collins, C.; Nieuwland, J.; Prinsen, E.; Sundaresan, V.; et al. Arabidopsis CYCD3 D-Type Cyclins Link Cell Proliferation and Endocycles and Are Rate-Limiting for Cytokinin Responses. Proc. Natl. Acad. Sci. USA 2007, 104, 14537–14542. [Google Scholar] [CrossRef]
- Hu, Y.; Xie, Q.; Chua, N.H. The Arabidopsis Auxin-Inducible Gene ARGOS Controls Lateral Organ Size. Plant Cell 2003, 15, 1951–1961. [Google Scholar] [CrossRef]
- Kim, J.H.; Kende, H. A Transcriptional Coactivator, AtGIF1, Is Involved in Regulating Leaf Growth and Morphology in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 13374–13379. [Google Scholar] [CrossRef]
- Horiguchi, G.; Kim, G.T.; Tsukaya, H. The Transcription Factor AtGRF5 and the Transcription Coactivator AN3 Regulate Cell Proliferation in Leaf Primordia of Arabidopsis thaliana: Control of Leaf Shape and Size by AtGRF5 and AN3. Plant J. 2005, 43, 68–78. [Google Scholar] [CrossRef]
- Kim, J.H. Biological Roles and an Evolutionary Sketch of the GRF–GIF Transcriptional Complex in Plants. BMB Rep. 2019, 52, 227–238. [Google Scholar] [CrossRef]
- Kim, J.H.; Tsukaya, H. Regulation of Plant Growth and Development by the GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR Duo. J. Exp. Bot. 2015, 66, 6093–6107. [Google Scholar] [CrossRef]
- Debernardi, J.M.; Mecchia, M.A.; Vercruyssen, L.; Smaczniak, C.; Kaufmann, K.; Inze, D.; Rodriguez, R.E.; Palatnik, J.F. Post-Transcriptional Control of GRF Transcription Factors by MicroRNA MiR396 and GIF Co-Activator Affects Leaf Size and Longevity. Plant J. 2014, 79, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Vercruyssen, L.; Tognetti, V.B.; Gonzalez, N.; Van Dingenen, J.; De Milde, L.; Bielach, A.; De Rycke, R.; Van Breusegem, F.; Inzé, D. GROWTH REGULATING FACTOR5 Stimulates Arabidopsis Chloroplast Division, Photosynthesis, and Leaf Longevity. Plant Physiol. 2015, 167, 817–832. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.H.; Ko, J.H.; Lee, S.; Lee, Y.; Pak, J.H.; Kim, J.H. The Arabidopsis GRF-INTERACTING FACTOR Gene Family Performs an Overlapping Function in Determining Organ Size as Well as Multiple Developmental Properties. Plant Physiol. 2009, 151, 655–668. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.H.; Kim, J.H. Spatio-Temporal Distribution Patterns of GRF-INTERACTING FACTOR Expression and Leaf Size Control. Plant Signal. Behav. 2014, 9, e29697. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.H.; Wynn, A.N.; Franks, R.G.; Hwang, Y.; Lim, J.; Kim, J.H. The Arabidopsis thaliana GRF-INTERACTING FACTOR Gene Family Plays an Essential Role in Control of Male and Female Reproductive Development. Dev. Biol. 2014, 386, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Kawade, K.; Horiguchi, G.; Usami, T.; Hirai, M.Y.; Tsukaya, H. ANGUSTIFOLIA3 Signaling Coordinates Proliferation between Clonally Distinct Cells in Leaves. Curr. Biol. 2013, 23, 788–792. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, H.; Kalve, S.; Wolabu, T.W.; Nakashima, J.; Golz, J.F.; Tadege, M. Control of Leaf Blade Outgrowth and Floral Organ Development by LEUNIG, ANGUSTIFOLIA 3 and WOX Transcriptional Regulators. New Phytol. 2019, 223, 2024–2038. [Google Scholar] [CrossRef]
- Vercruyssen, L.; Verkest, A.; Gonzalez, N.; Heyndrickx, K.S.; Eeckhout, D.; Han, S.K.; Jegu, T.; Archacki, R.; Van Leene, J.; Andriankaja, M.; et al. ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complex to regulate transcription during Arabidopsis leaf development. Plant Cell 2014, 26, 210–229. [Google Scholar] [CrossRef]
- Martin-Trillo, M.; Cubas, P. TCP genes: A family snapshot ten years later. Trends Plant Sci. 2010, 15, 31–39. [Google Scholar] [CrossRef]
- Efroni, I.; Blum, E.; Goldshmidt, A.; Eshed, Y. A Protracted and Dynamic Maturation Schedule Underlies Arabidopsis Leaf Development. Plant Cell 2008, 20, 2293–2306. [Google Scholar] [CrossRef]
- Nag, A.; King, S.; Jack, T. MiR319a Targeting of TCP4 Is Critical for Petal Growth and Development in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 22534–22539. [Google Scholar] [CrossRef] [PubMed]
- Schommer, C.; Debernardi, J.M.; Bresso, E.G.; Rodriguez, R.E.; Palatnik, J.F. Repression of Cell Proliferation by MiR319-Regulated TCP4. Mol. Plant 2014, 7, 1533–1544. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.E.; Mecchia, M.A.; Debernardi, J.M.; Schommer, C.; Weigel, D.; Palatnik, J.F. Control of Cell Proliferation in Arabidopsis thaliana by MicroRNA MiR396. Development 2010, 137, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Kim, J.; Jun, S.E.; Park, S.; Timilsina, R.; Kwon, D.S.; Kim, Y.; Park, S.J.; Hwang, J.Y.; Nam, H.G.; et al. ORESARA15, a PLATZ Transcription Factor, Mediates Leaf Growth and Senescence in Arabidopsis. New Phytol. 2018, 220, 609–623. [Google Scholar] [CrossRef]
- Meng, L.S.; Wang, Y.B.; Yao, S.Q.; Liu, A. Arabidopsis AINTEGUMENTA mediates salt tolerance by trans-repressing SCABP8. J. Cell Sci. 2015, 128, 2919–2927. [Google Scholar] [CrossRef]
- Lee, S.J.; Lee, B.H.; Jung, J.H.; Park, S.K.; Song, J.T.; Kim, J.H. GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR Specify Meristematic Cells of Gynoecia and Anthers. Plant Physiol. 2018, 176, 717–729. [Google Scholar] [CrossRef]
- Vanhaeren, H.; Inze, D.; Gonzalez, N. Plant Growth beyond Limits. Trends Plant Sci. 2018, 176, 717–729. [Google Scholar] [CrossRef]
- Tsukaya, H. Leaf Development. In The Arabidopsis Book; American Society of Plant Biologists: Rockville, MD, USA, 2013; Volume 11, p. e0163. [Google Scholar]
- Sugimoto-Shirasu, K.; Roberts, K. “Big it up”: Endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 2003, 6, 544–553. [Google Scholar] [CrossRef]
- De Veylder, L.; Larkin, J.C.; Schnittger, A. Molecular control of function endoreduplication in development and physiology. Trends Plant Sci. 2011, 16, 624–634. [Google Scholar] [CrossRef]
- Hisanaga, T.; Kawade, K.; Tsukaya, H. Compensation: A key to clarifying the organ-level regulation of lateral organ size in plants. J. Exp. Bot. 2015, 66, 1055–1063. [Google Scholar] [CrossRef]
- Autran, D.; Belcram, J.C.; Beemster, G.T.; Kronenberger, J.; Grandjean, O.; Inzé, D.; Traas, J. Cell numbers and leaf development in Arabidopsis: A functional analysis of the STRUWWELPETER gene. EMBO J. 2002, 21, 6036–6049. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Pérez, J.M.; Candela, H.; Micol, J.L. Understanding synergy in genetic interactions. Trends Genet. 2009, 25, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Powell, A.E.; Lenhard, M. Control of Organ Size in Plants. Curr. Biol. 2012, 22, R360–R367. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.; Xu, Q.; Wang, Z.; Zhuoma, Q. AINTEGUMENTA Negatively Regulates Age-Dependent Leaf Senescence Downstream of AUXIN RESPONSE FACTOR 2 in Arabidopsis thaliana. Plant Biotechnol. 2016, 33, 71–76. [Google Scholar] [CrossRef]
- Ichihashi, Y.; Kawade, K.; Usami, T.; Horiguchi, G.; Takahashi, T.; Tsukaya, H. Key Proliferative Activity in the Junction between the Leaf Blade and Leaf Petiole of Arabidopsis. Plant Physiol. 2011, 157, 1151–1162. [Google Scholar] [CrossRef]
- Kang, J.; Mizukami, Y.; Wang, H.; Fowke, L.; Dengler, N.G. Modification of Cell Proliferation Patterns Alters Leaf Vein Architecture in Arabidopsis thaliana. Planta 2007, 226, 1207–1218. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Jun, S.E.; Okushima, Y.; Nam, J.; Umeda, M.; Kim, G.T. Kip-Related Protein 3 Is Required for Control of Endoreduplication in the Shoot Apical Meristem and Leaves of Arabidopsis. Mol. Cells 2013, 35, 47–53. [Google Scholar] [CrossRef]
- Kim, G.T.; Tsukaya, H.; Uchimiya, H. The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev. 1998, 12, 2381–2391. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jun, S.E.; Kim, J.H.; Hwang, J.Y.; Huynh Le, T.T.; Kim, G.-T. ORESARA15 Acts Synergistically with ANGUSTIFOLIA3 and Separately from AINTEGUMENTA to Promote Cell Proliferation during Leaf Growth. Int. J. Mol. Sci. 2020, 21, 241. https://doi.org/10.3390/ijms21010241
Jun SE, Kim JH, Hwang JY, Huynh Le TT, Kim G-T. ORESARA15 Acts Synergistically with ANGUSTIFOLIA3 and Separately from AINTEGUMENTA to Promote Cell Proliferation during Leaf Growth. International Journal of Molecular Sciences. 2020; 21(1):241. https://doi.org/10.3390/ijms21010241
Chicago/Turabian StyleJun, Sang Eun, Jin Hee Kim, Ji Young Hwang, Thien Tu Huynh Le, and Gyung-Tae Kim. 2020. "ORESARA15 Acts Synergistically with ANGUSTIFOLIA3 and Separately from AINTEGUMENTA to Promote Cell Proliferation during Leaf Growth" International Journal of Molecular Sciences 21, no. 1: 241. https://doi.org/10.3390/ijms21010241
APA StyleJun, S. E., Kim, J. H., Hwang, J. Y., Huynh Le, T. T., & Kim, G.-T. (2020). ORESARA15 Acts Synergistically with ANGUSTIFOLIA3 and Separately from AINTEGUMENTA to Promote Cell Proliferation during Leaf Growth. International Journal of Molecular Sciences, 21(1), 241. https://doi.org/10.3390/ijms21010241