Abstract
Background: Although the scientific literature regarding sports genomics has grown during the last decade, some genes, such as peroxisome proliferator activated receptors (PPARs), have not been fully described in terms of their role in achieving extraordinary sports performance. Therefore, the purpose of this systematic review was to determine which elite sports performance constraints are positively influenced by PPARs and their coactivators. Methods: The Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines were used, with a combination of PPAR and sports keywords. Results: In total, 27 studies that referred to PPARs in elite athletes were included, where the Ala allele in PPARG rs1801282 was associated with strength and power elite athlete status in comparison to subelite athlete status. The C allele in PPARA rs4253778 was associated with soccer, and the G allele PPARA rs4253778 was associated with endurance elite athlete status. Other elite status endurance alleles were the Gly allele in PPARGC1A rs8192678 and the C allele PPARD rs2016520. Conclusions: PPARs can be used for estimating the potential to achieve elite status in human physical performance in strength and power, team, and aerobic sports disciplines. Carrying specific PPAR alleles can provide a partial benefit to achieving elite sports status, but does not preclude achieving elite status if they are absent.
1. Introduction
The scientific literature on exercise genomics has shown clear evidence that genetic markers are associated with endurance [1], power athlete status [2,3], trainability [4], and even psychological factors [5], and peroxisome proliferator activated receptors (PPARs) and/or their coactivators are often listed. While both sports performance and genomics are highly multifactorial domains, it is beneficial to summarize what phenotypic domains can be attributed to PPARs (and their coactivators) and where the analysis of phenotypic domains is redundant. Moreover, better knowledge, via functional genomics, of how PPARs (and their coactivators) may affect the individual response to physical activity or environmental factors is highly relevant not only for active individuals (athletes), but also for people who are undergoing a health treatment program that includes a physical intervention [6]. In this context, data from athletes can serve as a basis for hypotheses regarding the effectiveness of physical activity programs under extreme physiological conditions or for sedentary individuals, where the clear objective is to achieve health improvement. A recent review on the role of PPAR polymorphisms in trainability summarized several studies showing genotype/allele specific changes in health related markers [4].
PPARs are a subfamily of nuclear hormone receptors that form heterodimers with retinoid X receptors and regulate the transcription of several genes involved in lipid metabolism, energy utilization, and storage [7]. PPARs also regulate genes for glucose metabolism, carcinogenesis, and inflammation [8,9]. There are three isoforms of PPARs (PPARα, PPARβ/δ, and PPARγ, encoded by the PPARA, PPARD, and PPARG genes, respectively) that differ in their distribution and function [10]. For example, PPARγ is predominantly active in fat cells where it affects differentiation and growth; among other things, it is also an interesting target in pharmacotherapy for diabetes mellitus type 2 (DM2) [11]. An increased level of PPAR expression occurs in tissues that catabolize high amounts of fatty acids, such as the liver, kidney, brown adipose tissue, heart, and skeletal muscle [12,13]. In addition, muscle specific PPARβ/δ overexpression is considered to be a part of skeletal muscle plasticity. Therefore, the role of PPARs in elite aerobic performance is highly suspected [1,14].
The peroxisome proliferator activated receptor γ coactivator 1 (PGC1) family of transcriptional coactivators, consisting of three members, PGC1α, PGC1β, and the PGC-1 related coactivator (PRC), encoded by the PPARGC1A, PPARGC1Β, and PPRC1 genes, respectively, provides important links between these transcription factors and the physiological signals controlling cellular functions related to cellular and mitochondrial energy metabolism [15,16]. PGC1α is the most frequently studied and positively regulates mitochondrial biogenesis and respiration and many other metabolic processes, including adaptive thermogenesis, gluconeogenesis, and insulin signaling [17].
The links between PPARs (and their coactivators) and muscle morphology [18], oxygen uptake [19,20], power output [21], endurance performance [18], and human trainability [4] have already been associated with elite sports status in individual studies and, in the case of PPARGC1A Gly428Ser, by systematic review with meta-analyses [22]. Therefore, there has been an increase of PPAR analyses in the athletic population in recent years, where PPARA, PPARG, PPARD, and their transcriptional coactivators’ PPARGC1A and PPARGC1B gene polymorphisms contribute to the observed phenotypes. For example, it has been shown that prolonged endurance exercise increases the transcriptional activity of PPARGC1A in active subjects [23]. In contrast, a recent systematic review on genes related to the level of endurance performance in mice considered at least three PPAR gene variants (and their coactivators) to be associated with endurance capacity [24]. Previous literature reviews [2,3] focused on all possible genes that might have an association with strength and power athletes’ status and suggested that PPARs have important roles that require detailed analyses. So far, only PPARGC1A has been reviewed in relation to power athlete status [22], and endurance athlete status and other PPARs and their coactivators have not.
Since the scientific literature in sports genomics has grown during the last decade, some genes, such as PPARs and/or their coactivators, have not been adequately described in terms of their role in athlete training and achieving extraordinary sports performance. Therefore, the purpose of this systematic review was to determine which PPARs and their coactivators are positively or negatively associated with elite sports performance constraints. We hypothesize that PPARs and/or their coactivators might determine aerobic performance and team sports elite athlete status, but not speed and strength oriented elite athlete status.
2. Results
The literature search resulted in a total of 4916 articles, after removing duplicates. The number of eligible articles was further reduced to 79 (including 31 reviews) after screening article titles and abstracts according to the inclusion criteria that the articles include PPARs and/or their coactivators’ gene polymorphisms at the elite athlete level (Figure 1). Of these studies, 18 were rejected following the full-text screening, and three were rejected based on the methodological quality criteria. Finally, 27 studies (Figure 1) were included in the analysis.
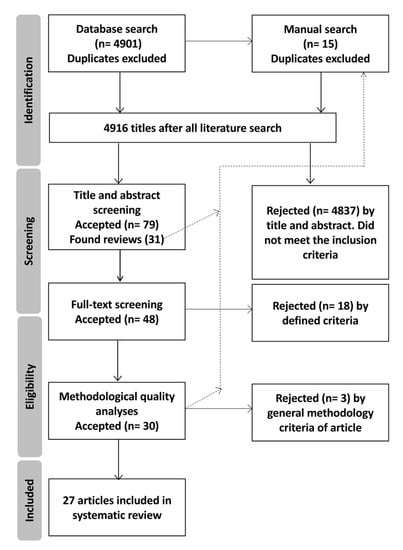
Figure 1.
Flowchart of the review for the articles included in the tables. The dotted line demonstrates the stages where a manual search of the reference lists of the selected articles was performed.
In total, 27 studies were included due to referring to PPARs with elite athlete status, where five PPARs were summarized as the main result of qualitative synthesis (Table 1). Thus, PPARs and their coactivators determined aerobic, speed, strength, and team sports elite athlete status. In total, 11 studies found differences between elite and subelite athletes or among elite athletes from different disciplines (Table 2). The comparison between PPARs in elite athletes and control groups only was reported in 14 studies (Table 3) and supported the main conclusions of this study. One study was a single case study and one without a control group (Table 3). The Ala allele in PPARG rs1801282 and the C allele in PPARA rs4253778 were associated with strength and power elite athlete status in comparison to subelite athletes’ status (Table 1 and Table 2). The G allele PPARA rs4253778, Gly allele in PPARGC1A rs8192678, and the PPARD rs2016520 C allele were associated with endurance elite athlete status in comparison to subelite athlete status (Table 1 and Table 2). The C allele in PPARA rs4253778 was associated with mixed endurance/strength-power (soccer) such as the PPARD A/C/C haplotype in rs2016520, rs2267668, and rs1053049, however only in comparison to control groups (Table 1, Table 2 and Table 3). In contrast, the G allele in PPARA rs4253778 was associated with mixed endurance/strength-power (soccer) with other elite athletes from combat sports and motorcycling.

Table 1.
Alleles and genotypes related to elite athlete status vs. subelite status in different types of disciplines and in comparison to controls. * The minority report results specific for the reported population.

Table 2.
PPAR alleles and genotypes in elite and subelite athletes and their differences among disciplines. TGS, total genetic score.

Table 3.
Results for PPAR alleles and genotypes in elite athletes and controls.
3. Discussion
The main finding of this review was that PPARs and their coactivator gene polymorphisms were related to the ability to achieve elite sports status for endurance, strength, power, and team sports oriented athletes. This consideration was specifically important for the C allele in PPARA rs4253778, the G allele PPARA rs4253778, the Gly allele in PPARGC1A rs8192678, and the C allele PPARD rs2016520, as those alleles have been found in higher frequencies in elite athletes than in subelite athletes (not just controls) and in studies including a large number of PPARs in the optimal genotype score [30,31,33] or haplotype [44]. Other findings (Table 1 and Table 3), where the genotype frequency differed between elite athletes and controls, were questionable; however, they still supported the hypothesis that PPAR alleles could influence extreme physical fitness phenotypes. Such an example was given by three studies devoted to the PPARGC1B gene in which two of them showed no association with athletes’ status [30,50]. However, the study of Ahmetov [18] examined the total genotype score of 15 genetic variants, where the PPARGC1B C allele was shown to be more common in a group of long endurance athletes compared to sedentary controls.
Although this study identified four alleles that were beneficial for elite athletes, missing the allele or the dominance of endurance or power genotypes does not mean that an athlete cannot achieve elite status, e.g., Eynon [51] reported a case study showing that athletes with the ACTN3 R577X heterozygote variation and five out of six “endurance oriented” genotypes (including PPARs) could be successful in a long 10,000 m and short 400 m run; similarly, elite long jumpers without the power associated ACTN3 genotype X577X have been reported [52]. On the other hand, Gonzales reported the presence of the Gly/Gly PPARGC1A rs8192678 genotype in a world-champion cross-country runner [38], but admitted that this genotype was not present in other elite runners. Therefore, our results can identify the potential to achieve elite sports levels, since those genotypes are also related to training response [4], but cannot play a role in whole talent identification.
The presence of the Ala allele PPARG rs1801282 and the C allele PPARA rs4253778 in elite athletes might be related to the molecular mechanisms required to sustain high anaerobic training loads [53]. Although PPARG rs1801282 Ala allele carriers have been found in individuals with better reactions to aerobic training in the typical population [54,55,56,57], their association in elite athletes might be related to the sustainability of periodic training, which requires tissue recovery and frequent training. Anaerobic training is accompanied by an increase in inflammatory markers, which are regulated by PPARs [8,9]. One study reported the association of the G allele in PPARA rs4253778 with power oriented sports (combat sports) in a comparison between elite athletes and controls (Table 2) [28], which might be explained by the mixed requirements of this sports discipline. However, this increased frequency has not been reported between elite and subelite athletes.
Most documented genetic predispositions to elite performance have been found in endurance athletes, where the G allele of PPARA rs4253778, the C allele of PPARD rs2016520, the Gly allele, and the Gly/Gly genotype of PPARGC1A rs8192678 have been associated with this status as candidate genes and as the crucial part of total genetic scores [30,31,33]. This confirmed the observations that the rs8192678 Gly allele may be a key element associated with the efficiency of aerobic metabolism; however, the question of how the rs8192678 Gly and Ser variants affect cardiorespiratory capacity remains unknown, although engagement of the PGC-1α coactivator in the regulation of energy metabolism, oxidative metabolism, mitochondrial biogenesis, and function has been proven, as have changes in muscle fiber types [42]. Moreover, the PPARGC1A rs8192678 Gly/Gly genotype has been associated with more significant increases in anaerobic threshold [54], more slow muscle fibers [55], more mitochondria activity, and a greater VO2 peak after aerobic training than the PPARGC1A rs8192678 Ser allele genotype. Another aspect is that plasmids bearing Gly or Ser at position 482 in the PGC-1α protein showed that the PPARGC1A 482Ser variant was less efficient as a coactivator of the myocyte enhancer factor 2C (MEF2C), which is a transcription factor regulating glucose transportation in skeletal muscle [58]. The described structure of the PPARs and their coactivators, therefore, targets many aspects necessary for elite athletic performance and might be used for training method selection or nutritional strategies [4]. Our results in the association of PPARGC1A Gly428Ser rs8192678 with endurance elite status seemed to be controversial with respect to previous findings [22], resulting in that this genotype was somewhat related to the power oriented athletes. This difference might be due to the contradictory finding in the original studies and that previous meta-analyses did not separate the comparisons between elite and subelite vs. comparisons between elite and control groups.
4. Materials and Methods
4.1. Review Process
The review was performed according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) [59] guidelines using the review protocol assigned in PROSPERO under Database No. CRD42018082236. The final article eligibility was assessed using the adapted “Strengthening the Reporting of Observational Studies in Epidemiology” (STROBE) checklist [60] (Table S1).
4.2. Literature Search
To find articles related to the role of PPAR polymorphisms in elite sports, we conducted a systematic computerized literature search on 20 August 2019, in PubMed (1940 to search date), Scopus (1823 to search date), and the Web of Science (1974 to search date). A combination of the following search terms was used: (PPAR) OR (peroxisome AND proliferator AND activated AND receptor) AND (sports) OR (physical AND activity) OR (endurance) OR (exercise) OR (performance) OR (movement). The search did not include comments, proceedings, editorial letters, conference abstracts, nor dissertations. Reviews were included for a manual search of their reference lists. A manual search of the reference lists of included articles was also performed (Figure 1).
4.3. Literature Selection
After identifying potential articles, the titles and abstracts were reviewed by two independent reviewers (P.S., M.P.) to select relevant articles for full-text screening according to the following inclusion criteria:
- Genotyping in PPARA, PPARG, PPARD, PPARGC1A, PPARGC1B, and genes.
- The population of athletes.
- Cross-sectional, cohort, case-control, intervention, control trial, or GWAS.
When the inclusion of articles was questionable, the reviewers agreed after a discussion. The full-text analyses of the relevant articles were performed by three independent reviewers (P.S., M.P., A.M.-S.) who also completed the data extraction form (Table S2). During the full-text screening, the following exclusion criteria were used:
- (1)
- the full text was not available in English;
- (2)
- the study did not contain an appropriate description of athlete performance status;
- (3)
- the study did not include a specification of the selected sports discipline;
- (4)
- the study did not report PPAR frequencies for elite athletes;
- (5)
- the study was not reproducible by the methodological quality criteria.
4.4. Qualitative Synthesis
The result of the qualitative synthesis was based on the comparison of the type of participants in the original studies, where the highest importance was considered for comparison of elite athletes to subelite athletes. Then, the comparison between elite athletes and controls was considered as a supportive level of meaningful. Elite sports status was determined during full-text screening, where we used the original status definition of the author if it was under elite status determination [61]. The synthesis summarized three categories of sports disciplines by the dominant metabolic demand for the disciplines: strength and power oriented athletes, endurance oriented athletes, and mixed type of activity according to previous definitions [25,30,46], where team sports such as soccer or ice-hockey were considered as mixed strength-power and endurance disciplines.
5. Conclusions
PPARs could be used for estimating the potential to achieve elite status in human physical performance in strength, power, team, and aerobic sports disciplines. Carrying specific PPARs alleles could provide a partial benefit for achieving elite sports status, but did not preclude achieving elite status if they were absent. The Ala allele in PPARG rs1801282 supported the achievement of elite athlete status in strength and power disciplines. The C allele in PPARA rs4253778 supported the achievement of elite athlete status in mixed strength and endurance soccer, and the G allele PPARA rs4253778 supported achievement in endurance athletes. The Gly allele in PPARGC1A rs8192678 and the C allele PPARD rs2016520 supported the achievement of elite athlete status in endurance sports disciplines.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/1/162/s1 link.
Author Contributions
P.S., M.P. and A.Z. conceived of and designed the review; P.S., M.P. and A.M.-S. performed the review; P.S., M.P. and A.M.-S. analyzed the data; P.S., M.P., A.M.-S. and J.C. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Charles University, grant number UNCE/HUM/03 and The Grant Agency of Czech Republic no GA19-12150S.
Acknowledgments
This study was supported by the UNCE/HUM/032, GACR GA19-12150S grant at Charles University.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| GWAS | genome-wide association study |
| PRISMA | Preferred Reporting Items for Systematic Reviews and Meta-Analyses |
| STROBE | Strengthening the Reporting of Observational Studies in Epidemiology |
| PPAR | peroxisome proliferator activated receptor |
| MEF2C | myocyte enhancer factor 2C |
| PROSPERO | international database of prospectively registered systematic reviews in health. |
| ACTN3 | alpha-actinin-3 |
| DM2 | diabetes mellitus type 2 |
References
- Semenova, E.A.; Fuku, N.; Ahmetov, I.I. Genetic profile of elite endurance athletes. In Sports, Exercise, and Nutritional Genomics; Elsevier Academic Press: London, UK, 2019; pp. 73–104. [Google Scholar]
- Maciejewska-Skrendo, A.; Sawczuk, M.; Cięszczyk, P.; Ahmetov, I.I. Genes and power athlete status. In Sports, Exercise, and Nutritional Genomics; Elsevier Academic Press: London, UK, 2019; pp. 41–72. [Google Scholar]
- Maciejewska-Skrendo, A.; Cięszczyk, P.; Chycki, J.; Sawczuk, M.; Smółka, W. Genetic Markers Associated with Power Athlete Status. J. Hum. Kinet. 2019, 68, 17. [Google Scholar] [CrossRef] [PubMed]
- Petr, M.; Stastny, P.; Zajac, A.; Tufano, J.J.; Maciejewska-Skrendo, A. The Role of Peroxisome Proliferator-Activated Receptors and Their Transcriptional Coactivators Gene Variations in Human Trainability: A Systematic Review. Int. J. Mol. Sci. 2018, 19, 1472. [Google Scholar] [CrossRef] [PubMed]
- Valeeva, E.V.; Ahmetov, I.I.; Rees, T. Psychogenetics and sports. Sports, Exercise, and Nutritional Genomics; Elsevier: Amsterdam, The Netherlands, 2019; pp. 147–165. [Google Scholar]
- Kersten, S.; Desvergne, B.; Wahli, W. Roles of PPARs in health and disease. Nature 2000, 405, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Pozzi, A.; Ibanez, M.R.; Gatica, A.E.; Yang, S.; Wei, S.; Mei, S.; Falck, J.R.; Capdevila, J.H. Peroxisomal proliferator-activated receptor-α-dependent inhibition of endothelial cell proliferation and tumorigenesis. J. Biol. Chem. 2007, 282, 17685–17695. [Google Scholar] [CrossRef]
- Dubuquoy, L.; Dharancy, S.; Nutten, S.; Pettersson, S.; Auwerx, J.; Desreumaux, P. Role of peroxisome proliferator-activated receptor γ and retinoid X receptor heterodimer in hepatogastroenterological diseases. Lancet 2002, 360, 1410–1418. [Google Scholar] [CrossRef]
- Cabrero, A.; Laguna, J.; Vazquez, M. Peroxisome proliferator-activated receptors and the control of inflammation. Curr. Drug Targets-Inflamm. Allergy 2002, 1, 243–248. [Google Scholar] [CrossRef]
- Leonardini, A.; Laviola, L.; Perrini, S.; Natalicchio, A.; Giorgino, F. Cross-talk between PPAR and insulin signaling and modulation of insulin sensitivity. PPAR Res. 2009. [Google Scholar] [CrossRef]
- Yessoufou, A.; Wahli, W. Multifaceted roles of peroxisome proliferator-activated receptors (PPARs) at the cellular and whole organism levels. Swiss Med Wkly. 2010, 140. [Google Scholar] [CrossRef]
- Kliewer, S.; Forman, B.; Blumberg, B.; Ong, E.; Borgmeyer, U.; Mangelsdorf, D.; Umesono, K.; Evans, R.M. Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proc. Natl. Acad. Sci. USA 1994, 91, 7355–7359. [Google Scholar] [CrossRef]
- Manickam, R.; Wahli, W. Roles of peroxisome proliferator-activated receptor β/δ in skeletal muscle physiology. Biochimie 2017, 136, 42–48. [Google Scholar] [CrossRef]
- Cagnin, S.; Chemello, F.; Ahmetov, I.I. Genes and response to aerobic training. In Sports, Exercise, and Nutritional Genomics; Elsevier Academic Press: London, UK, 2019; pp. 169–188. [Google Scholar]
- Gleyzer, N.; Scarpulla, R.C. PGC-1-related coactivator (PRC), a sensor of metabolic stress, orchestrates a redox-sensitive program of inflammatory gene expression. J. Biol. Chem. 2011, 286, 39715–39725. [Google Scholar] [CrossRef] [PubMed]
- Handschin, C.; Spiegelman, B.M. Peroxisome proliferator-activated receptor γ coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr. Rev. 2006, 27, 728–735. [Google Scholar] [CrossRef] [PubMed]
- Franks, P.W.; Christophi, C.A.; Jablonski, K.A.; Billings, L.K.; Delahanty, L.M.; Horton, E.S.; Knowler, W.C.; Florez, J.C.; Diabetes Prevention Program Research Group. Common variation at PPARGC1A/B and change in body composition and metabolic traits following preventive interventions: The Diabetes Prevention Program. Diabetologia 2014, 57, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Ahmetov, I.I.; Williams, A.G.; Popov, D.V.; Lyubaeva, E.V.; Hakimullina, A.M.; Fedotovskaya, O.N.; Mozhayskaya, I.A.; Vinogradova, O.L.; Astratenkova, I.V.; Montgomery, H.E.; et al. The combined impact of metabolic gene polymorphisms on elite endurance athlete status and related phenotypes. Hum. Gen. 2009, 126, 751–761. [Google Scholar] [CrossRef] [PubMed]
- Ahmetov, I.I.; Popov, D.V.; Mozhaiskaia, I.A.; Missina, S.S.; Astratenkova, I.V.; Vinogradova, O.L.; Rogozkin, V.A. Association of regulatory genes polymorphisms with aerobic and anaerobic performance of athletes. Rossiǐskii fiziologicheskiǐ zhurnal imeni IM Sechenova/Rossiǐskaia akademiia nauk 2007, 93, 837–843. [Google Scholar]
- Franks, P.W.; Barroso, I.; Luan, J.; Ekelund, U.; Crowley, V.E.F.; Brage, S.; Sandhu, M.S.; Jakes, R.W.; Middelberg, R.P.; Harding, A.H.; et al. PGC-1α Genotype Modifies the Association of Volitional Energy Expenditure with V̇O2max. Med. Sci. Sports. Exerc. 2003, 35, 1998–2004. [Google Scholar] [CrossRef]
- Petr, M.; Št’Astný, P.; Pecha, O.; Šteffl, M.; Šeda, O.; Kohlíková, E. PPARA intron polymorphism associated with power performance in 30-s anaerobic wingate test. PLoS ONE 2014, 9, e107171. [Google Scholar] [CrossRef]
- Tharabenjasin, P.; Pabalan, N.; Jarjanazi, H. Association of PPARGC1A Gly428Ser (rs8192678) polymorphism with potential for athletic ability and sports performance: A meta-analysis. PLoS ONE 2019, 14, e0200967. [Google Scholar] [CrossRef]
- Mathai, A.S.; Bonen, A.; Benton, C.R.; Robinson, D.L.; Graham, T.E. Rapid exercise-induced changes in PGC-1α mRNA and protein in human skeletal muscle. J. Appl. Physiol. 2008, 105, 1098–1105. [Google Scholar] [CrossRef]
- Nezhad, F.Y.; Verbrugge, S.A.J.; Schönfelder, M.; Becker, L.; De Angelis, M.H.; Wackerhage, H. Genes whose gain or loss-of-function increases endurance performance in Mice: A systematic literature review. Front. Physiol. 2019, 10, 262. [Google Scholar] [CrossRef]
- Ahmetov, I.I.; Mozhayskaya, I.A.; Flavell, D.M.; Astratenkova, I.V.; Komkova, A.I.; Lyubaeva, E.V.; Tarakin, P.P.; Shenkman, B.S.; Vdovina, A.B.; Netreba, A.I.; et al. PPARα gene variation and physical performance in Russian athletes. Eur. J. Appl. Physiol. 2006, 97, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Ahmetov, I.I.; Astratenkova, I.V.; Rogozkin, V.A. Association of a PPARD polymorphism with human physical performance. Mol. Biol. 2007, 41, 776–780. [Google Scholar] [CrossRef]
- Ahmetov, I.I.; Mozhayskaya, I.A.; Lyubaeva, E.V.; Vinogradova, O.L.; Rogozkin, V.A. PPARG Gene polymorphism and locomotor activity in humans. Bull. Exp. Biol. Med. 2008, 146, 630–632. [Google Scholar] [CrossRef] [PubMed]
- Cieszczyk, P.; Sawczuk, M.; Maciejewska, A.; Ficek, K.; Eider, A. Variation in peroxisome proliferator activated receptor α gene in elite combat athletes. Eur. J. Sport. Sci. 2011, 11, 119–123. [Google Scholar] [CrossRef]
- Cocci, P.; Pistolesi, L.; Guercioni, M.; Belli, L.; Carli, D.; Palermo, F.A. Genetic Variants and Mixed Sport Disciplines: A Comparison among Soccer, Combat and Motorcycle Athletes. Ann. Appl. Sport. Sci. 2019, 7, 1–9. [Google Scholar] [CrossRef]
- Drozdovska, S.B.; Dosenko, V.E.; Ahmetov, I.I.; Ilyin, V.N. The association of gene polymorphisms with athlete status in Ukrainians. Biol. Sport 2013, 30, 163–167. [Google Scholar] [CrossRef]
- Eynon, N.; Ruiz, J.R.; Meckel, Y.; Morán, M.; Lucia, A. Mitochondrial biogenesis related endurance genotype score and sports performance in athletes. Mitochondrion 2011, 11, 64–69. [Google Scholar] [CrossRef]
- Eynon, N.; Meckel, Y.; Sagiv, M.; Yamin, C.; Amir, R.; Sagiv, M.; Goldhammer, E.; Duarte, J.A.; Oliveira, J. Do PPARGC1A and PPARα polymorphisms influence sprint or endurance phenotypes? Scand. J. Med. Sci. Sports 2010, 20, e145–e150. [Google Scholar] [CrossRef]
- Egorova, E.S.; Borisova, A.V.; Mustafina, L.J.; Arkhipova, A.A.; Gabbasov, R.T.; Druzhevskaya, A.M.; Astratenkova, I.V.; Ahmetov, I.I. The polygenic profile of Russian football players. J. Sports. Sci. 2014, 32, 1286–1293. [Google Scholar] [CrossRef]
- Ginevičiene, V.; Pranckevičiene, E.; Milašius, K.; Kučinskas, V. Gene variants related to the power performance of the Lithuanian athletes. Cen. Eur. J. Biol. 2011, 6, 48–57. [Google Scholar] [CrossRef]
- Eynon, N.; Alves, A.J.; Yamin, C.; Meckel, Y. PPARA intron 1 A/C polymorphism and elite athlete status. Eur. J. Sport Sci. 2011, 11, 177–181. [Google Scholar] [CrossRef]
- Gineviciene, V.; Jakaitiene, A.; Tubelis, L.; Kucinskas, V. Variation in the ACE, PPARGC1A and PPARA genes in Lithuanian football players. Eur. J. Sport Sci. 2014, 14, S289–S295. [Google Scholar] [CrossRef] [PubMed]
- Gineviciene, V.; Jakaitiene, A.; Aksenov, M.O.; Aksenova, A.V.; Druzhevskaya, A.M.; Astratenkova, I.V.; Egorova, E.S.; Gabdrakhmanova, L.J.; Tubelis, L.; Kucinskas, V.; et al. Association analysis of ACE, ACTN3 and PPARGC1A gene polymorphisms in two cohorts of European strength and power athletes. Biol. Sport 2016, 33, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Freire, M.; Santiago, C.; Verde, Z.; Lao, J.I.; Olivan, J.; Gallego, F.G.; Lucia, A. Unique among unique. Is it genetically determined? Br. J. Sports Med. 2009, 43, 307–309. [Google Scholar] [CrossRef] [PubMed]
- Grealy, R.; Herruer, J.; Smith, C.L.E.; Hiller, D.; Haseler, L.J.; Griffiths, L.R. Evaluation of a 7-gene genetic profile for athletic endurance phenotype in ironman championship triathletes. PLoS ONE 2015, 10, e0145171. [Google Scholar] [CrossRef] [PubMed]
- Lucia, A.; Gomez-Gallego, F.; Barroso, I.; Rabadan, M.; Bandres, F.; San Juan, A.F.; Chicharro, J.L.; Ekelund, U.; Brage, S.; Earnest, C.P.; et al. PPARGC1A genotype (Gly482Ser) predicts exceptional endurance capacity in European men. J. Appl. Physiol. (1985) 2005, 99, 344–348. [Google Scholar] [CrossRef]
- Maciejewska, A.; Sawczuk, M.; Cieszczyk, P. Variation in the PPARalpha gene in Polish rowers. J. Sci. Med. Sport 2011, 14, 58–64. [Google Scholar] [CrossRef]
- Maciejewska, A.; Sawczuk, M.; Cieszczyk, P.; Mozhayskaya, I.A.; Ahmetov, I.I. The PPARGC1A gene Gly482Ser in Polish and Russian athletes. J. Sports Sci. 2012, 30, 101–113. [Google Scholar] [CrossRef]
- Maciejewska-Karlowska, A.; Sawczuk, M.; Cieszczyk, P.; Zarebska, A.; Sawczyn, S. Association between the Pro12Ala Polymorphism of the Peroxisome Proliferator-Activated Receptor Gamma Gene and Strength Athlete Status. PLoS ONE 2013, 8, e67172. [Google Scholar] [CrossRef]
- Maciejewska-Karlowska, A.; Hanson, E.D.; Sawczuk, M.; Cieszczyk, P.; Eynon, N. Genomic haplotype within the Peroxisome Proliferator-Activated Receptor Delta (PPARD) gene is associated with elite athletic status. Scand. J. Med. Sci. Sports 2014, 24, e148–e155. [Google Scholar] [CrossRef]
- Muniesa, C.A.; González-Freire, M.; Santiago, C.; Lao, J.I.; Buxens, A.; Rubio, J.C.; Martín, M.A.; Arenas, J.; Gomez-Gallego, F.; Lucia, A. World-class performance in lightweight rowing: Is it genetically influenced? A comparison with cyclists, runners and non-athletes. Br. J. Sports Med. 2010, 44, 898–901. [Google Scholar] [CrossRef] [PubMed]
- Peplonska, B.; Adamczyk, J.G.; Siewierski, M.; Safranow, K.; Maruszak, A.; Sozanski, H.; Gajewski, A.K.; Zekanowski, C. Genetic variants associated with physical and mental characteristics of the elite athletes in the Polish population. Scand. J. Med. Sci. Sports 2017, 27, 788–800. [Google Scholar] [CrossRef] [PubMed]
- Santiago, C.; Ruiz, J.R.; Muniesa, C.A.; González-Freire, M.; Gómez-Gallego, F.; Lucia, A. Does the polygenic profile determine the potential for becoming a world-class athlete? Insights from the sports of rowing. Scand. J. Med. Sci. Sports 2010, 20, e188–e194. [Google Scholar] [CrossRef] [PubMed]
- Tsianos, G.I.; Evangelou, E.; Boot, A.; Carola Zillikens, M.; Van Meurs, J.B.J.; Uitterlinden, A.G.; Ioannidis, J.P. Associations of polymorphisms of eight muscle—Or metabolism-related genes with performance in Mount Olympus marathon runners. J. Appl. Physiol. 2010, 108, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Tural, E.; Kara, N.; Agaoglu, S.A.; Elbistan, M.; Tasmektepligil, M.Y.; Imamoglu, O. PPAR-α and PPARGC1A gene variants have strong effects on aerobic performance of Turkish elite endurance athletes. Mol. Biol. Rep. 2014, 41, 5799–5804. [Google Scholar] [CrossRef] [PubMed]
- Yvert, T.; Miyamoto-Mikami, E.; Murakami, H.; Miyachi, M.; Kawahara, T.; Fuku, N. Lack of replication of associations between multiple genetic polymorphisms and endurance athlete status in Japanese population. Physiol. Rep. 2016, 4, e13003. [Google Scholar] [CrossRef]
- Eynon, N.; Birk, R.; Meckel, Y.; Lucia, A.; Nemet, D.; Eliakim, A. Physiological variables and mitochondrial-related genotypes of an athlete who excels in both short and long-distance running. Mitochondrion 2011, 11, 774–777. [Google Scholar] [CrossRef][Green Version]
- Lucia, A.; Oliván, J.; Gómez-Gallego, F.; Santiago, C.; Montil, M.; Foster, C. Citius and longius (faster and longer) with no α-actinin-3 in skeletal muscles? Br. J. Sports Med. 2007, 41, 616–617. [Google Scholar] [CrossRef]
- Aksenov, M.O.; Ilyin, A.B. Training process design in weightlifting sports customized to genetic predispositions. Teoriya i Praktika Fizicheskoy Kultury 2017, 6, 75–77. [Google Scholar]
- Stefan, N.; Thamer, C.; Staiger, H.; Machicao, F.; Machann, J.; Schick, F.; Venter, C.; Niess, A.; Laakso, M.; Fritsche, A.; et al. Genetic variations in PPARD and PPARGC1A determine mitochondrial function and change in aerobic physical fitness and insulin sensitivity during lifestyle intervention. J. Clin. Endocrin. Metabol. 2007, 92, 1827–1833. [Google Scholar] [CrossRef]
- Steinbacher, P.; Feichtinger, R.G.; Kedenko, L.; Kedenko, I.; Reinhardt, S.; Schönauer, A.L.; Leitner, I.; Sänger, A.M.; Stoiber, W.; Kofler, B.; et al. The single nucleotide polymorphism Gly482Ser in the PGC-1α gene impairs exercise-induced slow-twitch muscle fibre transformation in humans. PLoS ONE 2015, 10, e0123881. [Google Scholar] [CrossRef] [PubMed]
- Ring-Dimitriou, S.; Kedenko, L.; Kedenko, I.; Feichtinger, R.G.; Steinbacher, P.; Stoiber, W.; Forster, H.; Felder, T.K.; Muller, E.; Kolfer, B. Does genetic variation in PPARGC1A affect exercise-induced changes in ventilatory thresholds and metabolic syndrome? J. Exerc. Physiol. Online 2014, 17, 1–18. [Google Scholar]
- Hautala, A.J.; Leon, A.S.; Skinner, J.S.; Rao, D.C.; Bouchard, C.; Rankinen, T. Peroxisome proliferator-activated receptor-delta polymorphisms are associated with physical performance and plasma lipids: The HERITAGE Family Study. Am. J. Physiol. Heart Circ. Physiol. 2007, 292, H2498–H2505. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.-L.; Lu, W.-S.; Yan, L.; Wu, M.-C.; Xu, M.-T.; Chen, L.-H.; Cheng, H. Association between peroxisome proliferator-activated receptor-gamma coactivator-1alpha gene polymorphisms and type 2 diabetes in southern Chinese population: Role of altered interaction with myocyte enhancer factor 2C. Chin. Med. J. 2007, 120, 1878–1885. [Google Scholar] [CrossRef]
- Moher, D.; Schulz, K.F.; Simera, I.; Altman, D.G. Guidance for developers of health research reporting guidelines. PLoS Med. 2010, 7, e1000217. [Google Scholar] [CrossRef]
- Von Elm, E.; Altman, D.G.; Egger, M.; Pocock, S.J.; Gøtzsche, P.C.; Vandenbroucke, J.P. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) Statement: Guidelines for reporting observational studies. Prev. Med. 2007, 45, 247–251. [Google Scholar] [CrossRef]
- Swann, C.; Moran, A.; Piggott, D. Defining elite athletes: Issues in the study of expert performance in sports psychology. Psych. Sport Exerc. 2015, 16, 3–14. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).