Evaluation of Protein Kinase Inhibitors with PLK4 Cross-Over Potential in a Pre-Clinical Model of Cancer
Abstract
1. Introduction
2. Results
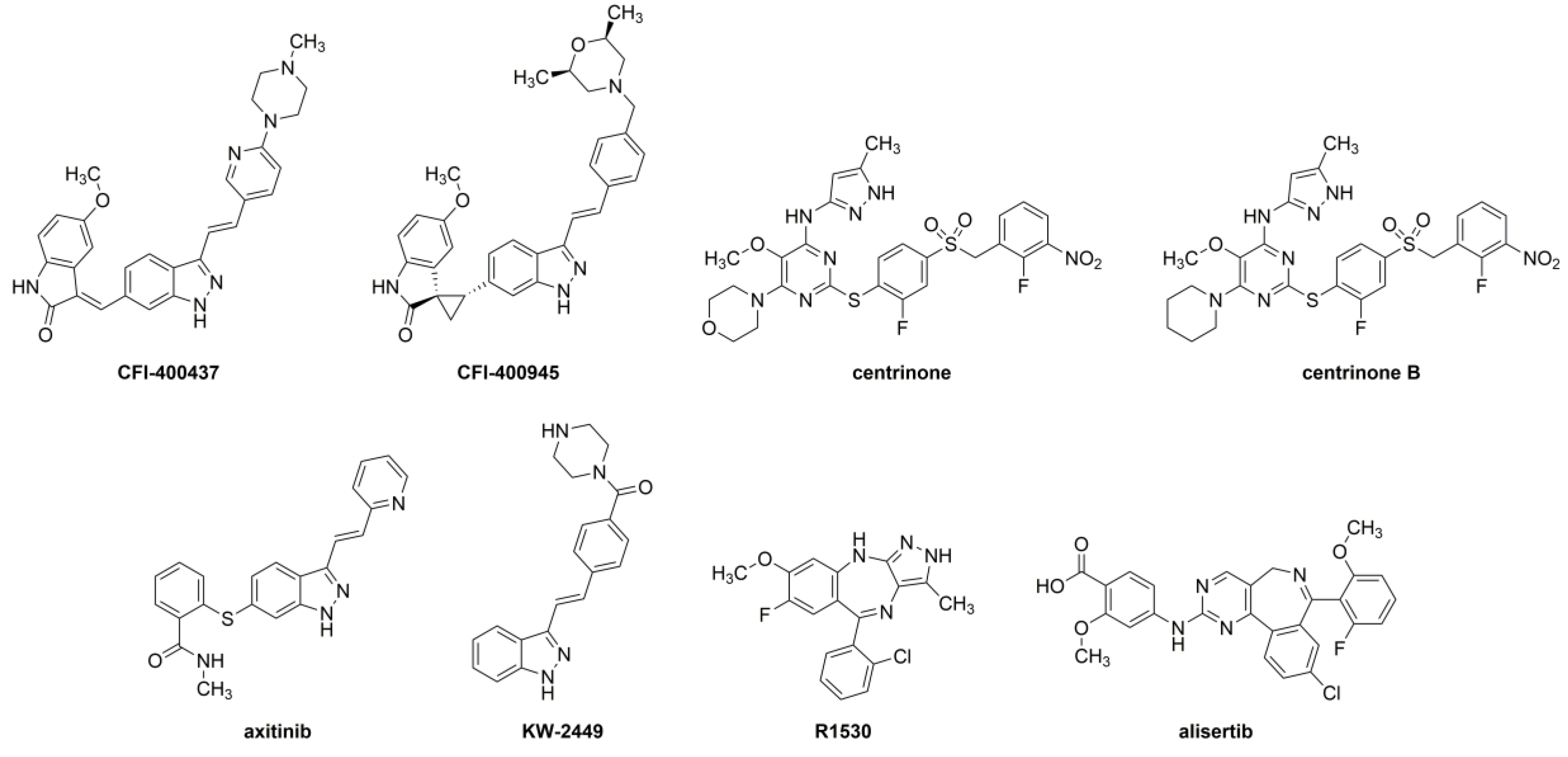
2.1. Structural Analysis of the Inhibitors
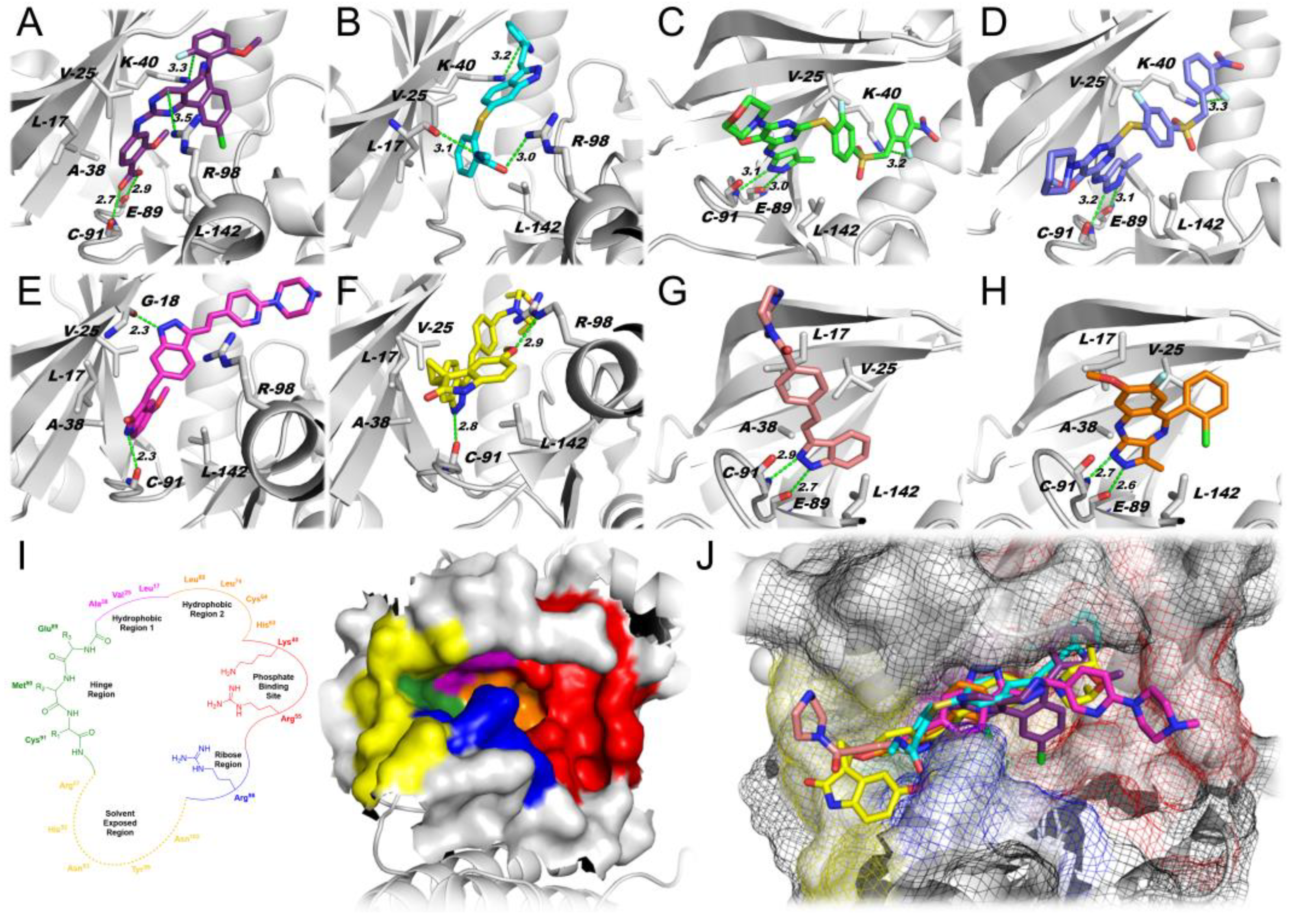
2.1.1. Docking Simulations
2.1.2. Physical Property Analysis
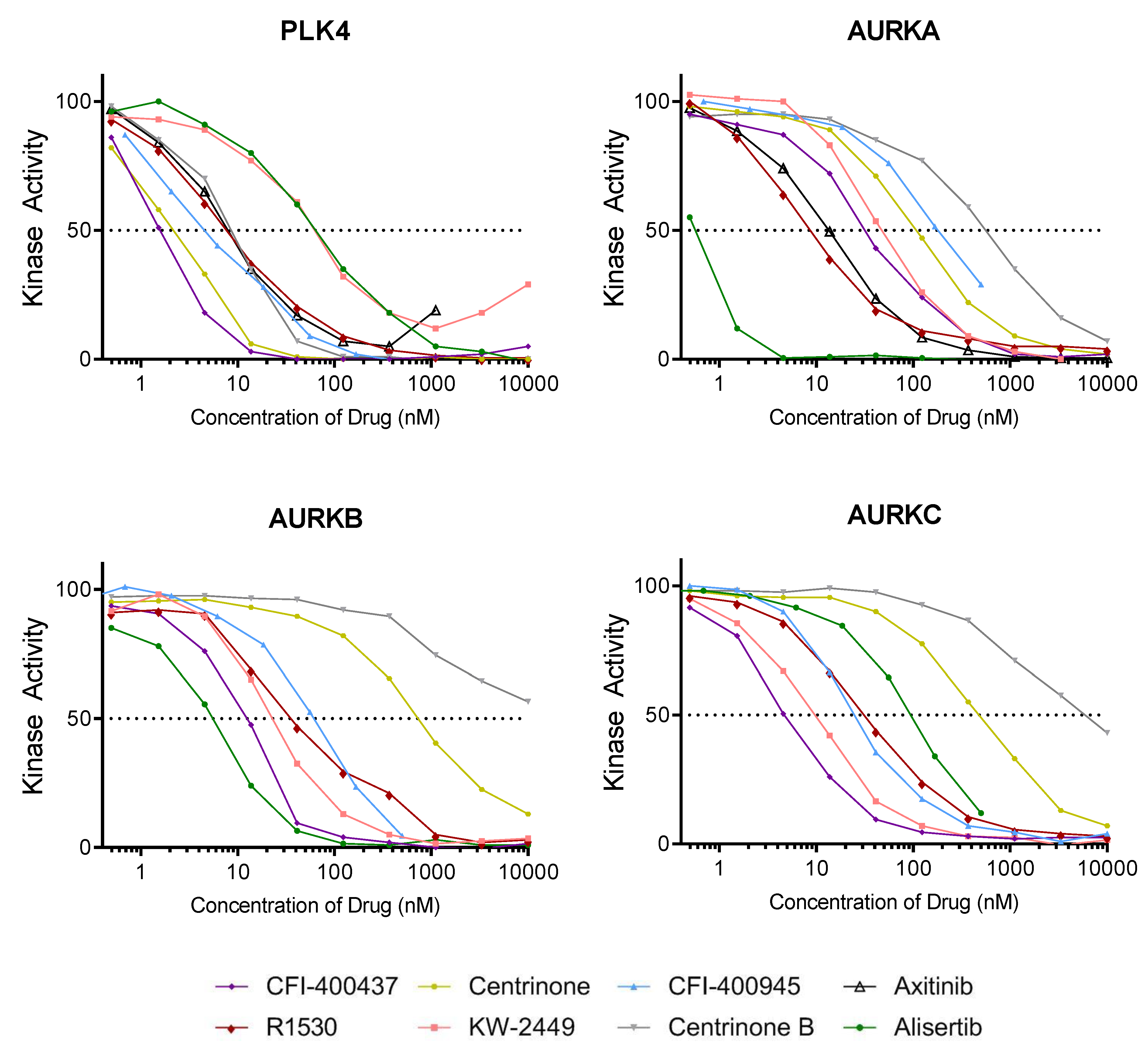
2.2. Kinase Assays
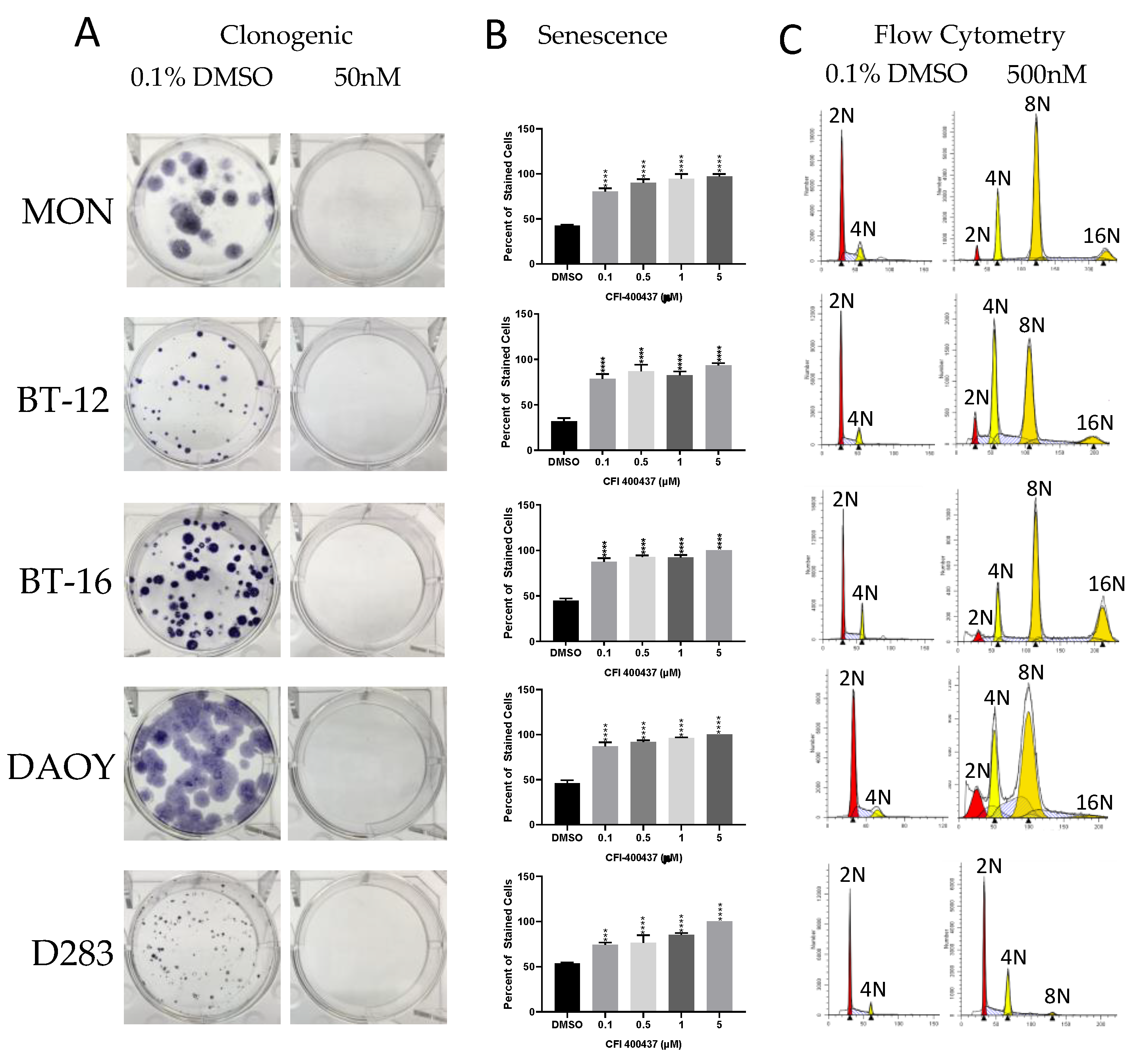
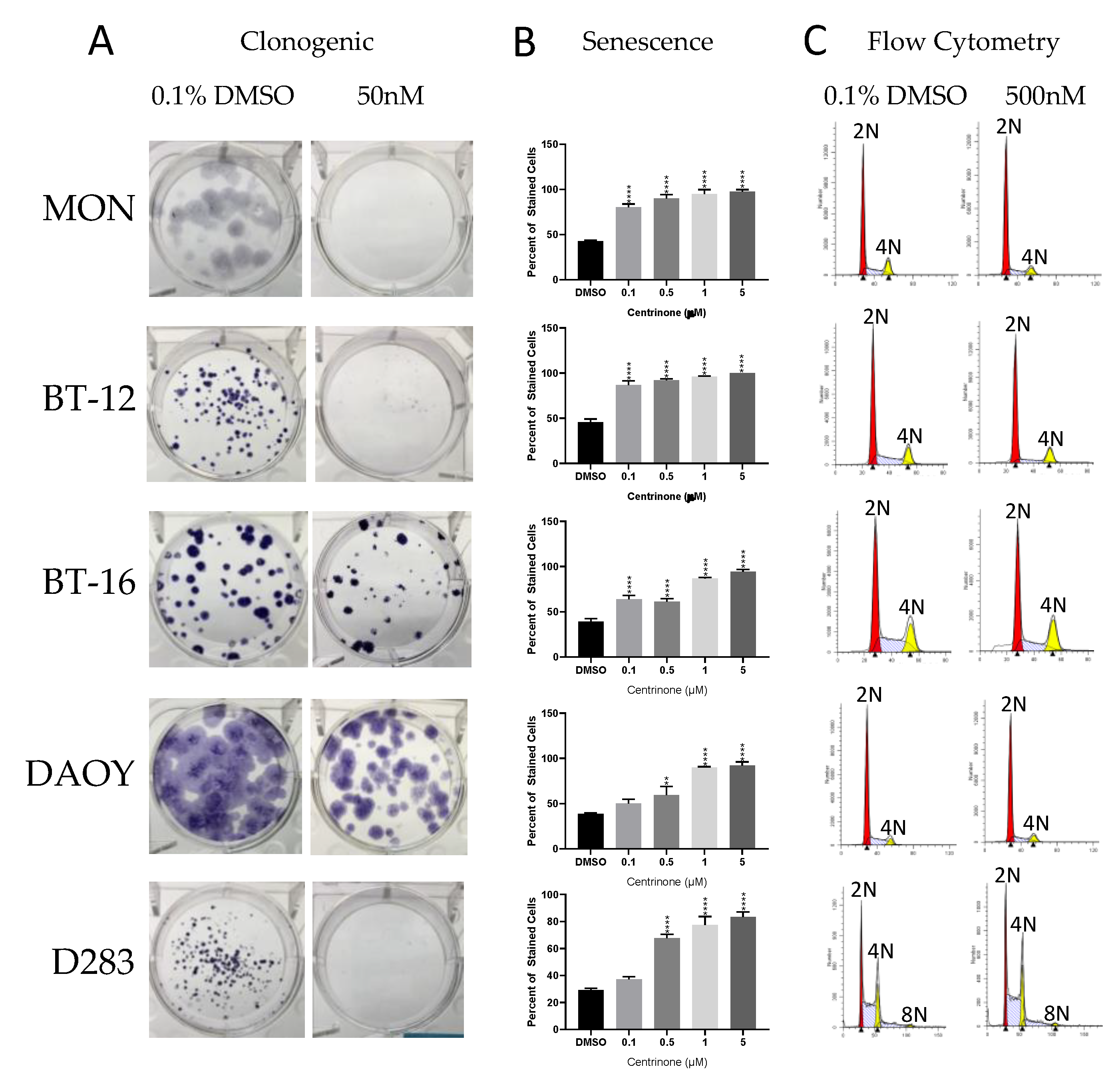
2.3. Cell-Based Studies
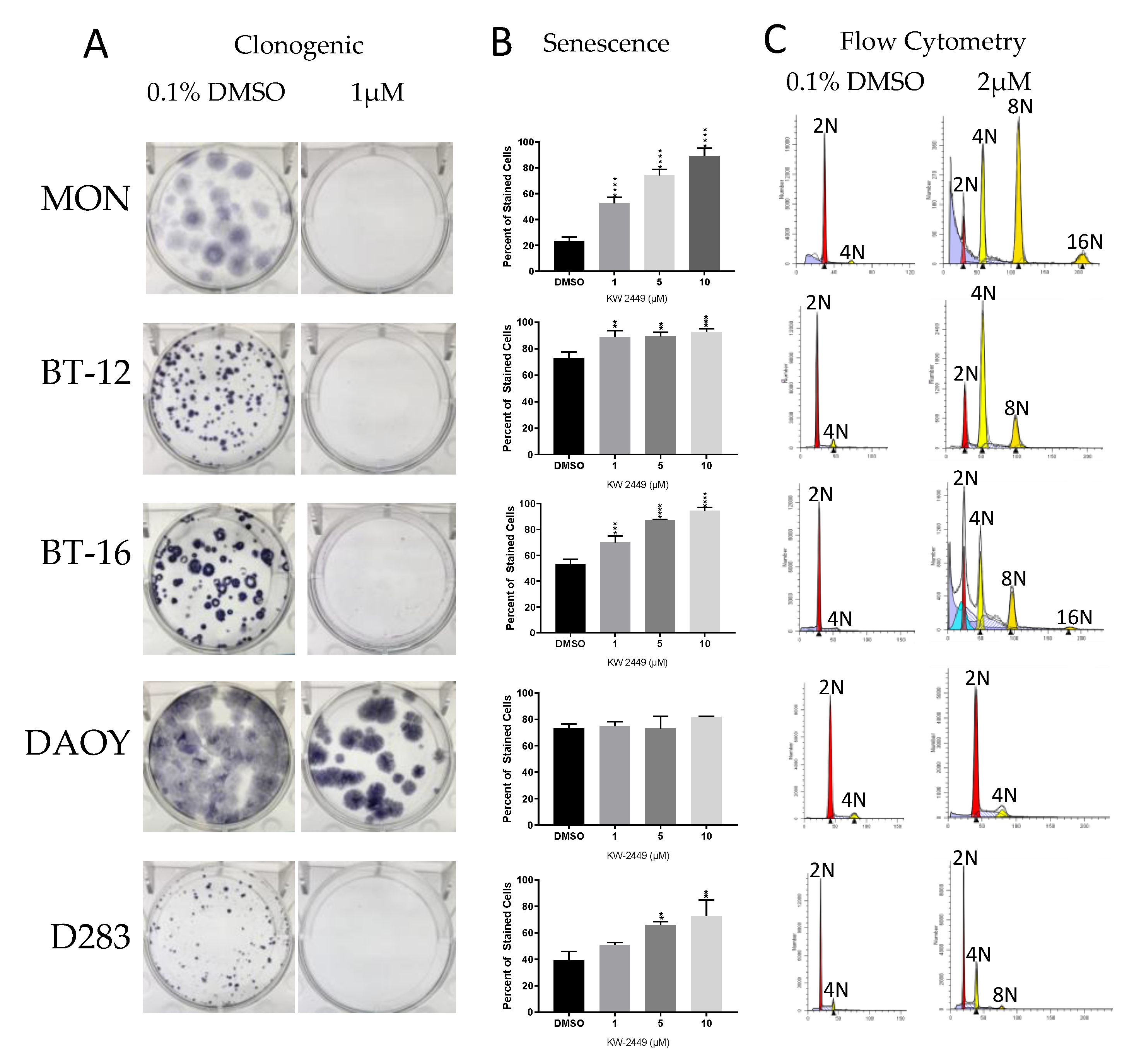
2.4. KW-2449
2.4.1. KW-2449 Potential for Cardio Toxicity and Drug-Drug Interaction Risk
2.4.2. KW-2449 Kinase Inhibition Screen
3. Discussion
3.1. Polyploidy and Aurora Kinases (AURKs)
3.2. Considerations for Future Development of a Brain Penetrant PLK4i and its Therapeutic Use
4. Materials and Methods
4.1. Inhibitors Tested
4.2. Structural Analysis of the Inhibitors
4.3. Kinase Assay
4.4. Cell Culture
4.5. Cell-Based Assays
4.5.1. Proliferation Assay
4.5.2. Viability Assay
4.5.3. Clonogenic Assay
4.5.4. Beta-Galactosidase Assay
4.5.5. Cell Cycle Analysis
4.6. Characterization of KW-2449
4.6.1. Kinase Activity Screening
4.6.2. Drug Safety and Toxicology
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AT/RT | Atypical Teratoid Rhabdoid Tumor |
| AURK | Aurora Kinase |
| AURKA | Aurora Kinase A |
| AURKB | Aurora Kinase B |
| AURKC | Aurora Kinase C |
| B3LYP | Becke-3-Lee Yang Parr |
| BBB | Blood Brain Barrier |
| CNS MPO | Central Nervous System Multi-Parameter Optimization |
| CYP450 | Cytochrome P450 |
| DFT | Density Functional Theory |
| EBT | Embryonal Brain Tumor |
| G3-MB | Group 3 MB |
| G4-MB | Group 4 MB |
| GBM | Glioblastoma multiforme |
| hERG | Human Ether-a-go-go-related gene |
| IC50 | Half Maximal Inhibitory Concentration |
| LogP | Lipophilicity |
| MB | Medulloblastoma |
| MRT | Malignant Rhabdoid Tumor |
| MW | Molecular Weight |
| PB | Polo-box |
| PBD | Protein Data Bank |
| PI | Propidium Iodide |
| PK | Protein Kinase |
| PKI | Protein Kinase Inhibitor |
| PLK | Polo-like Kinase |
| PLK4 | Polo-like kinase 4 |
| PLK4i | Polo-like kinase 4 inhibitor |
| PSA | Polar Surface Area |
| RT | Rhabdoid Tumor |
| SAR | Structure-Activity Relationship |
| SHH | Sonic hedgehog |
| STM | Signal Transduction Modifiers |
| TPSA | Total Polar Surface Area |
| TRK | Tropomyosin Receptor Kinase |
References
- Bhullar, K.S.; Lagaron, N.O.; McGowan, E.M.; Parmar, I.; Jha, A.; Hubbard, B.P.; Rupasinghe, H.P.V. Kinase-targeted cancer therapies: Progress, challenges and future directions. Mol. Cancer 2018, 17, 48. [Google Scholar] [CrossRef] [PubMed]
- Gross, S.; Rahal, R.; Stransky, N.; Lengauer, C.; Hoeflich, K.P. Targeting cancer with kinase inhibitors. J. Clin. Investig. 2015, 125, 1780–1789. [Google Scholar] [CrossRef]
- Nigg, E.A.; Holland, A.J. Once and only once: Mechanisms of centriole duplication and their deregulation in disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Levine, M.S.; Bakker, B.; Boeckx, B.; Moyett, J.; Lu, J.; Vitre, B.; Spierings, D.C.; Lansdorp, P.M.; Cleveland, D.W.; Lambrechts, D.; et al. Centrosome amplification is sufficient to promote spontaneous tumorigenesis in mammals. Dev. Cell 2017, 40, 313–322.e315. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.Y. A clinical overview of centrosome amplification in human cancers. Int. J. Biol. Sci. 2011, 7, 1122–1144. [Google Scholar] [CrossRef] [PubMed]
- Matthay, K.K.; Brisse, H.; Couanet, D.; Couturier, J.; Benard, J.; Mosseri, V.; Edeline, V.; Lumbroso, J.; Valteau-Couanet, D.; Michon, J. Central nervous system metastases in neuroblastoma: Radiologic, clinical, and biologic features in 23 patients. Cancer 2003, 98, 155–165. [Google Scholar] [CrossRef]
- Fukasawa, K. Centrosome amplification, chromosome instability and cancer development. Cancer Lett. 2005, 230, 6–19. [Google Scholar] [CrossRef]
- Conduit, P.T.; Wainman, A.; Raff, J.W. Centrosome function and assembly in animal cells. Nat. Rev. Mol. Cell Biol. 2015, 16, 611–624. [Google Scholar] [CrossRef]
- Prakash, A.; Garcia-Moreno, J.F.; Brown, J.A.L.; Bourke, E. Clinically applicable inhibitors impacting genome stability. Molecules 2018, 23, 1166. [Google Scholar] [CrossRef] [PubMed]
- Godinho, S.A.; Pellman, D. Causes and consequences of centrosome abnormalities in cancer. Philos. Trans. R. Soc. Lond. Ser. B. Biol. Sci. 2014, 369, 20130467. [Google Scholar] [CrossRef]
- Johnson, E.F.; Stewart, K.D.; Woods, K.W.; Giranda, V.L.; Luo, Y. Pharmacological and functional comparison of the polo-like kinase family: Insight into inhibitor and substrate specificity. Biochemistry 2007, 46, 9551–9563. [Google Scholar] [CrossRef]
- Slevin, L.K.; Nye, J.; Pinkerton, D.C.; Buster, D.W.; Rogers, G.C.; Slep, K.C. The structure of the plk4 cryptic polo box reveals two tandem polo boxes required for centriole duplication. Structure (London, England: 1993) 2012, 20, 1905–1917. [Google Scholar] [CrossRef]
- Klebba, J.E.; Buster, D.W.; McLamarrah, T.A.; Rusan, N.M.; Rogers, G.C. Autoinhibition and relief mechanism for polo-like kinase 4. Proc. Natl. Acad. Sci. USA 2015, 112, E657–E666. [Google Scholar] [CrossRef]
- Sillibourne, J.E.; Bornens, M. Polo-like kinase 4: The odd one out of the family. Cell Division 2010, 5, 25. [Google Scholar] [CrossRef]
- Holland, A.J.; Cleveland, D.W. Polo-like kinase 4 inhibition: A strategy for cancer therapy? Cancer Cell 2014, 26, 151–153. [Google Scholar] [CrossRef]
- Holland, A.J.; Fachinetti, D.; Zhu, Q.; Bauer, M.; Verma, I.M.; Nigg, E.A.; Cleveland, D.W. The autoregulated instability of polo-like kinase 4 limits centrosome duplication to once per cell cycle. Genes Dev. 2012, 26, 2684–2689. [Google Scholar] [CrossRef]
- Guderian, G.; Westendorf, J.; Uldschmid, A.; Nigg, E.A. Plk4 trans-autophosphorylation regulates centriole number by controlling betatrcp-mediated degradation. J. Cell Sci. 2010, 123, 2163–2169. [Google Scholar] [CrossRef]
- Holland, A.J.; Lan, W.; Niessen, S.; Hoover, H.; Cleveland, D.W. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J. Cell Biol. 2010, 188, 191–198. [Google Scholar] [CrossRef]
- Sillibourne, J.E.; Tack, F.; Vloemans, N.; Boeckx, A.; Thambirajah, S.; Bonnet, P.; Ramaekers, F.C.; Bornens, M.; Grand-Perret, T. Autophosphorylation of polo-like kinase 4 and its role in centriole duplication. Mol. Biol. Cell 2010, 21, 547–561. [Google Scholar] [CrossRef]
- Gonczy, P. Centrosomes and cancer: Revisiting a long-standing relationship. Nat. Rev. Cancer 2015, 15, 639–652. [Google Scholar] [CrossRef]
- Bettencourt-Dias, M.; Hildebrandt, F.; Pellman, D.; Woods, G.; Godinho, S.A. Centrosomes and cilia in human disease. Trends Genet. 2011, 27, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Dincer, T.; Yorgancioglu-Budak, G.; Olmez, A.; Er, I.; Dodurga, Y.; Ozdemir, O.M.; Toraman, B.; Yildirim, A.; Sabir, N.; Akarsu, N.A.; et al. Analysis of centrosome and DNA damage response in plk4 associated seckel syndrome. Eur. J. Hum. Genet. 2017, 25, 1118–1125. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.A.; Ahmad, I.; Klingseisen, A.; Hussain, M.S.; Bicknell, L.S.; Leitch, A.; Nurnberg, G.; Toliat, M.R.; Murray, J.E.; Hunt, D.; et al. Mutations in plk4, encoding a master regulator of centriole biogenesis, cause microcephaly, growth failure and retinopathy. Nat. Genet. 2014, 46, 1283–1292. [Google Scholar] [CrossRef] [PubMed]
- Shaheen, R.; Al Tala, S.; Almoisheer, A.; Alkuraya, F.S. Mutation in plk4, encoding a master regulator of centriole formation, defines a novel locus for primordial dwarfism. J. Med. Genet. 2014, 51, 814–816. [Google Scholar] [CrossRef] [PubMed]
- Macmillan, J.C.; Hudson, J.W.; Bull, S.; Dennis, J.W.; Swallow, C.J. Comparative expression of the mitotic regulators sak and plk in colorectal cancer. Ann. Surg. Oncol. 2001, 8, 729–740. [Google Scholar] [CrossRef]
- Marina, M.; Saavedra, H.I. Nek2 and plk4: Prognostic markers, drivers of breast tumorigenesis and drug resistance. Front. Biosci. (Landmark Ed.) 2014, 19, 352–365. [Google Scholar] [CrossRef]
- Mason, J.M.; Lin, D.C.; Wei, X.; Che, Y.; Yao, Y.; Kiarash, R.; Cescon, D.W.; Fletcher, G.C.; Awrey, D.E.; Bray, M.R.; et al. Functional characterization of cfi-400945, a polo-like kinase 4 inhibitor, as a potential anticancer agent. Cancer Cell 2014, 26, 163–176. [Google Scholar] [CrossRef]
- Kawakami, M.; Mustachio, L.M.; Zheng, L.; Chen, Y.; Rodriguez-Canales, J.; Mino, B.; Kurie, J.M.; Roszik, J.; Villalobos, P.A.; Thu, K.L.; et al. Polo-like kinase 4 inhibition produces polyploidy and apoptotic death of lung cancers. Proc. Natl. Acad. Sci. USA 2018, 115, 1913–1918. [Google Scholar] [CrossRef]
- Denu, R.A.; Shabbir, M.; Nihal, M.; Singh, C.K.; Longley, B.J.; Burkard, M.E.; Ahmad, N. Centriole overduplication is the predominant mechanism leading to centrosome amplification in melanoma. Mol. Cancer Res. 2018, 16, 517–527. [Google Scholar] [CrossRef]
- Hudnall, S.D.; Meng, H.; Lozovatsky, L.; Li, P.; Strout, M.; Kleinstein, S.H. Recurrent genetic defects in classical hodgkin lymphoma cell lines. Leuk. Lymphoma 2016, 57, 2890–2900. [Google Scholar] [CrossRef]
- Roberto, G.M.; Engel, E.E.; Scrideli, C.A.; Tone, L.G.; Brassesco, M.S. Downregulation of mir-10b* is correlated with altered expression of mitotic kinases in osteosarcoma. Pathol. Res. Pract. 2018, 214, 213–216. [Google Scholar] [CrossRef]
- Shinmura, K.; Kurabe, N.; Goto, M.; Yamada, H.; Natsume, H.; Konno, H.; Sugimura, H. Plk4 overexpression and its effect on centrosome regulation and chromosome stability in human gastric cancer. Mol. Biol. Rep. 2014, 41, 6635–6644. [Google Scholar] [CrossRef]
- Lohse, I.; Mason, J.; Cao, P.M.; Pintilie, M.; Bray, M.; Hedley, D.W. Activity of the novel polo-like kinase 4 inhibitor cfi-400945 in pancreatic cancer patient-derived xenografts. Oncotarget 2017, 8, 3064–3071. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, Z.; Huang, K.; Liu, Y.; Wei, C.; Zhou, J.; Zhang, W.; Wang, Q.; Liang, H.; Zhang, A.; et al. Plk4 is a determinant of temozolomide sensitivity through phosphorylation of ikbke in glioblastoma. Cancer Lett. 2019, 443, 91–107. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhang, C.Z.; Cai, M.; Fu, J.; Chen, G.G.; Yun, J. Downregulation of polo-like kinase 4 in hepatocellular carcinoma associates with poor prognosis. PLoS ONE 2012, 7, e41293. [Google Scholar] [CrossRef]
- Sredni, S.T.; Bailey, A.W.; Suri, A.; Hashizume, R.; He, X.; Louis, N.; Gokirmak, T.; Piper, D.R.; Watterson, D.M.; Tomita, T. Inhibition of polo-like kinase 4 (plk4): A new therapeutic option for rhabdoid tumors and pediatric medulloblastoma. Oncotarget 2017, 8, 111190–111212. [Google Scholar] [CrossRef]
- Sredni, S.T.; Suzuki, M.; Yang, J.P.; Topczewski, J.; Bailey, A.W.; Gokirmak, T.; Gross, J.N.; de Andrade, A.; Kondo, A.; Piper, D.R.; et al. A functional screening of the kinome identifies the polo-like kinase 4 as a potential therapeutic target for malignant rhabdoid tumors, and possibly, other embryonal tumors of the brain. Pediatric Blood Cancer 2017, 64, e26551. [Google Scholar] [CrossRef]
- Sredni, S.T.; Tomita, T. The polo-like kinase 4 gene (plk4) is overexpressed in pediatric medulloblastoma. Child’s Nerv. Syst. ChNS Off. J. Int. Soc. Pediatric Neurosurg. 2017, 33, 1031. [Google Scholar] [CrossRef] [PubMed]
- Bailey, A.W.; Suri, A.; Chou, P.M.; Pundy, T.; Gadd, S.; Raimondi, S.L.; Tomita, T.; Sredni, S.T. Polo-like kinase 4 (plk4) is overexpressed in central nervous system neuroblastoma (cns-nb). Bioengineering 2018, 5, 96. [Google Scholar] [CrossRef] [PubMed]
- Nemes, K.; Fruhwald, M.C. Emerging therapeutic targets for the treatment of malignant rhabdoid tumors. Expert Opin. Ther. Targets 2018, 22, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Sredni, S.T.; Tomita, T. Rhabdoid tumor predisposition syndrome. Pediatric Dev. Pathol. Off. J. Soc. Pediatric Pathol. Paediatr. Pathol. Soc. 2015, 18, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Fruhwald, M.C.; Biegel, J.A.; Bourdeaut, F.; Roberts, C.W.; Chi, S.N. Atypical teratoid/rhabdoid tumors-current concepts, advances in biology, and potential future therapies. Neuro-Oncology 2016, 18, 764–778. [Google Scholar] [CrossRef]
- Grupenmacher, A.T.; Halpern, A.L.; Bonaldo Mde, F.; Huang, C.C.; Hamm, C.A.; de Andrade, A.; Tomita, T.; Sredni, S.T. Study of the gene expression and microrna expression profiles of malignant rhabdoid tumors originated in the brain (at/rt) and in the kidney (rtk). Child’s Nerv. Syst. ChNS Off. J. Int. Soc. Pediatric Neurosurg. 2013, 29, 1977–1983. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; de Blank, P.M.; Kruchko, C.; Petersen, C.M.; Liao, P.; Finlay, J.L.; Stearns, D.S.; Wolff, J.E.; Wolinsky, Y.; Letterio, J.J.; et al. Alex’s lemonade stand foundation infant and childhood primary brain and central nervous system tumors diagnosed in the united states in 2007-2011. Neuro-Oncology 2015, 16, x1–x36. [Google Scholar] [CrossRef]
- Torchia, J.; Picard, D.; Lafay-Cousin, L.; Hawkins, C.E.; Kim, S.K.; Letourneau, L.; Ra, Y.S.; Ho, K.C.; Chan, T.S.; Sin-Chan, P.; et al. Molecular subgroups of atypical teratoid rhabdoid tumours in children: An integrated genomic and clinicopathological analysis. Lancet. Oncol. 2015, 16, 569–582. [Google Scholar] [CrossRef]
- Torchia, J.; Golbourn, B.; Feng, S.; Ho, K.C.; Sin-Chan, P.; Vasiljevic, A.; Norman, J.D.; Guilhamon, P.; Garzia, L.; Agamez, N.R.; et al. Integrated (epi)-genomic analyses identify subgroup-specific therapeutic targets in cns rhabdoid tumors. Cancer Cell 2016, 30, 891–908. [Google Scholar] [CrossRef] [PubMed]
- Johann, P.D.; Erkek, S.; Zapatka, M.; Kerl, K.; Buchhalter, I.; Hovestadt, V.; Jones, D.T.W.; Sturm, D.; Hermann, C.; Segura Wang, M.; et al. Atypical teratoid/rhabdoid tumors are comprised of three epigenetic subgroups with distinct enhancer landscapes. Cancer Cell 2016, 29, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Batora, N.V.; Sturm, D.; Jones, D.T.; Kool, M.; Pfister, S.M.; Northcott, P.A. Transitioning from genotypes to epigenotypes: Why the time has come for medulloblastoma epigenomics. Neuroscience 2014, 264, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Dubuc, A.M.; Remke, M.; Korshunov, A.; Northcott, P.A.; Zhan, S.H.; Mendez-Lago, M.; Kool, M.; Jones, D.T.; Unterberger, A.; Morrissy, A.S.; et al. Aberrant patterns of h3k4 and h3k27 histone lysine methylation occur across subgroups in medulloblastoma. Acta Neuropathol. 2013, 125, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Northcott, P.A.; Kool, M.; Pfister, S.M. The role of chromatin remodeling in medulloblastoma. Brain Pathol. (Zurich, Switzerland) 2013, 23, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Remke, M.; Ramaswamy, V.; Taylor, M.D. Medulloblastoma molecular dissection: The way toward targeted therapy. Curr. Opin. Oncol. 2013, 25, 674–681. [Google Scholar] [CrossRef] [PubMed]
- Parsons, D.W.; Li, M.; Zhang, X.; Jones, S.; Leary, R.J.; Lin, J.C.; Boca, S.M.; Carter, H.; Samayoa, J.; Bettegowda, C.; et al. The genetic landscape of the childhood cancer medulloblastoma. Science 2011, 331, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Huether, R.; Dong, L.; Chen, X.; Wu, G.; Parker, M.; Wei, L.; Ma, J.; Edmonson, M.N.; Hedlund, E.K.; Rusch, M.C.; et al. The landscape of somatic mutations in epigenetic regulators across 1000 paediatric cancer genomes. Nat. Commun. 2014, 5, 3630. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, V.; Taylor, M.D. Medulloblastoma: From myth to molecular. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2017, 35, 2355–2363. [Google Scholar] [CrossRef]
- Sampson, P.B.; Liu, Y.; Forrest, B.; Cumming, G.; Li, S.W.; Patel, N.K.; Edwards, L.; Laufer, R.; Feher, M.; Ban, F.; et al. The discovery of polo-like kinase 4 inhibitors: Identification of (1r,2s).2-(3-((e).4-(((cis).2,6-dimethylmorpholino)methyl)styryl). 1h.Indazol-6-yl)-5′-methoxyspiro[cyclopropane-1,3′-indolin]-2′-one (cfi-400945) as a potent, orally active antitumor agent. J. Med. Chem. 2015, 58, 147–169. [Google Scholar] [CrossRef] [PubMed]
- Sampson, P.B.; Liu, Y.; Patel, N.K.; Feher, M.; Forrest, B.; Li, S.W.; Edwards, L.; Laufer, R.; Lang, Y.; Ban, F.; et al. The discovery of polo-like kinase 4 inhibitors: Design and optimization of spiro[cyclopropane-1,3′[3h]indol]-2′(1′h).Ones as orally bioavailable antitumor agents. J. Med. Chem. 2015, 58, 130–146. [Google Scholar] [CrossRef]
- Liu, J.J.; Higgins, B.; Ju, G.; Kolinsky, K.; Luk, K.C.; Packman, K.; Pizzolato, G.; Ren, Y.; Thakkar, K.; Tovar, C.; et al. Discovery of a highly potent, orally active mitosis/angiogenesis inhibitor r1530 for the treatment of solid tumors. ACS Med. Chem. Lett. 2013, 4, 259–263. [Google Scholar] [CrossRef]
- Peart, P.A.; Tovar, J.D. Poly(cyclopropenone)s: Formal inclusion of the smallest huckel aromatic into pi-conjugated polymers. J. Org. Chem. 2010, 75, 5689–5696. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.L.; Anzola, J.V.; Davis, R.L.; Yoon, M.; Motamedi, A.; Kroll, A.; Seo, C.P.; Hsia, J.E.; Kim, S.K.; Mitchell, J.W.; et al. Cell biology. Reversible centriole depletion with an inhibitor of polo-like kinase 4. Science 2015, 348, 1155–1160. [Google Scholar] [CrossRef] [PubMed]
- Laufer, R.; Forrest, B.; Li, S.W.; Liu, Y.; Sampson, P.; Edwards, L.; Lang, Y.; Awrey, D.E.; Mao, G.; Plotnikova, O.; et al. The discovery of plk4 inhibitors: (e)-3-((1h-indazol-6-yl)methylene)indolin-2-ones as novel antiproliferative agents. J. Med. Chem. 2013, 56, 6069–6087. [Google Scholar] [CrossRef]
- Wilmes, L.J.; Pallavicini, M.G.; Fleming, L.M.; Gibbs, J.; Wang, D.; Li, K.L.; Partridge, S.C.; Henry, R.G.; Shalinsky, D.R.; Hu-Lowe, D.; et al. Ag-013736, a novel inhibitor of vegf receptor tyrosine kinases, inhibits breast cancer growth and decreases vascular permeability as detected by dynamic contrast-enhanced magnetic resonance imaging. Magn. Reson. Imaging 2007, 25, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Hu-Lowe, D.D.; Zou, H.Y.; Grazzini, M.L.; Hallin, M.E.; Wickman, G.R.; Amundson, K.; Chen, J.H.; Rewolinski, D.A.; Yamazaki, S.; Wu, E.Y.; et al. Nonclinical antiangiogenesis and antitumor activities of axitinib (ag-013736), an oral, potent, and selective inhibitor of vascular endothelial growth factor receptor tyrosine kinases 1, 2, 3. Clin. Cancer Res. 2008, 14, 7272–7283. [Google Scholar] [CrossRef] [PubMed]
- Pratz, K.W.; Cortes, J.; Roboz, G.J.; Rao, N.; Arowojolu, O.; Stine, A.; Shiotsu, Y.; Shudo, A.; Akinaga, S.; Small, D.; et al. A pharmacodynamic study of the flt3 inhibitor kw-2449 yields insight into the basis for clinical response. Blood 2009, 113, 3938–3946. [Google Scholar] [CrossRef]
- Shiotsu, Y.; Kiyoi, H.; Ishikawa, Y.; Tanizaki, R.; Shimizu, M.; Umehara, H.; Ishii, K.; Mori, Y.; Ozeki, K.; Minami, Y.; et al. Kw-2449, a novel multikinase inhibitor, suppresses the growth of leukemia cells with flt3 mutations or t315i-mutated bcr/abl translocation. Blood 2009, 114, 1607–1617. [Google Scholar] [CrossRef]
- Wetmore, C.; Boyett, J.; Li, S.; Lin, T.; Bendel, A.; Gajjar, A.; Orr, B.A. Alisertib is active as single agent in recurrent atypical teratoid rhabdoid tumors in 4 children. Neuro-Oncology 2015, 17, 882–888. [Google Scholar] [CrossRef] [PubMed]
- Melichar, B.; Adenis, A.; Lockhart, A.C.; Bennouna, J.; Dees, E.C.; Kayaleh, O.; Obermannova, R.; DeMichele, A.; Zatloukal, P.; Zhang, B.; et al. Safety and activity of alisertib, an investigational aurora kinase a inhibitor, in patients with breast cancer, small-cell lung cancer, non-small-cell lung cancer, head and neck squamous-cell carcinoma, and gastro-oesophageal adenocarcinoma: A five-arm phase 2 study. Lancet. Oncol. 2015, 16, 395–405. [Google Scholar] [PubMed]
- Liu, Z.; Lei, Q.; Wei, W.; Xiong, L.; Shi, Y.; Yan, G.; Gao, C.; Ye, T.; Wang, N.; Yu, L. Synthesis and biological evaluation of (e)-4-(3-arylvinyl-1h-indazol-6-yl)pyrimidin-2-amine derivatives as plk4 inhibitors for the treatment of breast cancer. RSC Adv. 2017, 7, 27737–27746. [Google Scholar] [CrossRef]
- Escudier, B.; Gore, M. Axitinib for the management of metastatic renal cell carcinoma. Drugs R&D 2011, 11, 113–126. [Google Scholar]
- Gross-Goupil, M.; Francois, L.; Quivy, A.; Ravaud, A. Axitinib: A review of its safety and efficacy in the treatment of adults with advanced renal cell carcinoma. Clin. Med. Insights. Oncol. 2013, 7, 269–277. [Google Scholar] [CrossRef]
- Cohen, E.E.W.; Rosen, L.S.; Vokes, E.E.; Kies, M.S.; Forastiere, A.A.; Worden, F.P.; Kane, M.A.; Sherman, E.; Kim, S.; Bycott, P.; et al. Axitinib is an active treatment for all histologic subtypes of advanced thyroid cancer: Results from a phase ii study. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2008, 26, 4708–4713. [Google Scholar] [CrossRef]
- Schiller, J.H.; Larson, T.; Ou, S.I.; Limentani, S.A.; Sandler, A.B.; Vokes, E.E.; Kim, S.; Liau, K.F.; Bycott, P.W.; Olszanski, A.J. Efficacy and safety of axitinib (ag-013736; ag) in patients (pts) with advanced non-small cell lung cancer (nsclc): A phase ii trial. J. Clin. Oncol. 2007, 25, 7507. [Google Scholar]
- Fruehauf, J.P.; Lutzky, J.; McDermott, D.F.; Brown, C.K.; Pithavala, Y.K.; Bycott, P.W.; Shalinsky, D.; Liau, K.F.; Niethammer, A.; Rixe, O. Axitinib (ag-013736) in patients with metastatic melanoma: A phase ii study. J. Clin. Oncol. 2008, 26, 9006. [Google Scholar] [CrossRef]
- Sells, T.B.; Chau, R.; Ecsedy, J.A.; Gershman, R.E.; Hoar, K.; Huck, J.; Janowick, D.A.; Kadambi, V.J.; LeRoy, P.J.; Stirling, M.; et al. Mln8054 and alisertib (mln8237): Discovery of selective oral aurora a inhibitors. ACS Med. Chem. Lett. 2015, 6, 630–634. [Google Scholar] [CrossRef] [PubMed]
- Venkataraman, S.; Alimova, I.; Tello, T.; Harris, P.S.; Knipstein, J.A.; Donson, A.M.; Foreman, N.K.; Liu, A.K.; Vibhakar, R. Targeting aurora kinase a enhances radiation sensitivity of atypical teratoid rhabdoid tumor cells. J. Neuro-Oncology 2012, 107, 517–526. [Google Scholar] [CrossRef]
- Pardridge, W.M. The blood-brain barrier: Bottleneck in brain drug development. NeuroRx J. Am. Soc. Exp. Neurother. 2005, 2, 3–14. [Google Scholar] [CrossRef]
- Gunosewoyo, H.; Yu, L.; Munoz, L.; Kassiou, M. Kinase targets in cns drug discovery. Future Med. Chem. 2017, 9, 303–314. [Google Scholar] [CrossRef]
- Heffron, T.P. Small molecule kinase inhibitors for the treatment of brain cancer. J. Med. Chem. 2016, 59, 10030–10066. [Google Scholar] [CrossRef]
- Chico, L.K.; Van Eldik, L.J.; Watterson, D.M. Targeting protein kinases in central nervous system disorders. Nat. Rev. Drug Discov. 2009, 8, 892–909. [Google Scholar] [CrossRef]
- Wager, T.T.; Hou, X.; Verhoest, P.R.; Villalobos, A. Central nervous system multiparameter optimization desirability: Application in drug discovery. ACS Chem. Neurosci. 2016, 7, 767–775. [Google Scholar] [CrossRef]
- Wager, T.T.; Hou, X.; Verhoest, P.R.; Villalobos, A. Moving beyond rules: The development of a central nervous system multiparameter optimization (cns mpo) approach to enable alignment of druglike properties. ACS Chem. Neurosci. 2010, 1, 435–449. [Google Scholar] [CrossRef]
- Levinson, N.M. The multifaceted allosteric regulation of aurora kinase a. Biochem. J. 2018, 475, 2025–2042. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.R.; Koretke, K.K.; Birkeland, M.L.; Sanseau, P.; Patrick, D.R. Evolutionary relationships of aurora kinases: Implications for model organism studies and the development of anti-cancer drugs. BMC Evol. Biol. 2004, 4, 39. [Google Scholar] [CrossRef] [PubMed]
- Lordier, L.; Chang, Y.; Jalil, A.; Aurade, F.; Garcon, L.; Lecluse, Y.; Larbret, F.; Kawashima, T.; Kitamura, T.; Larghero, J.; et al. Aurora b is dispensable for megakaryocyte polyploidization, but contributes to the endomitotic process. Blood 2010, 116, 2345–2355. [Google Scholar] [CrossRef] [PubMed]
- Noronha, S.; Alt, L.A.C.; Scimeca, T.E.; Zarou, O.; Obrzut, J.; Zanotti, B.; Hayward, E.A.; Pillai, A.; Mathur, S.; Rojas, J.; et al. Preclinical evaluation of the aurora kinase inhibitors amg 900, azd1152-hqpa, and mk-5108 on sw-872 and 93t449 human liposarcoma cells. In Vitro Cell. Dev. Biol. Anim. 2018, 54, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Ducat, D.; Zheng, Y. Aurora kinases in spindle assembly and chromosome segregation. Exp. Cell Res. 2004, 301, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Hauf, S.; Cole, R.W.; LaTerra, S.; Zimmer, C.; Schnapp, G.; Walter, R.; Heckel, A.; van Meel, J.; Rieder, C.L.; Peters, J.M. The small molecule hesperadin reveals a role for aurora b in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 2003, 161, 281–294. [Google Scholar] [CrossRef]
- Mitcheson, J.S.; Chen, J.; Lin, M.; Culberson, C.; Sanguinetti, M.C. A structural basis for drug-induced long qt syndrome. Proc. Natl. Acad. Sci. USA 2000, 97, 12329–12333. [Google Scholar] [CrossRef]
- Lynch, T.; Price, A. The effect of cytochrome P450 metabolism on drug response, interactions, and adverse effects. Am. Fam. Physician. 2007, 76, 391–396. [Google Scholar]
- Kemper, E.M.; Leenders, W.; Kusters, B.; Lyons, S.; Buckle, T.; Heerschap, A.; Boogerd, W.; Beijnen, J.H.; van Tellingen, O. Development of luciferase tagged brain tumour models in mice for chemotherapy intervention studies. Eur. J. Cancer (Oxford, England: 1990) 2006, 42, 3294–3303. [Google Scholar] [CrossRef]
- Bovée, J.V.M.G.; van Royen, M.; Bardoel, A.F.J.; Rosenberg, C.; Cornelisse, C.J.; Cleton-Jansen, A.-M.; Hogendoorn, P.C.W. Near-haploidy and subsequent polyploidization characterize the progression of peripheral chondrosarcoma. Am. J. Pathol. 2000, 157, 1587–1595. [Google Scholar] [CrossRef]
- Duensing, A.; Duensing, S. Centrosomes, polyploidy and cancer. Adv. Exp. Med. Biol. 2010, 676, 93–103. [Google Scholar] [PubMed]
- Mosieniak, G.; Sikora, E. Polyploidy: The link between senescence and cancer. Curr. Pharm. Des. 2010, 16, 734–740. [Google Scholar] [CrossRef]
- Davoli, T.; de Lange, T. The causes and consequences of polyploidy in normal development and cancer. Annu. Rev. Cell Dev. Biol. 2011, 27, 585–610. [Google Scholar] [CrossRef]
- Hau, P.M.; Siu, W.Y.; Wong, N.; Lai, P.B.; Poon, R.Y. Polyploidization increases the sensitivity to DNA-damaging agents in mammalian cells. FEBS Lett. 2006, 580, 4727–4736. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Wang, C.; He, B.; Yang, M.; Tong, M.; Long, Z.; Liu, B.; Peng, F.; Xu, L.; Zhang, Y.; et al. Aurora-a kinase: A potent oncogene and target for cancer therapy. Med. Res. Rev. 2016, 36, 1036–1079. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Qu, M.; Fang, F.; Fan, Z.; Chen, L.; Yue, W.; Xie, X.; Pei, X. Small molecule supplements improve cultured megakaryocyte polyploidization by modulating multiple cell cycle regulators. BioMed Res. Int. 2017, 2017, 2320519. [Google Scholar] [CrossRef]
- Harrington, E.A.; Bebbington, D.; Moore, J.; Rasmussen, R.K.; Ajose-Adeogun, A.O.; Nakayama, T.; Graham, J.A.; Demur, C.; Hercend, T.; Diu-Hercend, A.; et al. Vx-680, a potent and selective small-molecule inhibitor of the aurora kinases, suppresses tumor growth in vivo. Nat. Med. 2004, 10, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Versteege, I.; Medjkane, S.; Rouillard, D.; Delattre, O. A key role of the hsnf5/ini1 tumour suppressor in the control of the g1-s transition of the cell cycle. Oncogene 2002, 21, 6403–6412. [Google Scholar] [CrossRef]
- Singh, A.; Lun, X.; Jayanthan, A.; Obaid, H.; Ruan, Y.; Strother, D.; Chi, S.N.; Smith, A.; Forsyth, P.; Narendran, A. Profiling pathway-specific novel therapeutics in preclinical assessment for central nervous system atypical teratoid rhabdoid tumors (cns atrt): Favorable activity of targeting egfr- erbb2 signaling with lapatinib. Mol. Oncol. 2013, 7, 497–512. [Google Scholar] [CrossRef] [PubMed]
| Best Score Found (per Simulation) | |||||||
|---|---|---|---|---|---|---|---|
| Docking 1 | Docking 2 | Docking 3 | Docking 4 | Docking 5 | Docking 6 | Mean | |
| centrinone B | 83.58 | 78.20 | 79.18 | 83.800 | 79.86 | 81.40 | 81.00 |
| centrinone | 80.10 | 85.33 | 73.78 | 79.86 | 81.74 | 70.87 | 78.61 |
| alisertib | 65.55 | 68.66 | 69.23 | 69.50 | 69.78 | 69.32 | 68.67 |
| axitinib | 64.46 | 63.49 | 64.41 | 65.92 | 62.97 | 65.55 | 64.47 |
| CFI-400945 | 59.99 | 62.26 | 61.15 | 60.61 | 58.18 | 63.14 | 60.89 |
| R-1530 | 55.23 | 62.02 | 62.05 | 62.01 | 62.03 | 62.01 | 60.89 |
| CFI-400437 | 58.74 | 59.94 | 60.68 | 61.31 | 60.15 | 61.10 | 60.32 |
| KW-2449 | 51.85 | 57.68 | 58.51 | 59.05 | 59.12 | 58.49 | 57.45 |
| A | CFI-400945 | CFI-400437 | Centrinone | |||||
| CNS MPO Calculator | CNS MPO Calculator | CNS MPO Calculator | ||||||
| Property | Value | Property | Value | Property | Value | |||
| clogP | 5.05 | clogP | 4.07 | clogP | 4.03 | |||
| LogD7.4 | 4.98 | LogD7.4 | 3.98 | LogD7.4 | 4.03 | |||
| TPSA | 79.48 | TPSA | 86.38 | TPSA | 204.84 | |||
| MW | 534.65 | MW | 492.57 | MW | 633.65 | |||
| HBD | 2 | HBD | 2 | HBD | 2 | |||
| pKa | 6.6 | pKa | 7.2 | pKa | 2.3 | |||
| Centrinone B | R1530 | KW-2449 | ||||||
| CNS MPO Calculator | CNS MPO Calculator | CNS MPO Calculator | ||||||
| Property | Value | Property | Value | Property | Value | |||
| clogP | 4.31 | clogP | 3.82 | clogP | 2.56 | |||
| LogD7.4 | 4.31 | LogD7.4 | 3.82 | LogD7.4 | 2.14 | |||
| TPSA | 195.61 | TPSA | 62.3 | TPSA | 61.02 | |||
| MW | 631.68 | MW | 356.78 | MW | 332.40 | |||
| HBD | 2 | HBD | 2 | HBD | 2 | |||
| pKa | 4.2 | pKa | 5.6 | pKa | 8.3 | |||
| Axitinib | Alisertib | B | Compound | logBB | ||||
| CNS MPO Calculator | CNS MPO Calculator | CFI-400945 | 0.88 | |||||
| Property | Value | Property | Value | R1530 | 0.3 | |||
| clogP | 3.65 | clogP | 5.82 | KW-2449 | −0.07 | |||
| LogD7.4 | 3.65 | LogD7.4 | 2.9 | Centrinone B | −0.18 | |||
| TPSA | 95.97 | TPSA | 105.93 | Centrinone | −0.27 | |||
| MW | 386.47 | MW | 518.92 | Axitinib | −0.34 | |||
| HBD | 2 | HBD | 2 | CFI-400437 | −0.73 | |||
| pKa | 4.3 | pKa | 2.1 | Alisertib | −0.96 | |||
| PLK4 | AURKA | AURKB | AURKC | |
|---|---|---|---|---|
| CFI-400437 | 1.55 | 37.2 | 13.1 | 4.88 |
| centrinone | 2.71 | 108 | 680 | 493 |
| CFI-400945 | 4.85 | 188 | 70.7 | 106 |
| axitinib | 6.51 | 13.1 | 16.9 | 31.2 |
| R1530 | 7.06 | 6.22 | 42.6 | 30.9 |
| centrinone B | 8.69 | 623 | 10,000 | 5810 |
| KW-2449 | 52.6 | 45.8 | 23.8 | 23.2 |
| alisertib | 62.7 | 0.55 | 6.7 | 9.43 |
| SelectScreen IC50 (nM) | MON | BT-12 | BT-16 | DAOY | D283 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MTT | PB | MTT | PB | MTT | PB | MTT | PB | MTT | PB | ||
| CFI-400437 | 1.55 | 722 | 640 | 61.6 | 604 | 1920 | >10,000 | 684 | 768 | >10,000 | 6304 |
| centrinone | 2.71 | 1840 | 5000 | 3875 | 3400 | 1557 | 2220 | 1250 | 4500 | 6950 | 6650 |
| CFI-400945 | 4.85 | 67.4 | 5130 | 7250 | 8470 | 3300 | 5830 | 47.1 | 94 | 71 | 8400 |
| axitinib | 6.51 | >10,000 | 4500 | >10,000 | >10,000 | 5950 | >10,000 | 1147 | 1430 | 0.394 | 0.918 |
| R-1530 | 7.06 | 3200 | 3750 | >10,000 | >10,000 | 3895 | >10,000 | 829 | 1852 | >10,000 | >10,000 |
| centrinone-B | 8.69 | >10,000 | >10,000 | 5188 | 6320 | 5467 | >10,000 | >10,000 | >10,000 | 1240 | 1740 |
| KW-2449 | 52.6 | 2452 | 1743 | >10,000 | >10,000 | >10,000 | 1840 | >10,000 | >10,000 | 8050 | >10,000 |
| alisertib | 62.7 | 358 | 213 | >10,000 | >10,000 | >10,000 | >10,000 | 340 | 31 | 9533 | 9133 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suri, A.; Bailey, A.W.; Tavares, M.T.; Gunosewoyo, H.; Dyer, C.P.; Grupenmacher, A.T.; Piper, D.R.; Horton, R.A.; Tomita, T.; Kozikowski, A.P.; et al. Evaluation of Protein Kinase Inhibitors with PLK4 Cross-Over Potential in a Pre-Clinical Model of Cancer. Int. J. Mol. Sci. 2019, 20, 2112. https://doi.org/10.3390/ijms20092112
Suri A, Bailey AW, Tavares MT, Gunosewoyo H, Dyer CP, Grupenmacher AT, Piper DR, Horton RA, Tomita T, Kozikowski AP, et al. Evaluation of Protein Kinase Inhibitors with PLK4 Cross-Over Potential in a Pre-Clinical Model of Cancer. International Journal of Molecular Sciences. 2019; 20(9):2112. https://doi.org/10.3390/ijms20092112
Chicago/Turabian StyleSuri, Amreena, Anders W. Bailey, Maurício T. Tavares, Hendra Gunosewoyo, Connor P. Dyer, Alex T. Grupenmacher, David R. Piper, Robert A. Horton, Tadanori Tomita, Alan P. Kozikowski, and et al. 2019. "Evaluation of Protein Kinase Inhibitors with PLK4 Cross-Over Potential in a Pre-Clinical Model of Cancer" International Journal of Molecular Sciences 20, no. 9: 2112. https://doi.org/10.3390/ijms20092112
APA StyleSuri, A., Bailey, A. W., Tavares, M. T., Gunosewoyo, H., Dyer, C. P., Grupenmacher, A. T., Piper, D. R., Horton, R. A., Tomita, T., Kozikowski, A. P., Roy, S. M., & Sredni, S. T. (2019). Evaluation of Protein Kinase Inhibitors with PLK4 Cross-Over Potential in a Pre-Clinical Model of Cancer. International Journal of Molecular Sciences, 20(9), 2112. https://doi.org/10.3390/ijms20092112








