Phylogenetic, Molecular, and Functional Characterization of PpyCBF Proteins in Asian Pears (Pyrus pyrifolia)
Abstract
:1. Introduction
2. Results
2.1. Identifications and Characterizations of PpyCBF Subfamily
2.2. Strong Induction of PpyCBF Transcription by Various Abiotic Stresses and ABA Treatment
2.3. Increased Transcripts of PpyCBFs Induced by Low Temperature and ABA during Pear Bud Endodormancy
2.4. Overexpressions of PpyCBF2 and PpyCBF3 Positively Regulate Abiotic Stress Tolerances in Transgenic Arabidopsis
2.5. PpyCBF Transcriptional Activation of 6X C-Repeat Binding Sites and Stress-Responsive Genes
2.6. PpyCBFs Can Also Bind at the TCGAC Binding Site in the PpyCOR15A Promoter
3. Discussion
4. Materials and Methods
4.1. Identification and Characterization of PpyCBFs
4.2. Plant Materials and Abiotic Stress Treatments
4.3. Analysis of Stress Tolerance of Transgenic Plants
4.4. Histochemical Analysis of H2O2 and O2•−
4.5. RNA Extraction and cDNA Synthesis
4.6. qRT-PCR Analysis
4.7. Site-Directed Mutagenesis of Gene Promoters
4.8. Transient Expression and Luciferase Measurement
4.9. Yeast One-Hybrid Assay
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CORs | Cold Regulons |
| HMM | Hidden Markov Model |
| MEGA | Molecular Evolutionary Genetics Analysis |
| TF | Transcription factor |
| Y1H | Yeast one hybrid |
| SnRK2 | Snf1-Related kinase 2 |
| CTAB | Cetyltrimethyl Ammonium Bromide |
References
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed]
- Licausi, F.; Ohme Takagi, M.; Perata, P. APETALA 2/Ethylene Responsive Factor (AP 2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef]
- Jaglo, K.R.; Kleff, S.; Amundsen, K.L.; Zhang, X.; Haake, V.; Zhang, J.Z.; Deits, T.; Thomashow, M.F. Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol. 2001, 127, 910–917. [Google Scholar] [CrossRef]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration-and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef] [PubMed]
- Thomashow, M.F. Molecular basis of plant cold acclimation: Insights gained from studying the CBF cold response pathway. Plant Physiol. 2010, 154, 571–577. [Google Scholar] [CrossRef]
- Welling, A.; Palva, E.T. Involvement of CBF transcription factors in winter hardiness in birch. Plant Physiol. 2008, 147, 1199–1211. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.K.; Agarwal, P.; Reddy, M.; Sopory, S.K. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006, 25, 1263–1274. [Google Scholar] [CrossRef]
- Mizoi, J.; Shinozaki, K.; Yamaguchi Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 86–96. [Google Scholar] [CrossRef]
- Siddiqua, M.; Nassuth, A. Vitis CBF1 and Vitis CBF4 differ in their effect on Arabidopsis abiotic stress tolerance, development and gene expression. Plant Cell Environ. 2011, 34, 1345–1359. [Google Scholar] [CrossRef] [PubMed]
- Novillo, F.; Alonso, J.M.; Ecker, J.R.; Salinas, J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 3985–3990. [Google Scholar] [CrossRef]
- Benedict, C.; Skinner, J.S.; Meng, R.; Chang, Y.; Bhalerao, R.; Huner, N.P.; Finn, C.E.; Chen, T.H.; Hurry, V. The CBF1-dependent low temperature signalling pathway, regulon and increase in freeze tolerance are conserved in Populus spp. Plant Cell Environ. 2006, 29, 1259–1272. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Siddiqua, M.; Braybrook, S.; Nassuth, A. Three grape CBF/DREB1 genes respond to low temperature, drought and abscisic acid. Plant Cell Environ. 2006, 29, 1410–1421. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Tattersall, E.A.; Siddiqua, M.K.; Cramer, G.R.; Nassuth, A. CBF4 is a unique member of the CBF transcription factor family of Vitis vinifera and Vitis riparia. Plant Cell Environ. 2008, 31, 1–10. [Google Scholar] [CrossRef]
- Kidokoro, S.; Watanabe, K.; Ohori, T.; Moriwaki, T.; Maruyama, K.; Mizoi, J.; Myint Phyu Sin Htwe, N.; Fujita, Y.; Sekita, S.; Shinozaki, K. Soybean DREB 1/CBF type transcription factors function in heat and drought as well as cold stress responsive gene expression. Plant J. 2015, 81, 505–518. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Liu, X.D.; Chi, X.J.; Wu, C.A.; Li, Y.Z.; Song, L.L.; Liu, X.M.; Wang, Y.F.; Wang, F.W.; Zhang, C. Dwarf apple MbDREB1 enhances plant tolerance to low temperature, drought, and salt stress via both ABA-dependent and ABA-independent pathways. Planta 2011, 233, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Magome, H.; Yamaguchi, S.; Hanada, A.; Kamiya, Y.; Oda, K. dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J. 2004, 37, 720–729. [Google Scholar] [CrossRef]
- Haake, V.; Cook, D.; Riechmann, J.; Pineda, O.; Thomashow, M.F.; Zhang, J.Z. Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol. 2002, 130, 639–648. [Google Scholar] [CrossRef]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought-and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Dong, C.H.; Zhu, J.K. Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Curr. Opin. Plant Biol. 2007, 10, 290–295. [Google Scholar] [CrossRef]
- Norén, L.; Kindgren, P.; Stachula, P.; Rühl, M.; Eriksson, M.E.; Hurry, V.; Strand, Å. Circadian and plastid signaling pathways are integrated to ensure correct expression of the CBF and COR genes during photoperiodic growth. Plant Physiol. 2016, 171, 1392–1406. [Google Scholar]
- Rubio, S.; Noriega, X.; Pérez, F.J. Abscisic acid (ABA) and low temperatures synergistically increase the expression of CBF/DREB1 transcription factors and cold-hardiness in grapevine dormant buds. Ann. Bot. 2018, 20, 1–9. [Google Scholar] [CrossRef]
- Li, J.; Yan, X.; Yang, Q.; Ma, Y.; Yang, B.; Tian, J.; Teng, Y.; Bai, S. PpCBFs selectively regulate PpDAMs and contribute to the pear bud endodormancy process. Plant Mol. Biol. 2019, 1–12. [Google Scholar] [CrossRef]
- Li, J.; Xu, Y.; Niu, Q.; He, L.; Teng, Y.; Bai, S. Abscisic Acid (ABA) promotes the induction and maintenance of Pear (Pyrus pyrifolia White Pear Group) flower bud endodormancy. Int. J. Mol. Sci. 2018, 19, 310. [Google Scholar] [CrossRef]
- Xiong, L.; Wang, R.G.; Mao, G.; Koczan, J.M. Identification of drought tolerance determinants by genetic analysis of root response to drought stress and abscisic acid. Plant Physiol. 2006, 142, 1065–1074. [Google Scholar] [CrossRef] [PubMed]
- Hetherington, A.M. Guard cell signaling. Cell 2001, 107, 711–714. [Google Scholar] [CrossRef]
- Niu, Q.; Li, J.; Cai, D.; Qian, M.; Jia, H.; Bai, S.; Hussain, S.; Liu, G.; Teng, Y.; Zheng, X. Dormancy-associated MADS-box genes and microRNAs jointly control dormancy transition in pear (Pyrus pyrifolia white pear group) flower bud. J. Exp. Bot. 2015, 67, 239–257. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Chen, P.; Yan, Y.; Bao, C.; Li, X.; Wang, L.; Shen, X.; Li, H.; Liu, X.; Niu, C. An atypical R2R3 MYB transcription factor increases cold hardiness by CBF dependent and CBF independent pathways in apple. New Phytologist. 2018, 218, 201–218. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Li, R.; Qu, F.; You, C.; Wang, X.; Hao, Y. An apple NAC transcription factor negatively regulates cold tolerance via CBF-dependent pathway. J. Plant Physiol. 2018, 221, 74–80. [Google Scholar] [CrossRef]
- Jin, C.; Li, K.Q.; Xu, X.Y.; Zhang, H.P.; Chen, H.X.; Chen, Y.H.; Hao, J.; Wang, Y.; Huang, X.S.; Zhang, S.L. A novel NAC transcription factor, PbeNAC1, of Pyrus betulifolia confers cold and drought tolerance via Interacting with PbeDREBs and activating the expression of stress-responsive genes. Front. Plant Sci. 2017, 8, 1049. [Google Scholar] [CrossRef]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef]
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Tsz fung, F.C. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010, 61, 672–685. [Google Scholar] [CrossRef]
- Zhan, X.; Zhu, J.K.; Lang, Z. Increasing freezing tolerance: Kinase regulation of ICE1. Dev. Cell 2015, 32, 257–258. [Google Scholar] [CrossRef]
- Zhou, M.; Shen, C.; Wu, L.; Tang, K.; Lin, J. CBF dependent signaling pathway: A key responder to low temperature stress in plants. Crit. Rev. Biotechnol. 2011, 31, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y. The pear industry and research in China. Acta Hortic. 2011, 909, 161–170. [Google Scholar] [CrossRef]
- Teng, Y. Advances in the research on phylogeny of the genus Pyrus and the origin of pear cultivars native to East Asia. J. Fruit Sci. 2017, 34, 370–378. [Google Scholar]
- Ryu, J.Y.; Hong, S.Y.; Jo, S.H.; Woo, J.C.; Lee, S.; Park, C.M. Molecular and functional characterization of cold-responsive C-repeat binding factors from Brachypodium distachyon. BMC Plant Biol. 2014, 14, 15. [Google Scholar] [CrossRef] [PubMed]
- Kitashiba, H.; Ishizaka, T.; Isuzugawa, K.; Nishimura, K.; Suzuki, T. Expression of a sweet cherry DREB1/CBF ortholog in Arabidopsis confers salt and freezing tolerance. J. Plant Physiol. 2004, 161, 1171–1176. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Wang, F.; Wu, Z.; Li, Y.; Shi, G.; Hu, J.; Hou, X. Components of the Arabidopsis CBF cold-response pathway are conserved in non-heading Chinese cabbage. Plant Mol. Biol. Rep. 2011, 29, 525–532. [Google Scholar] [CrossRef]
- Wang, Q.J.; Xu, K.Y.; Tong, Z.G.; Wang, S.H.; Gao, Z.H.; Zhang, J.Y.; Zong, C.W.; Qiao, Y.S.; Zhang, Z. Characterization of a new dehydration responsive element binding factor in central arctic cowberry. Plant Cell Tissue Organ Cult. 2010, 101, 211–219. [Google Scholar] [CrossRef]
- Takemura, Y.; Kuroki, K.; Jiang, M.; Matsumoto, K.; Tamura, F. Identification of the expressed protein and the impact of change in ascorbate peroxidase activity related to endodormancy breaking in Pyrus pyrifolia. Plant Physiol. Biochem. 2015, 86, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Achard, P.; Renou, J.P.; Berthomé, R.; Harberd, N.P.; Genschik, P. Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr. Biol. 2008, 18, 656–660. [Google Scholar] [CrossRef] [PubMed]
- Uno, Y.; Furihata, T.; Abe, H.; Yoshida, R.; Shinozaki, K.; Yamaguchi Shinozaki, K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 2000, 97, 11632–11637. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.; Yan, X.; Li, J.; Yang, Q.; Jamil, W.; Teng, Y.; Bai, S. Genome wide identification and predicted functional analyses of NAC transcription factors in Asian pears. BMC Plant Biol. 2018, 18, 214. [Google Scholar] [CrossRef] [PubMed]
- Park, H.C.; Kim, M.L.; Kang, Y.H.; Jeon, J.M.; Yoo, J.H.; Kim, M.C.; Park, C.Y.; Jeong, J.C.; Moon, B.C.; Lee, J.H. Pathogen-and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol. 2004, 135, 2150–2161. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.R.; Wakeel, A.; Muhammad, N.; Liu, B.; Wu, M.; Liu, Y.; Ali, I.; Zaidi, S.H.R.; Azhar, W.; Song, G. Involvement of ethylene signaling in zinc oxide nanoparticle mediated biochemical changes in Arabidopsis thaliana leaves. Environ. Sci. Nano 2019, 6, 341–355. [Google Scholar] [CrossRef]
- Yang, Q.; Niu, Q.; Li, J.; Zheng, X.; Ma, Y.; Bai, S.; Teng, Y. PpHB22, a member of HD-Zip proteins, activates PpDAM1 to regulate bud dormancy transition in ‘Suli’ pear (Pyrus pyrifolia White Pear Group). Plant Physiol. Biochem. 2018, 127, 355–365. [Google Scholar] [CrossRef]
- Tao, R.; Bai, S.; Ni, J.; Yang, Q.; Zhao, Y.; Teng, Y. The blue light signal transduction pathway is involved in anthocyanin accumulation in ‘Red Zaosu’ pear. Planta 2018, 1–12. [Google Scholar] [CrossRef] [PubMed]
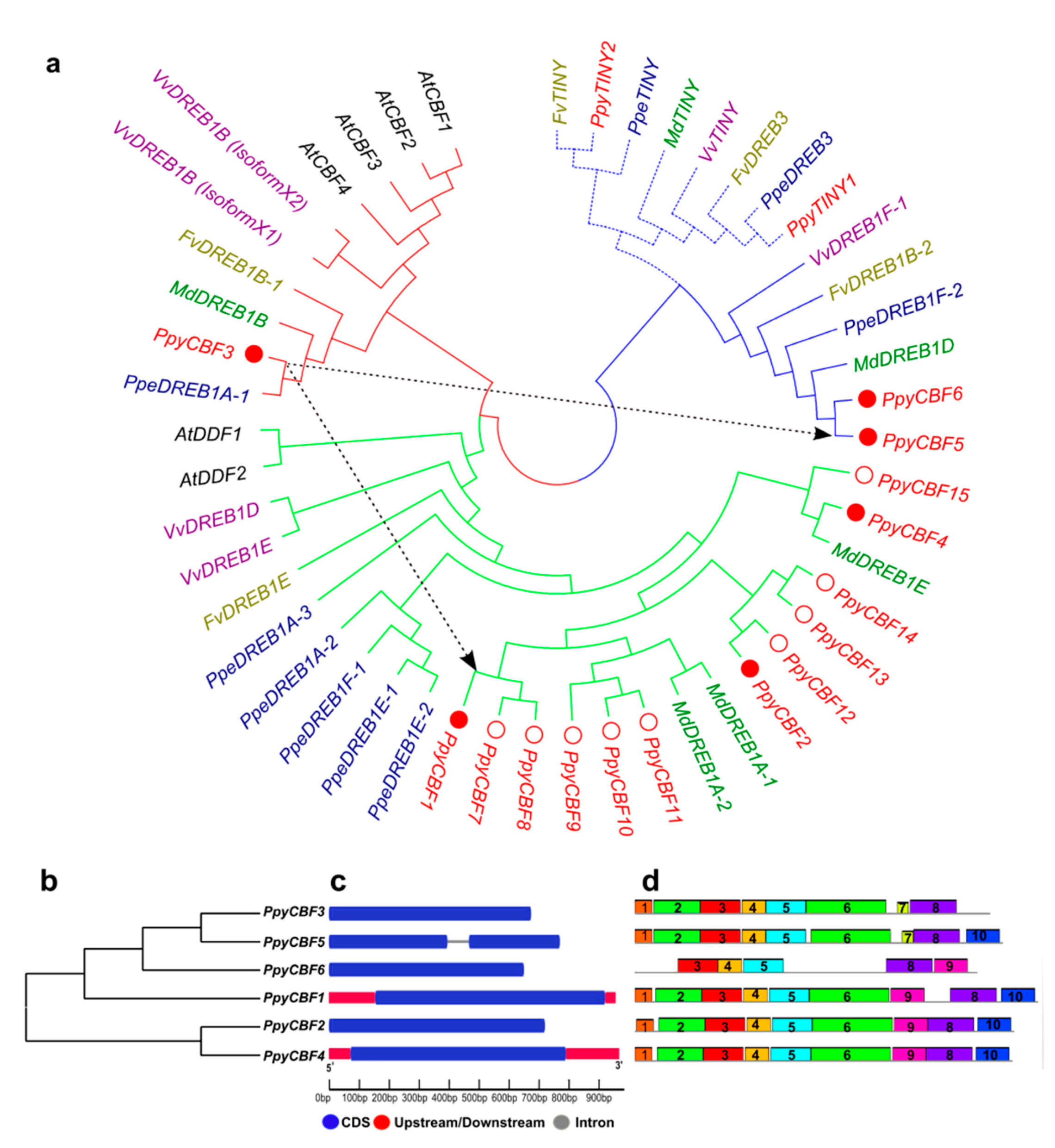


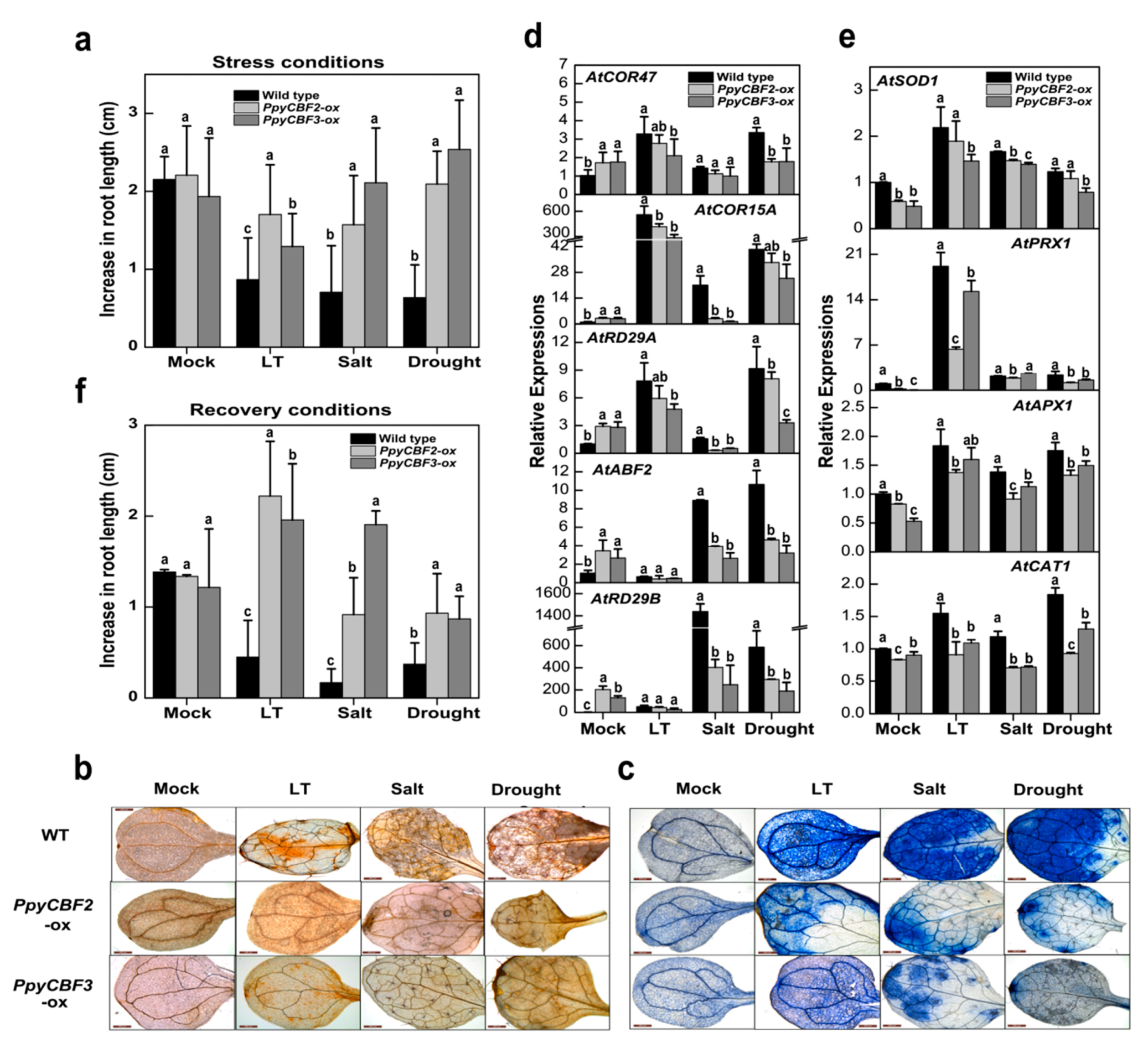
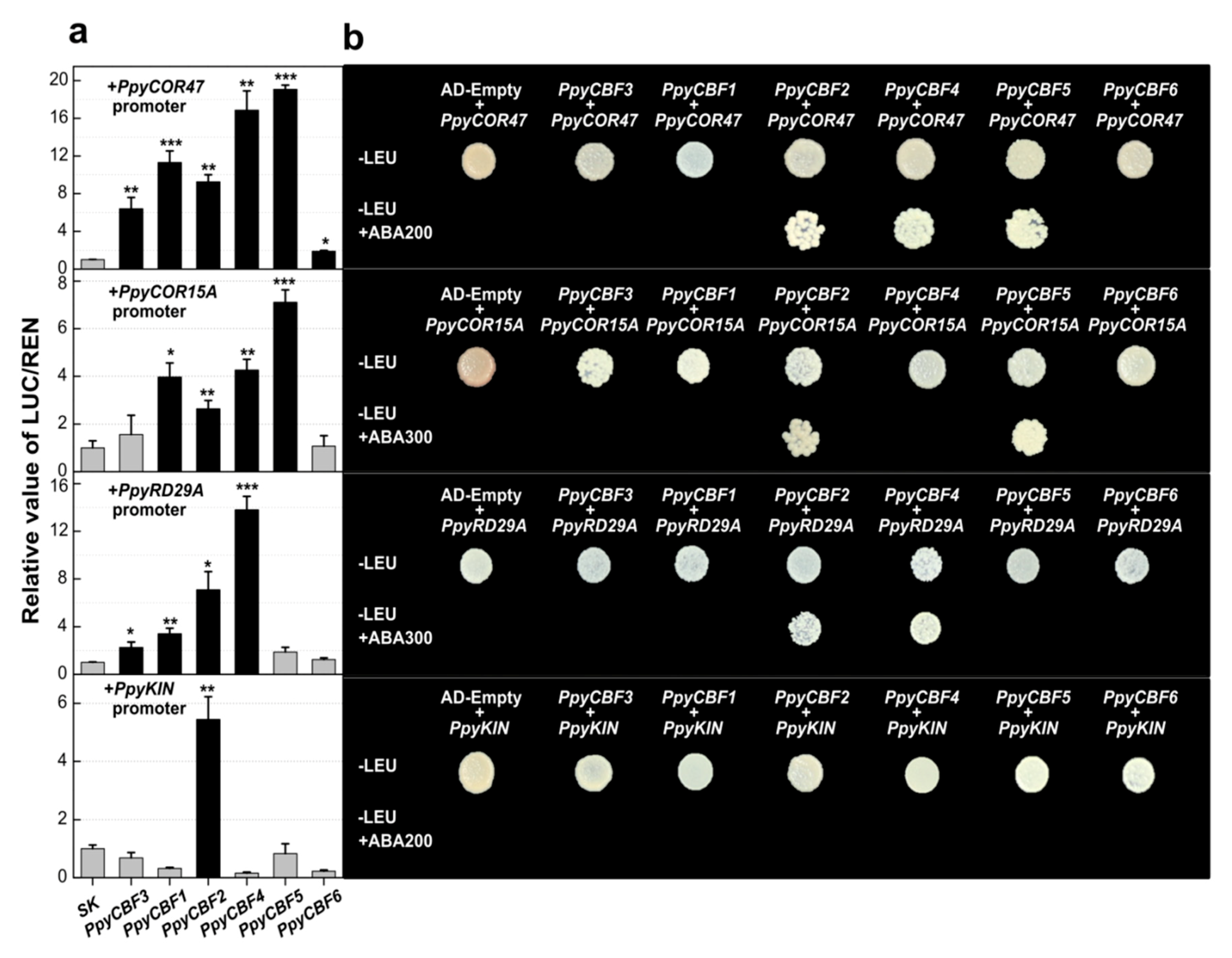
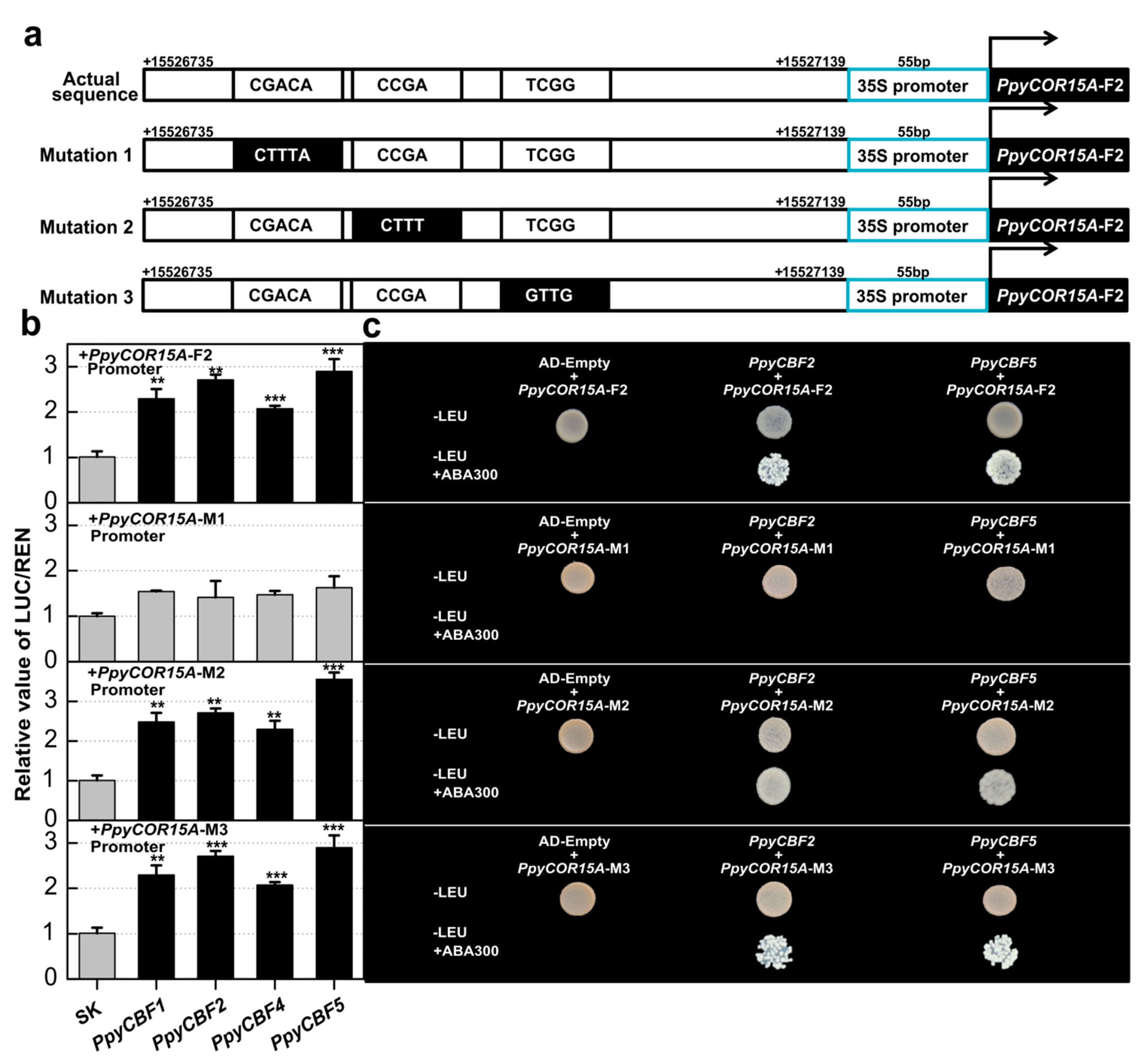
| TFs family | Functions | cis-Element | Sequences | PpyCBF3 | PpyCBF1 | PpyCBF2 | PpyCBF4 | PpyCBF5 | PpyCBF6 |
|---|---|---|---|---|---|---|---|---|---|
| ABI3/VP1 | ABA responsive | ABRE | CATGC | 1 | 4 | 1 | 4 | 1 | 1 |
| AP2/EREBP | Cold, drought, NaCl | CRT/DRE | CCGAC | 6 | 4 | 1 | 4 | 8 | 3 |
| AP2/RAV | Photoperiodism, flowering | B3 | CAACA | 10 | 8 | 5 | 7 | 9 | 8 |
| ARF | Auxin response | SURE | GAGACA | 3 | 2 | 2 | 2 | 2 | 1 |
| bHLH | Iron toxicity | IRO2 | CACGTGG | 0 | 0 | 2 | 2 | 0 | 2 |
| bZIP | ABA, NaCl, drought, heat | G-box1 | CACGTG | 0 | 1 | 2 | 2 | 0 | 3 |
| bZIP | Salt, Pathogen | GT-1-like box | GAAAAA | 3 | 3 | 7 | 3 | 4 | 4 |
| ERF | Defense responses | GCC box | AGCCG | 7 | 1 | 0 | 4 | 9 | 0 |
| GATA | Light response | GATA box | GATA | 14 | 16 | 16 | 11 | 12 | 15 |
| MADS | Plant development | MIKC | CC[A/T]5 | 1 | 0 | 1 | 3 | 1 | 2 |
| MYB like | Light response | I BOX | AAACCA | 1 | 0 | 2 | 1 | 0 | 0 |
| MYB/SANT | Gibberellin response | GARC | AACAAA | 6 | 3 | 6 | 4 | 2 | 3 |
| MYC-like bHLH | Cold stress | ICE1-like | CATTTG | 1 | 1 | 4 | 1 | 2 | 1 |
| NAC | Cold, drought, NaCl | NAC | CATGT | 2 | 3 | 3 | 2 | 3 | 3 |
| TCP/PCF1 | Oxidative stress | Site 2 | TGGGC | 3 | 1 | 3 | 1 | 1 | 2 |
| WRKY | Bacterial blight | PRE2 | ACGCTG | 1 | 0 | 0 | 0 | 2 | 0 |
| WRKY | Bacterial blight | PRE4 | TGCGCT | 1 | 0 | 0 | 0 | 2 | 1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmad, M.; Li, J.; Yang, Q.; Jamil, W.; Teng, Y.; Bai, S. Phylogenetic, Molecular, and Functional Characterization of PpyCBF Proteins in Asian Pears (Pyrus pyrifolia). Int. J. Mol. Sci. 2019, 20, 2074. https://doi.org/10.3390/ijms20092074
Ahmad M, Li J, Yang Q, Jamil W, Teng Y, Bai S. Phylogenetic, Molecular, and Functional Characterization of PpyCBF Proteins in Asian Pears (Pyrus pyrifolia). International Journal of Molecular Sciences. 2019; 20(9):2074. https://doi.org/10.3390/ijms20092074
Chicago/Turabian StyleAhmad, Mudassar, Jianzhao Li, Qinsong Yang, Wajeeha Jamil, Yuanwen Teng, and Songling Bai. 2019. "Phylogenetic, Molecular, and Functional Characterization of PpyCBF Proteins in Asian Pears (Pyrus pyrifolia)" International Journal of Molecular Sciences 20, no. 9: 2074. https://doi.org/10.3390/ijms20092074
APA StyleAhmad, M., Li, J., Yang, Q., Jamil, W., Teng, Y., & Bai, S. (2019). Phylogenetic, Molecular, and Functional Characterization of PpyCBF Proteins in Asian Pears (Pyrus pyrifolia). International Journal of Molecular Sciences, 20(9), 2074. https://doi.org/10.3390/ijms20092074






