Quantification of the Lamin A/C Transcript Variants in Cancer Cell Lines by Targeted Absolute Quantitative Proteomics and Correlation with mRNA Expression
Abstract
1. Introduction
2. Results
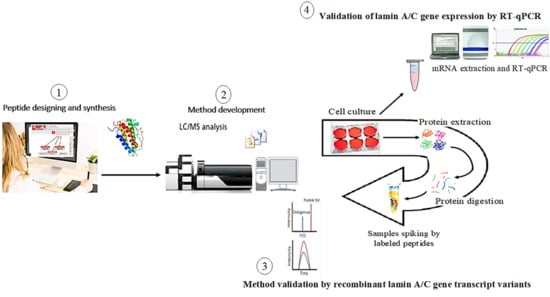
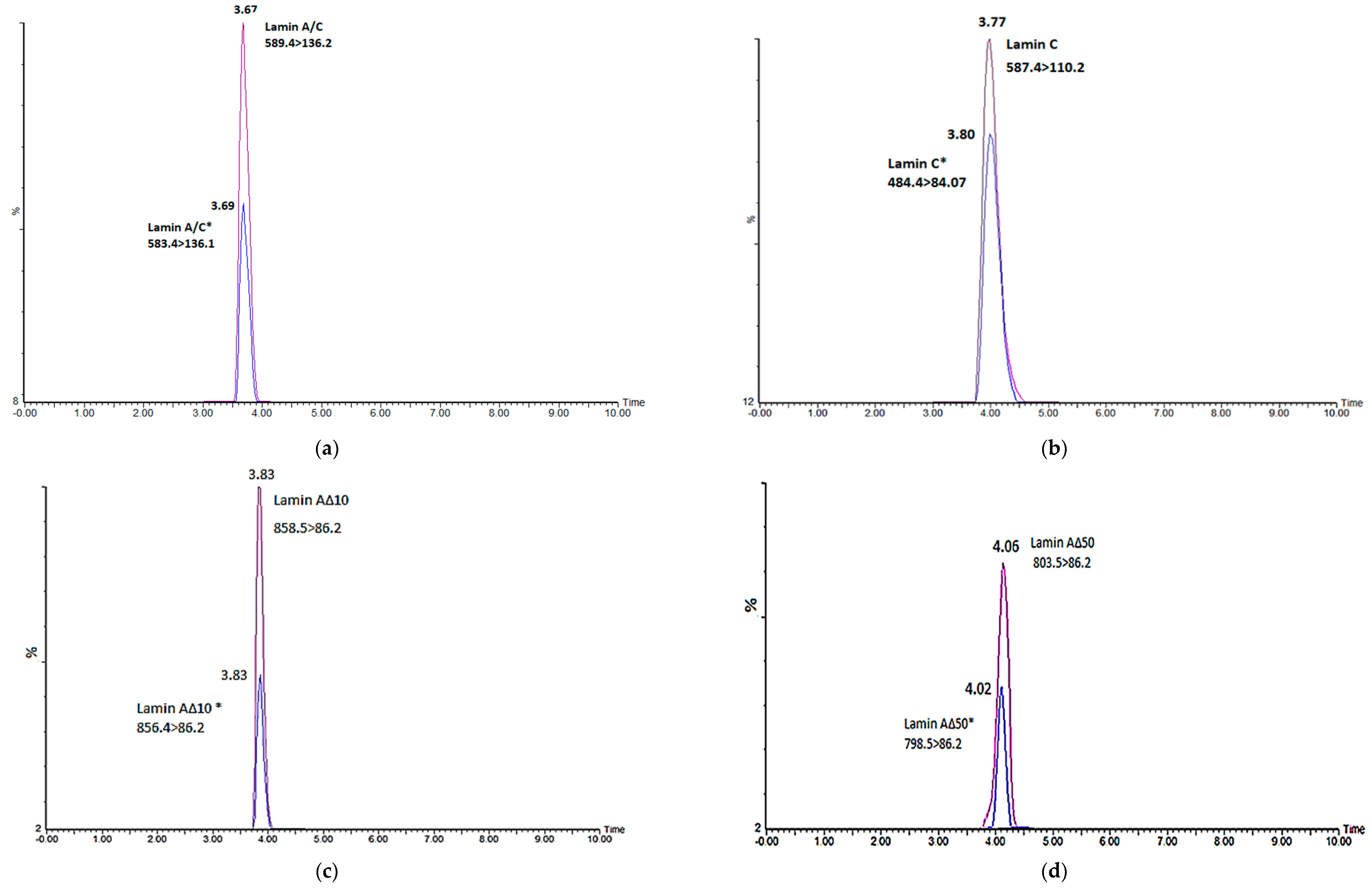
2.1. Method Development for Lamin A/C Gene Transcript Variants and Mass Spectrometry Optimization
2.2. Data Validation
2.2.1. Intra- and Inter-Day Accuracy and Precision
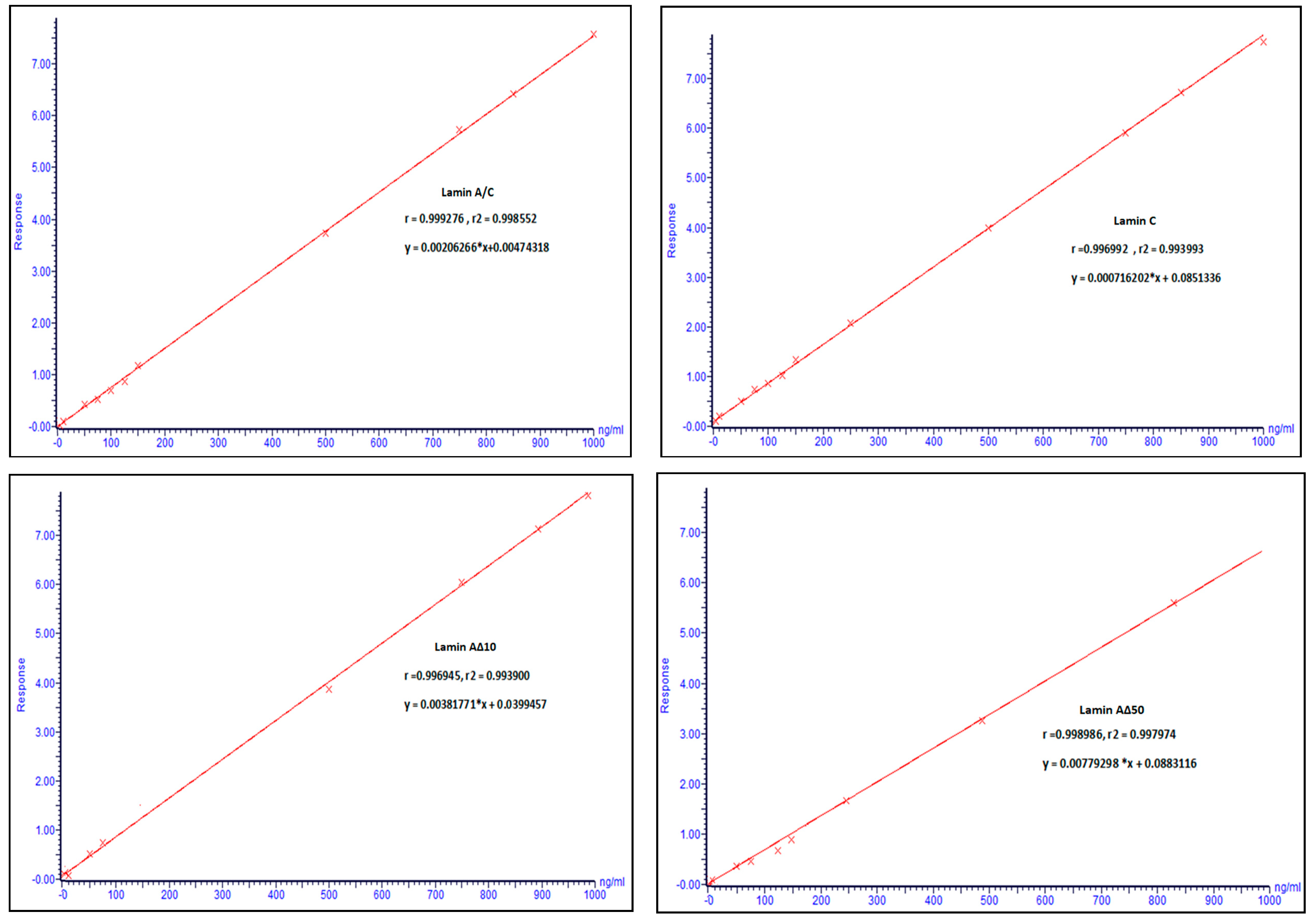
2.2.2. Linearity, Sensitivity, and Stability
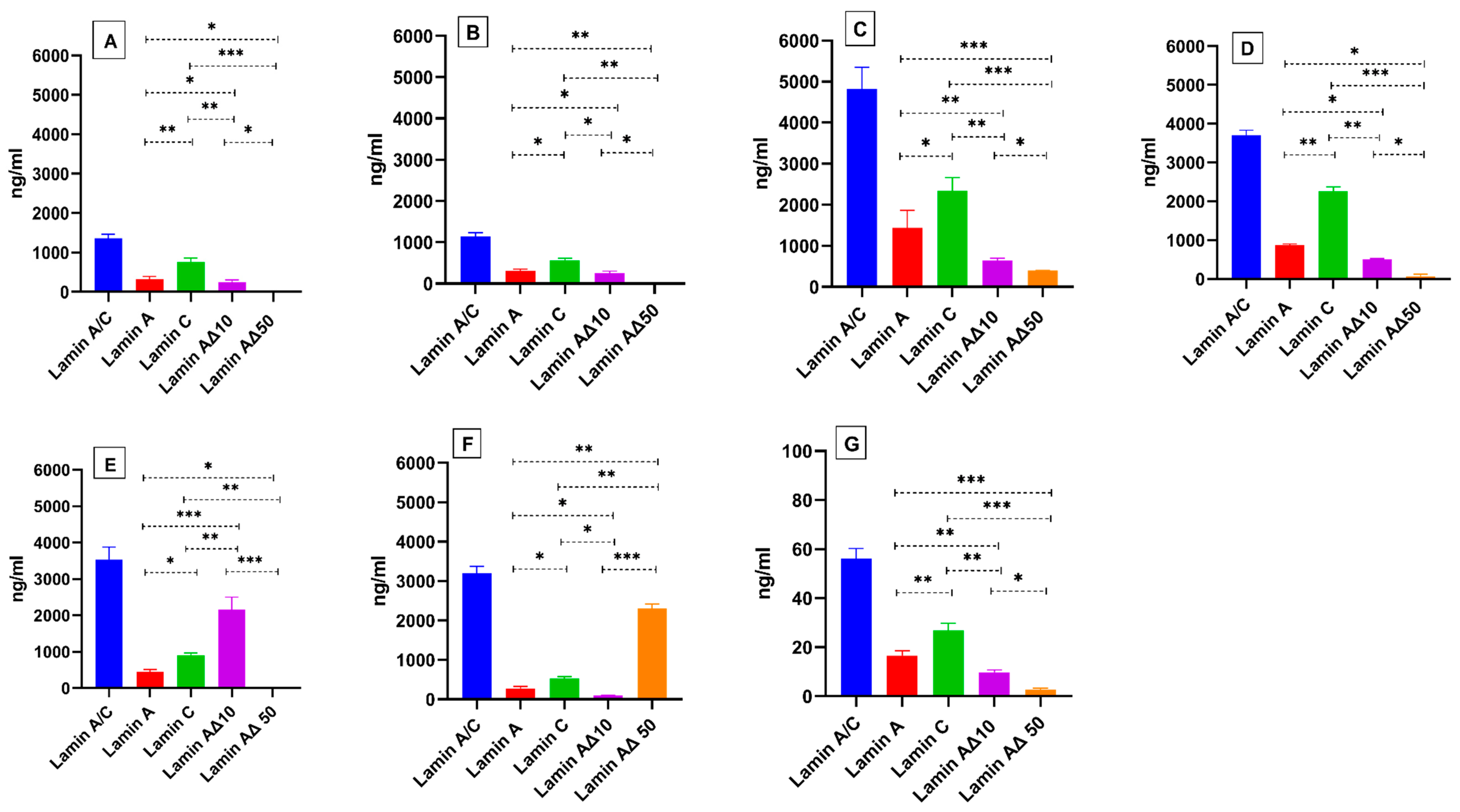
2.3. Validation of Mass Spectrometric Method by Recombinant Lamin A/C Gene Transcript Variants
2.4. Validation of Lamin A/C Gene Expression by RT-qPCR
3. Discussion
4. Materials and Methods
4.1. Chemicals and Materials
4.2. Establishing a Multiplex Assay for the Simultaneous Quantification of the Four Lamin A/C Gene Transcript Variants
4.2.1. Selection of Signature Peptides Specific for Lamin A/C Transcript Variants
4.2.2. Establishing a Mass Spectrometric Method for Lamin A/C Gene Transcript Variants
LC/MS Conditions
Preparation of Calibration Standards and Quality Control (QC) Samples
Validation of the LC/MS Method
Stability
4.3. Validation of the LC/MS Method by Recombinant Lamin A/C Gene Transcript Variants
4.4. Validation of Lamin A/C Gene Expression by RT-qPCR
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| °C | Degrees Celsius |
| µg/mL | microgram/milliliter |
| C-terminus | Carboxyl terminus |
| CV | Coefficient of Variation |
| DNA | Deoxyribonucleic acid |
| FBS | Fetal bovine serum |
| GFP | Green Fluorescent Protein |
| HQC | High Quality control |
| IS | Internal Standard |
| L/mL/µL | Liter/milliliter/microliter |
| LC–MS/MS | Liquid Chromatography Mass Spectrometry MS |
| TV2 | lamin C |
| TV3 | lamin AΔ10 |
| TV4 | lamin AΔ50 |
| LQC | Low Quality control |
| M/mM/µM | Molar/millimolar/micromolar |
| mRNA | messenger RNA |
| ng/mL | nanogram/milliliter |
| PBS | Phosphate buffered saline |
| PCR | Polymerase chain reaction |
| QC | Quality control |
| qPCR | Quantitative PCR |
| RE | Relative Error |
| REC% | recovery percent |
| RNA | Ribonucleic acid |
References
- Agrelo, R.; Setien, F.; Espada, J.; Artiga, M.J.; Rodriguez, M.; Pérez-Rosado, A.; Sanchez-Aguilera, A.; Fraga, M.F.; Piris, M.A.; Esteller, M. Inactivation of the LAMIN A/C gene by CpG island promoter hypermethylation in hematologic malignancies, and its association with poor survival in nodal diffuse large B-cell lymphoma. J. Clin. Oncol. 2005, 23, 3940–3947. [Google Scholar] [CrossRef] [PubMed]
- Olkowicz, M.; Jablonska, P.; Rogowski, J.; Smolenski, R.T. Simultaneous accurate quantification of HO-1, CD39, and CD73 in human calcified aortic valves using multiple enzyme digestion–filter aided sample pretreatment (MED-FASP) method and targeted proteomics. Talanta 2018, 182, 492–499. [Google Scholar] [CrossRef] [PubMed]
- Borroni, A.P.; Emanuelli, A.; Shah, P.A.; Ilić, N.; Apel-Sarid, L.; Paolini, B.; Manikoth Ayyathan, D.; Koganti, P.; Levy-Cohen, G.; Blank, M. Smurf2 regulates stability and the autophagic–lysosomal turnover of lamin A and its disease-associated form progerin. Aging Cell 2018, 17, e12732. [Google Scholar] [CrossRef] [PubMed]
- Domon, B.; Aebersold, R. Mass spectrometry and protein analysis. Science 2006, 312, 212–217. [Google Scholar] [CrossRef]
- Benson, E.K.; Lee, S.W.; Aaronson, S.A. Role of progerin-induced telomere dysfunction in HGPS premature cellular senescence. J. Cell Sci. 2010, 123, 2605–2612. [Google Scholar] [CrossRef]
- Capo-Chichi, C.D.; Cai, K.Q.; Smedberg, J.; Ganjei-Azar, P.; Godwin, A.K.; Xu, X.X. Loss of A-type Lamin expression compromises nuclear envelope integrity in breast cancer. Chin. J. Cancer 2011, 30, 415. [Google Scholar] [CrossRef] [PubMed]
- Worman, H.J. Cell signaling abnormalities in cardiomyopathy caused by lamin A/C gene mutations. Biochem. Soc. Trans. 2018, 46, 37–42. [Google Scholar] [CrossRef]
- Wu, Z.; Wu, L.; Weng, D.; Xu, D.; Geng, J.; Zhao, F. Reduced expression of lamin A/C correlates with poor histological differentiation and prognosis in primary gastric carcinoma. J. Exp. Clin. Cancer Res. 2009, 28, 8. [Google Scholar] [CrossRef]
- Domínguez-Gerpe, L.; Araújo-Vilar, D. Prematurely aged children: Molecular alterations leading to Hutchinson-Gilford progeria and Werner syndromes. Curr. Aging Sci. 2008, 1, 202–212. [Google Scholar] [CrossRef]
- Lukasova, E.; Kovarik, A.; Kozubek, S. Consequences of lamin B1 and lamin B receptor downregulation in senescence. Cells 2018, 7, 11. [Google Scholar] [CrossRef]
- Abdel Rahman, A.M.; Kamath, S.D.; Gagné, S.; Lopata, A.L.; Helleur, R. Comprehensive proteomics approach in characterizing and quantifying allergenic proteins from northern shrimp: Toward better occupational asthma prevention. J. Proteome Res. 2013, 12, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.M.; Kamath, S.D.; Lopata, A.L.; Robinson, J.J.; Helleur, R.J. Biomolecular characterization of allergenic proteins in snow crab (Chionoecetes opilio) and de novo sequencing of the second allergen arginine kinase using tandem mass spectrometry. J. Proteomics 2011, 74, 231–241. [Google Scholar] [CrossRef]
- Hsiao, Y.C.; Chi, L.M.; Chien, K.Y.; Chiang, W.F.; Chen, S.F.; Chuang, Y.N.; Lin, S.Y.; Wu, C.C.; Chang, Y.T.; Chu, L.J.; et al. Development of a multiplexed assay for oral cancer candidate biomarkers using peptide immunoaffinity enrichment and targeted mass spectrometry. Mol. Cell. Proteomics 2017, 16, 1829–1849. [Google Scholar] [CrossRef] [PubMed]
- Jacob, M.; Malkawi, A.; Albast, N.; Al Bougha, S.; Lopata, A.; Dasouki, M.; Rahman, A.M.A. A targeted metabolomics approach for clinical diagnosis of inborn errors of metabolism. Anal. Chim. Acta 2018, 1025, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zheng, W.; Wang, W.; Shen, H.; Liu, L.; Lou, W.; Wang, X.; Yang, P. A new panel of pancreatic cancer biomarkers discovered using a mass spectrometry-based pipeline. Br. J. Cancer 2017, 117, 1846. [Google Scholar] [CrossRef]
- Al-Saaidi, R.A.; Rasmussen, T.B.; Birkler, R.I.; Palmfeldt, J.; Beqqali, A.; Pinto, Y.M.; Nissen, P.H.; Baandrup, U.; Mølgaard, H.; Hey, T.M.; et al. The clinical outcome of LMNA missense mutations can be associated with the amount of mutated protein in the nuclear envelope. Eur. J. Heart Fail. 2018, 20, 1404–1412. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Tang, M.; Liu, Y.; Wang, J.; Wu, Z. Comparative Proteomics of Chromium-Transformed Beas-2B Cells by 2D-DIGE and MALDI-TOF/TOF MS. Biol. Trace Elem. Res. 2018, 185, 78–88. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Aljada, A.; Doria, J.; Saleh, A.M.; Al-Matar, S.H.; Al-Gabbani, S.; Shamsa, H.B.; Al-Bawab, A.; Ahmed, A.A. Altered LAMIN A/C splice variant expression as a possible diagnostic marker in breast cancer. Cell. Oncol. 2016, 39, 161–174. [Google Scholar] [CrossRef]
- Meshorer, E.; Misteli, T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat. Rev. Mol. Cell Biol. 2006, 7, 540. [Google Scholar] [CrossRef]
- Stadelmann, B.; Khandjian, E.; Hirt, A.; Lüthy, A.; Weil, R.; Wagner, H.P. Repression of nuclear lamin A and C gene expression in human acute lymphoblastic leukemia and non-Hodgkin’s lymphoma cells. Leukemia Res. 1990, 14, 815–821. [Google Scholar] [CrossRef]
- Scaffidi, P.; Misteli, T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat. Med. 2005, 11, 440. [Google Scholar] [CrossRef] [PubMed]
- Zhong, N.; Radu, G.; Ju, W.; Brown, W.T. Novel progerin-interactive partner proteins hnRNP E1, EGF, Mel 18, and UBC9 interact with lamin A/C. Biochem. Biophys. Res. Commun. 2005, 338, 855–861. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Worman, H.J. Expression of nuclear lamins in human tissues and cancer cell lines and transcription from the promoters of the LAMIN A/C and B1 genes. Exp. Cell Res. 1997, 236, 378–384. [Google Scholar] [CrossRef]
- Briand, N.; Collas, P. Laminopathy-causing Lamin A mutations reconfigure Lamina-associated domains and local spatial chromatin conformation. Nucleus 2018, 9, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Burtner, C.R.; Kennedy, B.K. Progeria syndromes and ageing: What is the connection? Nat. Rev. Mol. Cell Biol. 2010, 11, 567. [Google Scholar] [CrossRef]
- Mattout, A.; Dechat, T.; Adam, S.A.; Goldman, R.D.; Gruenbaum, Y. Nuclear Lamins, diseases and aging. Curr. Opin. Cell Biol. 2006, 18, 335–341. [Google Scholar] [CrossRef]
- Dixon, J.R. The international conference on harmonization good clinical practice guideline. Qual. Assur. 1999, 6, 65–74. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef]
- Peng, J.; Elias, J.E.; Thoreen, C.C.; Licklider, L.J.; Gygi, S.P. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: The yeast proteome. J. Proteome Res. 2003, 2, 43–50. [Google Scholar] [CrossRef]
- Rahman, A.M.; Kamath, S.; Lopata, A.L.; Helleur, R.J. Analysis of the allergenic proteins in black tiger prawn (Penaeus monodon) and characterization of the major allergen tropomyosin using mass spectrometry. Rapid Commun. Mass Spectrum. 2010, 24, 2462–2470. [Google Scholar] [CrossRef] [PubMed]
- González-Cruz, R.D.; Sadick, J.S.; Fonseca, V.C.; Darling, E.M. Nuclear lamin protein C is linked to lineage-specific, whole-cell mechanical properties. Cell. Mol. Bioeng. 2018, 11, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Debes, J.D.; Sebo, T.J.; Heemers, H.V.; Kipp, B.R.; De Anna, L.H.; Lohse, C.M.; Tindall, D.J. p300 modulates nuclear morphology in prostate cancer. Cancer Res. 2005, 65, 708–712. [Google Scholar] [PubMed]
- Machiels, B.M.; Broers, J.L.; Raymond, Y.; Ley, L.D.; Kuijpers, H.; Caberg, N.; Ramaekers, F. Abnormal A-type lamin organization in a human lung carcinoma cell line. Eur. J. Cell Biol. 1995, 67, 328–335. [Google Scholar] [PubMed]
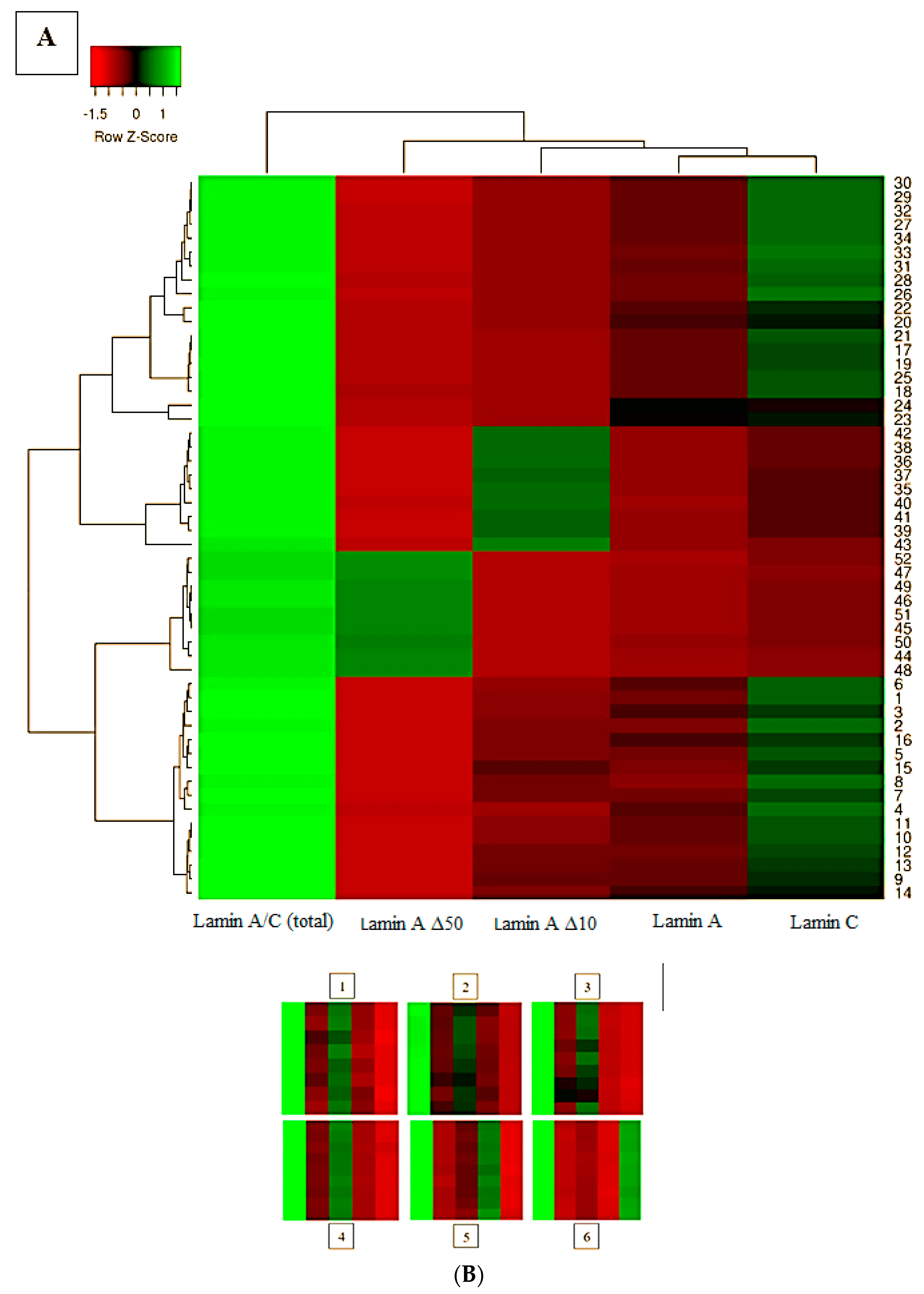
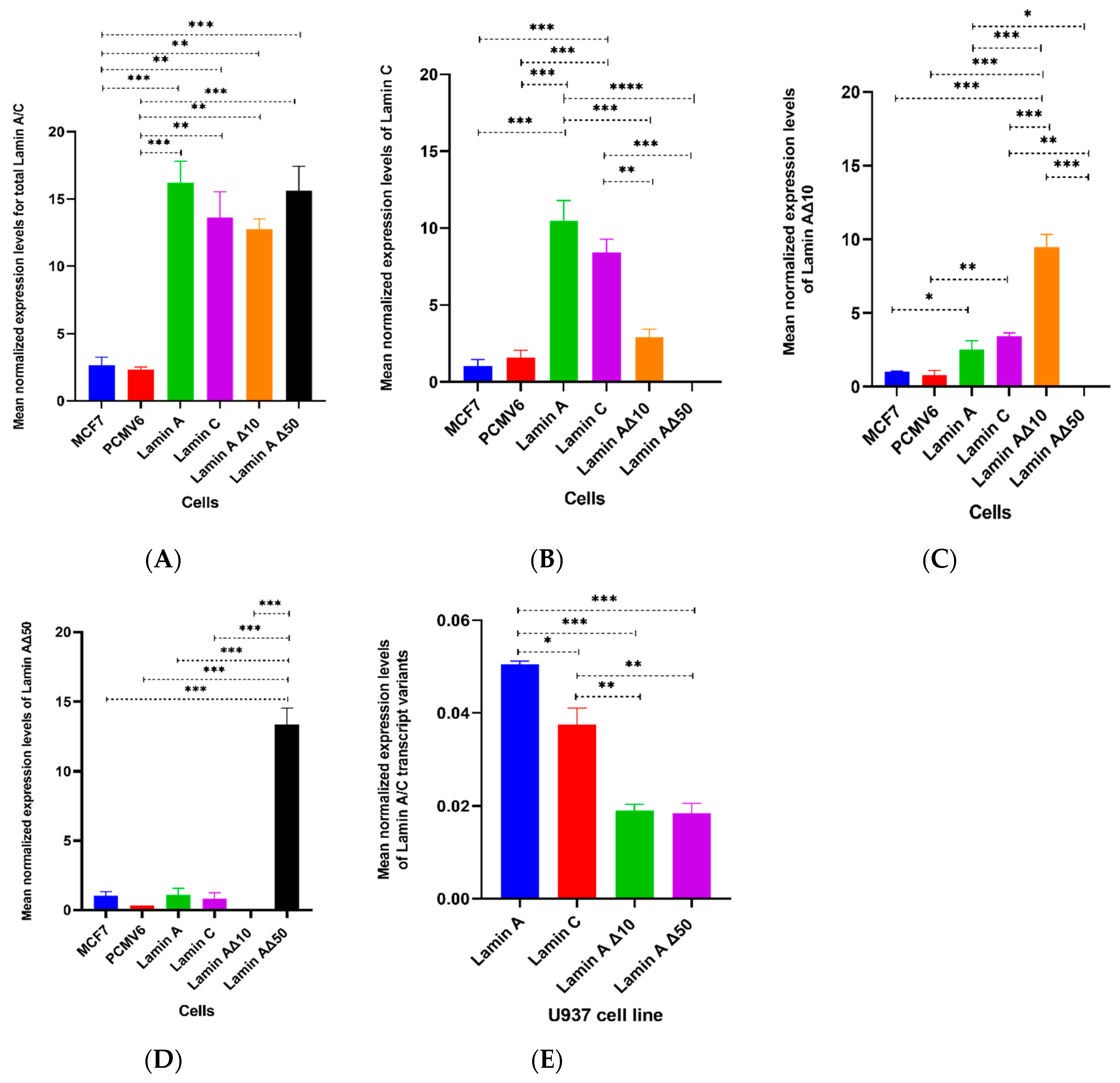
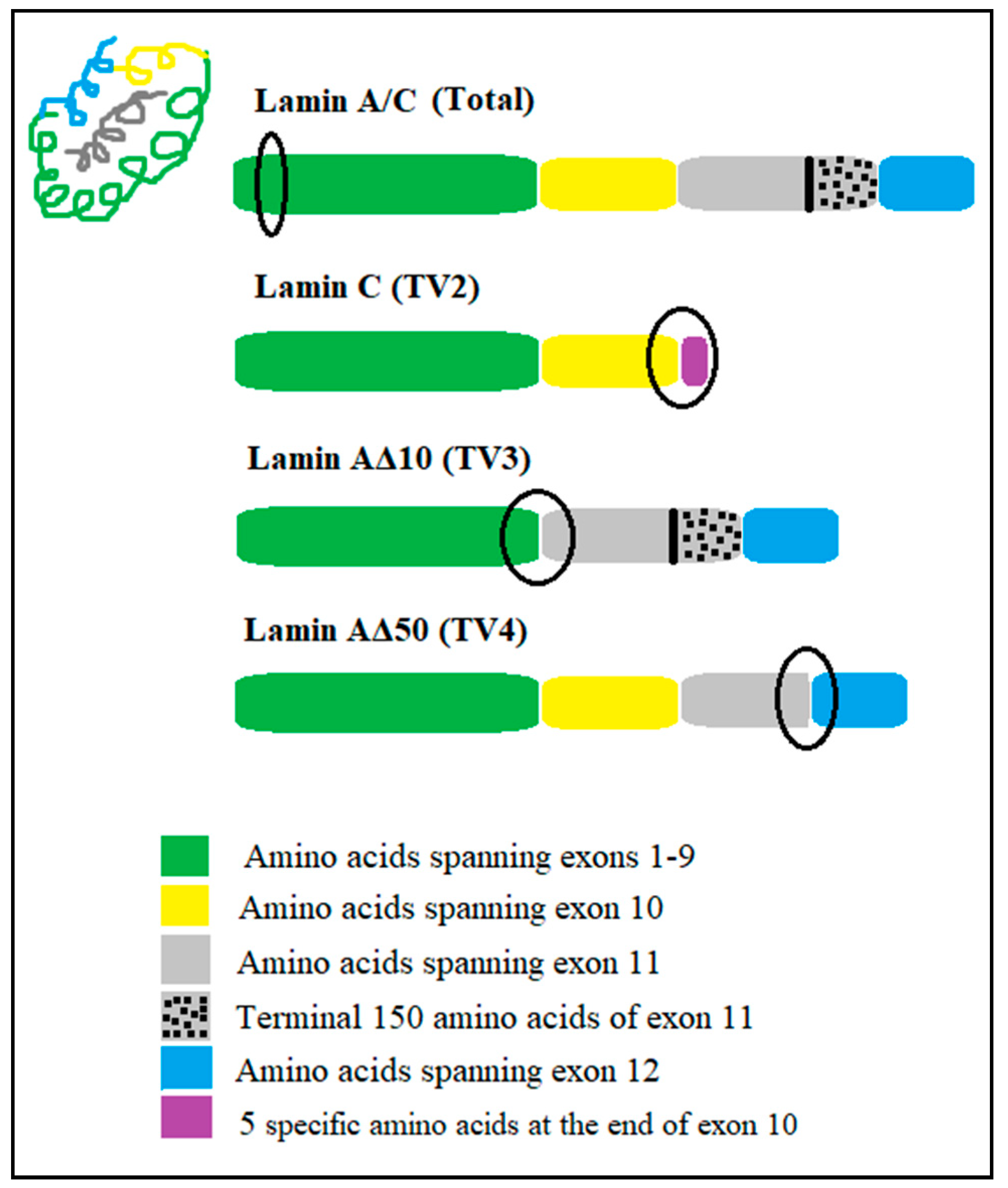
| Total Time (min) | Flow Rate (mL/min) | % A | % B |
|---|---|---|---|
| Initial | 0.200 | 90.0 | 10.0 |
| 1.00 | 0.200 | 90.0 | 10.0 |
| 1.01 | 0.200 | 90.0 | 10.0 |
| 6.00 | 0.200 | 10.0 | 90.0 |
| 6.20 | 0.200 | 10.0 | 90.0 |
| 6.50 | 0.200 | 90.0 | 10.0 |
| 10.00 | 0.200 | 90.0 | 10.0 |
| Compound | Parent or Precursor (m/z) | Daughter or Product (m/z) | Cone Voltage (V) | Collision Energy (V) | Retention Time |
|---|---|---|---|---|---|
| lamin A/C a | 583.4000 | 70.1100 | 44 | 68 | 3.771 |
| 91.1000 | 44 | 84 | 3.775 | ||
| 136.1000 | 44 | 44 | 3.741 | ||
| lamin A/C b | 589.5000 | 86.2000 | 46 | 54 | 3.741 |
| 90.2000 | 46 | 90 | 3.741 | ||
| 136.2000 | 46 | 42 | 3.740 | ||
| lamin C a | 484.4000 | 72.1100 | 16 | 50 | 3.801 |
| 84.0700 | 16 | 50 | 3.801 | ||
| 110.6000 | 16 | 40 | 3.804 | ||
| lamin C b | 587.4000 | 80.3000 | 16 | 46 | 3.805 |
| 102.2000 | 16 | 40 | 3.801 | ||
| 110.2000 | 16 | 54 | 3.801 | ||
| lamin A Δ10 a | 856.4000 | 86.2000 | 52 | 54 | 3.861 |
| 110.2000 | 52 | 62 | 3.861 | ||
| 173.2000 | 52 | 42 | 3.861 | ||
| lamin A Δ10 b | 858.5000 | 70.2000 | 8 | 96 | 3.861 |
| 86.2000 | 8 | 54 | 3.861 | ||
| 110.2000 | 8 | 72 | 3.861 | ||
| lamin A Δ50 a | 798.5000 | 60.2000 | 26 | 108 | 4.095 |
| 70.2000 | 26 | 82 | 4.094 | ||
| 86.2000 | 26 | 48 | 4.094 | ||
| lamin A Δ50 b | 803.5000 | 60.2000 | 26 | 96 | 4.095 |
| 70.2000 | 26 | 78 | 4.095 | ||
| 86.2000 | 26 | 40 | 4.094 |
| Intra-Day Validation (n = 6) | |||||||||||||
| Day Validation | The Signature Peptide of Transcript Variant | 150 ng/mL | 300 ng/mL | 600 ng/mL | |||||||||
| Mean | SD | Precision (CV%) | Accuracy (%) | Mean | SD | Precision (CV%) | Accuracy (%) | Mean | SD | Precision (CV%) | Accuracy (%) | ||
| Day1 | lamin A/C | 153.00 | 10.61 | 6.94 | 102.00 | 312.00 | 8.03 | 7.72 | 104.00 | 602.18 | 10.16 | 1.68 | 100.36 |
| lamin C | 144.48 | 26.55 | 14.38 | 96.32 | 268.46 | 15.72 | 5.85 | 89.48 | 594.90 | 7.39 | 1.24 | 99.15 | |
| lamin AΔ10 | 161.35 | 12.34 | 7.64 | 107.56 | 312.78 | 8.80 | 2.81 | 104.26 | 631.75 | 36.65 | 5.80 | 105.29 | |
| lamin AΔ50 | 146.15 | 10.31 | 7.05 | 97.43 | 307.75 | 9.28 | 3.01 | 102.58 | 600.61 | 11.30 | 1.88 | 100.10 | |
| Day2 | lamin A/C | 143.92 | 8.88 | 6.17 | 95.94 | 274.38 | 13.75 | 5.01 | 91.46 | 526.73 | 28.82 | 5.47 | 87.79 |
| lamin C | 155.87 | 9.39 | 6.03 | 103.91 | 290.53 | 4.60 | 1.58 | 96.84 | 620.95 | 14.92 | 2.40 | 103.49 | |
| lamin AΔ10 | 138.07 | 16.23 | 11.75 | 92.04 | 293.57 | 12.30 | 4.19 | 97.86 | 580.78 | 22.46 | 3.87 | 96.80 | |
| lamin AΔ50 | 156.90 | 5.56 | 3.54 | 104.60 | 305.72 | 6.49 | 2.12 | 101.91 | 602.60 | 4.56 | 0.76 | 100.43 | |
| Day3 | lamin A/C | 152.75 | 21.68 | 14.19 | 101.83 | 295.10 | 29.19 | 9.89 | 98.37 | 608.78 | 76.04 | 12.49 | 101.46 |
| lamin C | 139.45 | 5.45 | 3.91 | 92.97 | 272.02 | 21.50 | 7.90 | 90.67 | 586.33 | 11.21 | 1.91 | 97.72 | |
| lamin AΔ10 | 151.10 | 8.61 | 5.69 | 100.73 | 295.50 | 12.99 | 4.40 | 98.50 | 586.33 | 10.69 | 1.82 | 97.72 | |
| lamin AΔ50 | 151.65 | 2.30 | 1.52 | 101.10 | 302.75 | 13.00 | 4.29 | 100.92 | 602.73 | 11.29 | 1.87 | 100.46 | |
| Inter-Day Validation (n = 18) | |||||||||||||
| The Signature Peptide of Transcript Variant | 150 ng/mL | 300 ng/mL | 600 ng/mL | ||||||||||
| Mean | SD | Precision (CV%) | Accuracy (%) | Mean | SD | Precision (CV%) | Accuracy (%) | Mean | SD | Precision (CV%) | Accuracy (%) | ||
| lamin A/C | 149.89 | 14.61 | 9.75 | 99.93 | 290.49 | 21.70 | 7.47 | 96.83 | 579.23 | 58.67 | 10.13 | 96.54 | |
| lamin C | 146.60 | 17.09 | 11.66 | 97.73 | 277.01 | 17.72 | 6.40 | 92.34 | 599.96 | 18.94 | 3.16 | 99.99 | |
| lamin AΔ10 | 159.60 | 15.50 | 9.71 | 106.40 | 300.62 | 14.00 | 4.66 | 100.21 | 599.62 | 33.60 | 5.60 | 99.94 | |
| lamin AΔ50 | 148.98 | 6.88 | 4.62 | 99.32 | 305.41 | 9.59 | 3.14 | 101.80 | 601.98 | 9.06 | 1.51 | 100.33 | |
| lamin A/C | 149.89 | 14.61 | 9.75 | 99.93 | 290.49 | 21.70 | 7.47 | 96.83 | 579.23 | 58.67 | 10.13 | 96.54 | |
| The Signature Peptide of Transcript Variant | Linearity (n = 3) | Sensitivity (n = 3) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Slope | SD | Intercept | R | LLOQ (nM) | Mean | SD | Precision (CV%) | Accuracy (%) | |
| lamin A/C | 0.0026 | 0.0005 | 0.0089 | 0.9969 | 50.00 | 52.37 | 7.06 | 13.48 | 104.73 |
| lamin C | 0.0003 | 0.0002 | −0.0223 | 0.9726 | 50.00 | 48.47 | 6.05 | 12.48 | 96.93 |
| lamin AΔ10 | 0.0032 | 0.0008 | −0.0428 | 0.9917 | 50.00 | 60.20 | 8.80 | 14.62 | 120.40 |
| lamin AΔ50 | 0.0032 | 0.0018 | −0.0091 | 0.9933 | 50.00 | 44.23 | 5.26 | 11.89 | 88.47 |
| Stability (%) | |||||
|---|---|---|---|---|---|
| Storage Temp (°C) | ng/mL | The Signature Peptide of Transcript Variant | |||
| lamin A/C | lamin C | lamin AΔ10 | lamin AΔ50 | ||
| Fresh (−80 °C) | 150 | 100 | 100 | 100 | 100 |
| 300 | 100 | 100 | 100 | 100 | |
| 600 | 100 | 100 | 100 | 100 | |
| 2 weeks (−20 °C) | 150 | 100.65 | 101.89 | 99.17 | 100.21 |
| 300 | 99.97 | 97.45 | 99.76 | 99.59 | |
| 600 | 99.28 | 100.09 | 99.64 | 99.94 | |
| 4 weeks (−20 °C) | 150 | 98.06 | 98.27 | 98.76 | 96.99 |
| 300 | 97.46 | 96.45 | 98.26 | 96.44 | |
| 600 | 99.22 | 99.86 | 100.07 | 99.31 | |
| 1 week (4 °C) | 150 | 97.59 | 98.95 | 97.56 | 96.97 |
| 300 | 98.25 | 96.3 | 96.14 | 95.87 | |
| 600 | 97.85 | 99.07 | 100.03 | 99.802 | |
| 1 week (RT) | 150 | 87.75 | 86.56 | 79.52 | 80.37 |
| 300 | 86.99 | 92.55 | 93.58 | 83.69 | |
| 600 | 95.6 | 96.77 | 98.99 | 95.65 | |
| Transcript Variant | Sequence Position | NCBI Reference | Signature Peptide Sequence |
|---|---|---|---|
| lamin A/C (Total) | AA78–89; exon 1 | NM_170707.3 | AAYEAELGDAR AAYEAELGDAR (A = d3-Ala) * |
| lamin C | AA547–572; in the junction region between the end of exon 10 with its specific 5 amino acids | NM_005572.3 | SVTVVEDDEDEDGDDLLHHHHVSGSR SVTVVEDDEDEDGDDLLHHHHVSGSR (V = d8-Val) * |
| lamin AΔ10 | AA529–553; in the junction between exon 9 and exon 11 | NM_170708.3 | TALINSTGEGSHCSSSGDPAEYNLR TALINSTGEGSHCSSSGDPAEYNLR (A = d3-Ala) * |
| lamin AΔ50 | AA599–615; in the junction region of the exonic cryptic site of exon 11 which lacks the terminal 150 amino acids and exon 12 | NM_001282626.1 | ASASGSGAQSPQNCSIM ASASGSGAQSPQNCSIM (A = d3-Ala) * |
| Probe | Primer | |
|---|---|---|
| Lamin A | 5′-CGCTGAGTACAACCT -3′ | F 5′- GACGAGGATGAGGATGGAGA-3′ R 5′-GAGTGACCGTGACACTGGAG-3′ |
| Lamin C | 5′-AGATGACCTGCTCCATCACC-3′ | F 5′-GTGGAAGGCACAGAACACCT-3′ R 5′-GCGGCGGCTACCACTCAC-3′ |
| Lamin AΔ10 | 5′-AGTACAACCTGCGCTCGCGC-3′ | F 5′-AACTCCACTGGGGAAGGCTCC-3′ R 5′-GCTCCTGAGCCGCTGGCAGA-3′ |
| Lamin AΔ50 | 5′-AGCATCATGTAATCTGGGACCT-3′ | F 5′-GCGTCAGGAGCCCTGAGC-3′ R 5′-GACGCAGGAAGCCTCCAC-3′ |
| Ubiquitin | 5′-CCCACCTCTGAGACGGAGCACCAG-3′ | F 5′-ACTACAACATCCAGAAAGAGTCCA-3′ R 5′- CCAGTCAGGGTCTTCACGAAG-3′ |
| RPL-13 | 5′-CGCAAGCGGATGAACACCAACCCT-3′ | F 5′-AACAAGTTGAAGTACCTGGCTTTC-3′ R 5′-TGGTTTTGTGGGGCAGCATA-3′ |
| Cyclophilin A | 5′-ACAGCTCAAAGGAGACGCGGCCCA-3′ | F 5′-CCCACCGTGTTCTTCGACAT-3′ R 5′-TTTCTGCTGTCTTTGGGACCTT-3′ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Qahtani, W.S.; Abduljabbar, M.; AlSuhaibani, E.S.; Abdel Rahman, A.; Aljada, A. Quantification of the Lamin A/C Transcript Variants in Cancer Cell Lines by Targeted Absolute Quantitative Proteomics and Correlation with mRNA Expression. Int. J. Mol. Sci. 2019, 20, 1902. https://doi.org/10.3390/ijms20081902
Al-Qahtani WS, Abduljabbar M, AlSuhaibani ES, Abdel Rahman A, Aljada A. Quantification of the Lamin A/C Transcript Variants in Cancer Cell Lines by Targeted Absolute Quantitative Proteomics and Correlation with mRNA Expression. International Journal of Molecular Sciences. 2019; 20(8):1902. https://doi.org/10.3390/ijms20081902
Chicago/Turabian StyleAl-Qahtani, Wedad Saeed, Mai Abduljabbar, Entissar S. AlSuhaibani, Anas Abdel Rahman, and Ahmad Aljada. 2019. "Quantification of the Lamin A/C Transcript Variants in Cancer Cell Lines by Targeted Absolute Quantitative Proteomics and Correlation with mRNA Expression" International Journal of Molecular Sciences 20, no. 8: 1902. https://doi.org/10.3390/ijms20081902
APA StyleAl-Qahtani, W. S., Abduljabbar, M., AlSuhaibani, E. S., Abdel Rahman, A., & Aljada, A. (2019). Quantification of the Lamin A/C Transcript Variants in Cancer Cell Lines by Targeted Absolute Quantitative Proteomics and Correlation with mRNA Expression. International Journal of Molecular Sciences, 20(8), 1902. https://doi.org/10.3390/ijms20081902