Functional Analysis of the Soybean GmCDPK3 Gene Responding to Drought and Salt Stresses
Abstract
1. Introduction
2. Results
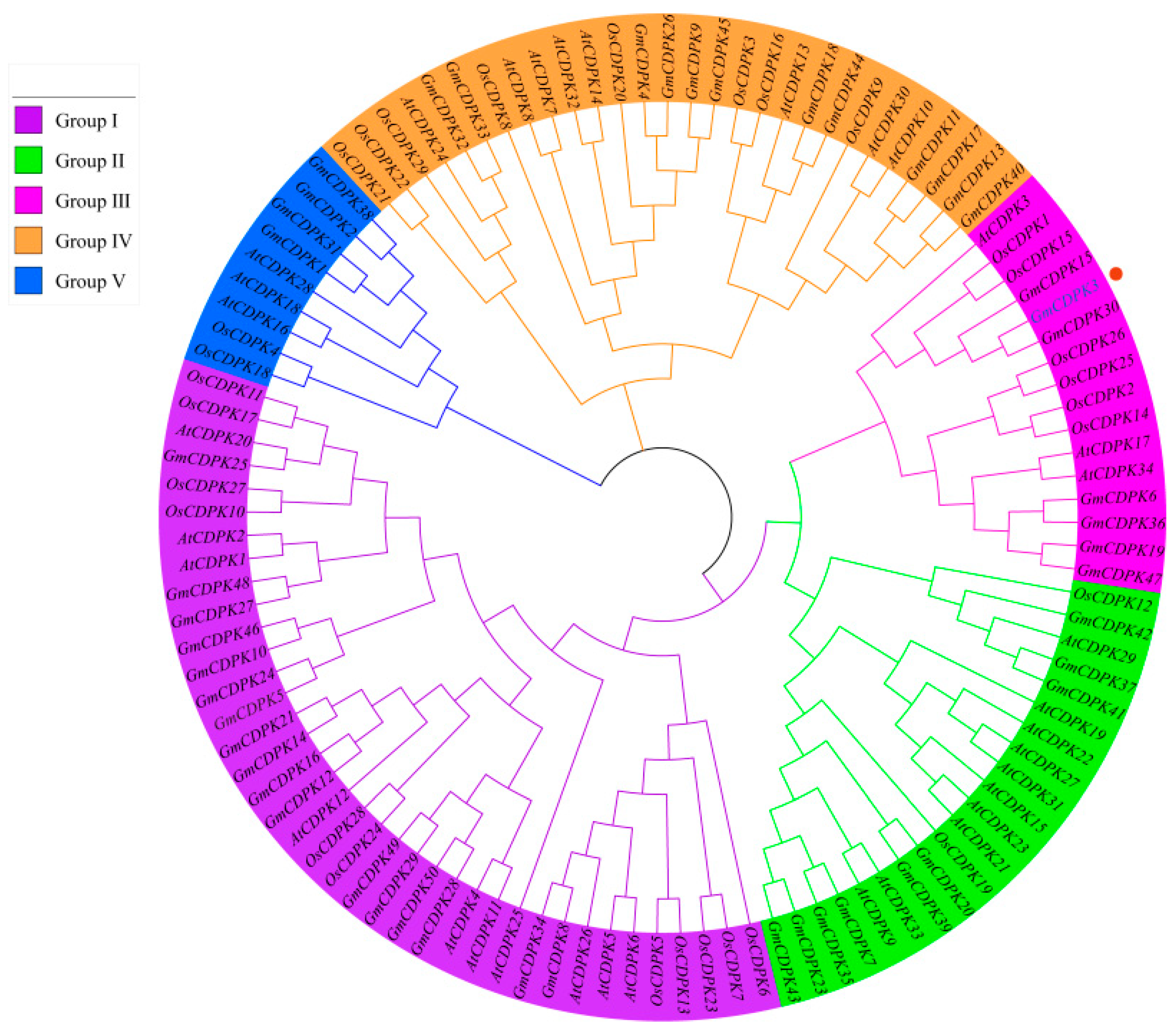
2.1. Phylogenetic Tree Inference in CDPKs
2.2. Analysis of Gene and Protein Structure of 17 Selected GmCDPK
2.3. GmCDPK Protein Tertiary Structure Homology Modeling
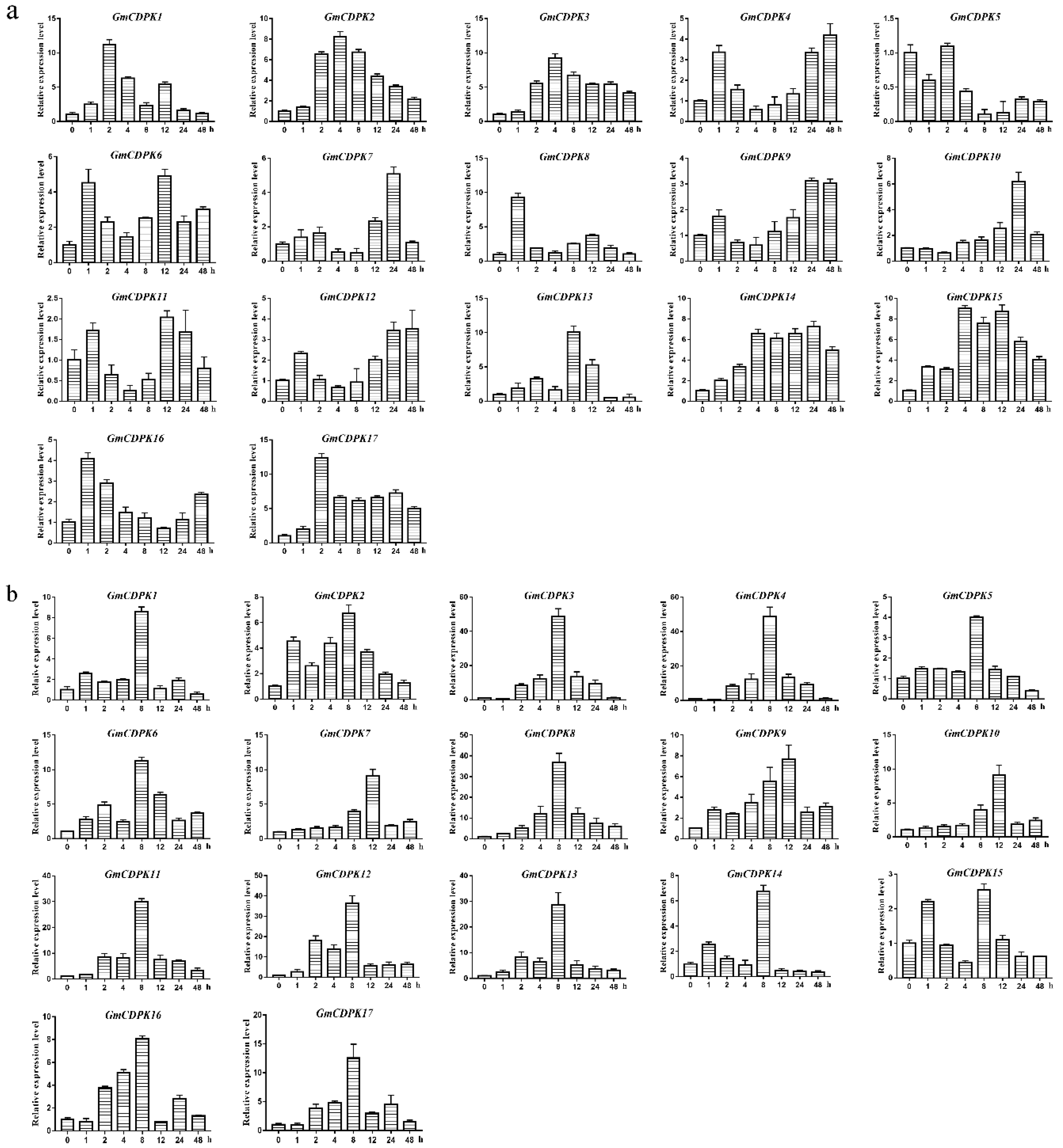
2.4. Expression of 17 GmCDPKs in Different Tissues and at Different Developmental Stages
2.5. Promoter Regions of 17 GmCDPKs Contain Various Stress Response Elements
2.6. Candidate Genes Involved in Drought and Salt Stresses
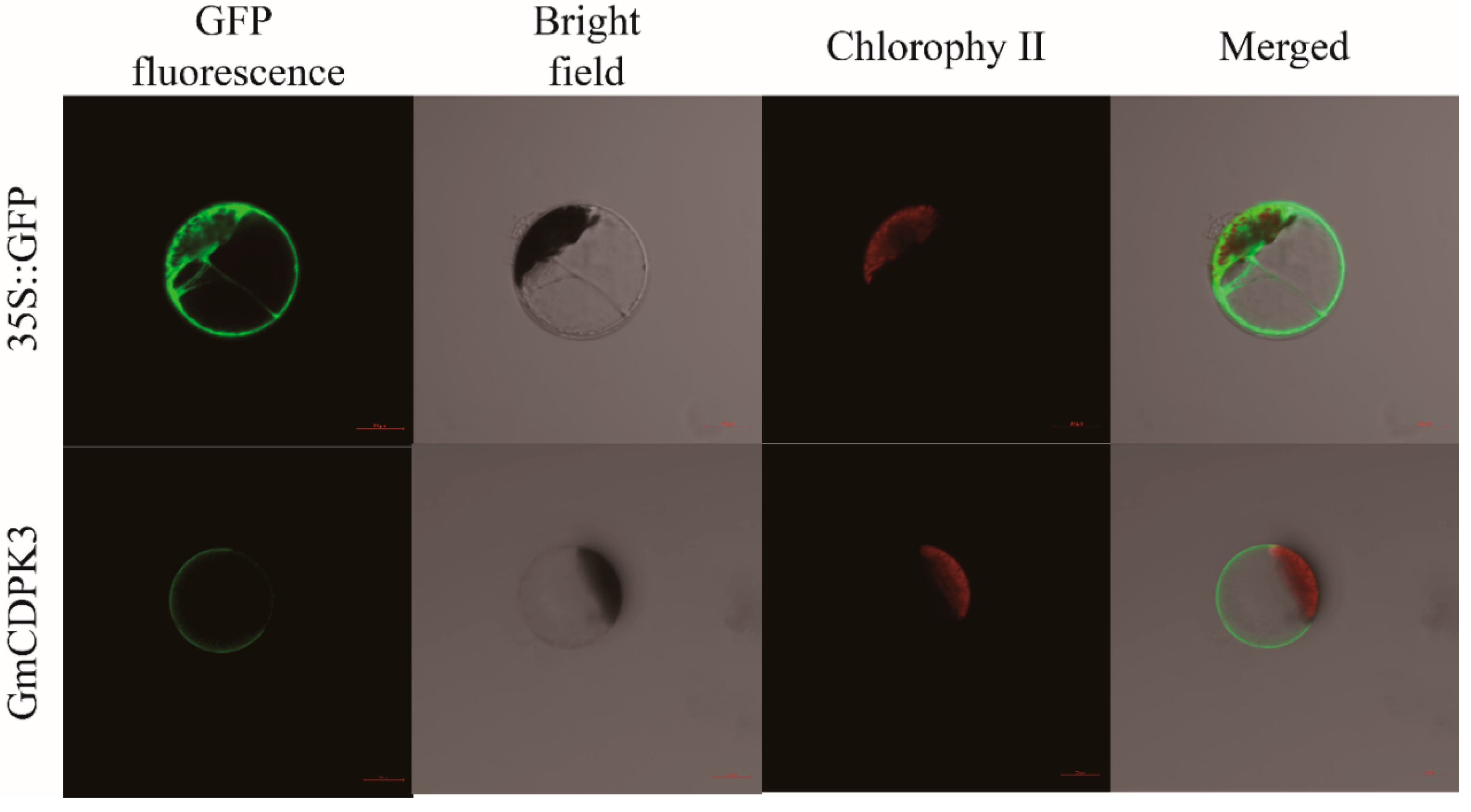
2.7. GmCDPK3 Localized on the Cell Membrane
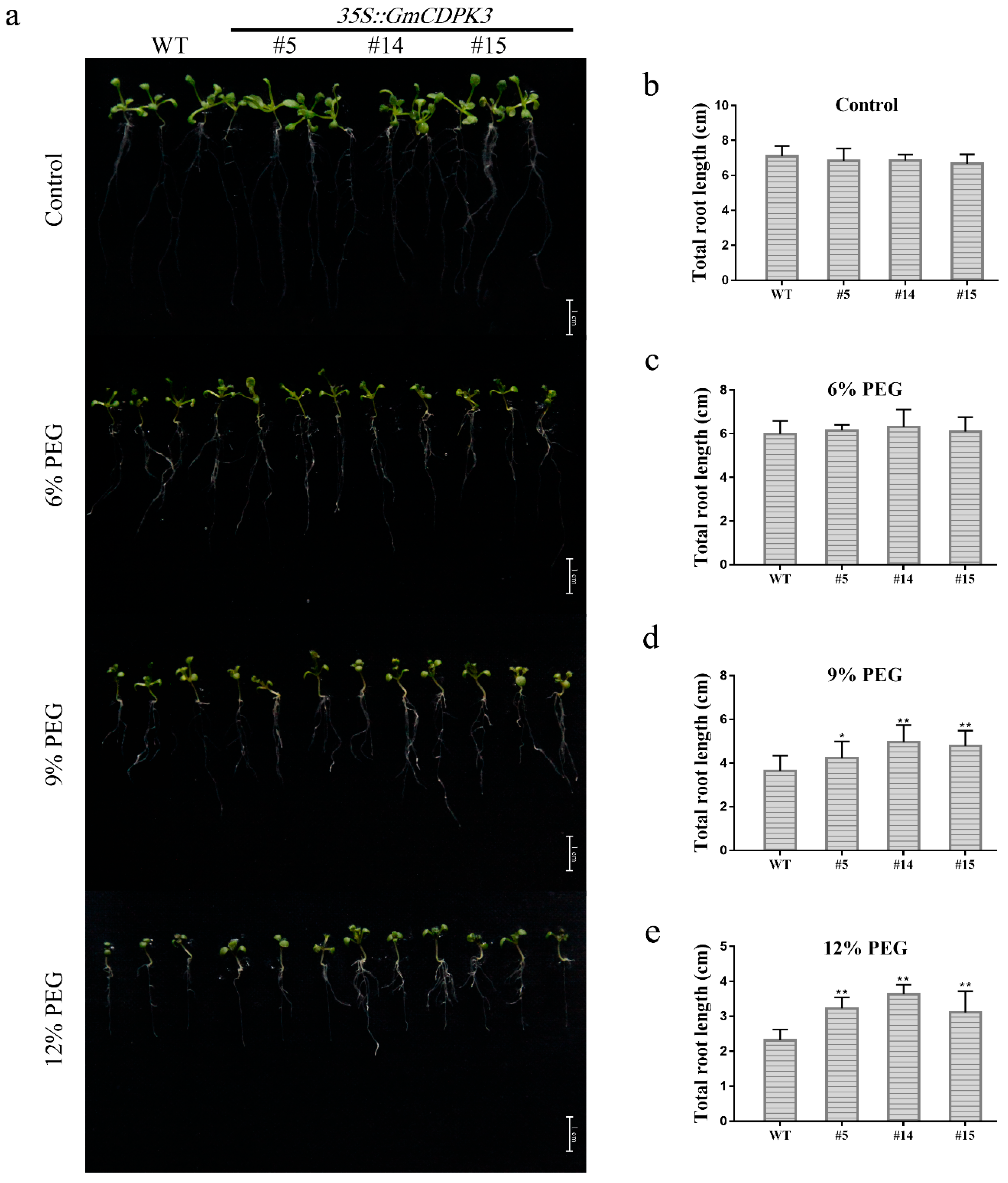
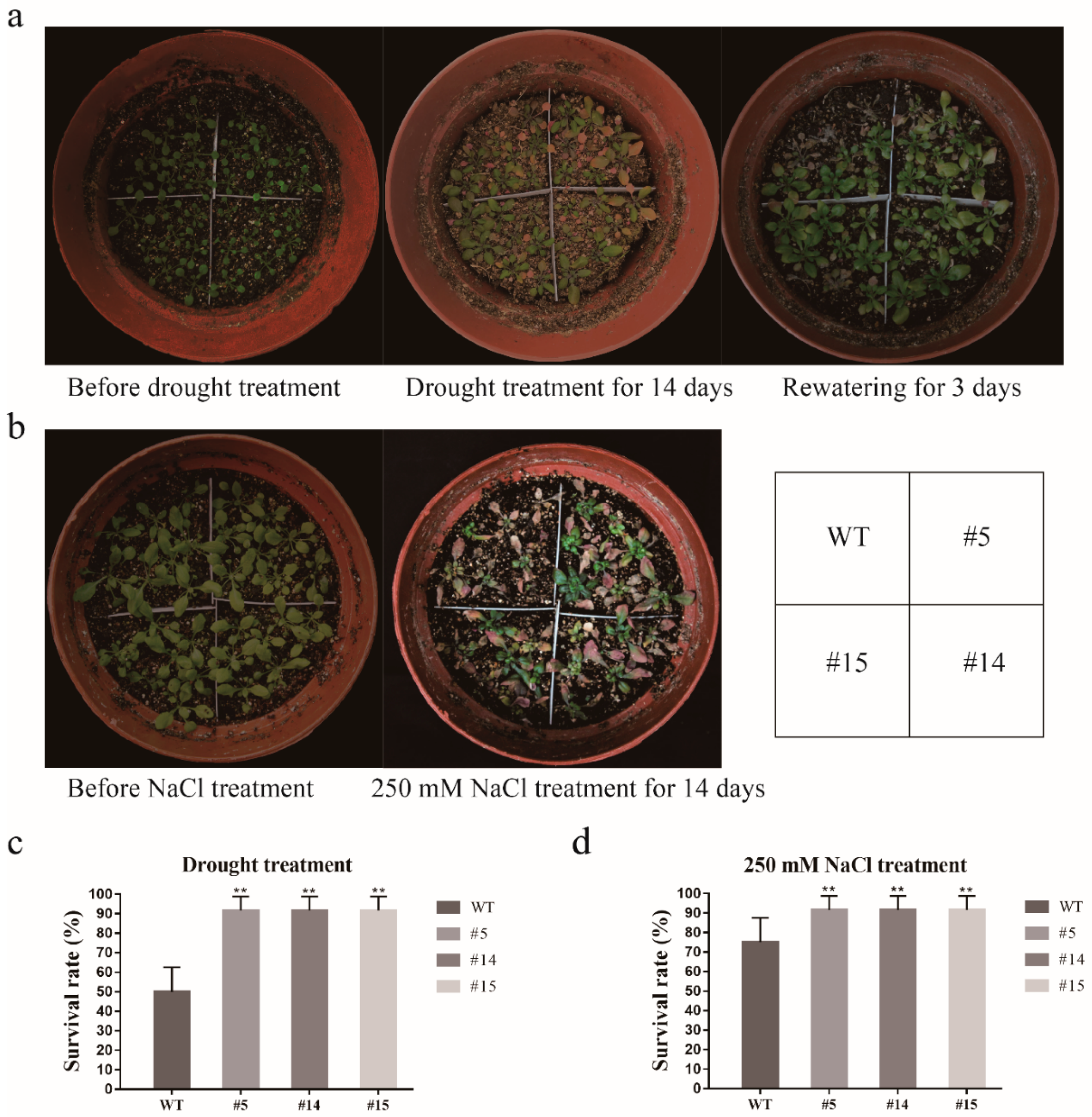
2.8. GmCDPK3 Conferred Drought Tolerance in Transgenic Arabidopsis
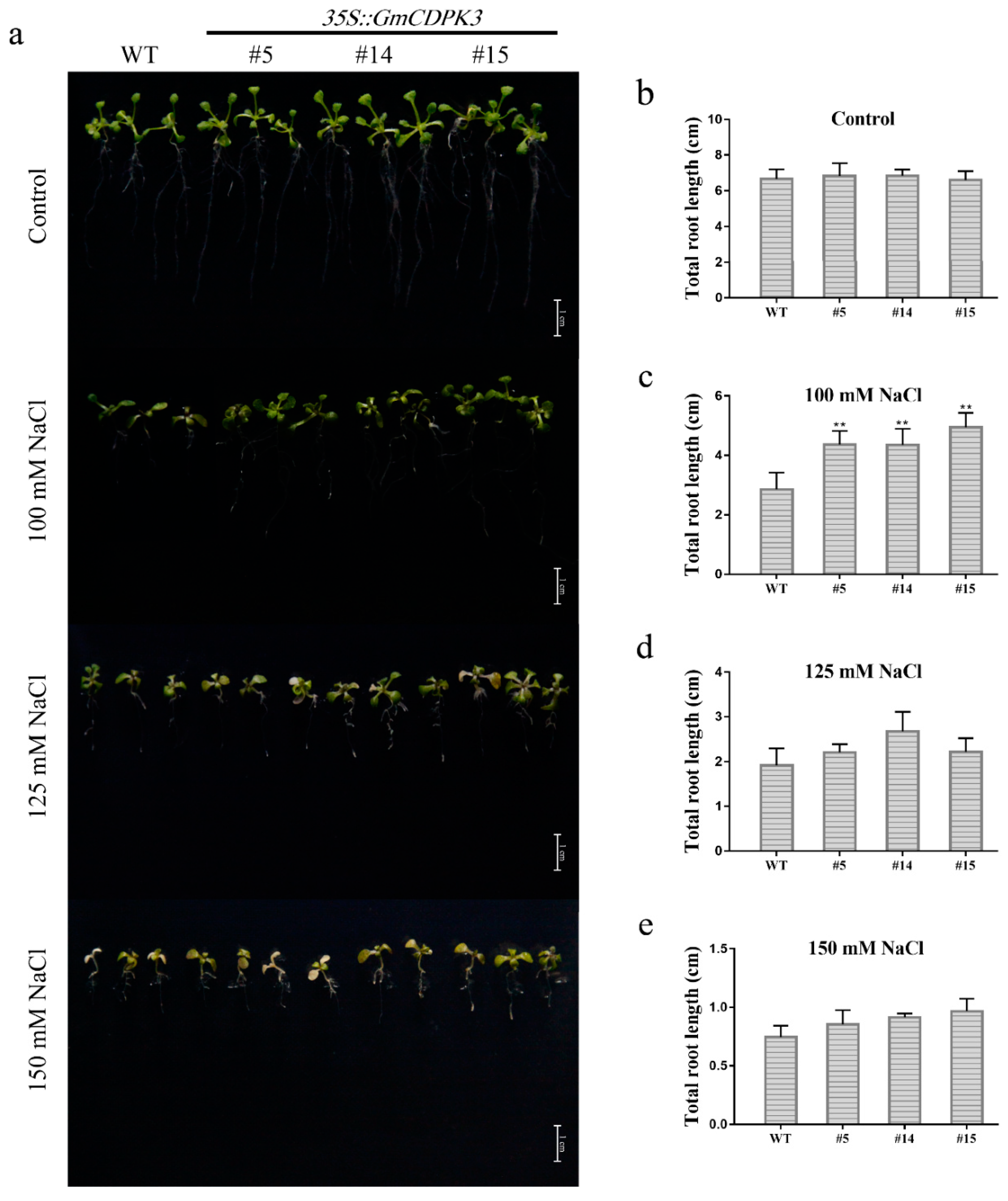
2.9. Salt Tolerance of GmCDPK3 in Arabidopsis
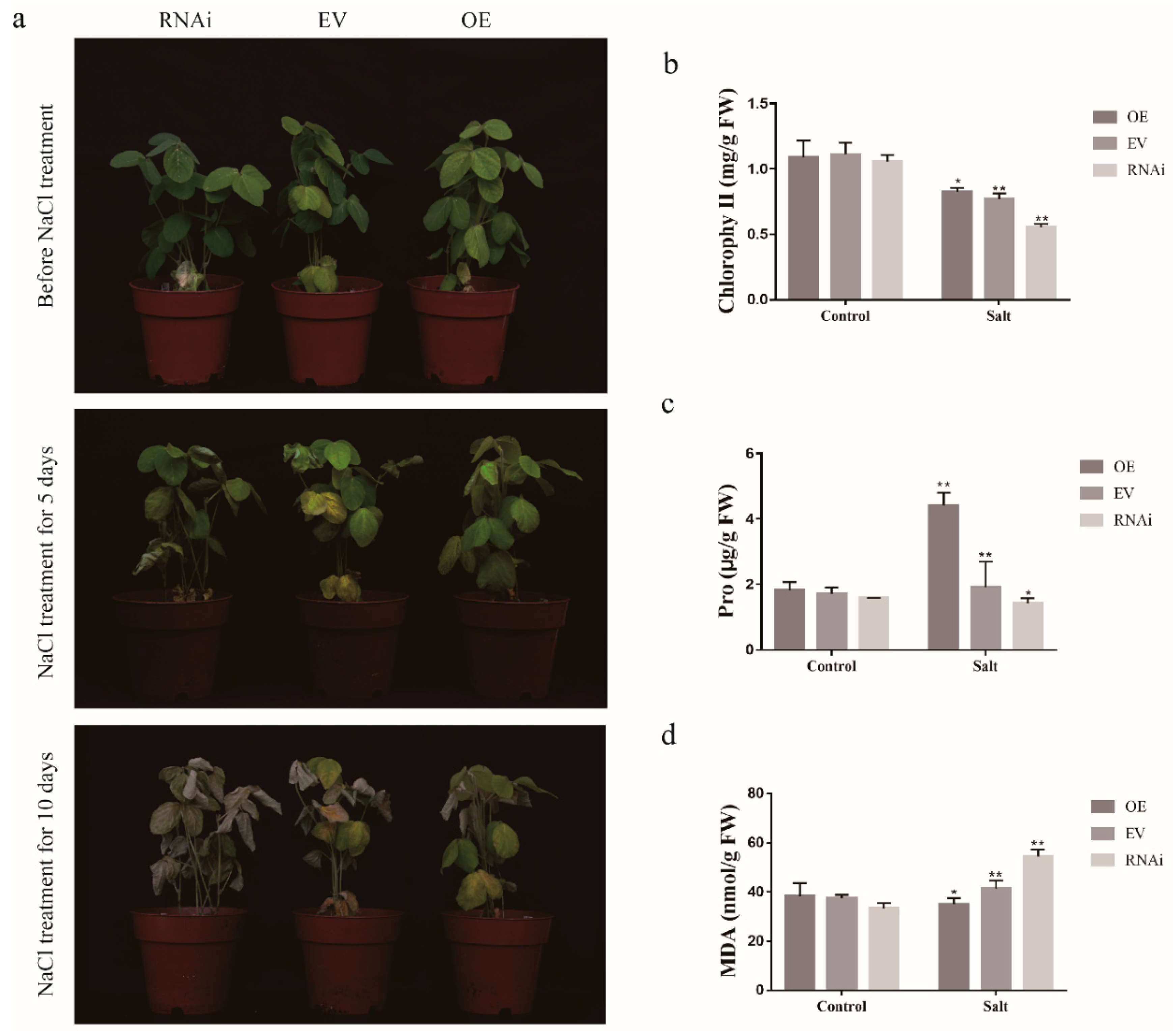
2.10. Positive Effect of GmCDPK3 in Transgenic Soybean Hairy Roots Under Drought and Salt Treatment
3. Discussion
4. Materials and Methods
4.1. Phylogenetic Tree Analysis and Gene Source
4.2. Sequence Analysis of 17 Drought and Salt-Tolerant GmCDPKs
4.3. Promoter Analysis of GmCDPKs
4.4. Planting of Plant Materials
4.5. RNA Extraction and qRT-PCR
4.6. Subcellular Localization of GmCDPK3
4.7. Drought and Salt Stress Assays of Transgenic Arabidopsis Plants
4.8. Vector Construction of GmCDPK3
4.9. Transformation of Soybean Hairy Roots
4.10. Trypan Blue Staining
4.11. Determination of Pro, MDA, and Chlorophyll Contents
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| A. rhizogenes | Agrobacterium rhizogenes |
| ABA | Abscisic acid |
| CDPK | Calcium-dependent protein kinase |
| qRT-PCR | Quantitative real-time PCR |
| pI | Isoelectric points |
| GFP | Green fluorescent protein |
| MDA | Malondialdehyde |
| PRO | Proline |
| PEG | Polyethylene glycol |
References
- Bohnert, H.J.; Nelson, D.E.; Jensen, R.G. Adaptations to environmental stresses. Plant. Cell. 1995, 7, 1099–1111. [Google Scholar] [CrossRef] [PubMed]
- Bray, E.A. Plant responses to water deficit. Trends Plant. Sci. 1997, 2, 48–54. [Google Scholar] [CrossRef]
- Zhu, J.K.; Hasegawa, P.M.; Bressan, R.A.; Bohnert, H.J. Molecular aspects of osmotic stress in plants. Crit. Rev. Plant. Sci. 1997, 16, 253–277. [Google Scholar] [CrossRef]
- Ghosh, A.; Greenberg, M.E. Calcium signaling in neurons: Molecular mechanisms and cellular consequences. Science 1995, 268, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Rudd, J.J.; Franklin Tong, V.E. Unravelling response-specificity in Ca2+ signaling pathways in plant cells. New Phytol. 2001, 151, 7–33. [Google Scholar] [CrossRef]
- Hamel, L.P.; Sheen, J.; Seguin, A. Ancient signals: Comparative genomics of green plant CDPKs. Trends Plant. Sci. 2014, 19, 79–89. [Google Scholar] [CrossRef]
- Harper, J.F.; Harmon, A. Plants, symbiosis and parasites: A calcium signalling connection. Nat. Rev. Mol. Cell Biol. 2005, 6, 555–566. [Google Scholar] [CrossRef]
- Valmonte, G.R.; Arthur, K.; Higgins, C.M.; MacDiarmid, R.M. Calcium-dependent protein kinases inplants: Evolution, expression and function. Plant. Cell Physiol. 2014, 55, 551–569. [Google Scholar] [CrossRef]
- Roberts, D.M.; Harmon, A.C. Calcium-modulated proteins: Targets of intracellular calcium signals in higher plants. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 1992, 43, 375–414. [Google Scholar] [CrossRef]
- Wu, Z.G.; Wu, S.J.; Wang, Y.C.; Zheng, L.L. Advances in studies of calcium-dependent protein kinase (CDPK) in plants. Acta Prataculturae Sinica. 2018, 27, 204–214. [Google Scholar]
- Cheng, S.H.; Willmann, M.R.; Chen, H.C.; Sheen, J. Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 2002, 129, 469–485. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Agarwal, P.; Arora, R.; Kapoor, S.; Tyagi, A.K. Expression analysis of calcium-dependent proteinkinase gene family during reproductive development and abiotic stress conditions in rice (Oryza sativa L. ssp. indica). Mol. Genet. Genom. 2007, 278, 493–505. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.W.; Liu, M.; Lu, L.; He, M.; Qu, W.Q.; Xu, Q.; Qi, X.H.; Chen, X.H. Genome-wide analysis and expression of the calcium-dependent protein kinase gene family in cucumber. Mol. Genet. Genom. 2015, 290, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Li, A.L.; Zhu, Y.F.; Tan, X.M.; Wang, X.; Wei, B.; Guo, H.Z.; Zhang, Z.L.; Chen, X.B.; Zhao, G.Y.; Kong, X.Y.; et al. Evolutionary and functional study of the CDPK gene family in wheat (Triticumaestivum L.). Plant Mol. Biol. 2008, 66, 429–443. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.L.; Che, Z.J.; Zeng, X.R.; Zhou, X.Q.; Hélder, M.S.; Wang, H.; Yu, D.Y. Genome-wide analysis of calcium-dependent protein kinases and their expression patterns in response to herbivore and wounding stresses in soybean. Funct. Integr. Genom. 2016, 16, 481–493. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.F.; Zhao, W.Y.; Fu, J.D.; Liu, Y.W.; Chen, M.; Zhou, Y.B.; Ma, Y.Z.; Xu, Z.S.; Xi, Y.J. Genome-Wide Analysis of CDPK Family in Foxtail Millet and Determination of SiCDPK24 Functions in Drought Stress. Front. Plant. Sci. 2018, 9, 651. [Google Scholar] [CrossRef] [PubMed]
- Asano, T.; Tanaka, N.; Yang, G.X.; Hayashi, N.; Komatsu, S. Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: Comprehensive analysis of the CDPKs gene family in rice. Plant. Cell Physiol. 2005, 46, 356–366. [Google Scholar] [CrossRef]
- Wang, J.P.; Xu, Y.P.; Munyampundu, J.P.; Liu, T.Y.; Cai, X.Z. Calcium-dependent protein kinase (CDPK) and CDPK-related kinase (CRK) gene families in tomato: Genome-wide identification and functional analyses in disease resistance. Mol. Genet. Genom. 2016, 291, 661–676. [Google Scholar] [CrossRef]
- Cai, H.Y.; Cheng, J.B.; Yan, Y.; Xiao, Z.L.; Li, J.Z.; Mou, S.L.; Qiu, A.L.; Lai, Y.; Guan, D.Y.; He, S.L. Genome-wide identification and expression analysis of calcium-dependent protein kinase and its closely related kinase genes in Capsicum annuum. Front. Plant. Sci. 2015, 6, 737. [Google Scholar] [CrossRef]
- Zhang, K.; Han, Y.T.; Zhao, F.L.; Hu, Y.; Gao, Y.R.; Ma, Y.F.; Zheng, Y.; Wang, Y.J.; Wen, Y.Q. Genome-wide identification and expression analysis of the CDPK gene family in grape, Vitis spp. BMC Plant. Biol. 2015, 15, 164. [Google Scholar] [CrossRef]
- Hettenhausen, C.; Sun, G.L.; He, Y.B.; Zhuang, H.F.; Sun, T.; Qi, J.F.; Wu, J.Q. Genome-wide identification of calcium-dependent protein kinases in soybean and analyses of their transcriptional responses to insect herbivory and drought stress. Sci. Rep. 2016, 6, 18973. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xu, C.; Zhu, Y.; Zhang, L.; Zhou, F.; Chen, H.; Lin, Y. The calcium-dependent kinase OsCPK24 functions in cold stress responses in rice. BMC Genom. Biol. 2018, 60, 173–188. [Google Scholar] [CrossRef]
- Bian, S.M.; Jin, D.H.; Li, R.H.; Xie, X.; Gao, G.; Sun, W.; Li, Y.; Zhai, L.; Li, X. Genome-Wide Analysis of CCA1-Like Proteins in Soybean and Functional Characterization of GmMYB138a. Int. J. Mol. Sci. 2017, 18, 2040. [Google Scholar] [CrossRef] [PubMed]
- Asano, T.; Hayashi, N.; Kobayashi, M.; Aoki, N.; Mitsuhara, I.; Ichikawa, H.; Komatus, S.; Hirochika, H.; Kikuchi, S.; Ohsuqi, R. A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant. J. 2012, 69, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.J.; Wei, F.J.; Wang, C.; Wu, J.J.; Ratnasekera, D.; Liu, W.X.; Wu, W.H. Arabidopsis calcium-dependent protein kinase CPK10 functions in abscisic susceptibility to early blight pathogen in potato via reactive oxygen species burst. New Phytol. 2012, 196, 223–237. [Google Scholar]
- Choi, H.I.; Park, H.J.; Park, J.H.; Kim, S.; Im, M.Y.; Seo, H.H.; Kim, Y.W.; Hwang, I.; Kim, S.Y. Arabidopsis calcium-dependent protein kinase AtCPK32 interacts with ABF4, a transcriptional regulator of abscisic acid-responsive gene expression, and modulates its activity. Plant. Physiol. 2005, 139, 1750–1761. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhou, X.; Chang, S.; Chu, Z.; Wang, H.; Han, S.; Wang, Y. Calcium-dependent protein kinase 21 phosphorylates 14–3-3 proteins in response to ABA signaling and salt stress in rice. Biochem. Biophys. Res. Commun. 2017, 493, 1450–1456. [Google Scholar] [CrossRef] [PubMed]
- Boyer, J.S. Plant productivity and environment. Science 1982, 218, 443–448. [Google Scholar] [CrossRef]
- Liu, F.; Ma, H.; Peng, L.; Du, Z.; Ma, B.; Liu, X. Effect of the inoculation of plant growth-promoting rhizobacteria on the photosynthetic characteristics of Sambucus williamsii Hance container seedlings under drought stress. AMB Expr. 2019, 9, 2–9. [Google Scholar] [CrossRef]
- Vurukonda, S.S.; Vardharajula, S.; Shrivastava, M.; SkZ, A. Enhancement of drought stress tolerance in crops by plant growth promoting rhizobacteria. Microbiol. Res. 2016, 184, 13–24. [Google Scholar] [CrossRef]
- Du, H.; Wang, N.L.; Cui, F.; Li, X.H.; Xiao, J.H.; Xiong, L.Z. Characterization of the β-Carotene Hydroxylase Gene DSM2 Conferring Drought and Oxidative Stress Resistance by Increasing Xanthophylls and Abscisic Acid Synthesis in Rice. Plant. Physiol. 2010, 154, 1304–1318. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.J.; Fan, L.; Yang, J.; Cao, R.Z.; Yang, C.Y.; Zhang, J.; Wang, D.M. A Glycine max sodium/hydrogen exchanger enhances salt tolerance through maintaining higher Na+ efflux rate and K+/Na+ ratio in Arabidopsis. BMC Plant. Biol. 2019, 19, 469. [Google Scholar] [CrossRef] [PubMed]
- Szabados, L.; Savour, A. Proline: A multifunctional amino acid. Trends Plant. Sci. 2009, 15, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, S.; Funk, D.; Szabados, L.; Rentsch, D. Proline metabolism and transport in plant development. Amino Acids 2010, 39, 949–962. [Google Scholar] [CrossRef]
- Dimitrios, T. Assessment of lipid peroxidation by measuring malondialdehyde (MDA) and relatives in biological samples: Analytical and biological challenges. Anal. Biochem. 2017, 524, 13–30. [Google Scholar]
- Baker, N.R. A possible role for photosystem II in environmental perturbations of photosynthesis. Physiol. Plant. 1991, 81, 563–570. [Google Scholar] [CrossRef]
- Kolb, E.; Legué, V.; Bogeat-Triboulot, M.B. Physical root-soil interactions. Phys. Biol. 2017, 16, 065004. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Xu, Z.S.; Chen, M.; Li, L.C.; Ma, Y.Z. Functions of the ERF transcription factor family in plants. Botany 2008, 86, 969–977. [Google Scholar] [CrossRef]
- Xu, Z.S.; Chen, M.; Li, L.C.; Ma, Y.Z. Functions and application of the AP2/ERF transcription factor family in crop improvement. Integr. Plant. Biol. 2011, 53, 570–585. [Google Scholar] [CrossRef]
- Liu, P.; Xu, Z.S.; Pan-Pan, L.; Hu, D.; Chen, M.; Li, L.C.; Ma, Y.Z. A wheat PI4K gene whose product possesses threonine autophophorylation activity confers tolerance to drought and salt in Arabidopsis. Exp. Bot. 2013, 64, 2915–2927. [Google Scholar] [CrossRef] [PubMed]
- Flowers, T.J. Improving crop salt tolerance. Exp. Bot. 2004, 55, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.T.; Zhao, M.J.; Wang, C.T.; Gao, Y.; Wang, Y.X.; Liu, Y.W.; Chen, M.; Chen, J.; Zhou, Y.B.; Xu, Z.S.; et al. Identification and characterization of GmMYB118 responses to drought and salt stress. BMC Plant. Biol. 2018, 18, 320. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Le, D.T.; Nishiyama, R.; Watanabe, Y.; Mochida, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S. Genome-wide expression profiling of soybean two component system genes in soybean root and shoot tissues under dehydration stress. DNA Res. 2011, 18, 17–29. [Google Scholar] [CrossRef]
- He, G.H.; Xu, J.Y.; Wang, Y.X.; Liu, J.M.; Li, P.S.; Chen, M.; Ma, Y.Z.; Xu, Z.S. Drought responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant. Biol. 2016, 16, 116. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for agrobacterium mediated transformation of Arabidopsis thaliana. Plant. J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Bu, Q.; Lv, T.; Shen, H.; Luong, P.; Wang, J.; Wang, Z.; Huang, Z.; Xiao, L.; Engineer, C.; Kim, T.H.; et al. Regulation of drought tolerance by the F-box protein MAX2 in Arabidopsis. Plant. Physiol. 2014, 164, 424–439. [Google Scholar] [CrossRef]
- Feng, C.Z.; Chen, Y.; Wang, C.; Kong, Y.H.; Wu, W.H.; Chen, Y.F. Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant. J. 2014, 80, 654–668. [Google Scholar] [CrossRef]
- Kereszt, A.; Li, D.X.; Indrasumunar, A.; Nguyen, C.D.; Nontachaiyapoom, S.; Kinkema, M.; Gresshoff, P.M. Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2007, 2, 948–952. [Google Scholar] [CrossRef]
- Wang, F.; Chen, H.W.; Li, Q.T.; Wei, W.; Li, W.; Zhang, W.K.; Ma, B.; Bi, Y.D.; Lai, Y.C.; Liu, X.L.; et al. GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant. J. 2015, 83, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.P.; Xu, Z.S.; Zheng, W.J.; Zhao, W.; Wang, Y.X.; Yu, T.F.; Chen, M.; Zhou, Y.B.; Min, D.H.; Ma, Y.Z.; et al. Genome-wide analysis of the RAV family in soybean and functional identification of GmRAV-03 involvement in salt and drought stresses and exogenous ABA treatment. Front. Plant. Sci. 2017, 8, 905. [Google Scholar] [CrossRef] [PubMed]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.; Liu, Y.-X.; Yu, Q.; Zhao, S.-P.; Zhao, J.-Y.; Ru, J.-N.; Cao, X.-Y.; Fang, Z.-W.; Chen, J.; Zhou, Y.-B.; et al. Functional Analysis of the Soybean GmCDPK3 Gene Responding to Drought and Salt Stresses. Int. J. Mol. Sci. 2019, 20, 5909. https://doi.org/10.3390/ijms20235909
Wang D, Liu Y-X, Yu Q, Zhao S-P, Zhao J-Y, Ru J-N, Cao X-Y, Fang Z-W, Chen J, Zhou Y-B, et al. Functional Analysis of the Soybean GmCDPK3 Gene Responding to Drought and Salt Stresses. International Journal of Molecular Sciences. 2019; 20(23):5909. https://doi.org/10.3390/ijms20235909
Chicago/Turabian StyleWang, Dan, Yuan-Xia Liu, Qian Yu, Shu-Ping Zhao, Juan-Ying Zhao, Jing-Na Ru, Xin-You Cao, Zheng-Wu Fang, Jun Chen, Yong-Bin Zhou, and et al. 2019. "Functional Analysis of the Soybean GmCDPK3 Gene Responding to Drought and Salt Stresses" International Journal of Molecular Sciences 20, no. 23: 5909. https://doi.org/10.3390/ijms20235909
APA StyleWang, D., Liu, Y.-X., Yu, Q., Zhao, S.-P., Zhao, J.-Y., Ru, J.-N., Cao, X.-Y., Fang, Z.-W., Chen, J., Zhou, Y.-B., Chen, M., Ma, Y.-Z., Xu, Z.-S., & Lan, J.-H. (2019). Functional Analysis of the Soybean GmCDPK3 Gene Responding to Drought and Salt Stresses. International Journal of Molecular Sciences, 20(23), 5909. https://doi.org/10.3390/ijms20235909






