Abstract
Watermelon is a good source of citrulline, a non-protein amino acid. Citrulline has several therapeutic and clinical implications as it produces nitric oxide via arginine. In plants, citrulline plays a pivotal role in nitrogen transport and osmoprotection. The purpose of this study was to identify single nucleotide polymorphism (SNP) markers associated with citrulline metabolism using a genome-wide association study (GWAS) and understand the role of citrulline in watermelon domestication. A watermelon collection consisting of 187 wild, landraces, and cultivated accessions was used to estimate citrulline content. An association analysis involved a total of 12,125 SNPs with a minor allele frequency (MAF)>0.05 in understanding the population structure and phylogeny in light of citrulline accumulation. Wild egusi types and landraces contained low to medium citrulline content, whereas cultivars had higher content, which suggests that obtaining higher content of citrulline is a domesticated trait. GWAS analysis identified candidate genes (ferrochelatase and acetolactate synthase) showing a significant association of SNPs with citrulline content. Haplotype networking indicated positive selection from wild to domesticated watermelon. To our knowledge, this is the first study showing genetic regulation of citrulline variation in plants by using a GWAS strategy. These results provide new insights into the citrulline metabolism in plants and the possibility of incorporating high citrulline as a trait in watermelon breeding programs.
1. Introduction
Non-protein amino acids present in legumes, fruits, seeds, and nuts are ubiquitous in the human diet. Besides containing several health-promoting bioactive compounds, fruits accumulate substantial amounts of free non-protein amino acids. With a diversity of phytochemicals such as carotenoids, flavonoids, and triterpenoids, watermelon (Citrullus lanatus var. vulgaris) fruits also accumulate a substantial amount of a non-protein amino acid—citrulline. Scientific studies have demonstrated several health benefits of citrulline, such as anti-atherosclerotic (hardening of the arteries) effects, reduction of aortic blood pressure and stiffness in individuals with hypertension and cardiovascular diseases, improving lipid profiles by lowering cholesterol, lowering inflammation, and increasing athletic performance [1,2,3,4,5]. Additionally, many recent reviews have covered the clinical impacts of citrulline administration on human health in detail [6,7,8]. Consumption of watermelon has been shown to increase plasma arginine concentrations in adults [9,10]. Watermelons are readily available in most parts of the world, including dry and hot areas of the African continent, where most fruits would not thrive. As watermelon is a nutrient-dense fruit, it is recommended as part of a healthy meal plan as per the U.S. Department of Agriculture’s MyPlate guidelines. In plants, citrulline has been suggested to have a functional role in the nitrogen transport and maintenance of cellular osmolarity during abiotic stresses in plants [11,12]. Members of the Cucurbitaceae family are generally considered to contain relatively large amounts of free citrulline, although watermelon accumulates the highest quantities [13]. Citrulline content in watermelons is spatially and developmentally regulated, with the highest values occurring at fruit maturity [14,15,16,17,18]. Unlike its investigation in plants, citrulline regulation has been extensively studied in the mammalian, prokaryotic, and yeast systems [19]. In the absence of functional nitric oxide synthase (NOS), citrulline in plants is synthesized as a metabolic intermediate during arginine biosynthesis by using carbamoyl phosphate and ornithine. Several studies reported the presence of genotypic variation for citrulline content in a selected set of cultivated watermelon varieties [11,16,18,20,21]. A moderate to high range of heritability [21,22] for citrulline content within cultivated watermelons implies its possible genetic improvement with selective breeding.
A complex compartmentalized network of genes coordinates several metabolic pathways to regulate amino acid metabolism in plants. To understand the molecular regulation and genetic inheritance of amino acids, these networks can be unraveled with the availability of whole-genome sequences and other functional resources. Over the last 2 decades, genome-wide association studies (GWASs) have continued to be the favorite tool to identify causal genetic loci of quantitative or qualitative traits in diverse germplasm collections exploiting evolutionarily conserved recombination events. Several GWAS studies have successfully identified candidate genes by using primary metabolites such as amino acid profiles [23,24,25]. As a naturally rich source of citrulline and with the advent of a newly available genome [26], watermelon could serve as an excellent model to study the evolutionary, biochemical, and molecular determinants of citrulline metabolism and regulation. We previously analyzed genome-wide diversity in watermelon by using a large set of SNPs from accessions collected around the world [27,28] to identify genetic loci controlling traits such as fruit firmness, trichome density and length, fruit length, width, rind thickness and soluble solids [29].
However, we lack information on the genetic basis for variability in citrulline content in watermelon fruits. Our studies with GWAS in watermelon will allow us to estimate population structure and linkage disequilibrium (LD), connecting the variation in the genome with the citrulline content in watermelon germplasm. To understand the evolutionary significance of citrulline in watermelon domestication and its possible genetic regulation, this current study involved (1) characterizing genotypic and phenotypic diversity for citrulline content in a watermelon diversity panel representing wild types, landraces and cultivars; (2) identifying candidate genes significantly associated with citrulline variation; and (3) validating the role of selected genes showing significant association by quantitative real-time-PCR analysis.
2. Results
2.1. Phenotypic and Geographic Variation in Citrulline Content
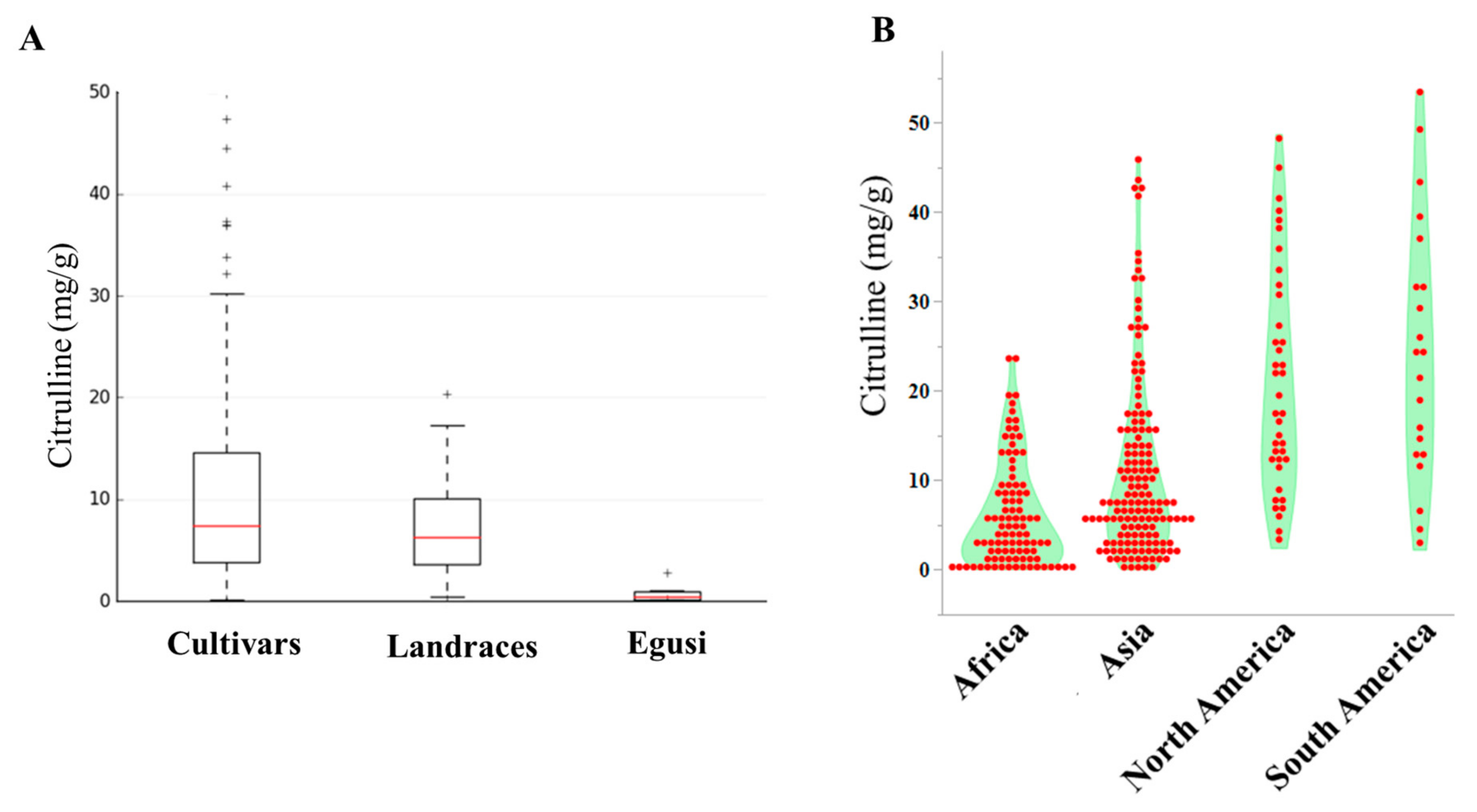
The current study examined the collection of 144 Citrullus. lanatus var. vulgaris (sweet watermelons) accessions, 34 semi-wild types (hereafter called landraces), and 9 accessions belonging to Citrullus. mucasospermus (egusi) (Table S1). Our collection contained plant introduction (PI) accessions from Africa, Asia, Europe, North America, and South America. African types were from Algeria, Botswana, Egypt, Ethiopia, Ghana, Kenya, Liberia, Mali, Nigeria, Senegal, South Africa, Sudan, Zaire, Zambia, and Zimbabwe. The detailed distribution and quantitative variation of citrulline content of accessions used in this study are in Figure S1 and Table S1. The mean, range, and distribution of citrulline content in cultivars, landraces, and egusi types are shown in Figure 1A. The means of citrulline content were 11.08, 7.35, and 0.8 mg/g in cultivars, landraces, and egusi types respectively, which indicates high citrulline content as a feature of cultivars. The average citrulline content in the accessions from North and South America was significantly higher (p ≤ 0.0001) than the accessions from Africa (Figure 1B). Among the selected accessions, mean citrulline content was 10.0 ± 0.04 mg/g (ranging from 0.10 to 47.3 mg/g). The dispersion of free citrulline across the accessions was leptokurtic (kurtosis = 3.27) and asymmetrical (skewness = 1.8). Accessions PI 559993, PI 426625, PI 560020, and PI 526238 had the lowest free citrulline in flesh and Garrisonian, Cole’s Early, and PI 442826 had the highest. Restricted maximum likelihood/best linear unbiased prediction (REML/BLUP) [30,31], variance component estimation revealed significant variation in citrulline content within accessions. The estimates of heritability based on REML analysis were high (83%) for citrulline content, whereas the genetic gain at 5% selection intensity was 9.5%.
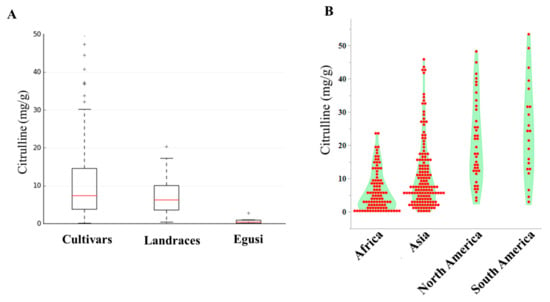
Figure 1.
Citrulline content in watermelon accessions. (A) Box plots showing the range, mean, and distribution of citrulline content in cultivars, landraces, and egusi types. (B) Violin-scaled contour map showing world geographic variation in citrulline content across the accessions.
2.2. Population Structure of Various Accessions Based on Citrulline Content
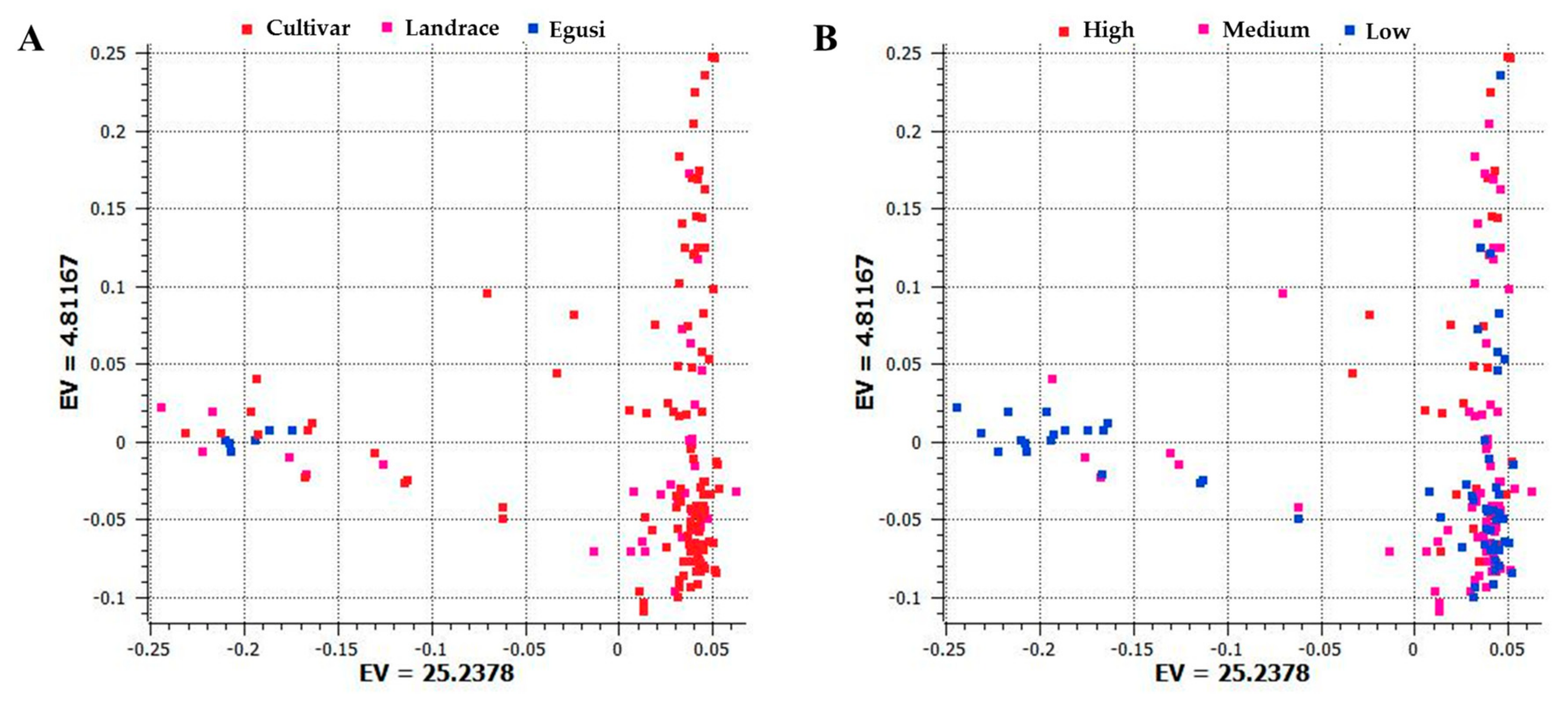
To examine divergence across accessions during evolution, analysis of population structure and phylogenetic relationships and PCA were carried out [27]. Using the SNP dataset, we constructed a PCA with the first and second principal components PC1 (25.24) and PC2 (4.81) that separated egusi, landraces, and sweet watermelons (Figure 2A). This PCA also separated low, medium, and high citrulline content types (Figure 2B). Sweet watermelon types contained the highest citrulline content as compared with their ancestral progenitors. Egusi types and landraces contained low to medium citrulline content, whereas cultivars showed increased content, which indicates that high citrulline content is a domesticated trait.
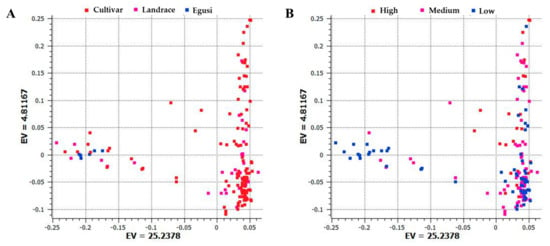
Figure 2.
Principal component analysis (PCA) based on the first two components showing the distribution of (A) cultivars, landraces, and egusi types; (B) and low, medium, and high citrulline content in 187 watermelon accessions by using 1410 single nucleotide polymorphisms (SNPs) generated by genotyping by sequencing. Each dot represents an accession. EV indicates the percentage of explained variance.
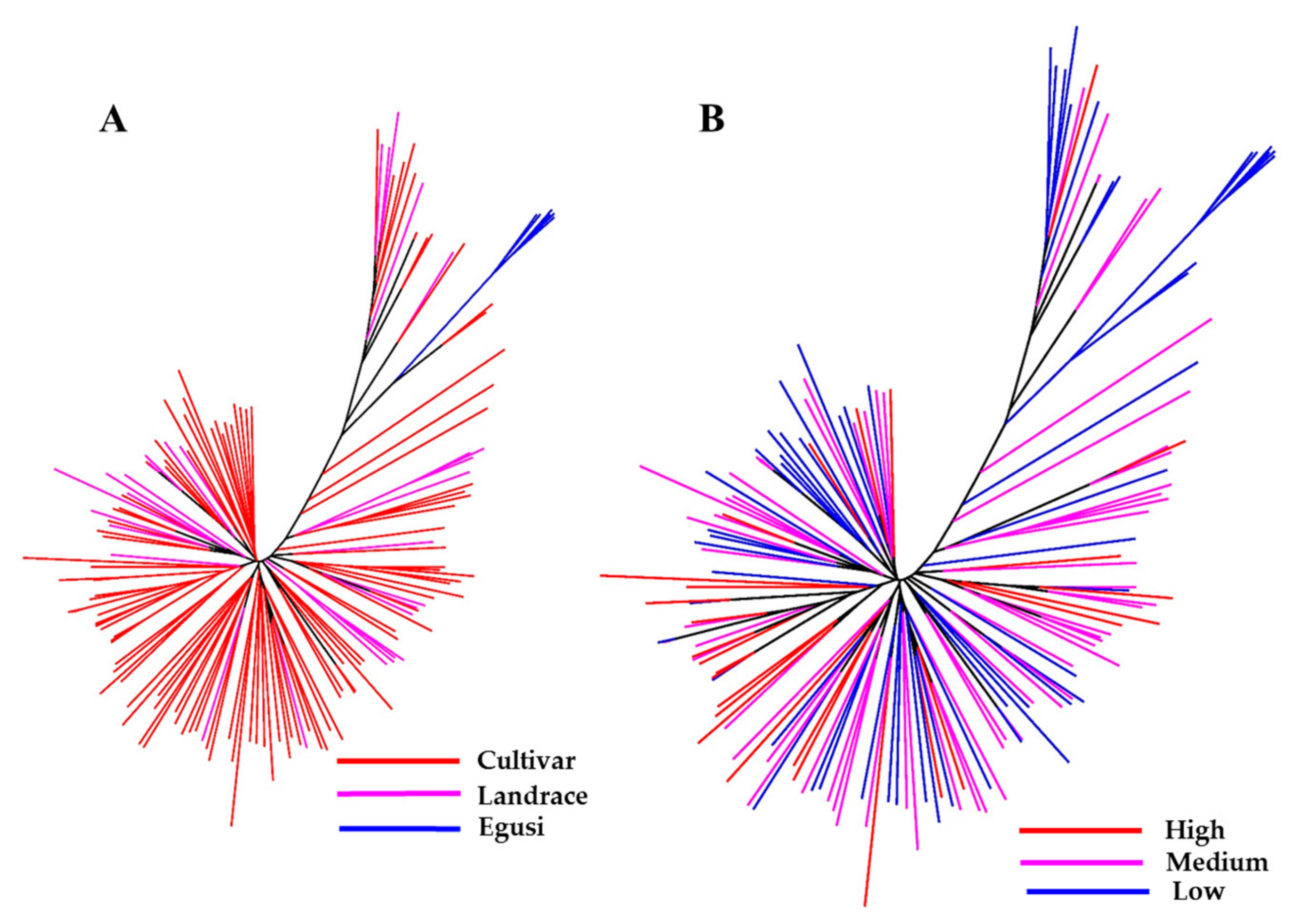
Using the final SNP dataset, we constructed an unrooted neighbor-joining (NJ) tree to infer phylogenetic relationships and understand the distribution of citrulline content across the C. lanatus accessions (Figure 3A,B). One group (colored blue in the tree) represents the entire egusi collections. The pink- and red-colored clades represent sweet watermelons and landraces (most from South Africa), many with a hard rind and white flesh resembling an intermediate between egusi and sweet watermelon. This study indicated that most low to medium citrulline-content types are egusi and landraces.
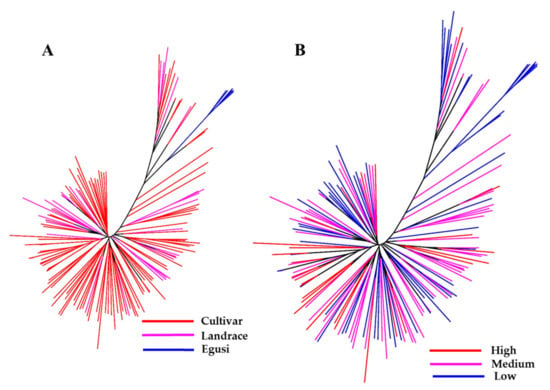
Figure 3.
Genetic relationship between a set of 187 watermelon accessions. Neighbor-joining (NJ) tree constructed with 1410 high-quality SNPs explains most of the genetic structure of watermelon germplasm by (A) type and (B) citrulline content. Accessions in blue-, pink-, and red-colored clades are egusi-types, sweet watermelons, and landraces, respectively. In (B), blue, pink, and red clades represent low, medium, and high citrulline content, respectively.
2.3. Genome-wide Association Study to Locate Quantitative Trait Loci for Citrulline Content
We used a GWAS with 12,125 SNPs to identify alleles that affect citrulline content (Figure 4); individual SNP associations along with the details of major and minor allele frequencies and magnitude of associations are in Table 1 and detailed annotations for all associated SNP markers are in Table S2. We found 12 SNPs associated with citrulline content (Table 1 and Table 2). Significantly associated SNPs for citrulline content were S02_33508197, S02_33508131, S02_28460679, S04_19161720, S04_10803195, S04_19161725, S06_30930976, S06_30991451, S07_12838412, S07_6258382, S09_9172194, and S10_19726131 and were found in ferrochelatase, F-box/LRR-repeat protein 2, Golgi SNAP receptor complex member 2, DNA polymerase I/DNA polymerase I, acetolactate synthase, BAG family molecular chaperone regulator 1, TLC ATP/ADP transporter, protein of unknown function, and phototropic-responsive NPH3 family protein genes, respectively. Biological roles, molecular processes, and the cellular location of these genes are in Table 2.
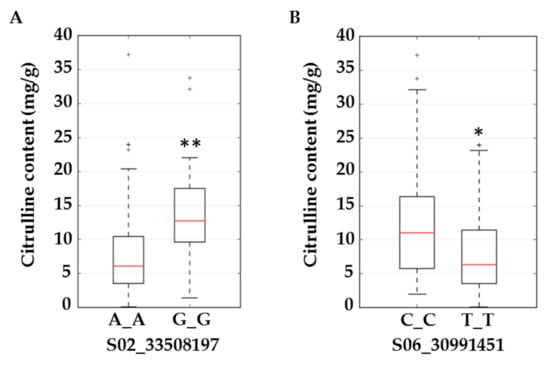
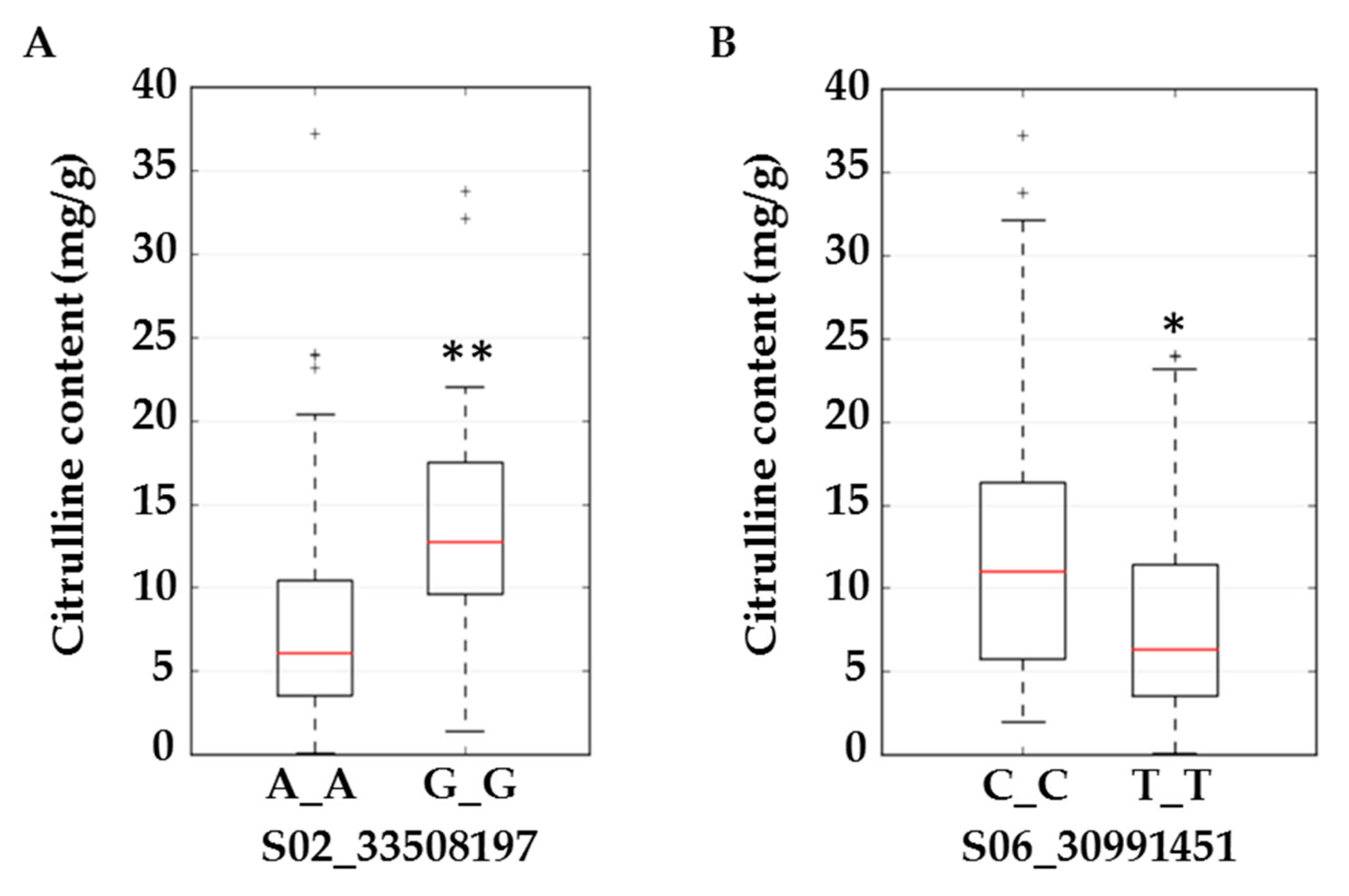
Figure 4.
Boxplots for citrulline content (mg/g) in flesh tissue at SNP S02_33508197 located in the intron of ferrochelatase (A) and S06_30991451 located in an exon of acetolactate synthase (B). Significant differences (based on the Kruskal–Wallis test) with p ≤ 0.01 and p ≤ 0.05 are marked with two (**) and one (*) asterisks respectively.

Table 1.
The significant non-synonymous SNPs associated with citrulline content in watermelon flesh.

Table 2.
Gene ontology classification of the genes associated with significant SNPs.
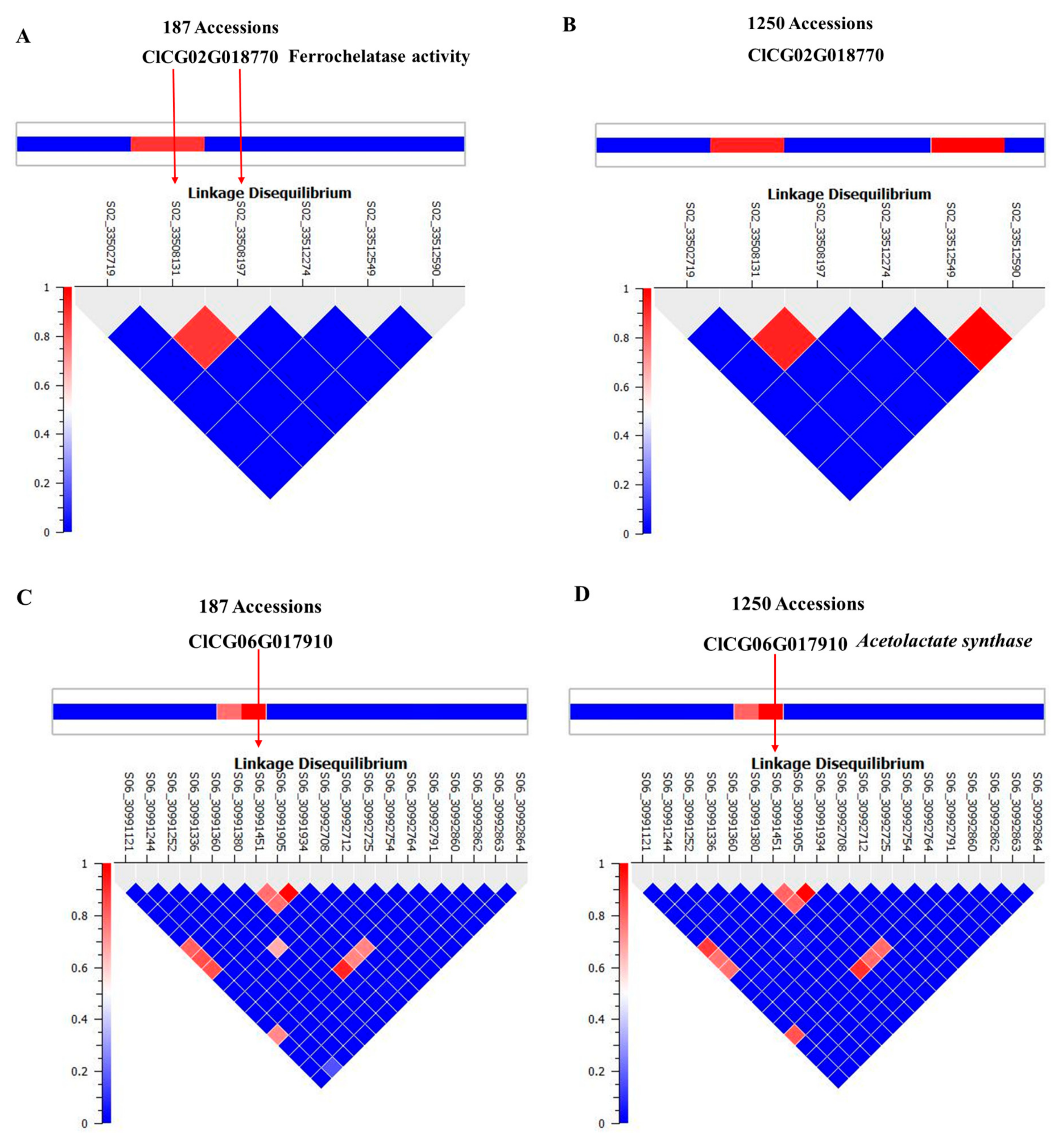
We selected ferrochelatase (FC) and acetolactate synthase (ALS) genes for further validation. Allelic effects of SNPs located in these genes are in Figure 4. S02_33508197 is located in the intron of FC. Allele frequencies for S02_33508197 were 0.83 for AA and 0.17 for the minor allele GG. Average citrulline content was 5 mg/g for the AA-containing genotype and 12.5 mg/g for the GG-containing genotypes (Figure 4A). S06_30991451 is located in an exon of ALS and a non-synonymous mutation causing N→S with allele frequencies for TT and CC of 0.79 and 0.21, respectively. Allelic effects for major (TT) and minor (CC) alleles can be noted from the box plot (Figure 4B). Strong LD is noted around the associated SNPs in Figure 5A,C. LD around these two genes was further confirmed from a robust set of 1250 accessions (Figure 5B,D) in a recently published study [26].
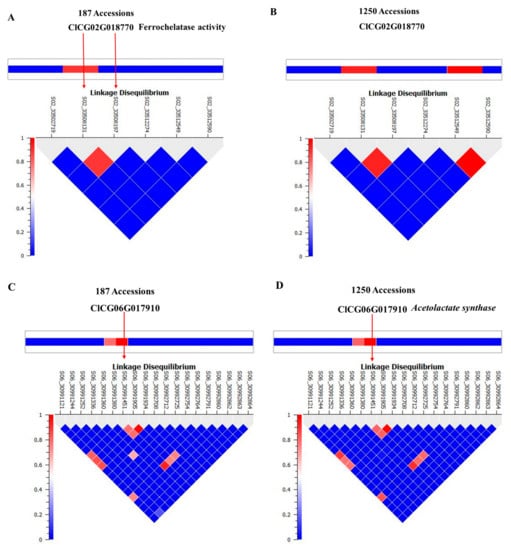
Figure 5.
Linkage disequilibrium structure and implicated genomic regions for SNPs aligned with: (A,B) ferrochelatase; (C,D) acetolactate synthase.
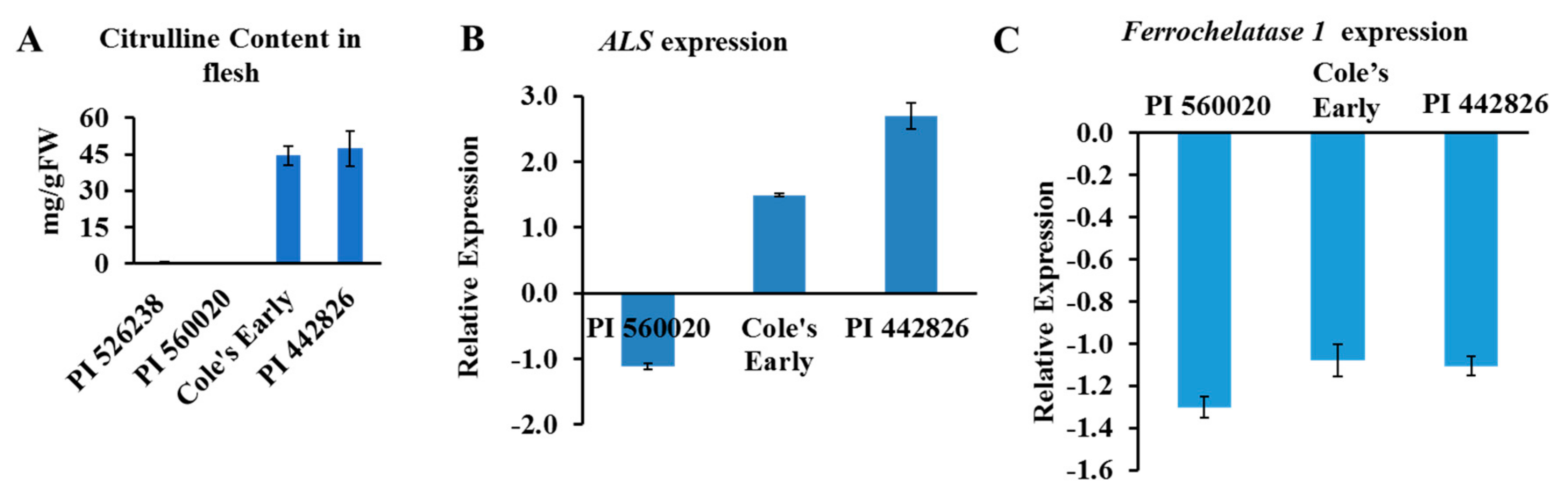
The watermelon genome has a single copy of ALS gene (a large subunit) and two genes that encode ALS small subunits (ClCG09G014670 and ClCG03G010140, putative ALS small subunits 1 and 2, respectively). The ALS enzyme facilitates the first step in the biosynthesis of branched-chain amino acids (BCAAs; valine, leucine, and isoleucine) in microbes and plants. Moreover, ALS enzyme is inhibited by a group of imidazolinone and sulfonylurea herbicides [32,33,34], thereby preventing biosynthesis of BCAAs. We used real-time quantitative PCR to validate the association of the ALS gene(s) in selected high and low citrulline-content watermelon accessions (Figure 6A). The relative expression of ALS mRNA was significantly upregulated in flesh tissues of high citrulline-content watermelon accessions and downregulated in PI560020, with low citrulline content (Figure 6B). In summary, the expression of ALS showed strong association with citrulline content in watermelon.
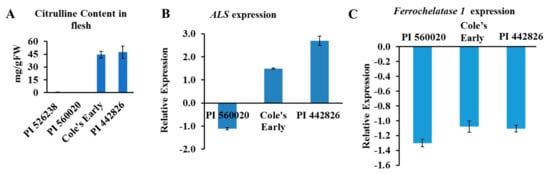
Figure 6.
(A) Citrulline content and expression of candidate genes in flesh of selected watermelon accessions. Data are means ± SD (n = 3). (B) Expression of acetolactate synthase (ALS) and (C) Ferrochelatase 1 genes in flesh. PI 526238 accession, with low citrulline content, was used as a calibrator for relative expression in (A). Gene expression was normalized to that of Actin 8. Data are means ± SD (n = 3).
The watermelon genome database search revealed two genes coding for FC enzyme: ClCG02G018770 (appeared in GWAS) and ClCG08G016940. The BLAST search against the Arabidopsis thaliana genome revealed that the watermelon ClCG08G016940 and ClCG02G018770 genes are orthologues of AT5G26030 and AT2G30390, labeled FC1 and FC2, respectively.
The mRNA abundance for FC1 was quantified by qRT-PCR, and we wondered whether FC1 transcript level was associated with citrulline level in watermelon. Reduced FC1 transcript abundance was seen in all three accessions (Figure 6C). However, the reason as to why the FC1 expression was low in accessions with both high and low citrulline content is elusive. We hypothesize that FC1 may act in a spatiotemporal and growth dependent manner. Our data suggest that the ALS and FC genes may have a strong association with citrulline accumulation. However, future analysis is required to characterize the functions of FC and ALS proteins in citrulline-BCAA metabolism and NO turnover in watermelon.
2.4. Haplotyping and Network Analysis of Acetolactate Synthase and Ferrochelatase
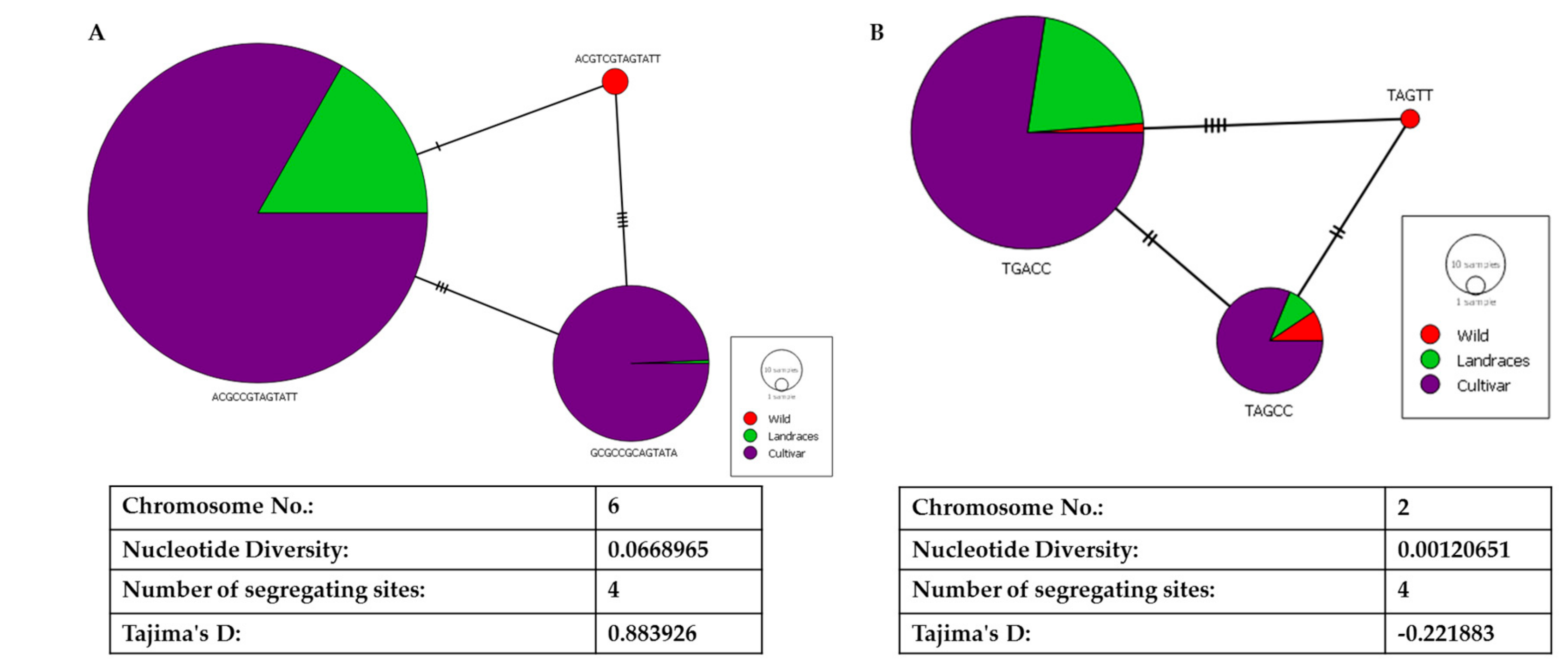
With the four segregating sites in the LD block in ALS, we could build a network of haplotypes for egusi (wild), landraces, and cultivated watermelon (Figure 7A). Haplotype ACGTCGTAGTATT had undergone a single nucleotide change to form a landrace haplotype (ACGCCGTAGTATT). GCGCCGCAGTATA has three segregating sites as compared with the landrace haplotype and four segregating sites as compared with the wild haplotype. We noted Tajima’s D as 0.88 and nucleotide diversity of 0.07, indicating a high degree of positive selection around this gene. Contrastingly, for the other causal gene FC, we noted reduced nucleotide diversity of 0.01 and negative Tajima’s D (−0.221) indicating purifying selection in evolution (Figure 7B).
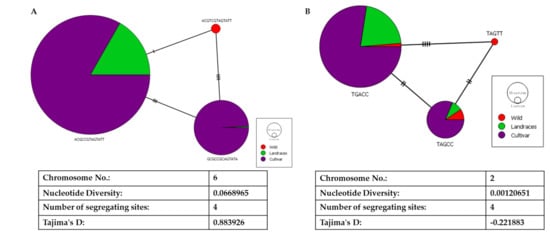
Figure 7.
Haplotyping and network analysis of (A) acetolactate synthase and (B) ferrochelatase on chromosomes 6 and 2, respectively.
3. Discussion
The current research aimed to analyze citrulline content evolution by using GBS-generated SNPs across the global collections of sweet watermelon, landraces, and egusi types. Watermelons were cultivated in the northeast of Africa around 5000 years ago [35]. Pictures on Egyptian frescos and seeds found in the grave of King Tutankhamun indicated that the flesh of watermelons was still white and tasted bland [36]. These bland vegetable-kind watermelons were cultivated for thousands of years before sweet watermelons arose [37]. Depictions of cut watermelons throughout the centuries (Giovanni Stanchi 1608 to c. 1675) deviated from the modern watermelons. When explored through flesh of landraces, one can find the evidence for these anthropological records. Because of this, in the current study, landraces are named as semi-wild as the sweet watermelons are quite different. Modern watermelon provides a large amount of water and nutrients, such as sugars, carotenoids, lycopene, minerals, and amino acids, including citrulline. Although citrulline is the most abundant free amino acid in watermelon [12], because of the crop’s commercial value, genetic mapping efforts have mostly focused on sugar content [38,39]. During domestication, dessert watermelons were mostly selected for qualitative traits such as sweetness, flesh color, and rind pattern [40]. Although primitive dessert watermelon landraces, with naturally low sugar content, were expected to be valuable sources of bioactive compounds [41], our study demonstrates that citrulline content is, in general, lower in landraces and egusi types. Modern cultivars from the Americas that are rich in citrulline may have acquired the trait inadvertently during the selection for sweetness or the trait might have an unknown role in adaptation. A positive correlation of sugar content with citrulline was demonstrated in modern watermelon cultivars [18,21,22]. The distinctness of American, European and Asian ecotypes has been described in watermelons [29,42,43], but an independent parallel phenotypic evolution for citrulline during domestication cannot be ruled out [44].
3.1. Genetic Characterization of ALS and FC Locus Haplogroups
We genotyped 187 watermelon accessions with a mix of cultivars, landraces and egusi types predominantly collected from Africa, Europe, North America, and Asia with ~12K SNPs. A subset of this data allowed for estimating genomic diversity across the LD around the associated locus among various groups, constructing an NJ tree, resolving population structure, estimating chromosome-wise LD patterns and understanding the extent of population differentiation and haplotype networking in terms of citrulline content in watermelons. This study revealed high nucleotide diversity and Tajima’s D for the ALS locus, which showed a strong association with citrulline content. In this study sweet watermelon in South Africa, and most landraces of South Africa clustered with sweet watermelon, which strengthens the argument that the Kalahari Desert could be the center of origin [45,46,47]. Alternatively, egusi types of northeast Africa could be the progenitors of landrace watermelon types because they share segregating sites in the both haplotype networks, thereby indicating a stepwise evolutionary pattern and probably is the second event of domestication. Similar to our observation in this study of haplotype networking in the ALS and FC loci between egusi and sweet watermelon, Chomicki and Renner [48] reclassified egusi (var. mucasospermus) and sweet watermelons (var. vulgaris) as two different species naming egusi as C. mucasospermus. Knowledge of causal genes underlying domestication traits and distribution of functionally diverse alleles in different watermelon populations may be required to better demonstrate the evolutionary route of dessert watermelons [26]. The current SNP-based analysis revealed citrulline accumulation as an important step for the domestication process of sweet watermelons. In this study, we identified 105 private SNPs segregating in sweet watermelons that was not found in related wild species. Such SNPs are valuable because they can be of adaptive importance and would be of immense use for generating passport information for enhancing nutritional traits such as citrulline.
3.2. Plausible Role of Candidate Genes Identified in Citrulline Biosynthesis
In eukaryotic cells, FC proteins are located in the mitochondrial inner membrane and facilitate the final step in the heme biosynthetic pathway, inserting a ferrous iron into protoporphyrin IX to yield the heme [49]. In animals, arginine can be a substrate for citrulline biosynthesis via the action of NOS [50,51,52]. However, in plants, arginine-dependent NOS-like proteins are not functionally validated; therefore, the synthesis of citrulline directly from arginine in plants remains debated [18,53,54]. In animals, NOS proteins are soluble hemoproteins that facilitate the conversion of l-arginine to citrulline and NO [55]. NO, a crucial signaling molecule also called a gaseous hormone in plants and animals regulates many physiological and biochemical processes and stress responses in plants (reviewed by Domingos et al. [56]). In animals, NO preferentially binds to hemoproteins in both ferrous and ferric states [57,58], which indicates a strong association of heme synthesis by FC and the arginine–citrulline cycle. We observed that both low and high citrulline content accessions showed reduced FC1 expression. However, the reason as to why the FC1 transcript levels were low in accessions with both high and low citrulline content is elusive. We hypothesize that FC1 expression may be regulated in a spatio-temporal and growth-dependent manner. Further analysis of FC1 expression in watermelon flesh at various fruit developmental stages may reveal the association of citrulline content and FC1 transcript abundance.
The ALS complex consists of three pairs of subunits, a large subunit responsible for catalysis, and a small subunit for feedback inhibition. All three genes encoding the putative ALS large subunit (ClCG06G017910), ALS small subunit_1 (ASsu1; ClCG09G014670) and ALS small subunit_2 (ASsu2; ClCG03G010140) proteins were found in the watermelon genome. There are no known direct metabolic links between citrulline and BCAA synthesis (BCAAs) in the canonical or non-canonical systems. Because of the sub-optimal levels of free citrulline in most plants, limited information is available on its relationship with other amino acids. Nonetheless, some studies suggest the possibility of unknown links between citrulline/arginine and BCAA pathways; for example, (1) arginine, a catabolic product of citrulline, is significantly and positively correlated with BCAAs in soybean seeds [59]; (2) levels of BCAAs were increased along with those of citrulline, arginine, and ornithine during conditional downregulation of target of rapamycin gene in Arabidopsis [60]; and (3) citrulline level was increased in leaves of the rice ALS mutant [61].
4. Materials and Methods
4.1. Plant Materials, Citrulline Extraction, and Quantification
Selfed seeds of 187 National Plant Germplasm System collections of C. lanatus var. vulgaris and C. lanatus var. mucasospermus were grown in the greenhouse. The authors are grateful to R. Jarret, Plant Genetic Resources Conservation Unit, USDA-ARS (Griffin, GA, USA) for providing the seeds of germplasm accessions. Citrulline content in the flesh determined by using three biological replicates for each accession (Table S1). For determining citrulline content, ~20 mg lyophilized flesh tissue for each biological replicate was homogenized by using 3 mm Demag stainless steel balls (Abbott Ball Co., CT, USA) and a Harbil model 5G-HD paint shaker, suspended in 20 mM cold HCl and centrifuged at 14,609× g for 20 min at 4 °C. The supernatant was filtered by using 0.45 uM 96-well filters (Pall Life Sciences, NY, USA). The filtrate was derivatized with an AccQ-Fluor reagent kit (Waters Corp., Milford, MA, USA) per the manufacturer’s protocol. UPLC-ESI-MS/MS analysis involved using a Waters Acquity H-class UPLC system coupled to a Waters Xevo TQ mass spectrometer with an electrospray ionization (ESI) probe, Waters ACQUITY UPLC Fluorescence (FLR) detector and Water’s AccQ•Tag Ultra column. The mobile phase consisted of water (0.1% formic acid v/v) (A) and acetonitrile (0.1% formic acid v/v) (B). The column heater was set to 60 °C, and the mobile phase flow rate was maintained at a constant rate of 0.6 mL/min. By using the Waters IntelliStart software, multiple reaction monitoring transitions for citrulline were optimized. The ESI source was operated at 150 °C, with desolvation temperature 450 °C, desolvation gas flow rate 900 L/h and capillary voltage 3.2 kV. Multiple reaction monitoring was performed in the positive mode. Instrument monitoring and data acquisition, integration, and quantification involved using Water’s MassLynx software.
4.2. Phylogenetic and Population Genomic Analyses
Phenotypic data were analyzed by using JMP software (JMP Pro 14 (SAS Institute, Cary, NC, SAS http://www.jmp.com/en_us/home.html)). The variance components were estimated by using REML-BLUP analysis. The broad-sense heritability was estimated as H2 = δ2g/( δ2g + δ2e/r), where δ2g and δ2e are estimated genotypic and error variances, respectively [62]. The genetic gain was calculated as Gs = K × H2 × (δ2p)−1/2, where K is the selection intensity at 5% (k = 2.056), H2 is heritability in a broad sense and (δ2p)−1/2 is the phenotypic standard deviation. Totals of 11,485 SNPs [27] and 16,292 SNPs [26] were combined and further filtered using MAF ≥ 0.01 to identify 12,125 SNPs that have a call rate of 70% in our GWAS panel. For population stratification analysis, we used more stringent cut off for SNP selection based on MAF of 0.1 and 90% call rate. Further informative SNPs were selected by removing SNPs located in a LD block by 50% cut off and also SNPs that deviated from Hardy–Weinberg equilibrium were discarded. A neighbor-joining (NJ) tree for watermelon accessions was constructed by using TASSEL 5.0 [63]. Archaeopteryx [64] was used to visualize and analyze the tree. For analyzing population structure, we used the principal components, or eigenvectors, of principal component analysis (PCA), and corresponding eigenvalues were estimated by using the EIGENSTRAT algorithm [65] with the SNP & Variation Suite (SVS v8.8.1; Golden Helix, Inc., Bozeman, MT, www.goldenhelix.com).
4.3. Association Analysis
For GWAS, the population structure Q matrix was replaced by the PC matrix. The PC matrix and identity by descent (IBD) was calculated from LD-pruned SNPs in SVS v8.1.5. GWAS involved a multiple-locus mixed linear model developed by the EMMAX method and implemented in SVS v8.1.5. We used a PC matrix (first two vectors) and the IBD matrix to correct for population stratification. Manhattan plots for associated SNPs were visualized in GenomeBrowse v1.0 (Golden Helix, Inc) (Figure S2). The SNP p-values from GWAS underwent false discovery rate (FDR) analysis.
4.4. Haplotype Network Analysis
To evaluate the haplotype frequency, we first specified the haplogroups and the frequency with which a determined haplotype appeared in each haplogroup. Then, we determined the genetic constitution as the proportion of SNPs that belong to a haplogroup. The haplotype frequency analysis involved estimating haplotype diversity. The number of haplotypes and the haplotype diversity values were calculated by using DnaSP 5.10 (http://www.ub.edu/dnasp/DnaSP_OS.html) [66]. Haplotype networking and LD calculation were performed by using publicly available SNP data for 1209 C. lanatus var. vulgaris and 51 C. mucasospermus (egusi) [21]. Adjacent SNP pairs within a chromosome underwent LD analysis by using expectation maximization [67]. All LD plots as well as LD measurements and haplotype frequency calculations involved use of SVS v8.8.1. This collection consisted 220 PIs from Africa, 352 from Asia, 497 from Europe, 121 from North America, and 16 from South America [26].
4.5. Total RNA Extraction and qRT-PCR
Total RNA was extracted from watermelon flesh (three biological replicates) by using TRIzolTM Reagent (Life Technologies, Carlsbad, CA, USA). On-column DNase I treatment was used to remove genomic DNA. The RNA concentrations and quality were determined by using NanoDrop 2000 (Thermofisher, Waltham, MA, USA). The first strand cDNAs were synthesized with 1 µg total RNA in a 20 µL reaction mixture with SuperScriptTM II Reverse Transcriptase (Thermo Fisher Scientific, USA) per the manufacturer’s instructions. cDNAs were then diluted by adding 180 µL sterile nuclease-free water. qRT-PCR was performed with PowerUpTM SYBR Green Master Mix (Applied Biosystems, Thermo Fisher Scientific, USA) and the StepOnePlusTM Real-Time PCR System (Applied Biosystems). PCR was programmed at 95 °C for 30 s, followed by 95 °C for 10 s and 60 °C for 60 s for 40 cycles. Gene expression was normalized to Actin 8 expression (housekeeping reference), and transcript abundance of target genes was calculated with the PI 526238 accession, with significantly low citrulline content, as a calibrator. The 2−ΔΔCT method was used to estimate relative transcript level [68]. The primers for qRT-PCR analysis are in Table S3.
5. Conclusions
This study demonstrated a wide variation in watermelon germplasm for citrulline content. Identifying such variability and introgressing it into high-yield lines will be key to long-term watermelon breeding for improved nutraceutical content. We found moderate to high (83%) broad-sense heritability, which indicates the possibility of successful citrulline enhancement with introgression breeding. Here we provide new insights into the domestication and population genetic structure based on citrulline content in geographically spread-out watermelon accessions. The SNP loci associated with citrulline content identified in this study would benchmark the efforts of development of molecular markers to enhance nutritional quality in elite watermelon cultivars.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/21/5392/s1.
Author Contributions
U.K.R., P.N., A.L. and V.J. designed the research program. S.B.A. collected the samples. U.K.R., V.L.A., P.N. analyzed the data and conducted the GWAS. V.J. performed metabolic analysis. S.S. performed gene expression analysis. C.L.-O. performed haplotyping and network analysis. G.P. performed field evaluation. U.K.R., S.S., P.N., A.L., and V.J. wrote the manuscript. All authors read and approved the manuscript.
Funding
This work is supported by the USDA-NIFA 2017-38821-26434, USDA-NIFA 2018-38821-27734, USDA-NIFA [Hatch Project No TEX09647/project accession no. 1011513] and National Watermelon Promotion Board.
Acknowledgments
We thank Diwas Silwal for assistance in sample processing for metabolic analysis.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Abbreviations
| ALS | Acetolactate synthase |
| ASsu1 | Acetolactate synthase small subunit_1 |
| ASsu1 | Acetolactate synthase small subunit_2 |
| BCAAs | branched-chain amino acid synthesis |
| FDR | false discovery rate |
| FC | Ferrochelatase |
| GWAS | genome-wide association studies |
| IBD | identity by descent |
| LD | linkage disequilibrium |
| NJ | neighbor-Joining |
| PI | Plant Introduction |
| SNP | single nucleotide polymorphism |
References
- Alsop, P.; Hauton, D. Oral nitrate and citrulline decrease blood pressure and increase vascular conductance in young adults: a potential therapy for heart failure. Eur. J. Appl. Physiol. 2016, 116, 1651–1661. [Google Scholar] [CrossRef] [PubMed]
- Moinard, C.; Maccario, J.; Walrand, S.; Lasserre, V.; Marc, J.; Boirie, Y.; Cynober, L. Arginine behaviour after arginine or citrulline administration in older subjects. Br. J. Nutr. 2016, 115, 399–404. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Morita, M.; Hayashi, T.; Ochiai, M.; Maeda, M.; Yamaguchi, T.; Ina, K.; Kuzuya, M. Oral supplementation with a combination of L-citrulline and L-arginine rapidly increases plasma L-arginine concentration and enhances NO bioavailability. Biochem. Biophys. Res. Commun. 2014, 454, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Bahri, S.; Zerrouk, N.; Aussel, C.; Moinard, C.; Crenn, P.; Curis, E.; Chaumeil, J.-C.; Cynober, L.; Sfar, S. Citrulline: from metabolism to therapeutic use. Nutrition 2013, 29, 479–484. [Google Scholar] [CrossRef]
- Schellekens, G.A.; de Jong, B.A.W.; van den Hoogen, F.H.J.; van de Putte, L.B.A.; van Venrooij, W.J. Pillars Article: Citrulline is an Essential Constituent of Antigenic Determinants Recognized by Rheumatoid Arthritis–specific Autoantibodies. J. Clin. Invest. 1998, 101, 273–281. [Google Scholar] [CrossRef]
- Allerton, T.D.; Proctor, D.N.; Stephens, J.M.; Dugas, T.R.; Spielmann, G.; Irving, B.A. l-Citrulline Supplementation: Impact on Cardiometabolic Health. Nutrients 2018, 10, 921. [Google Scholar] [CrossRef]
- Papadia, C.; Osowska, S.; Cynober, L.; Forbes, A. Citrulline in health and disease. Review on human studies. Clin. Nutr. 2018, 37, 1823–1828. [Google Scholar] [CrossRef]
- Breuillard, C.; Bonhomme, S.; Couderc, R.; Cynober, L.; De Bandt, J.P. In vitro anti-inflammatory effects of citrulline on peritoneal macrophages in Zucker diabetic fatty rats. Br. J. Nutr. 2015, 113, 120–124. [Google Scholar] [CrossRef]
- Collins, J.K.; Wu, G.; Perkins-Veazie, P.; Spears, K.; Claypool, P.L.; Baker, R.A.; Clevidence, B.A. Watermelon consumption increases plasma arginine concentrations in adults. Nutrition 2007, 23, 261–266. [Google Scholar] [CrossRef]
- Wu, G.; Collins, J.K.; Perkins-Veazie, P.; Siddiq, M.; Dolan, K.D.; Kelly, K.A.; Heaps, C.L.; Meininger, C.J. Dietary supplementation with watermelon pomace juice enhances arginine availability and ameliorates the metabolic syndrome in Zucker diabetic fatty rats. J. Nutr. 2007, 137, 2680–2685. [Google Scholar] [CrossRef]
- Akashi, K.; Miyake, C.; Yokota, A. Citrulline, a novel compatible solute in drought-tolerant wild watermelon leaves, is an efficient hydroxyl radical scavenger. FEBS Lett. 2001, 508, 438–442. [Google Scholar] [CrossRef]
- Joshi, V.; Fernie, A.R. Citrulline metabolism in plants. Amino Acids 2017, 49, 1543–1559. [Google Scholar] [CrossRef] [PubMed]
- Fish, W.W.; Bruton, B.D. Quantification of L-citrulline and other physiologic amino acids in watermelon and selected cucurbits. In Proceedings of the Cucurbitaceae 2010, Charleston, SC, USA, 14–18 November 2010; pp. 152–154. [Google Scholar]
- Rimando, A.M.; Perkins-Veazie, P.M. Determination of citrulline in watermelon rind. J. Chromatogr. A 2005, 1078, 196–200. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.R.; Fish, W.; Levi, A.; King, S.; Wehner, T.; Perkins-Veazie, P. L-citrulline levels in watermelon cultivars from three locations. Cucurbit. Genet. Coop. Rpt. 2010, 33, 36–39. [Google Scholar] [CrossRef]
- Davis, A.R.; Webber, C.L.; Fish, W.W.; Wehner, T.C.; King, S.; Perkins-Veazie, P. L-citrulline levels in watermelon cultigens tested in two environments. HortScience 2011, 46, 1572–1575. [Google Scholar] [CrossRef]
- Akashi, K.; Mifune, Y.; Morita, K.; Ishitsuka, S.; Tsujimoto, H.; Ishihara, T. Spatial accumulation pattern of citrulline and other nutrients in immature and mature watermelon fruits. J. Sci. Food Agric. 2017, 97, 479–487. [Google Scholar] [CrossRef]
- Joshi, V.; Joshi, M.; Silwal, D.; Noonan, K.; Rodriguez, S.; Penalosa, A. Systematized biosynthesis and catabolism regulate citrulline accumulation in watermelon. Phytochemistry 2019, 162, 129–140. [Google Scholar] [CrossRef]
- Curis, E.; Nicolis, I.; Moinard, C.; Osowska, S.; Zerrouk, N.; Bénazeth, S.; Cynober, L. Almost all about citrulline in mammals. Amino Acids 2005, 29, 177. [Google Scholar] [CrossRef]
- Yokota, A.; Kawasaki, S.; Iwano, M.; Nakamura, C.; Miyake, C.; Akashi, K. Citrulline and DRIP-1 protein (ArgE homologue) in drought tolerance of wild watermelon. Ann. Bot. 2002, 89, 825–832. [Google Scholar] [CrossRef]
- Wehner, T.C.; Naegele, R.P.; Perkins-Veazie, P. Heritability and Genetic Variance Components Associated with Citrulline, Arginine, and Lycopene Content in Diverse Watermelon Cultigens. HortScience 2017, 52, 936–940. [Google Scholar] [CrossRef]
- Fall, L.A.; Perkins-Veazie, P.; Ma, G.; McGregor, C. QTLs associated with flesh quality traits in an elite × elite watermelon population. Euphytica 2019, 215, 30. [Google Scholar] [CrossRef]
- Angelovici, R.; Lipka, A.E.; Deason, N.; Gonzalez-Jorge, S.; Lin, H.; Cepela, J.; Buell, R.; Gore, M.A.; Dellapenna, D. Genome-wide analysis of branched-chain amino acid levels in Arabidopsis seeds. Plant Cell 2013, 25, 4827–4843. [Google Scholar] [CrossRef] [PubMed]
- Angelovici, R.; Batushansky, A.; Deason, N.; Gonzalez-Jorge, S.; Gore, M.A.; Fait, A.; DellaPenna, D. Network-Guided GWAS Improves Identification of Genes Affecting Free Amino Acids. Plant Physiol. 2017, 173, 872–886. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Liu, H.; Chen, J.; Shi, T.; Zhang, C.; Sun, D.; He, Z.; Hao, Y.; Chen, W. Genome-Wide Association Studies of Free Amino Acid Levels by Six Multi-Locus Models in Bread Wheat. Front. Plant Sci. 2018, 9, 1196. [Google Scholar] [CrossRef]
- Wu, S.; Wang, X.; Reddy, U.; Sun, H.; Bao, K.; Gao, L.; Mao, L.; Patel, T.; Ortiz, C.; Abburi, V.L.; et al. Genome of ‘Charleston Gray’, the principal American watermelon cultivar, and genetic characterization of 1,365 accessions in the U.S. National Plant Germplasm System watermelon collection. Plant Biotechnol. J. 2019, 0, 1–13. [Google Scholar] [CrossRef]
- Nimmakayala, P.; Levi, A.; Abburi, L.; Abburi, V.L.; Tomason, Y.R.; Saminathan, T.; Vajja, V.G.; Malkaram, S.; Reddy, R.; Wehner, T.C.; et al. Single nucleotide polymorphisms generated by genotyping by sequencing to characterize genome-wide diversity, linkage disequilibrium, and selective sweeps in cultivated watermelon. BMC Genom. 2014, 15, 767. [Google Scholar] [CrossRef]
- Reddy, U.K.; Nimmakayala, P.; Levi, A.; Abburi, V.L.; Saminathan, T.; Tomason, Y.R.; Vajja, G.; Reddy, R.; Abburi, L.; Wehner, T.C.; et al. High-resolution genetic map for understanding the effect of genome-wide recombination rate on nucleotide diversity in watermelon. G3 2014, 4, 2219–2230. [Google Scholar] [CrossRef]
- Reddy, U.K.; Abburi, L.; Abburi, V.L.; Saminathan, T.; Cantrell, R.; Vajja, V.G.; Reddy, R.; Tomason, Y.R.; Levi, A.; Wehner, T.C. A genome-wide scan of selective sweeps and association mapping of fruit traits using microsatellite markers in watermelon. J. Hered. 2014, 106, 166–176. [Google Scholar] [CrossRef]
- Patterson, H.D.; Thompson, R. Recovery of inter-block information when block sizes are unequal. Biometrika 1971, 58, 545–554. [Google Scholar] [CrossRef]
- Henderson, C.R. Sire evaluation and genetic trends. J. Anim. Sci. 1973, 1973, 10–41. [Google Scholar] [CrossRef]
- Chaleff, R.S.; Mauvais, C. Acetolactate synthase is the site of action of two sulfonylurea herbicides in higher plants. Science 1984, 224, 1443–1445. [Google Scholar] [CrossRef] [PubMed]
- Shaner, D.L.; Anderson, P.C.; Stidham, M.A. Imidazolinones: potent inhibitors of acetohydroxyacid synthase. Plant Physiol. 1984, 76, 545–546. [Google Scholar] [CrossRef] [PubMed]
- McCourt, J.A.; Pang, S.S.; King-Scott, J.; Guddat, L.W.; Duggleby, R.G. Herbicide-binding sites revealed in the structure of plant acetohydroxyacid synthase. Proc. Natl. Acad. Sci. USA 2006, 103, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Paris, H.S. Origin and emergence of the sweet dessert watermelon, Citrullus lanatus. Ann. Bot. 2015, 116, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Paris, H.S.; Daunay, M.C.; Janick, J. Medieval iconography of watermelons in Mediterranean Europe. Ann. Bot. 2013, 112, 867–879. [Google Scholar] [CrossRef]
- Vergauwen, D.; De Smet, I. Watermelons versus Melons: A Matter of Taste. Trends Plant Sci. 2019, 24, 973–976. [Google Scholar] [CrossRef]
- Sandlin, K.; Prothro, J.; Heesacker, A.; Khalilian, N.; Okashah, R.; Xiang, W.; Bachlava, E.; Caldwell, D.G.; Taylor, C.A.; Seymour, D.K.; et al. Comparative mapping in watermelon [Citrullus lanatus (Thunb.) Matsum. et Nakai]. Theor. Appl. Genet. 2012, 125, 1603–1618. [Google Scholar] [CrossRef]
- Hashizume, T.; Shimamoto, I.; Hirai, M. Construction of a linkage map and QTL analysis of horticultural traits for watermelon [Citrullus lanatus (THUNB.) MATSUM & NAKAI] using RAPD, RFLP and ISSR markers. Theor. Appl. Genet. 2003, 106, 779–785. [Google Scholar] [CrossRef]
- Gusmini, G.; Wehner, T.C. Foundations of yield improvement in watermelon. Crop Sci. 2005, 45, 141–146. [Google Scholar] [CrossRef]
- Davis, A.R.; Levi, A.; Tetteh, A.; Wehner, T.; Russo, V.; Pitrat, M. Evaluation of watermelon and related species for resistance to race 1W powdery mildew. J. Am. Soc. Hortic. Sci. 2007, 132, 790–795. [Google Scholar] [CrossRef]
- Guo, S.; Zhang, J.; Sun, H.; Salse, J.; Lucas, W.J.; Zhang, H.; Zheng, Y.; Mao, L.; Ren, Y.; Wang, Z.; et al. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat. Genet. 2013, 45, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Zhao, H.; Kou, Q.; Jiang, J.; Guo, S.; Zhang, H.; Hou, W.; Zou, X.; Sun, H.; Gong, G.; et al. A high resolution genetic map anchoring scaffolds of the sequenced watermelon genome. PLoS ONE 2012, 7, e29453. [Google Scholar] [CrossRef] [PubMed]
- Gross, B.L.; Olsen, K.M. Genetic perspectives on crop domestication. Trends Plant Sci. 2010, 15, 529–537. [Google Scholar] [CrossRef]
- Hancock, J.F. Plant Evolution and the Origin of Crop Species; CABI: Wallingford, OX, UK, 2012. [Google Scholar]
- Meyer, R.S.; DuVal, A.E.; Jensen, H.R. Patterns and processes in crop domestication: an historical review and quantitative analysis of 203 global food crops. New Phytol. 2012, 196, 29–48. [Google Scholar] [CrossRef]
- Vavilov, N.I. Proiskhozhdenie i geografiya kul’turnykh rastenii (Origin and Geography of Cultivated Plants); Cambridge University Press: Cambridge, UK, 1987; ISBN 13:978-0521111591. [Google Scholar]
- Chomicki, G.; Renner, S.S. Watermelon origin solved with molecular phylogenetics including Linnaean material: another example of museomics. New Phytol. 2015, 205, 526–532. [Google Scholar] [CrossRef]
- Ferreira, G.C.; Franco, R.; Lloyd, S.G.; Moura, I.; Moura, J.J.; Huynh, B.H. Structure and function of ferrochelatase. J. Bioenerg. Biomembr. 1995, 27, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Noble, M.A.; Munro, A.W.; Rivers, S.L.; Robledo, L.; Daff, S.N.; Yellowlees, L.J.; Shimizu, T.; Sagami, I.; Guillemette, J.G.; Chapman, S.K. Potentiometric analysis of the flavin cofactors of neuronal nitric oxide synthase. Biochemistry 1999, 38, 16413–16418. [Google Scholar] [CrossRef]
- Stuehr, D.; Pou, S.; Rosen, G.M. Oxygen reduction by nitric-oxide synthases. J. Biol. Chem. 2001, 276, 14533–14536. [Google Scholar] [CrossRef]
- Förstermann, U.; Sessa, W.C. Nitric oxide synthases: regulation and function. Eur. Heart J. 2011, 33, 829–837. [Google Scholar] [CrossRef]
- Gas, E.; Flores-Perez, U.; Sauret-Gueto, S.; Rodriguez-Concepcion, M. Hunting for plant nitric oxide synthase provides new evidence of a central role for plastids in nitric oxide metabolism. Plant Cell 2009, 21, 18–23. [Google Scholar] [CrossRef]
- Fröhlich, A.; Durner, J. The hunt for plant nitric oxide synthase (NOS): is one really needed? Plant Sci. 2011, 181, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Hamza, I.; Dailey, H.A. One ring to rule them all: trafficking of heme and heme synthesis intermediates in the metazoans. Biochim. Biophys. Acta 2012, 1823, 1617–1632. [Google Scholar] [CrossRef] [PubMed]
- Domingos, P.; Prado, A.M.; Wong, A.; Gehring, C.; Feijo, J.A. Nitric oxide: A multitasked signaling gas in plants. Mol. Plant 2015, 8, 506–520. [Google Scholar] [CrossRef] [PubMed]
- Henry, Y.; Guissani, A. Interactions of nitric oxide with hemoproteins: roles of nitric oxide in mitochondria. Cell. Mol. Life Sci. 1999, 55, 1003–1014. [Google Scholar] [CrossRef]
- Kim-Shapiro, D.B.; Gladwin, M.T. Heme Protein Metabolism of NO and Nitrite. In Nitric Oxide; Ignarro, L.J., Freeman, B.A., Eds.; Academic Press, London Wall: London, UK, 2017; pp. 85–96. [Google Scholar] [CrossRef]
- Assefa, Y.; Purcell, L.C.; Salmeron, M.; Naeve, S.; Casteel, S.N.; Kovacs, P.; Archontoulis, S.; Licht, M.; Below, F.; Kandel, H.; et al. Assessing Variation in US Soybean Seed Composition (Protein and Oil). Front. Plant Sci. 2019, 10, 298. [Google Scholar] [CrossRef]
- Caldana, C.; Li, Y.; Leisse, A.; Zhang, Y.; Bartholomaeus, L.; Fernie, A.R.; Willmitzer, L.; Giavalisco, P. Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in A rabidopsis thaliana. Plant J. 2013, 73, 897–909. [Google Scholar] [CrossRef]
- Endo, M.; Shimizu, T.; Fujimori, T.; Yanagisawa, S.; Toki, S. Herbicide-resistant mutations in acetolactate synthase can reduce feedback inhibition and lead to accumulation of branched-chain amino acids. Food Nutr. Sci. 2013, 4, 522–528. [Google Scholar] [CrossRef][Green Version]
- Nyquist, W.E.; Baker, R. Estimation of heritability and prediction of selection response in plant populations. Crit. Rev. Plant Sci. 1991, 10, 235–322. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Han, M.V.; Zmasek, C.M. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinf. 2009, 10, 356. [Google Scholar] [CrossRef]
- Patterson, N.; Price, A.L.; Reich, D. Population structure and eigenanalysis. PLoS Genet. 2006, 2, e190. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Slatkin, M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid LDpopulation. Mol. Biol. Evol. 1995, 12, 921–927. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).