Two Novel er1 Alleles Conferring Powdery Mildew (Erysiphe pisi) Resistance Identified in a Worldwide Collection of Pea (Pisum sativum L.) Germplasms
Abstract
:1. Introduction
2. Results
2.1. Phenotypic Evaluation
2.2. PsMLO1 Sequence Analysis
2.3. Genetic Analysis and Mapping of er1-8 and er1-9
2.4. Development of Functional Markers for er1-8 and er1-9
2.5. Validation and Application of Functional Markers
3. Discussion
4. Materials and Methods
4.1. Plant Material and E. pisi Isolate
4.2. Phenotypic Evaluation
4.3. RNA Extraction and PsMLO1 Sequence Analysis
4.4. Genetic Analysis of Pea Germplasms Carrying Novel Alleles
4.5. Genetic Mapping of the Resistance Alleles er1-8 and er1-9
4.6. Development of Functional Markers for er1-8 and er1-9
4.7. Validation and Application of Functional Markers
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| SSR | Simple sequence repeat |
| SNP | Single nucleotide polymorphism |
| InDel | Insertion/deletion |
| KASPar | Kompetitive allele-specific PCR |
References
- Ali, S.M.; Sharma, B.; Ambrose, M.J. Current status and future strategy in breeding pea to improve resistance to biotic and abiotic stresses. Euphytica 1993, 1, 115–126. [Google Scholar]
- Wang, X.; Zhu, Z.; Duan, C.; Zong, X. Identification and Control Technology of Disease and Pest on Faba Bean and Pea; Chinese Agricultural Science and Technology Press: Beijing, China, 2007. [Google Scholar]
- Gritton, E.T.; Ebert, R.D. Interaction of planting date and powdery mildew on pea plant performance. Am. Soc. Horti. Sci. 1975, 100, 137–142. [Google Scholar]
- Peng, H.X.; Yao, G.; Jia, R.L.; Liang, H.Y. Identification of pea germplasm resistance to powdery mildew. J. Southwest Agric. Univ. 1991, 13, 384–386. (In Chinese) [Google Scholar]
- Smith, P.H.; Foster, E.M.; Boyd, L.A.; Brown, J.K.M. The early development of Erysiphe pisi on Pisum sativum L. Plant. Pathol. 1996, 45, 302–309. [Google Scholar] [CrossRef]
- Ghafoor, A.; McPhee, K. Marker assisted selection (MAS) for developing powdery mildew resistant pea cultivars. Euphytica 2012, 186, 593–607. [Google Scholar] [CrossRef]
- Fondevilla, S.; Rubiales, D. Powdery mildew control in pea, a review. Agron. Sustain. Dev. 2012, 32, 401–409. [Google Scholar] [CrossRef]
- Ondřej, M.; Dostálová, R.; Odstrčilová, L. Response of Pisum sativum germplasm resistant to Erysiphe pisi to inoculation with Erysiphe baeumleri, a new pathogen of peas. Plant. Prot. Sci. 2005, 41, 95–103. [Google Scholar] [CrossRef]
- Attanayakea, R.N.; Glaweab, D.A.; McPheec, K.E.; Dugand, F.M.; Chend, W. Erysiphe trifolii—A newly recognized powdery mildew pathogen of pea. Plant. Pathol. 2010, 59, 712–720. [Google Scholar] [CrossRef]
- Fondevilla, S.; Chattopadhyay, C.; Khare, N.; Rubiales, D. Erysiphe trifolii is able to overcome er1 and Er3, but not er2, resistance genes in pea. Eur. J. Plant. Pathol. 2013, 136, 557–563. [Google Scholar] [CrossRef]
- Harland, S.C. Inheritance of immunity to mildew in Peruvian forms of Pisum sativum. Heredity 1948, 2, 263–269. [Google Scholar] [CrossRef]
- Heringa, R.J.; Van Norel, A.; Tazelaar, M.F. Resistance to powdery mildew (Erysiphe polygoni D.C.) in peas (Pisum sativum L.). Euphytica 1969, 18, 163–169. [Google Scholar] [CrossRef]
- Fondevilla, S.; Torres, A.M.; Moreno, M.T.; Rubiales, D. Identification of a new gene for resistance to powdery mildew in Pisum fulvum, a wild relative of pea. Breed. Sci. 2007, 57, 181–184. [Google Scholar] [CrossRef]
- Humphry, M.; Reinstädler, A.; Ivanov, S.; Bisseling, T.; Panstruga, R. Durable broad-spectrum powdery mildew resistance in pea er1 plants is conferred by natural loss-of-function mutations in PsMLO1. Mol. Plant. Pathol. 2011, 12, 866–878. [Google Scholar] [CrossRef] [PubMed]
- Sarala, K. Linkage Studies in Pea (Pisum sativum L.) with Reference to Er Gene for Powdery Mildew Resistance and Other Genes. Ph.D. Thesis, Indian Agricultural Research Institute, New Delhi, India, 1993. [Google Scholar]
- Dirlewanger, E.; Isaac, P.G.; Ranade, S.; Belajouza, M.; Cousin, R.; Vienne, D. Restriction fragment length polymorphism analysis of loci associated with disease resistance genes and developmental traits in Pisum sativum L. Theor. Appl. Genet. 1994, 88, 17–27. [Google Scholar] [CrossRef]
- Timmerman, G.M.; Frew, T.J.; Weeden, N.F. Linkage analysis of er1, a recessive Pisum sativum gene for resistance to powdery mildew fungus (Erysiphe pisi DC). Theor. Appl. Genet. 1994, 88, 1050–1055. [Google Scholar] [CrossRef]
- Tiwari, K.R.; Penner, G.A.; Warkentin, T.D. Identification of coupling and repulsion phase RAPD markers for powdery mildew resistance gene er1 in pea. Genome 1998, 41, 440–444. [Google Scholar] [CrossRef]
- Janila, P.; Sharma, B. RAPD and SCAR markers for powdery mildew resistance gene er in pea. Plant. Breed. 2004, 123, 271–274. [Google Scholar] [CrossRef]
- Ek, M.; Eklund, M.; von Post, R.; Dayteg, C.; Henriksson, T.; Weibull, P.; Ceplitis, A.; Isaac, P.; Tuvesson, S. Microsatellite markers for powdery mildew resistance in pea (Pisum sativum L.). Hereditas 2005, 142, 86–91. [Google Scholar] [CrossRef]
- Pereira, G.; Marques, C.; Ribeiro, R.; Formiga, S.; Dâmaso, M.; Sousa, T.; Farinhó, M.; Leitão, J.M. Identification of DNA markers linked to an induced mutated gene conferring resistance to powdery mildew in pea (Pisum sativum L.). Euphytica 2010, 171, 327–335. [Google Scholar] [CrossRef]
- Tonguc, M.; Weeden, N.F. Identification and mapping of molecular markers linked to er1 gene in pea. J. Plant. Mol. Biol. Biotech. 2010, 1, 1–5. [Google Scholar]
- Srivastava, R.K.; Mishra, S.K.; Singh, K.; Mohapatra, T. Development of a coupling-phase SCAR marker linked to the powdery mildew resistance gene er1 in pea (Pisum sativum L.). Euphytica 2012, 86, 855–866. [Google Scholar] [CrossRef]
- Wang, Z.; Fu, H.; Sun, S.; Duan, C.; Wu, X.; Yang, X.; Zhu, Z. Identification of powdery mildew resistance gene in pea line X9002. Acta Agron. Sin. 2015, 41, 515–523, (In Chinese with English abstract). [Google Scholar] [CrossRef]
- Sun, S.; Wang, Z.; Fu, H.; Duan, C.; Wang, X.; Zhu, Z. Resistance to powdery mildew in the pea cultivar Xucai 1 is conferred by the gene er1. Crop. J. 2015, 3, 489–499. [Google Scholar] [CrossRef]
- Sun, S.; Deng, D.; Wang, Z.; Duan, C.; Wu, X.; Wang, X.; Zong, X.; Zhu, Z. A novel er1 allele and the development and validation of its functional marker for breeding pea (Pisum sativum L.) resistance to powdery mildew. Appl. Genet. 2016, 129, 909–919. [Google Scholar]
- Sun, S.; Fu, H.; Wang, Z.; Duan, C.; Zong, X.; Zhu, Z. Discovery of a novel er1 allele conferring powdery mildew resistance in Chinese pea (Pisum sativum L.) landraces. PLoS ONE 2016, 11, e0147624. [Google Scholar] [CrossRef]
- Katoch, V.; Sharma, S.; Pathania, S.; Banayal, D.K.; Sharma, S.K.; Rathour, R. Molecular mapping of pea powdery mildew resistance gene er2 to pea linkage group III. Mol. Breed. 2010, 25, 229–237. [Google Scholar] [CrossRef]
- Cobos, M.J.; Rubiales, D.; Fondevilla, S. Er3 gene conferring resistance to Erysiphe pisi is located in pea LGIV. In Proceedings of the Second International Legume Society Conference, Troia, Portugal, 11–14 October 2016. [Google Scholar]
- Tiwari, K.R.; Penner, G.A.; Warkentin, T.D. Inheritance of powdery mildew resistance in pea. Can. J. Plant. Sci. 1997, 77, 307–310. [Google Scholar] [CrossRef] [Green Version]
- Vaid, A.; Tyagi, P.D. Genetics of powdery mildew resistance in pea. Euphytica 1997, 96, 203–206. [Google Scholar] [CrossRef]
- Fondevilla, S.; Carver, T.L.W.; Moreno, M.T.; Rubiales, D. Macroscopic and histological characterisation of genes er1 and er2 for powdery mildew resistance in pea. Eur. J. Plant. Pathol. 2006, 115, 309–321. [Google Scholar] [CrossRef]
- Fondevilla, S.; Cubero, J.I.; Rubiales, D. Confirmation that the Er3 gene, conferring resistance to Erysiphe pisi in pea, is a different gene from er1 and er2 genes. Plant. Breed. 2011, 130, 281–282. [Google Scholar] [CrossRef]
- Bai, Y.; Pavan, S.; Zheng, Z.; Zappel, N.F.; Reinstadler, A.; Lotti, C.; DeGiovanni, C.; Ricciardi, L.; Lindhout, P.; Visser, R.; et al. Naturally occurring broad-spectrum powdery mildew resistance in a central American tomato accession is caused by loss of MLO1 function. Mol. Plant. Microbe Interact. 2008, 21, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Büschges, R.; Hollricher, K.; Panstruga, R.; Simons, G.; Wolter, M.; Frijters, A.; van Daelen, R.; van der Lee, T.; Diergaarde, P.; Groenendijk, J. The barley MLO gene, a novel control element of plant pathogen resistance. Cell 1997, 88, 695–705. [Google Scholar] [CrossRef]
- Consonni, C.; Humphry, M.E.; Hartmann, H.A.; Livaja, M.; Durner, J.; Westphal, L.; Vogel, J.; Lipka, V.; Kemmerling, B.; Schulze-Lefert, P.; et al. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 2006, 38, 716–720. [Google Scholar] [CrossRef] [PubMed]
- Devoto, A.; Hartmann, H.A.; Piffanelli, P.; Elliott, C.; Simmons, C.; Taramino, G.; Goh, C.S.; Cohen, F.E.; Emerson, B.C.; Schulze-Lefert, P.; et al. Molecular phylogeny and evolution of the plant-specific seven-transmembrane MLO family. J. Mol. Evol. 2003, 56, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Pavan, S.; Schiavulli, A.; Appiano, M.; Marcotrigiano, A.R.; Cillo, F.; Visser, R.G.F.; Bai, Y.; Lotti, C.; Luigi Ricciardi, L. Pea powdery mildew er1 resistance is associated to loss-of-function mutations at a MLO homologous locus. Appl. Genet. 2011, 123, 1425–1431. [Google Scholar] [CrossRef] [PubMed]
- Rispail, N.; Rubiales, D. Genome-wide identification and comparison of legume MLO gene family. Sci. Rep. 2016, 6, 32673. [Google Scholar] [CrossRef]
- Santo, T.; Rashkova, M.; Alabaca, C.; Leitao, J. The ENU–induced powdery mildew resistant mutant pea (Pisum sativum L.) lines S (er1mut1) and F (er1mut2) harbour early stop codons in the PsMLO1 gene. Mol. Breed. 2013, 32, 723–727. [Google Scholar] [CrossRef]
- Fu, H.; Sun, S.; Zhu, Z.; Duan, C.; Yang, X. Phenotypic and genotypic identification of powdery mildew resistance in pea cultivars or lines from Canada. J. Plant. Genet. Resour. 2014, 15, 1028–1033, (In Chinese with English abstract). [Google Scholar]
- Ma, Y.; Coyne, C.J.; Main, D.; Pavan, S.; Sun, S.; Zhu, Z.; Zong, X.; Leitão, J.; McGee, R.J. Development and validation of breeder-friendly KASPar markers for er1, a powdery mildew resistance gene in pea (Pisum sativum L.). Mol. Breed. 2017, 37, 151. [Google Scholar] [CrossRef]
- Sudheesh, S.; Lombardi, M.; Leonforte, A.; Cogan, N.O.I.; Materne, M.; Forster, J.W.; Kaur, S. Consensus Genetic Map Construction for Field Pea (Pisum sativum L.), Trait dissection of biotic and abiotic stress tolerance and development of a diagnostic marker for the er1 powdery mildew resistance gene. Plant. Mol. Biol. Rep. 2015, 33, 1391–1403. [Google Scholar] [CrossRef]
- Pavan, S.; Schiavulli, A.; Appiano, M.; Miacola, C.; Visser, R.G.F.; Bai, Y.L.; Lotti, C.; Ricciardi, L. Identification of a complete set of functional markers for the selection of er1 powdery mildew resistance in Pisum sativum L. Mol. Breed. 2013, 31, 247–253. [Google Scholar] [CrossRef]
- Peng, H.X.; Yao, G. On resistance to powdery mildew of pea varieties Chinese. Acta Phytopathol Sin. 1993, 23, 62. (In Chinese) [Google Scholar]
- Liu, A.A. Identification method of resistance of pea to powdery mildew using detached leaves. Acta Phytophylacica Sin. 2002, 29, 19–123, (In Chinese with English abstract). [Google Scholar]
- Zeng, L.; Li, M.Q.; Yang, X.M. Identification of resistance of peas resources to powdery mildew. Grassl. Turf. 2012, 32, 35–38, (In Chinese with English abstract). [Google Scholar]
- Wang, Z.; Bao, S.; Duan, C.; Zong, X.; Zhu, Z. Screening and molecular identification of resistance to powdery mildew in pea germplasm. Acta Agron. Sin. 2013, 39, 1030–1038, (In Chinese with English abstract). [Google Scholar] [CrossRef]
- Lu, J.; Yang, X.; Wang, C.; Yang, F.; Zhang, L. Screening for pea resources resistant to pea powdery mildew in field. Gansu Agr. Sci. Technol. 2015, 41, 154–158, (In Chinese with English abatract). [Google Scholar]
- Fu, H. Phenotyping and Genotyping Powdery Mildew Resistance in Pea. Ph.D. Thesis, Gansu Agricultural University, Gansu, China, 2014. (In Chinese with English abstract). [Google Scholar]
- Sun, S.; He, Y.; Dai, C.; Duan, C.; Zhu, Z. Two major er1 alleles confer powdery mildew resistance in three pea cultivars bred in Yunnan Province, China. The Crop. J. 2016, 4, 353–359. [Google Scholar] [CrossRef]
- Kusch, S.; Panstruga, R. Mlo–based resistance, an apparently universal “Weapon” to defeat powdery mildew disease. MPMI 2017, 30, 179–189. [Google Scholar] [CrossRef]
- Kim, E.; Magen, A.; Ast, G. Different levels of alternative splicing among eukaryotes. Nucl. Acids Res. 2007, 35, 125–131. [Google Scholar] [CrossRef]
- Reddy, A.S.N. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant. Biol. 2007, 58, 267–294. [Google Scholar] [CrossRef]
- Ner–Gaon, H.; Halachmi, R.; Savaldi–Goldstein, S.; Rubin, E.; Ophir, R.; Fluhr, R. Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant. J. 2004, 39, 877–885. [Google Scholar] [CrossRef] [PubMed]
- Barbazuk, W.B.; Fu, Y.; McGinnis, K.M. Genome-wide analyses of alternative splicing in plants, opportunities and challenges. Genome Res. 2008, 18, 1381–1392. [Google Scholar] [CrossRef] [PubMed]
- Rubiales, D.; Brown, J.K.M.; Martín, A. Hordeum chilense resistance to powdery mildew and its potential use in cereal breeding. Euphytica 1993, 67, 215–220. [Google Scholar] [CrossRef]
- Rana, J.C.; Banyal, D.K.; Sharma, K.D.; Sharma, M.K.; Gupta, S.K.; Yadav, S.K. Screening of pea germplasm for resistance to powdery mildew. Euphytica 2013, 189, 271–282. [Google Scholar] [CrossRef]
- Shure, M.; Wessler, S.; Fedoroff, N. Molecular-identification and isolation of the waxy locus in maize. Cell 1983, 35, 225–233. [Google Scholar] [CrossRef]
- Bordat, A.; Savois, V.; Nicolas, M.; Salse, J.; Chauveau, A.; Bourgeois, M.; Potier, J.; Houtin, H.; Rond, C.; Murat, F.; et al. Translational genomics in legumes allowed placing in silico 5460 unigenes on the pea functional map and identified candidate genes in Pisum sativum L. Genes Genome Genet. 2011, 1, 93–103. [Google Scholar] [CrossRef]
- Loridon, K.; McPhee, K.; Morin, J.; Dubreuil, P.; Pilet-Nayel, M.L.; Aubert, G.; Rameau, C.; Baranger, A.; Coyne, C.; Lejeune-Hénaut, I.; et al. Microsatellite marker polymorphism and mapping in pea (Pisum sativum L.). Theor. Appl. Genet. 2005, 111, 1022–1031. [Google Scholar] [CrossRef]
- Lander, E.S.; Daly, M.J.; Lincoln, S.E. Constructing genetic linkage maps with MAPMAKER/EXP Version 3.0, a tutorial and reference manual. In Institute for Biomedical Research Technical Report, 3rd ed.; Whitehead, A., Ed.; Whitehead Institute for Biomedical Research: Cambridge, MA, USA, 1993. [Google Scholar]
- Kosambi, D.D. The estimation of map distances from recombination values. Ann. Eugen. 1944, 12, 172–175. [Google Scholar] [CrossRef]
- Liu, R.H.; Meng, J.L. MapDraw, A Microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas 2003, 25, 317–321, (In Chinese with English abstract). [Google Scholar]
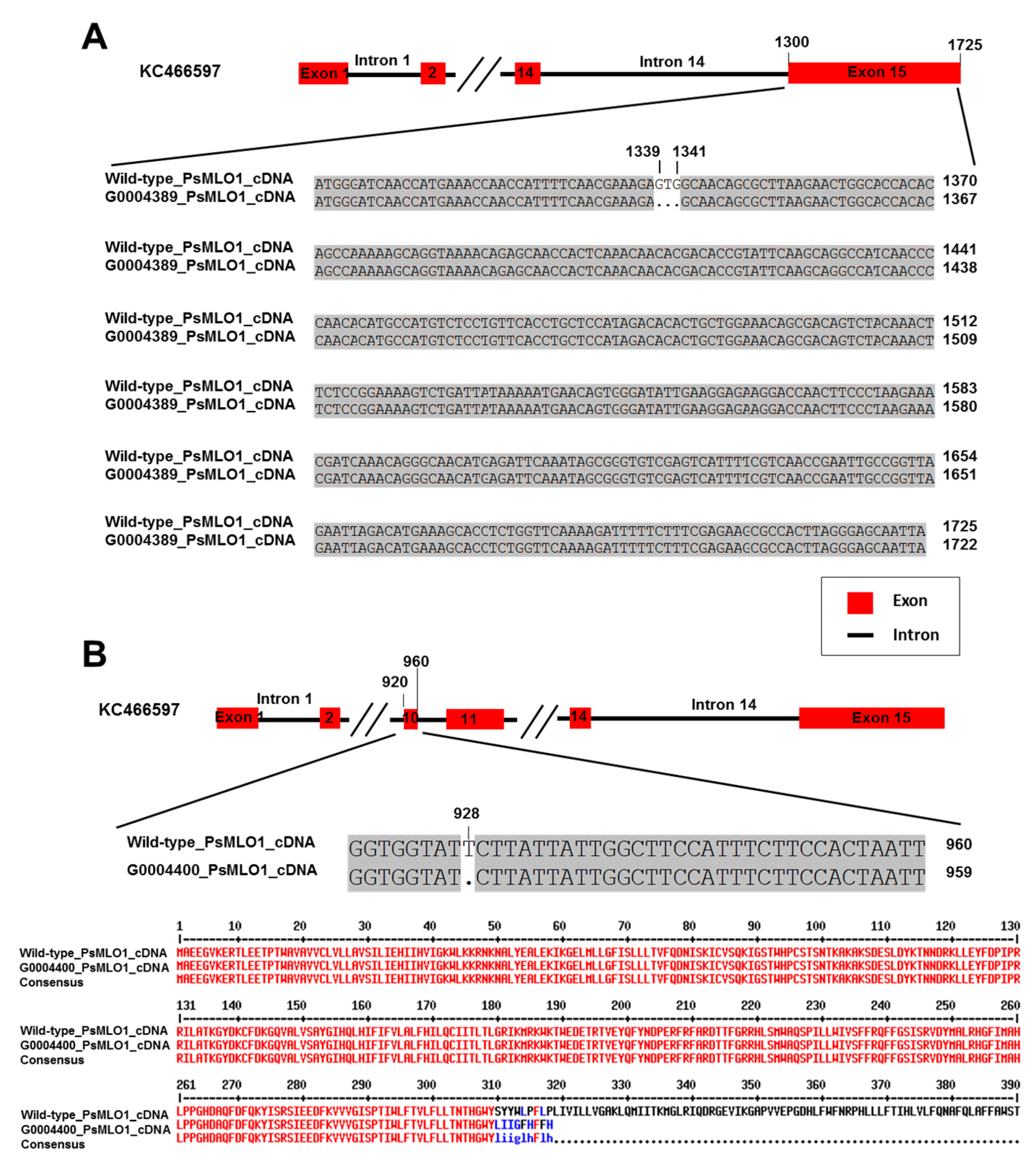
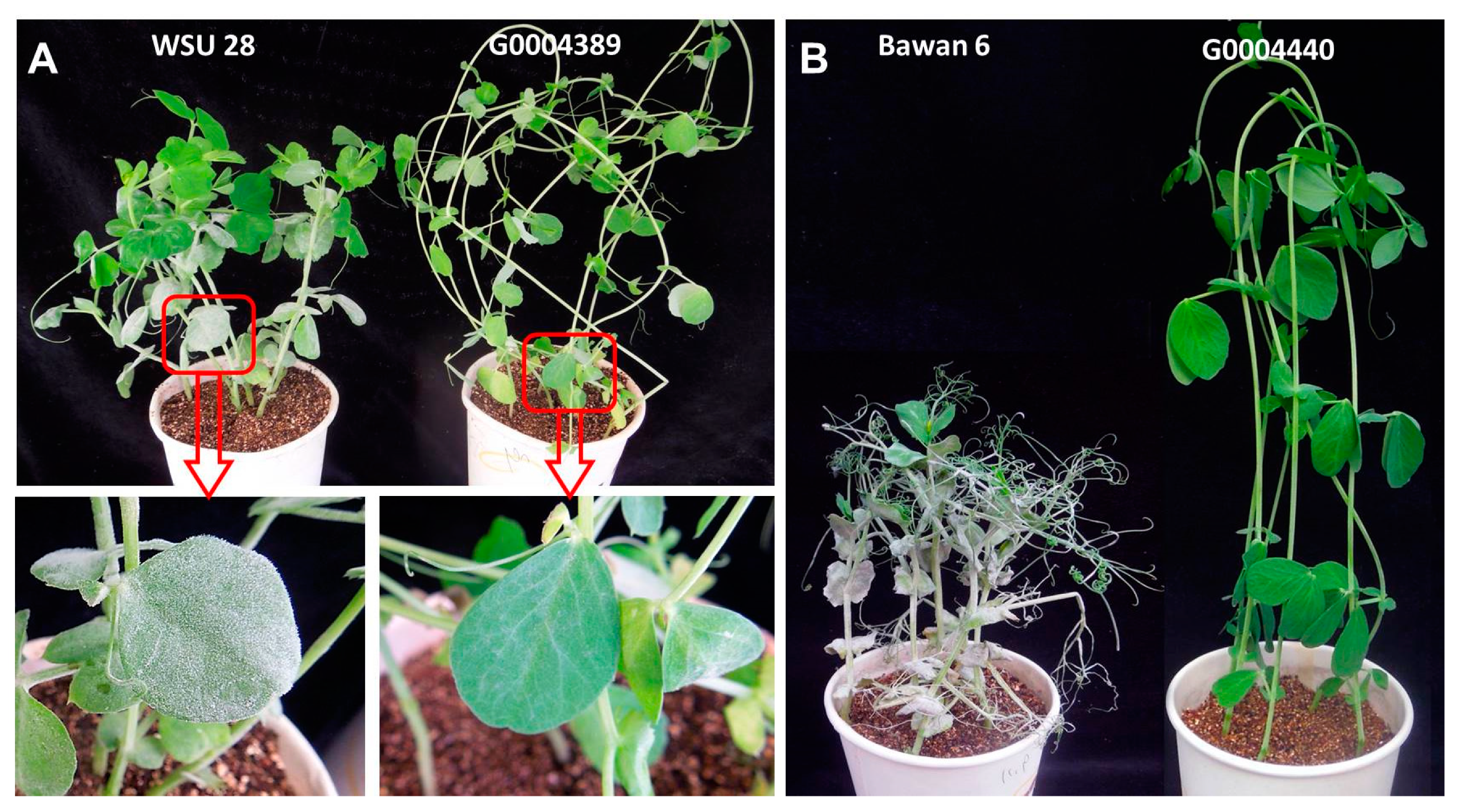
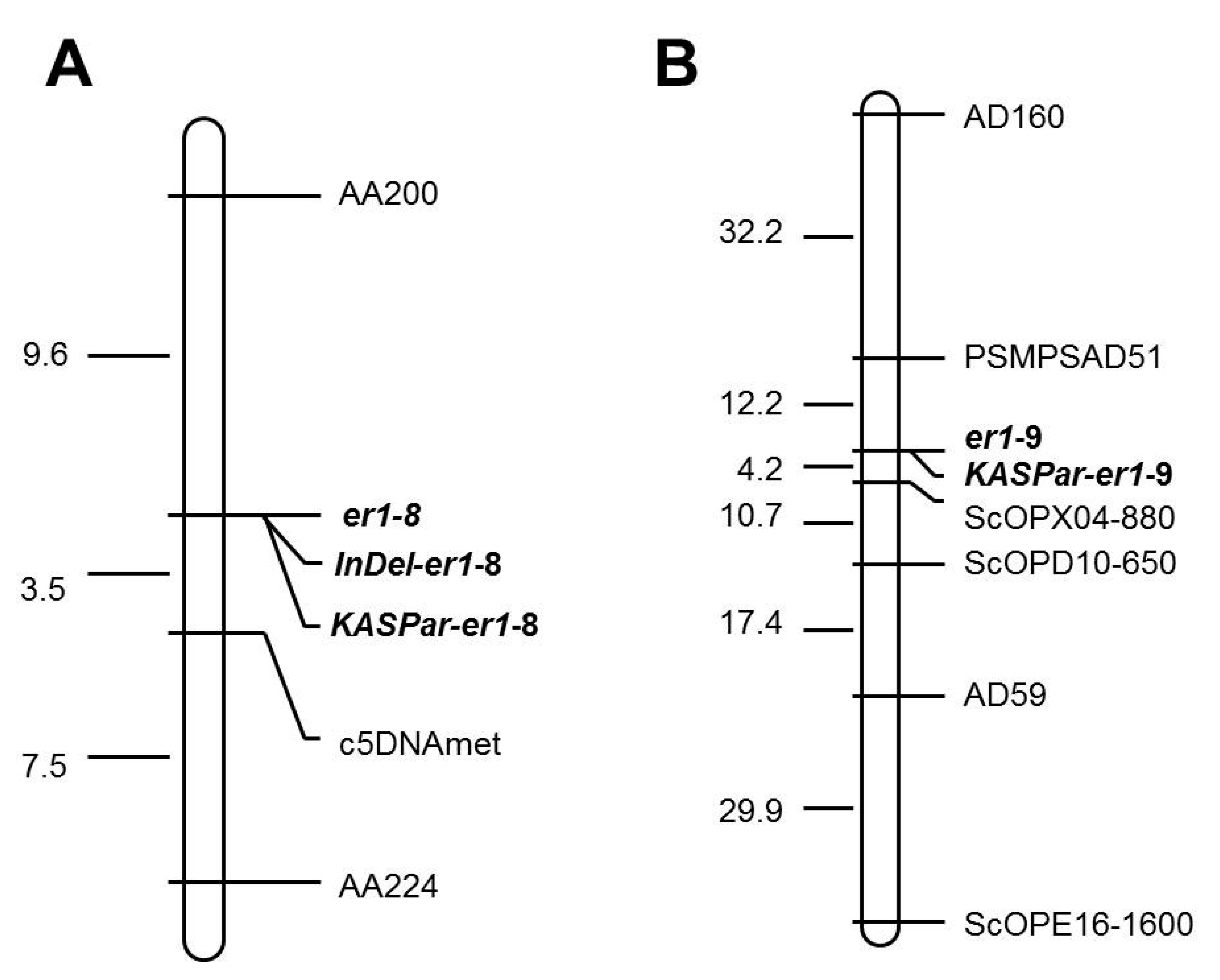
| No. | Accession No./Germplasm Name | Origin | Phenotype | er1 Allele | Reference |
|---|---|---|---|---|---|
| 1 | G0004389 | Afghanistan | I | er1-8 | This study |
| 2 | G0004382 | Australia | I | er1-1 | This study |
| 3 | G0004400 | Australia | I | er1-9 | This study |
| 4 | G0004417 | Australia | I | er1-2 | This study |
| 5 | G0004434 | Australia | I | er1-2 | This study |
| 6 | G0004448 | Australia | I | er1-2 | This study |
| 7 | G0004450 | Australia | I | er1-2 | This study |
| 8 | G0002102 | Canada | I | er1-6 | This study |
| 9 | G0006514 | Canada | R | er1-2 | This study |
| 10 | G0006515 | Canada | R | er1-2 | This study |
| 11 | G0006516 | Canada | I | er1-2 | This study |
| 12 | G0006519 | Canada | I | er1-2 | This study |
| 13 | G0003925 | Canada | I | er1-1 | [41] |
| 14 | Cooper | Canada | I | er1-1 | [41] |
| 15 | G0005576 | China, Chongqing | I | er1-2 | [27] |
| 16 | G0006273 | China, Gansu | I | er1-2 | [24] |
| 17 | 20012 | China, Gansu | I | er1-1 | This study |
| 18 | Jia2 | China, Gansu | I | er1-2 | This study |
| 19 | Texuan11 | China, Gansu | I | er1-2 | This study |
| 20 | Hehuan66 | China, Gansu | R | er1-1 | This study |
| 21 | Longwan 1 | China, Gansu | S | Er1 | [51] |
| 22 | PI391630 | China, Guangdong | I | er1-4 | [14] |
| 23 | Xucai1 | China, Hebei | I | er1-2 | [25] |
| 24 | G0003694 | China, Hebei | R | er1-6 | [27] |
| 25 | Bawan 6 | China, Hebei | S | Er1 | [24] |
| 26 | L0314 | China, Yunnan | I | er1-1 | [51] |
| 27 | L1332 | China, Yunnan | I | er1-2 | [51] |
| 28 | L1335 | China, Yunnan | I | er1-2 | [51] |
| 29 | G0001747 | China, Yunnan | R | er1-6 | This study |
| 30 | G0001752 | China, Yunnan | I | er1-6 | [27] |
| 31 | G0001763 | China, Yunnan | I | er1-6 | [27] |
| 32 | G0001764 | China, Yunnan | I | er1-6 | [27] |
| 33 | G0001767 | China, Yunnan | I | er1-6 | [27] |
| 34 | G0001768 | China, Yunnan | I | er1-6 | [27] |
| 35 | G0001773 | China, Yunnan | I | er1-6 | This study |
| 36 | G0001777 | China, Yunnan | I | er1-6 | [27] |
| 37 | G0001778 | China, Yunnan | I | er1-6 | [27] |
| 38 | G0001780 | China, Yunnan | I | er1-6 | [27] |
| 39 | G0003824 | China, Yunnan | R | er1-6 | [27] |
| 40 | G0003825 | China, Yunnan | I | er1-6 | [27] |
| 41 | G0003826 | China, Yunnan | I | er1-6 | [27] |
| 42 | G0003831 | China, Yunnan | R | er1-6 | [27] |
| 43 | G0003834 | China, Yunnan | R | er1-6 | [27] |
| 44 | G0003836 | China, Yunnan | R | er1-6 | [27] |
| 45 | G0003839 | China, Yunnan | R | er1-6 | This study |
| 46 | G0005117 | China, Yunnan | I | er1-6 | This study |
| 47 | G0003974 | China, Yunnan | I | er1-7 | This study |
| 48 | G0003975 | China, Yunnan | I | er1-7 | This study |
| 49 | Yunwan4 | China, Yunnan | R | er1-1 | This study |
| 50 | Yunwan18 | China, Yunnan | R | er1-2 | This study |
| 51 | Yunwan35 | China, Yunnan | I | er1-2 | This study |
| 52 | Yunwan37 | China, Yunnan | I | er1-6 | This study |
| 53 | L2157 | China, Yunnan | I | er1-2 | This study |
| 54 | G0002848 | Denmark | I | er1-2 | This study |
| 55 | G0002971 | England | I | er1-2 | This study |
| 56 | G0002859 | Germany | I | er1-2 | This study |
| 57 | G0002860 | Germany | I | er1-2 | This study |
| 58 | G0002883 | Germany | I | er1-2 | This study |
| 59 | G0003895 | ICRISAT | I | er1-7 | [26] |
| 60 | G0003897 | ICRISAT | I | er1-2 | This study |
| 61 | G0003899 | ICRISAT | I | er1-7 | [26] |
| 62 | G0003907 | ICRISAT | I | er1-2 | This study |
| 63 | G0003911 | ICRISAT | I | er1-2 | This study |
| 64 | G0003961 | India | I | er1-2 | This study |
| 65 | G0003967 | India | I | er1-7 | [26] |
| 66 | G0003958 | India | I | er1-7 | [26] |
| 67 | G0006285 | Japan | R | er1-2 | This study |
| 68 | G0004332 | Mexico | R | er1-1 | This study |
| 69 | G0004394 | Nepal | R | er1-7 | [26] |
| 70 | G0002980 | Unknown country | I | er1-2 | This study |
| 71 | G0003931 | Unknown country | I | er1-7 | [26] |
| 72 | G0003935 | Unknown country | I | er1-2 | This study |
| 73 | G0003936 | Unknown country | I | er1-7 | [26] |
| 74 | G0003942 | Unknown country | I | er1-1 | This study |
| 75 | G0003943 | Unknown country | I | er1-1 | This study |
| 76 | G0002128 | USA | I | er1-2 | This study |
| 77 | G0002129 | USA | I | er1-2 | This study |
| 78 | G0002131 | USA | I | er1-2 | This study |
| 79 | G0002132 | USA | I | er1-2 | This study |
| 80 | G0002134 | USA | I | er1-2 | This study |
| 81 | G0002137 | USA | I | er1-2 | This study |
| 82 | G0002183 | USA | I | er1-2 | This study |
| 83 | G0002235 | USA | I | er1-6 | This study |
| 84 | G0002250 | USA | I | er1-2 | This study |
| 85 | G0002602 | USA | I | er1-2 | This study |
| 86 | G0002608 | USA | I | er1-2 | This study |
| 87 | G0002847 | USA | I | er1-2 | This study |
| 88 | G0002960 | USA | I | er1-2 | This study |
| Country | No. of Pea Germplasm Accessions Contained er1 Alleles | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| er1-1 | er1-2 | er1-3 | er1-4 | er1-5 | er1-6 | er1-7 | er1-8 | er1-9 | Total | |
| USA | - | 12 | - | - | - | 1 | - | - | - | 13 |
| Canada | - | 4 | - | - | - | 1 | - | - | - | 5 |
| Germany | - | 3 | - | - | - | - | - | - | - | 3 |
| ICRISAT | - | 3 | - | - | - | - | - | - | - | 3 |
| India | - | 1 | - | - | - | - | - | - | - | 1 |
| Australia | 1 | 4 | - | - | - | - | - | - | 1 | 6 |
| England | - | 1 | - | - | - | - | - | - | - | 1 |
| Denmark | - | 1 | - | - | - | - | - | - | - | 1 |
| Nepal | - | - | - | - | - | - | - | - | - | 0 |
| Japan | - | 1 | - | - | - | - | - | - | - | 1 |
| Afghanistan | - | - | - | - | - | - | - | 1 | - | 1 |
| Mexico | 1 | - | - | - | - | - | - | - | - | 1 |
| China | 3 | 5 | - | - | - | 5 | 2 | - | - | 15 |
| Unknown country | 2 | 2 | - | - | - | - | - | - | - | 4 |
| Total | 7 | 37 | - | - | - | 7 | 2 | 1 | 1 | 55 |
| Markers | Primers | Sequence Information (5′-3′) | Annealing Tm |
|---|---|---|---|
| InDel-er1-8 | Forward | GTTTTGACTGATATGACAGATGGGA | 55 °C |
| Reverse | GTTTGTAGACTGTCGCTGTTTCC | ||
| KASPar-er1-8 | Forward-TGG | TGGCAACAGCGCTTAAGAACTGG | 65–57 °C touchdown |
| Forward | GAGCAACAGCGCTTAAGAACTGG | ||
| Common reverse | TGGTTGGTTTCATGGTTGATCCCATC | ||
| KASPar-er1-9 | Forward-T | TTTTGTTATATGGGCAGGGTGGTATT | 65–57 °C touchdown |
| Forward | TGTTATATGGGCAGGGTGGTATC | ||
| Common reverse | CAAAATGTAGATTATGCTTACAATTAGTGGA |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, S.; Deng, D.; Duan, C.; Zong, X.; Xu, D.; He, Y.; Zhu, Z. Two Novel er1 Alleles Conferring Powdery Mildew (Erysiphe pisi) Resistance Identified in a Worldwide Collection of Pea (Pisum sativum L.) Germplasms. Int. J. Mol. Sci. 2019, 20, 5071. https://doi.org/10.3390/ijms20205071
Sun S, Deng D, Duan C, Zong X, Xu D, He Y, Zhu Z. Two Novel er1 Alleles Conferring Powdery Mildew (Erysiphe pisi) Resistance Identified in a Worldwide Collection of Pea (Pisum sativum L.) Germplasms. International Journal of Molecular Sciences. 2019; 20(20):5071. https://doi.org/10.3390/ijms20205071
Chicago/Turabian StyleSun, Suli, Dong Deng, Canxing Duan, Xuxiao Zong, Dongxu Xu, Yuhua He, and Zhendong Zhu. 2019. "Two Novel er1 Alleles Conferring Powdery Mildew (Erysiphe pisi) Resistance Identified in a Worldwide Collection of Pea (Pisum sativum L.) Germplasms" International Journal of Molecular Sciences 20, no. 20: 5071. https://doi.org/10.3390/ijms20205071
APA StyleSun, S., Deng, D., Duan, C., Zong, X., Xu, D., He, Y., & Zhu, Z. (2019). Two Novel er1 Alleles Conferring Powdery Mildew (Erysiphe pisi) Resistance Identified in a Worldwide Collection of Pea (Pisum sativum L.) Germplasms. International Journal of Molecular Sciences, 20(20), 5071. https://doi.org/10.3390/ijms20205071






