Genome-Wide Identification and Characterization of Cucumber BPC Transcription Factors and Their Responses to Abiotic Stresses and Exogenous Phytohormones
Abstract
:1. Introduction
2. Results
2.1. Identification and Characterization of Cucumber BPC Genes
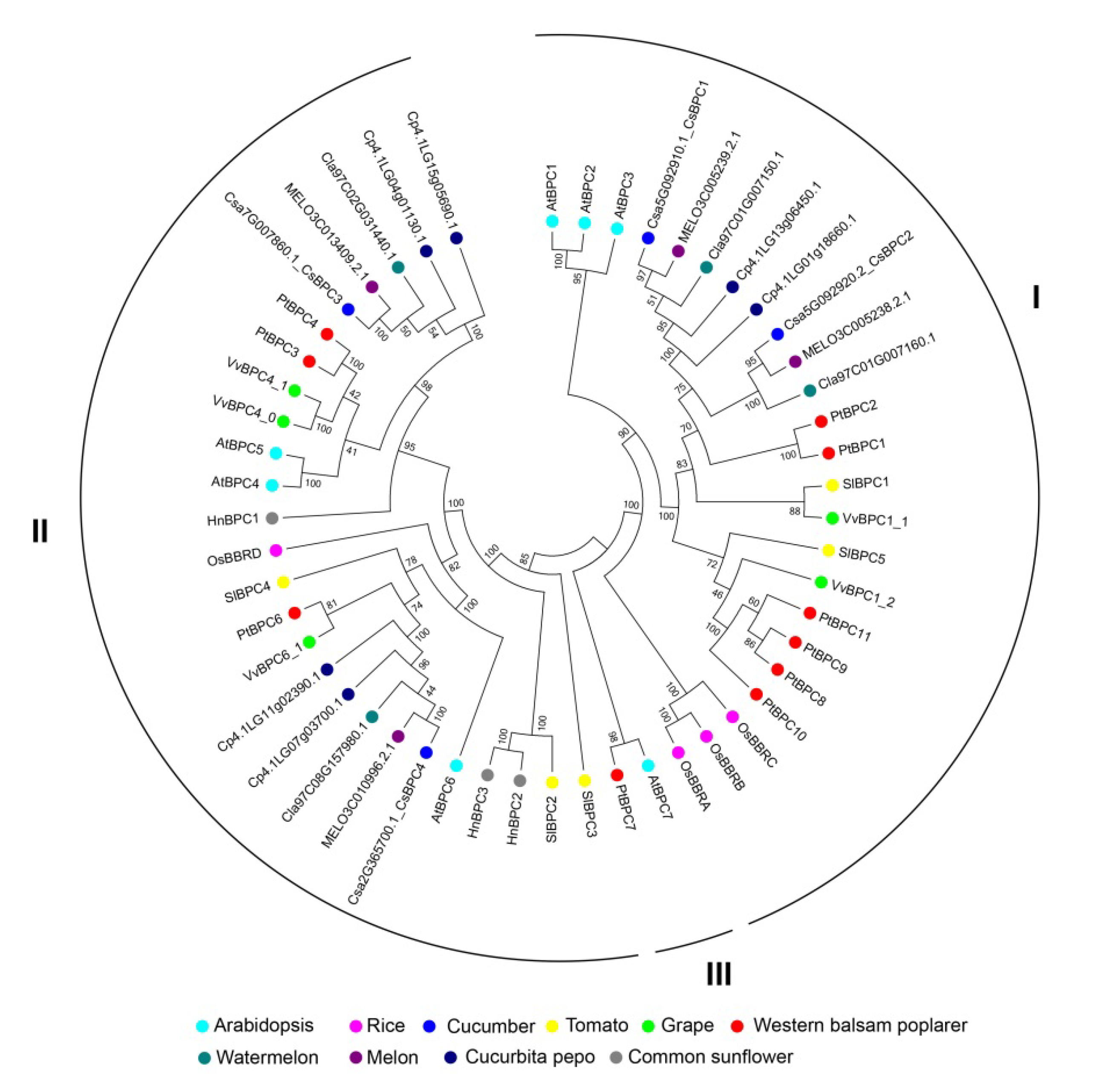
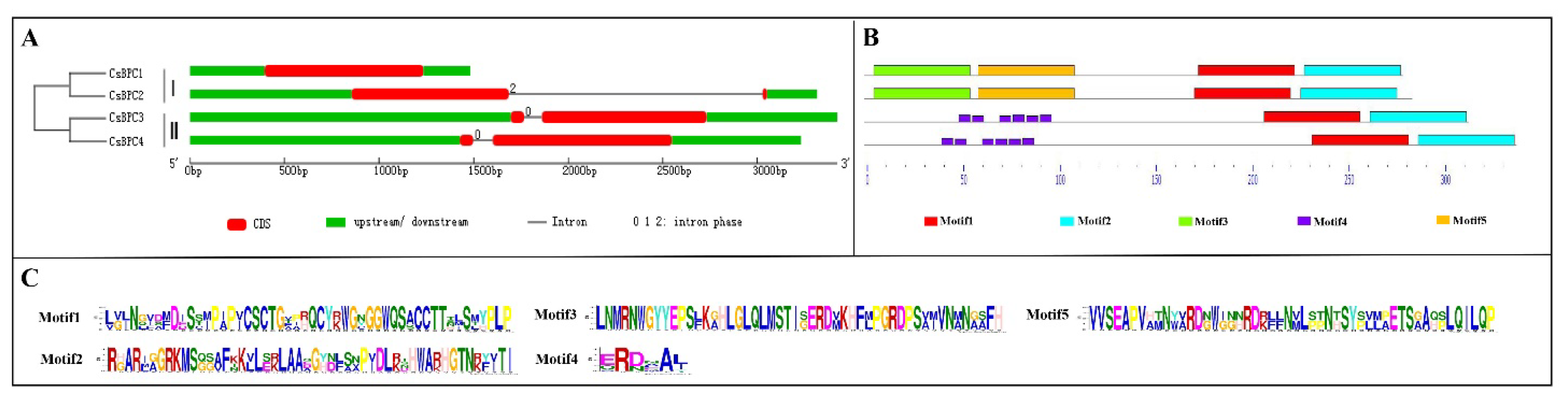
2.2. Phylogenetic Gene Structure and Motif Analysis
2.3. Expression Profiles in Different Tissues
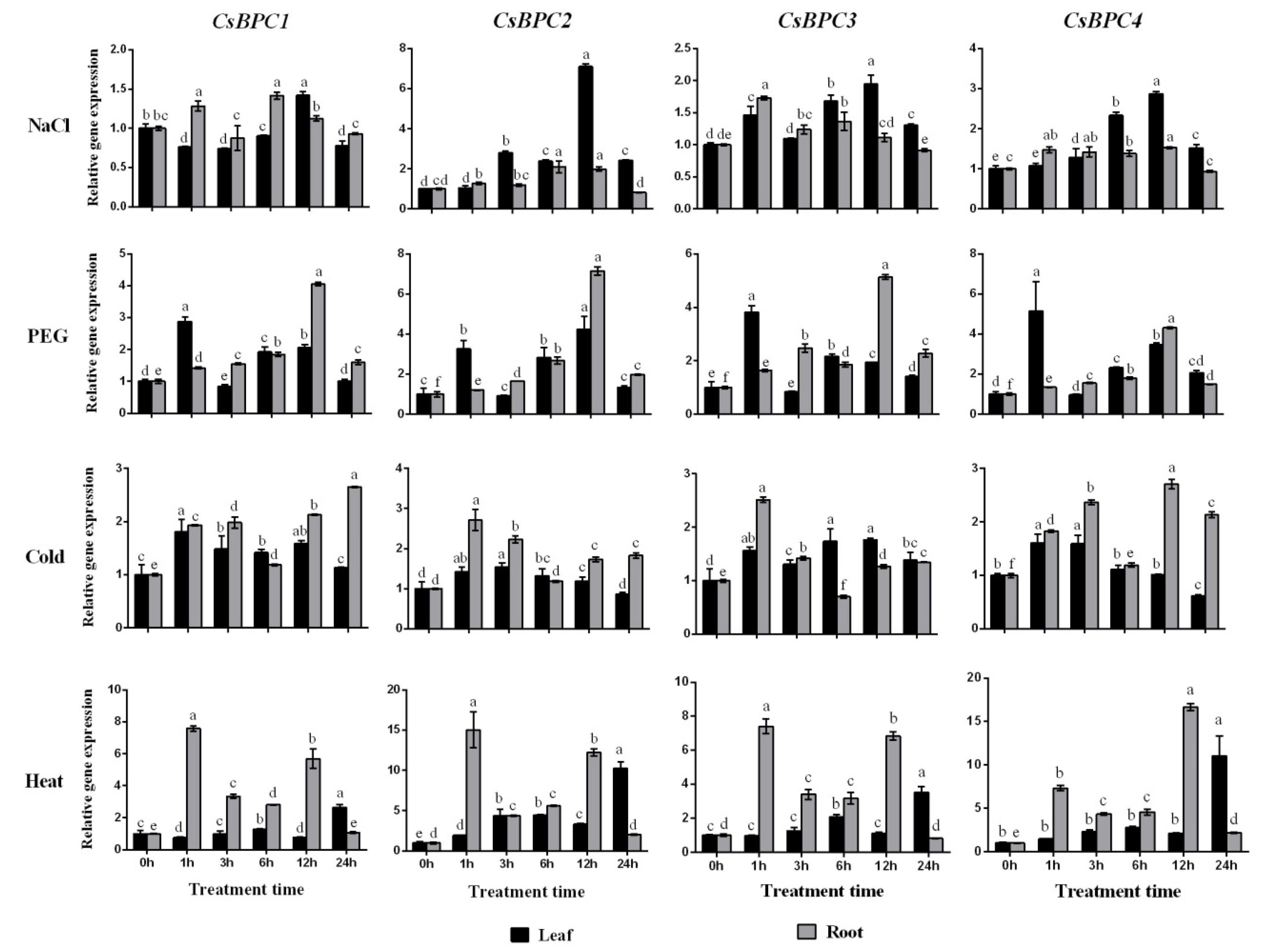
2.4. Expression Patterns of the CsBPC Genes under Different Abiotic Stress and Phytohormone Treatments
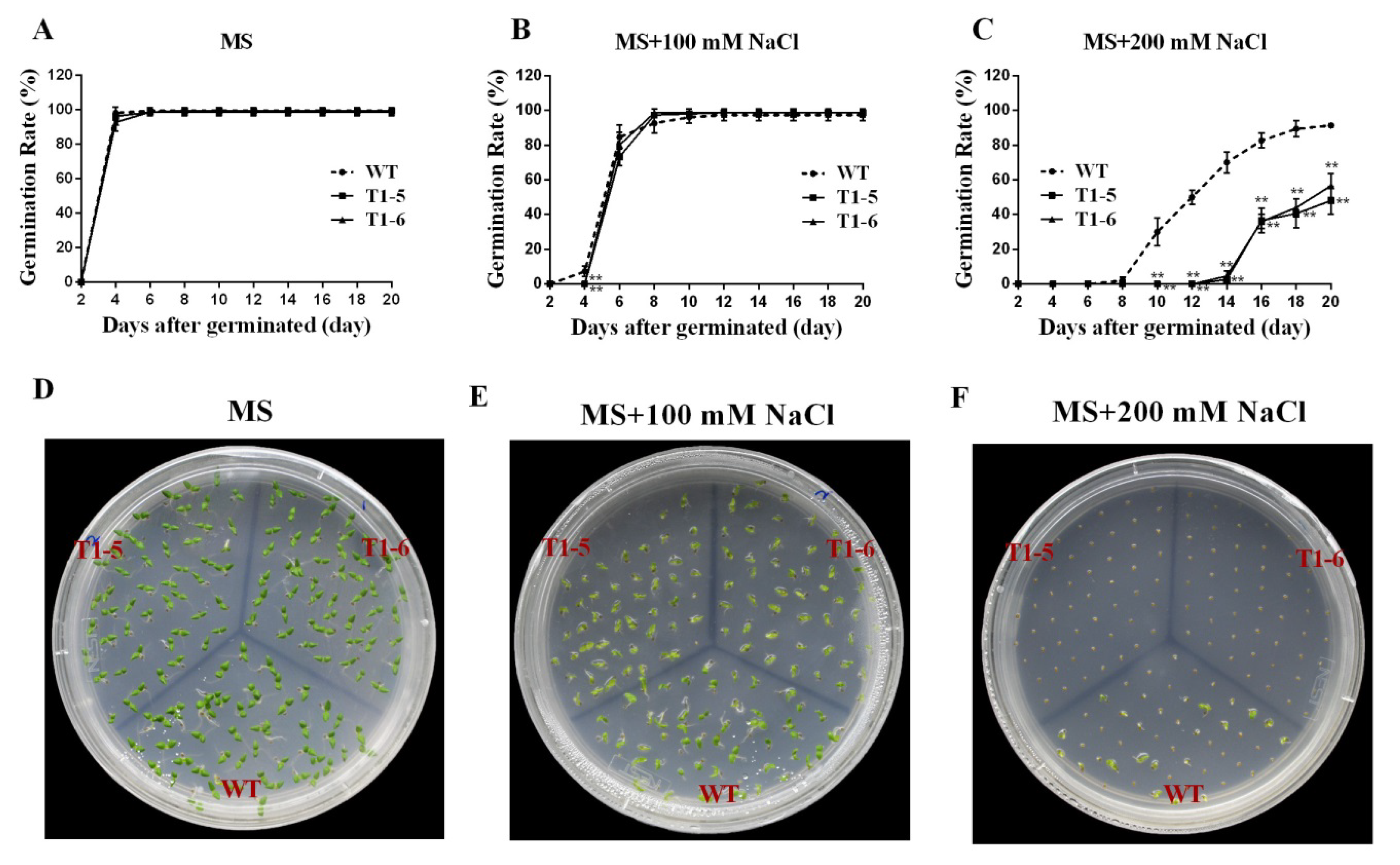
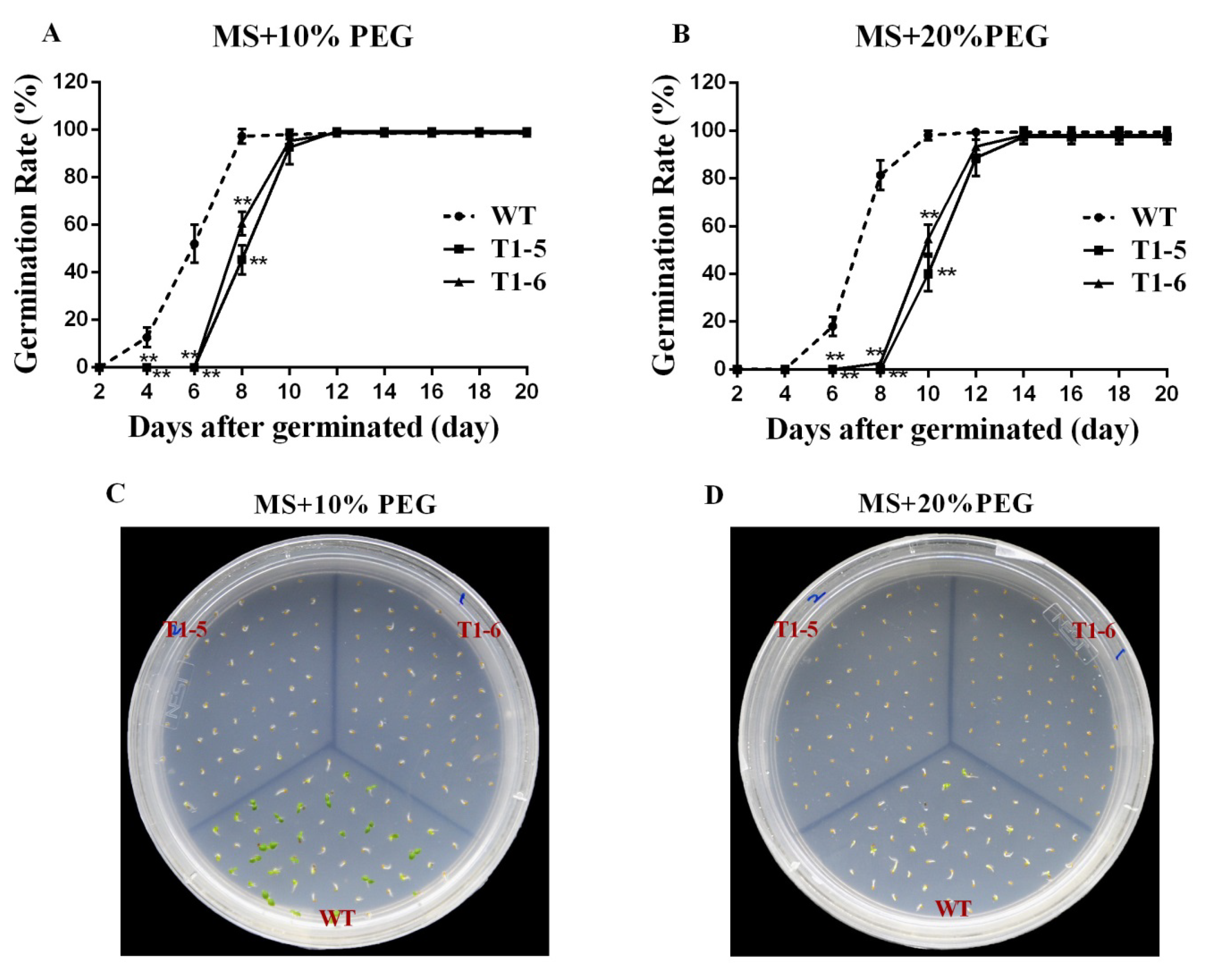
2.5. Germination Assays under Stress and ABA Treatments
3. Discussion
4. Materials and Methods
4.1. Identification of BPC Gene Family Members in Cucumber
4.2. Phylogenetic Gene Structure and Conserved Motif Analyses
4.3. Plant Material, Growth Conditions, and Treatments
4.4. Vector Construction and Tobacco Transformation
4.5. RNA Extraction and qRT-PCR Analysis
4.6. Seed Germination of Transgenic Tobacco Plants under Hyperosmotic Stress Conditions
4.7. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| wt | wild-type |
| qRT-PCR | quantitative real-time polymerase chain reaction |
| PEG | polyethylene glycol |
| MS | Murashige and Skoog |
| ABA | abscisic acid |
References
- Tanurdzic, M.; Banks, J.A. Sex-determining mechanisms in land plants. Plant. Cell 2004, 16, S61–S71. [Google Scholar] [CrossRef]
- Zhang, B.C.; Tolstikov, V.; Turnbull, C.; Hicks, L.M.; Fiehn, O. Divergent metabolome and proteome suggest functional independence of dual phloem transport systems in cucurbits. Proc. Natl. Acad. Sci. USA 2010, 107, 13532–13537. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Singh, A.P.; Chung, G.C. Rapid accumulation of hydrogen peroxide in cucumber roots due to exposure to low temperature appears to mediate decreases in water transport. J. Exp. Bot 2004, 55, 1733–1741. [Google Scholar] [CrossRef] [Green Version]
- Klobus, G.; Janicka-Russak, M. Modulation by cytosolic components of proton pump activities in plasma membrane and tonoplast from Cucumis sativus roots during salt stress. Physiol Plant. 2004, 121, 84–92. [Google Scholar] [CrossRef]
- Zhong, M.; Yuan, Y.H.; Shu, S.; Sun, J.; Guo, S.R.; Yuan, R.N.; Tang, Y.Y. Effects of exogenous putrescine on glycolysis and Krebs cycle metabolism in cucumber leaves subjected to salt stress. Plant. Growth Regul 2016, 79, 319–330. [Google Scholar] [CrossRef]
- Janoudi, A.K.; Widders, I.E.; Flore, J.A. Water deficits and environmental factors affect photosynthesis in leaves of cucumber (Cucumis sativus). J. Am. Soc. Hortic Sci. 1993, 118, 366–370. [Google Scholar] [CrossRef]
- Klein, E.; Katan, J.; Gamliel, A. Soil suppressiveness by organic amendment to Fusarium disease in cucumber: effect on pathogen and host. Phytoparasitica 2016, 44, 239–249. [Google Scholar] [CrossRef]
- Sangwan, I.; O’Brian, M.R. Identification of a soybean protein that interacts with GAGA element dinucleotide repeat DNA. Plant. Physiol. 2002, 129, 1788–1794. [Google Scholar] [CrossRef]
- Santi, L.; Wang, Y.M.; Stile, M.R.; Berendzen, K.; Wanke, D.; Roig, C.; Pozzi, C.; Muller, K.; Muller, J.; Rohde, W.; et al. The GA octodinucleotide repeat binding factor BBR participates in the transcriptional regulation of the homeobox gene Bkn3. Plant. J. 2003, 34, 813–826. [Google Scholar] [CrossRef]
- Meister, R.J.; Williams, L.A.; Monfared, M.M.; Gallagher, T.L.; Kraft, E.A.; Nelson, C.G.; Gasser, C.S. Definition and interactions of a positive regulatory element of the Arabidopsis INNER NOOUTER promoter. Plant. J. 2004, 37, 426–438. [Google Scholar] [CrossRef]
- Kooiker, M.; Airoldi, C.A.; Losa, A.; Manzotti, P.S.; Finzi, L.; Kater, M.M.; Colombo, L. BASIC PENTACYSTEINE1, a GA binding protein that induces conformational changes in the regulatory region of the homeotic Arabidopsis gene SEEDSTICK. Plant. Cell 2005, 17, 722–729. [Google Scholar] [CrossRef]
- Monfared, M.M.; Simon, M.K.; Meister, R.J.; Roig-Villanova, I.; Kooiker, M.; Colombo, L.; Fletcher, J.C.; Gasser, C.S. Overlapping and antagonistic activities of BASIC PENTACYSTEINE genes affect a range of developmental processes in Arabidopsis. Plant. J. 2011, 66, 1020–1031. [Google Scholar] [CrossRef]
- Simonini, S.; Roig-Villanova, I.; Gregis, V.; Colombo, B.; Colombo, L.; Kater, M.M. Basic pentacysteine proteins mediate MADS domain complex binding to the DNA for tissue-specific expression of target genes in Arabidopsis. Plant. Cell 2012, 24, 4163–4172. [Google Scholar] [CrossRef]
- Simonini, S.; Kater, M.M. Class I BASIC PENTACYSTEINE factors regulate HOMEOBOX genes involved in meristem size maintenance. J. Exp. Bot. 2014, 65, 1455–1465. [Google Scholar] [CrossRef] [Green Version]
- Hecker, A.; Brand, L.H.; Peter, S.; Simoncello, N.; Kilian, J.; Harter, K.; Gaudin, V.; Wanke, D. The Arabidopsis GAGA-Binding Factor BASIC PENTACYSTEINE6 Recruits the POLYCOMB-REPRESSIVE COMPLEX1 Component LIKE HETEROCHROMATIN PROTEIN1 to GAGA DNA Motifs. Plant. Physiol 2015, 168, 1013–1024. [Google Scholar] [CrossRef]
- Anusak, P.; Ditta, G.S.; Beth, S.; Liljegren, S.J.; Elvira, B.; Ellen, W.; Yanofsky, M.F. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 2003, 424, 85–88. [Google Scholar]
- Rebecca, F.; Anusak, P.; Raffaella, B.; Maarten, K.; Lorenzo, B.; Gary, D.; Yanofsky, M.F.; Kater, M.M.; Lucia, C. MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant. Cell 2003, 15, 2603–2611. [Google Scholar]
- Berger, N.; Dubreucq, B.; Roudier, F.; Dubos, C.; Lepiniec, L. Transcriptional regulation of Arabidopsis LEAFY COTYLEDON2 involves RLE, a cis-element that regulates trimethylation of histone H3 at lysine-27. Plant. Cell 2011, 23, 4065–4078. [Google Scholar] [CrossRef]
- Stone, S.L.; Kwong, L.W.; Yee, K.M.; Pelletier, J.; Lepiniec, L.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 2001, 98, 11806–11811. [Google Scholar] [CrossRef]
- Kroj, T.; Savino, G.; Valon, C.; Giraudat, J.; Parcy, F. Regulation of storage protein gene expression in Arabidopsis. Development 2003, 130, 6065–6073. [Google Scholar] [CrossRef]
- Mu, Y.; Liu, Y.M.; Bai, L.Q.; Li, S.Z.; He, C.X.; Yan, Y.; Yu, X.C.; Li, Y.S. Cucumber CsBPCs regulate the expression of CsABI3 during seed germination. Front. Plant. Sci. 2017, 8, 459. [Google Scholar] [CrossRef]
- Gong, R.; Cao, H.; Zhang, J.; Xie, K.; Wang, D.; Yu, S. Divergent functions of the GAGA-binding transcription factor family in rice. Plant. J. 2018, 94, 32–47. [Google Scholar] [CrossRef]
- Peng, L.T.; Shi, Z.Y.; Li, L.; Shen, G.Z.; Zhang, J.L. Overexpression of transcription factor OsLFL1 delays flowering time in Oryza sativa. J. Plant. Physiol 2008, 165, 876–885. [Google Scholar] [CrossRef]
- Peng, L.T.; Shi, Z.Y.; Li, L.; Shen, G.Z.; Zhang, J.L. Ectopic expression of OsLFL1 in rice represses Ehd1 by binding on its promoter. Biochem. Biophys. Res. Commun. 2007, 360, 251–256. [Google Scholar] [CrossRef]
- Shanks, C.M.; Hecker, A.; Cheng, C.Y.; Brand, L.; Collani, S.; Schmid, M.; Schaller, G.E.; Wanke, D.; Harter, K.; Kieber, J.J. Role of BASIC PENTACYSTEINE transcription factors in a subset of cytokinin signaling responses. Plant. J. 2018, 95, 458–473. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Gallagher, K.L. Transcription factors on the move. Curr. Opin. Plant. Biol. 2012, 15, 645–651. [Google Scholar] [CrossRef]
- Mao, K.; Dong, Q.; Li, C.; Liu, C.; Ma, F. Genome wide identification and characterization of apple bHLH transcription factors and expression analysis in response to drought and salt stress. Front. Plant. Sci. 2017, 8, 480. [Google Scholar] [CrossRef]
- Wang, Y.; Zhan, D.F.; Li, H.L.; Guo, D.; Zhu, J.H.; Peng, S.Q. Transcriptome-wide identification and characterization of MYB transcription factor genes in the laticifer cells of Hevea brasiliensis. Front. Plant. Sci. 2017, 8, 1974. [Google Scholar] [CrossRef]
- Xie, T.; Chen, C.; Li, C.; Liu, J.; Liu, C.; He, Y. Genome-wide investigation of WRKY gene family in pineapple: evolution and expression profiles during development and stress. BMC Genomics 2018, 19, 490. [Google Scholar] [CrossRef]
- Wanke, D.; Hohenstatt, M.L.; Dynowski, M.; Bloss, U.; Hecker, A.; Elgass, K.; Hummel, S.; Hahn, A.; Caesar, K.; Schleifenbaum, F.; et al. Alanine zipper-like coiled-coil domains are necessary for homotypic dimerization of plant GAGA-factors in the nucleus and nucleolus. PLoS ONE 2011, 6, e16070. [Google Scholar] [CrossRef]
- Theune, M.L.; Hummel, S.; Jaspert, N.; Lafos, M.; Wanke, D. Dimerization of the BASIC PENTACYSTEINE domain in plant GAGA-factors is mediated by disulfide bonds and required for DNA-binding. J. Adv. Plant. Biol. 2017, 1, 26–39. [Google Scholar] [CrossRef]
- Kucera, B.; Cohn, M.A.; Leubner-Metzger, G. Plant hormone interactions during seed dormancy release and germination. Seed Sci. Res. 2005, 15, 281–307. [Google Scholar] [CrossRef]
- Warpeha, K.M.; Montgomery, B.L. Light and hormone interactions in the seed-to-seedling transition. Environ. Exp. Bot. 2016, 121, 56–65. [Google Scholar] [CrossRef] [Green Version]
- Gupta, R.; Chakrabarty, S.K. Gibberellic acid in plant: still a mystery unresolved. Plant. Signal. Behav. 2013, 8. [Google Scholar] [CrossRef]
- Hedden, P.; Thomas, S.G. Gibberellin biosynthesis and its regulation. Biochem. J. 2012, 444, 11–25. [Google Scholar] [CrossRef] [Green Version]
- Perilli, S.; Moubayidin, L.; Sabatini, S. The molecular basis of cytokinin function. Curr. Opin. Plant. Biol. 2010, 13, 21–26. [Google Scholar] [CrossRef]
- Clouse, S.D. Brassinosteroids. The Arabidopsis Book 2011, 9, e0151. [Google Scholar] [CrossRef] [Green Version]
- Corbineau, F.; Xia, Q.; Bailly, C.; El-Maarouf-Bouteau, H. Ethylene, a key factor in the regulation of seed dormancy. Front. Plant. Sci. 2014, 5, 539. [Google Scholar] [CrossRef] [Green Version]
- Sakata, Y.; Komatsu, K.; Takezawa, D. ABA as a universal plant hormone. Progress Botany 2014, 75, 57–96. [Google Scholar]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant. Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef]
- Kakimoto, T. Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate: ATP/ADP isopentenyltransferases. Plant. Cell Physiol. 2001, 42, 677–685. [Google Scholar] [CrossRef]
- Buechel, S.; Leibfried, A.; To, J.P.C.; Zhao, Z.; Andersen, S.U.; Kieber, J.J.; Lohmann, J.U. Role of A-type ARABIDOPSIS RESPONSE REGULATORS in meristem maintenance and regeneration. Eur. J. Cell Biol. 2010, 89, 279–284. [Google Scholar] [CrossRef]
- Zhong, Z.; Andersen, S.U.; Karin, L.; Karel, D.; Andrej, M.; Schultheiss, S.J.; Lohmann, J.U. Hormonal control of the shoot stem-cell niche. Nature 2010, 465, 1089–1092. [Google Scholar]
- Bateman, A.; Birney, E.; Durbin, R.; Eddy, S.R.; Howe, K.L.; Sonnhammer, E.L. The Pfam protein families database. Nucleic Acids Res. 2000, 28, 263–266. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef]
- Yu, C.S.; Chen, Y.C.; Lu, C.H.; Hwang, J.K. Prediction of protein subcellular localization. Proteins 2006, 64, 643–651. [Google Scholar] [CrossRef]
- Wang, P.; Wang, S.; Chen, Y.; Xu, X.; Guang, X.; Zhang, Y. Genome-wide Analysis of the MADS-Box Gene Family in Watermelon. Comput. Biol. Chem. 2019, 80, 341–350. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Horsch, R.B.; Fry, J.E.; Hoffmann, N.L.; Eichholtz, D.; Rogers, S.G.; Fraley, R.T. A simple and general method for transferring genes into plants. Science 1985, 227, 1229–1231. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
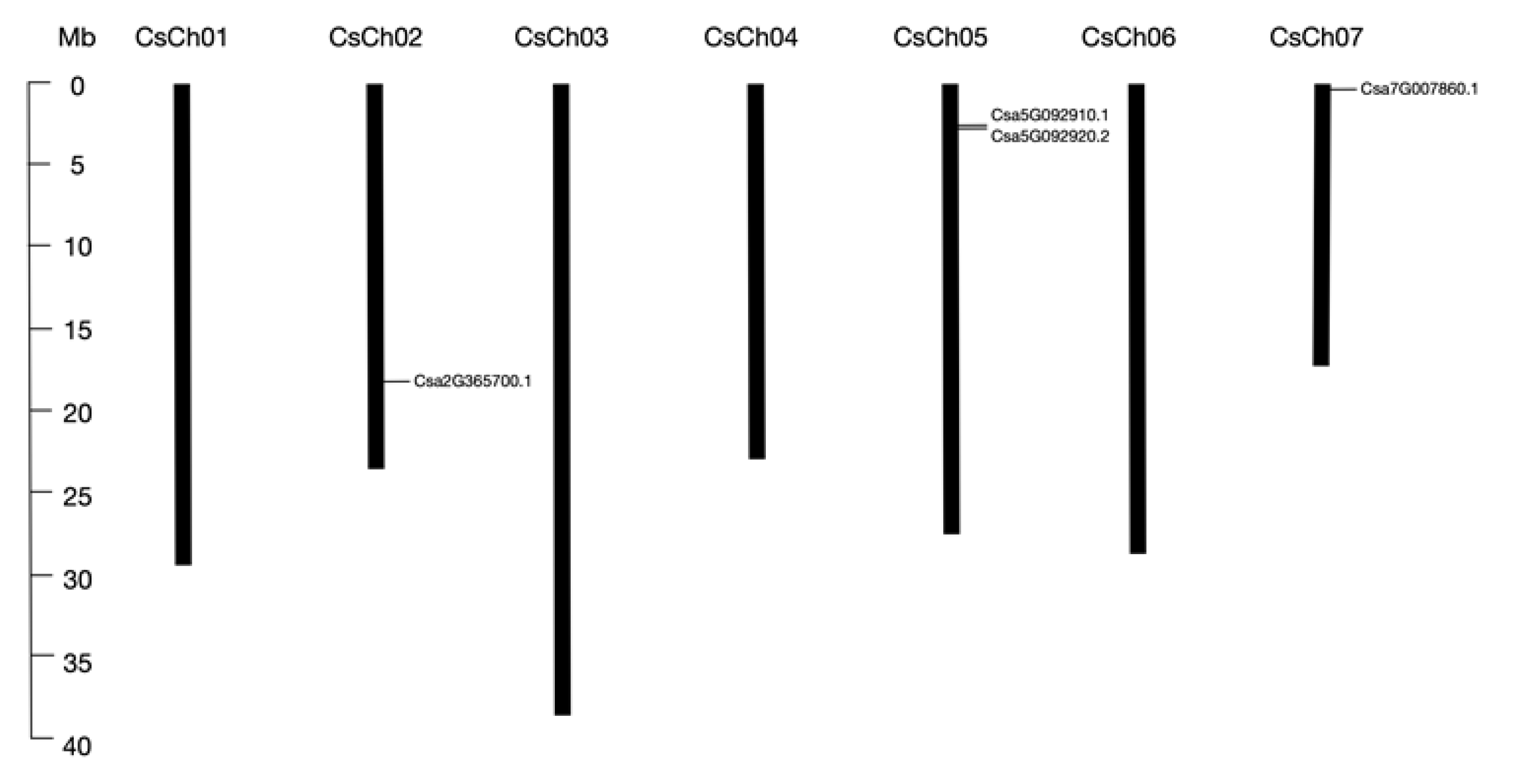



| Gene ID | Length (aa) | Molecular Weight (KD) | Chromosome | Location | pI | Strand Direction | Subcellular Location |
|---|---|---|---|---|---|---|---|
| Csa2G365700.1 | 338 | 37.8 | 2 | 17669793–17673023 | 9.52 | + | Nuclear |
| Csa5G092910.1 | 279 | 31.1 | 5 | 2693816–2695298 | 9.62 | - | Nuclear |
| Csa5G092920.2 | 284 | 31.7 | 5 | 2696954–2700269 | 9.84 | - | Nuclear |
| Csa7G007860.1 | 313 | 35 | 7 | 388479–391899 | 9.6 | + | Nuclear |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Miao, L.; Huang, B.; Gao, L.; He, C.; Yan, Y.; Wang, J.; Yu, X.; Li, Y. Genome-Wide Identification and Characterization of Cucumber BPC Transcription Factors and Their Responses to Abiotic Stresses and Exogenous Phytohormones. Int. J. Mol. Sci. 2019, 20, 5048. https://doi.org/10.3390/ijms20205048
Li S, Miao L, Huang B, Gao L, He C, Yan Y, Wang J, Yu X, Li Y. Genome-Wide Identification and Characterization of Cucumber BPC Transcription Factors and Their Responses to Abiotic Stresses and Exogenous Phytohormones. International Journal of Molecular Sciences. 2019; 20(20):5048. https://doi.org/10.3390/ijms20205048
Chicago/Turabian StyleLi, Shuzhen, Li Miao, Bin Huang, Lihong Gao, Chaoxing He, Yan Yan, Jun Wang, Xianchang Yu, and Yansu Li. 2019. "Genome-Wide Identification and Characterization of Cucumber BPC Transcription Factors and Their Responses to Abiotic Stresses and Exogenous Phytohormones" International Journal of Molecular Sciences 20, no. 20: 5048. https://doi.org/10.3390/ijms20205048
APA StyleLi, S., Miao, L., Huang, B., Gao, L., He, C., Yan, Y., Wang, J., Yu, X., & Li, Y. (2019). Genome-Wide Identification and Characterization of Cucumber BPC Transcription Factors and Their Responses to Abiotic Stresses and Exogenous Phytohormones. International Journal of Molecular Sciences, 20(20), 5048. https://doi.org/10.3390/ijms20205048






