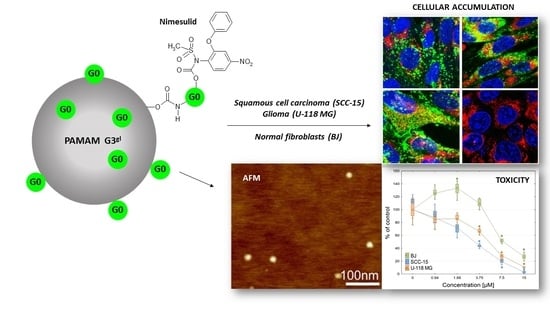
Mixed-Generation PAMAM G3-G0 Megamer as a Drug Delivery System for Nimesulide: Antitumor Activity of the Conjugate Against Human Squamous Carcinoma and Glioblastoma Cells
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
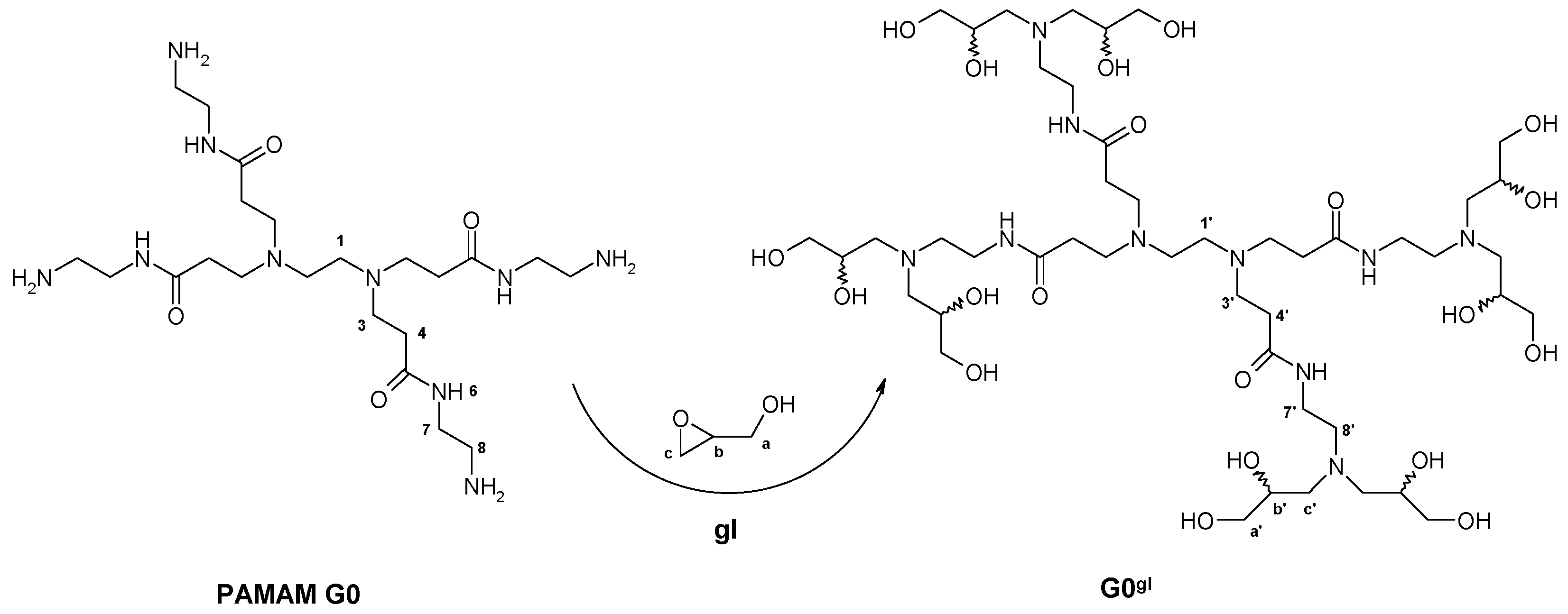
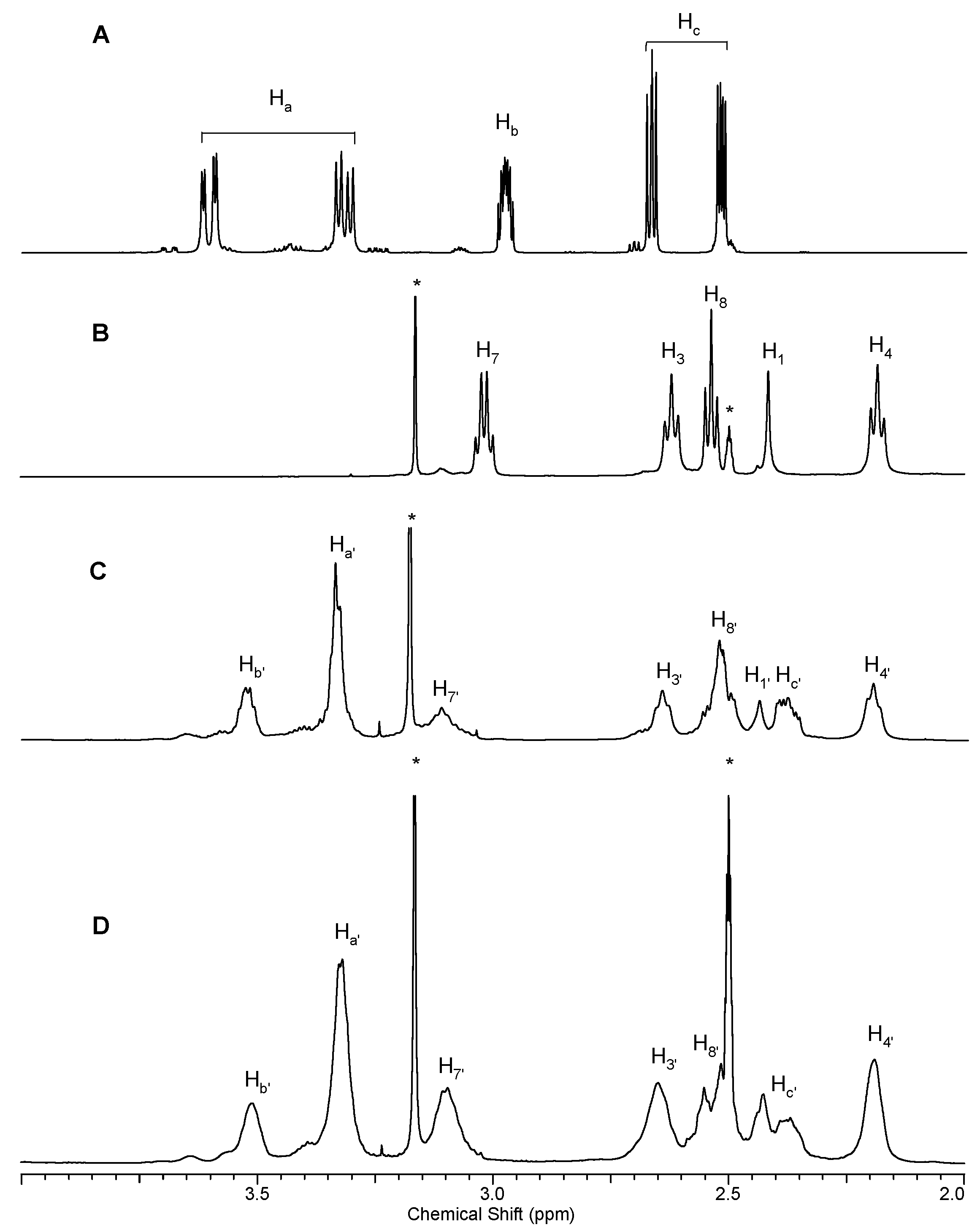
2.1.1. Reaction of PAMAM Dendrimers with Glycidol
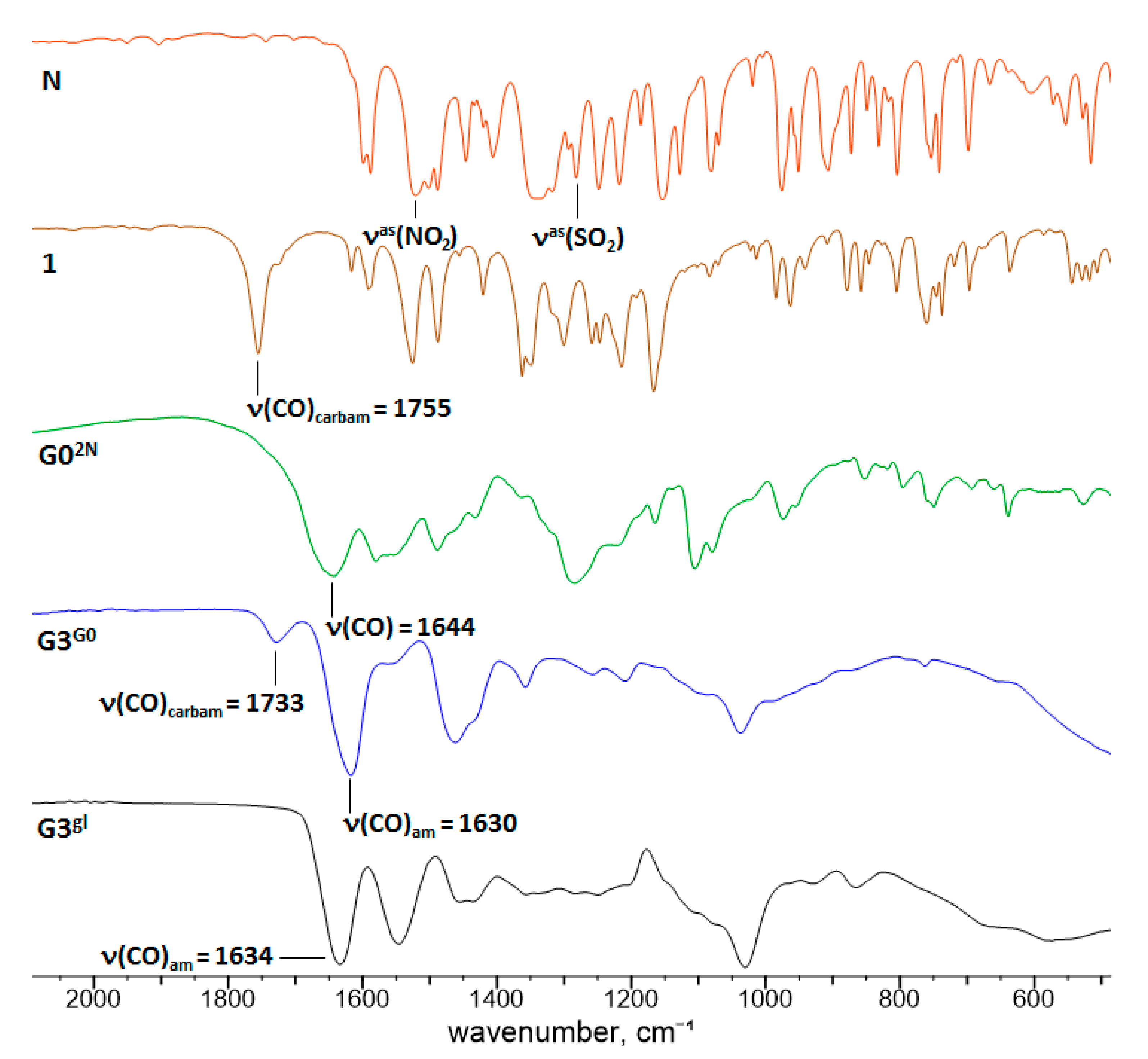
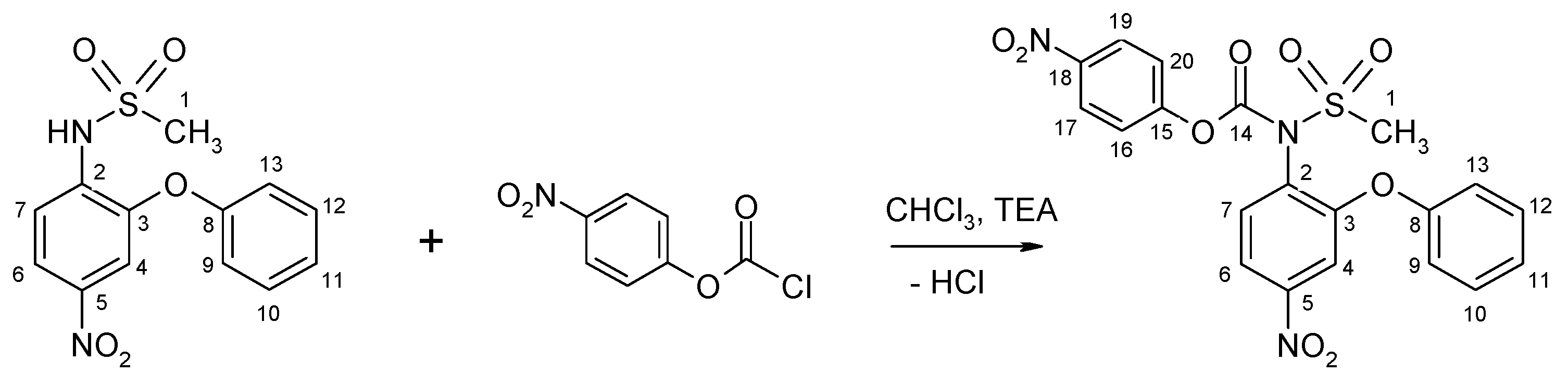
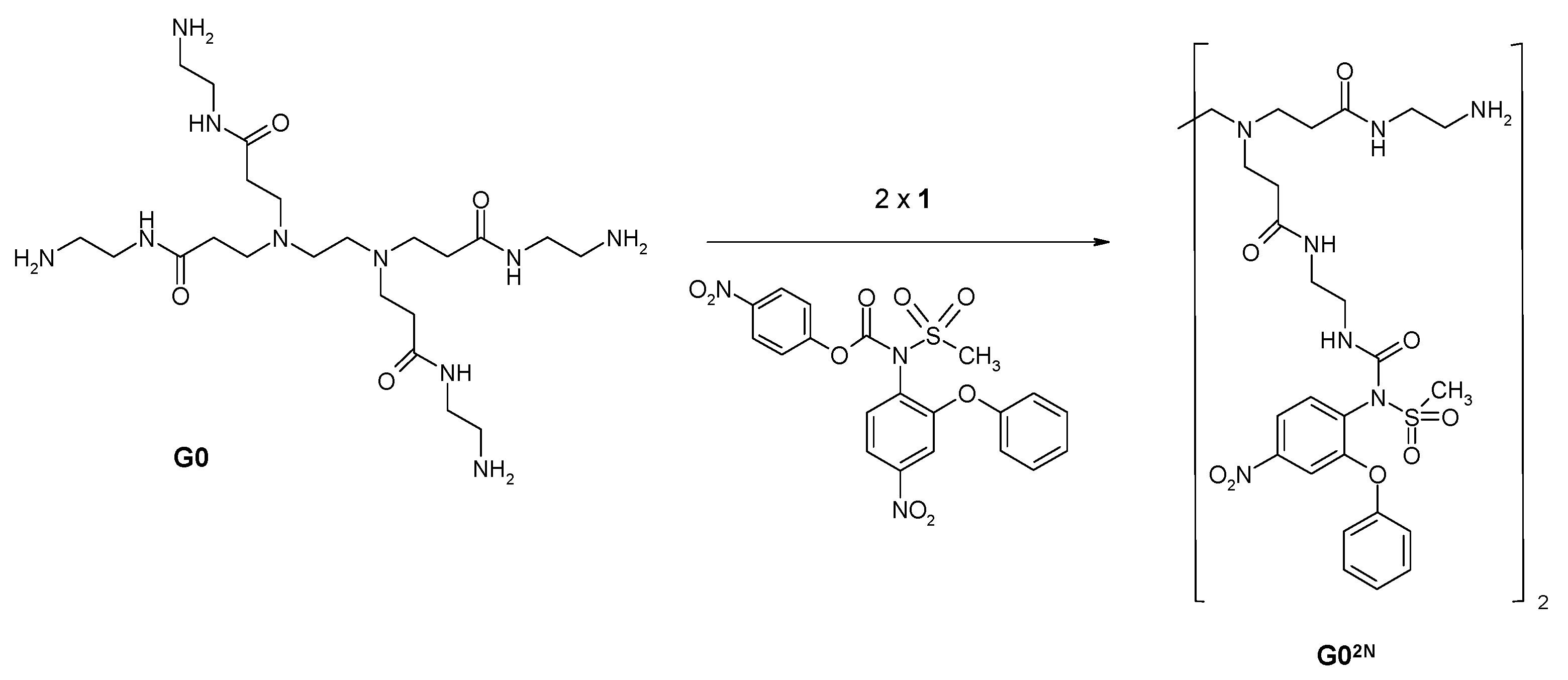
2.1.2. Nimesulide Activation and Attachment to PAMAM G0
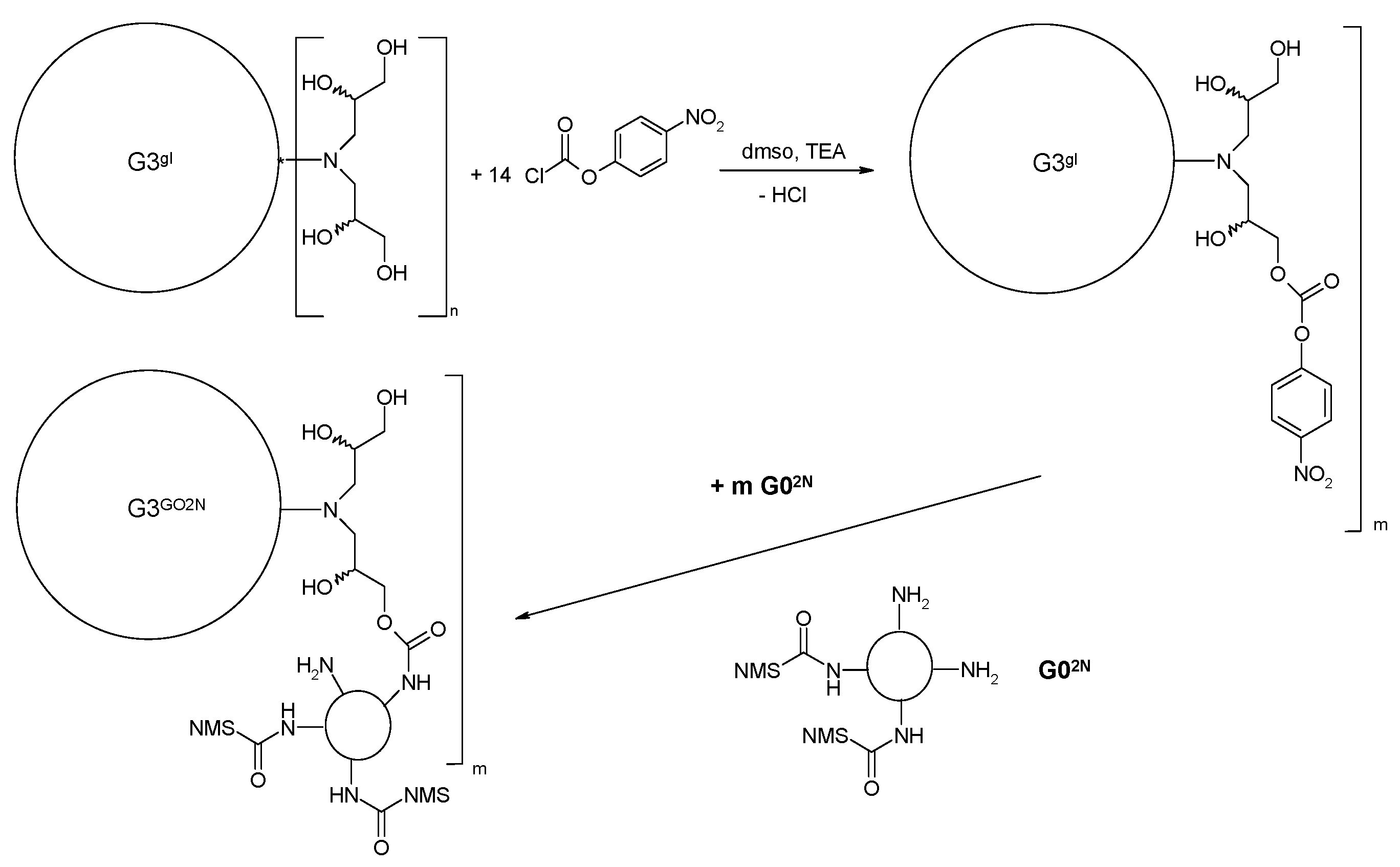
2.1.3. Synthesis of Megamer with Conjugated G0
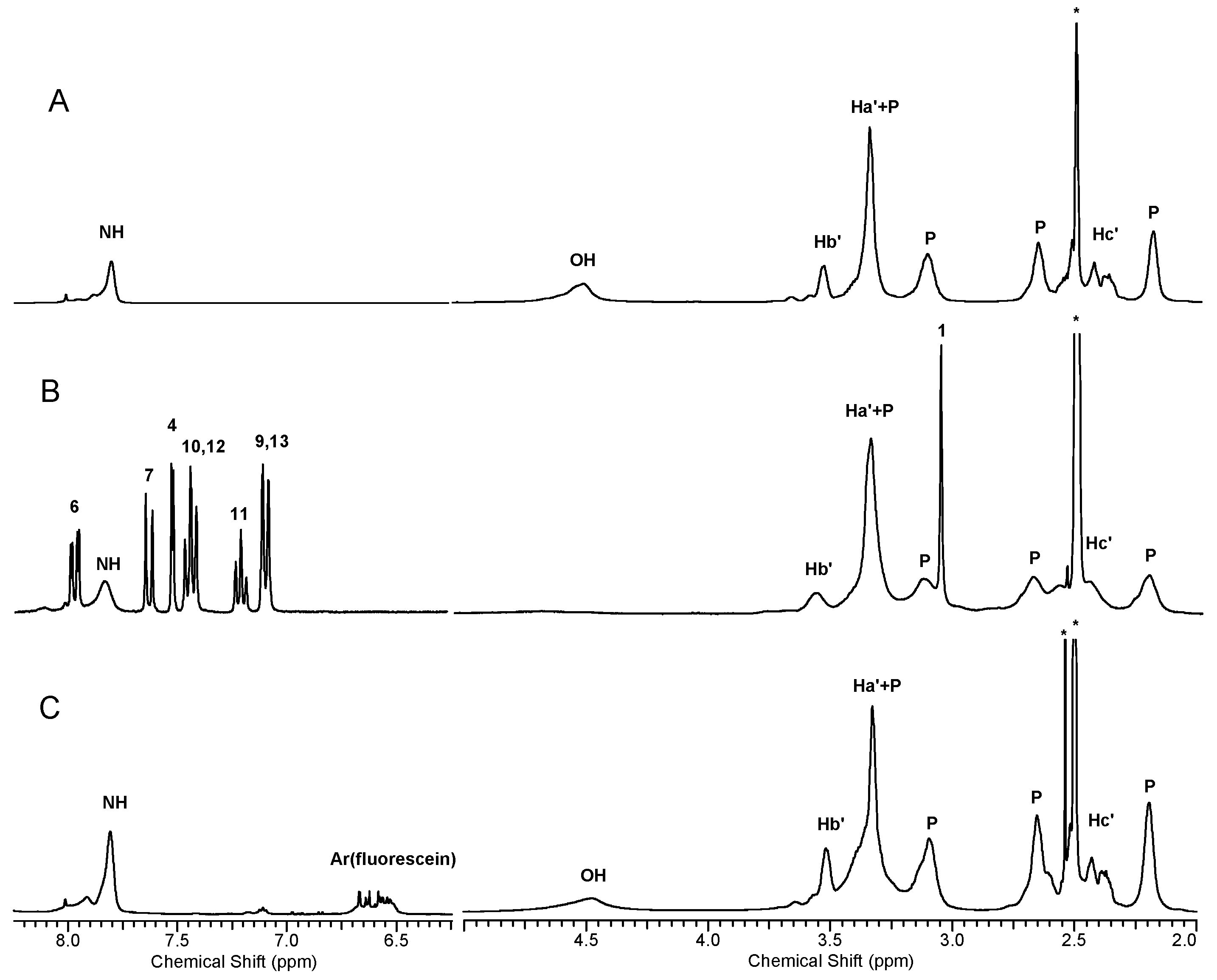
2.1.4. Molecular Weight and Size of Dendrimers and Megamers
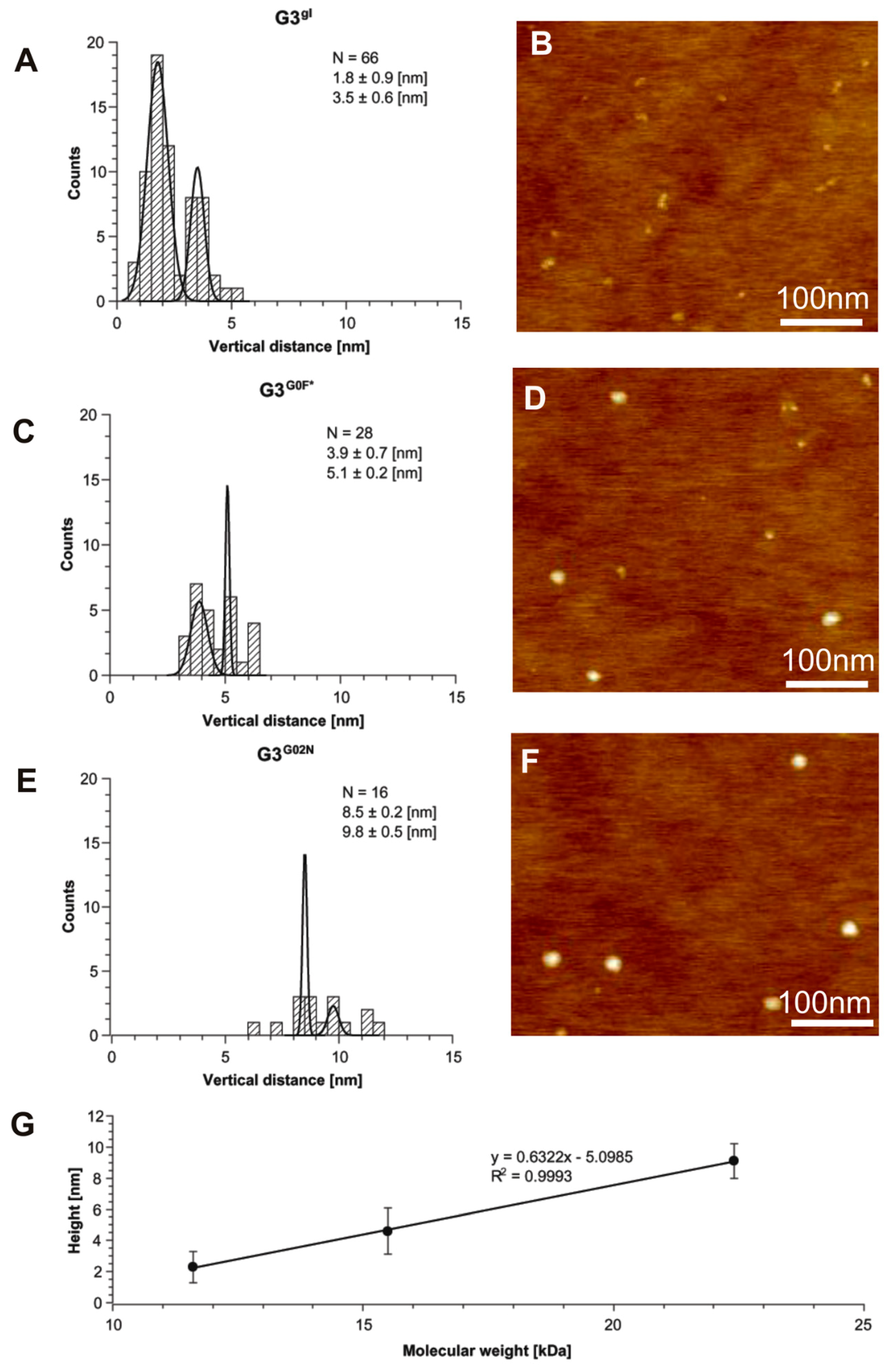
2.1.5. Structural and Height Analysis of Dendrimers Using AFM
2.2. Biology
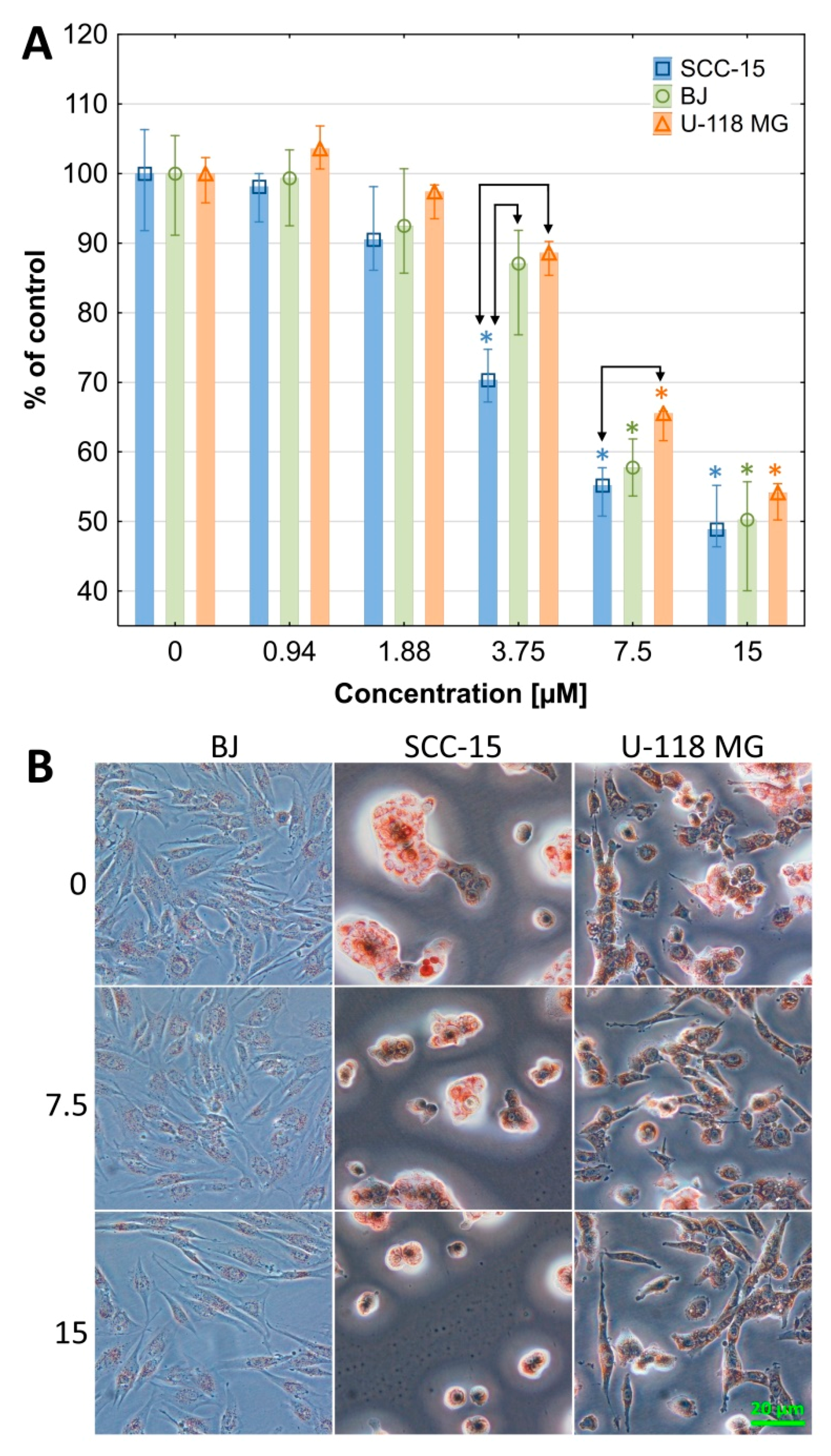
2.2.1. Cytotoxicity
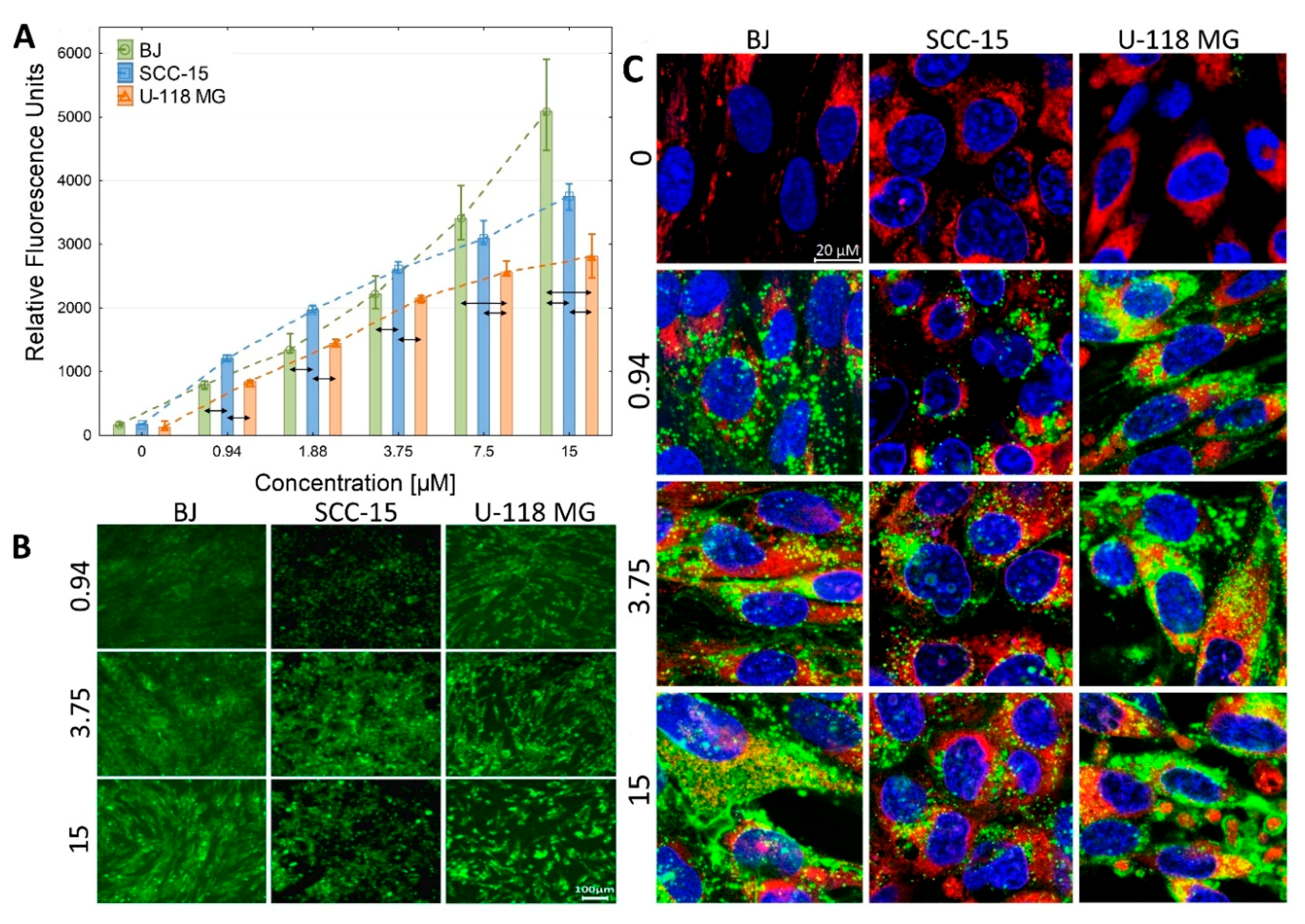
2.2.2. Cellular Accumulation
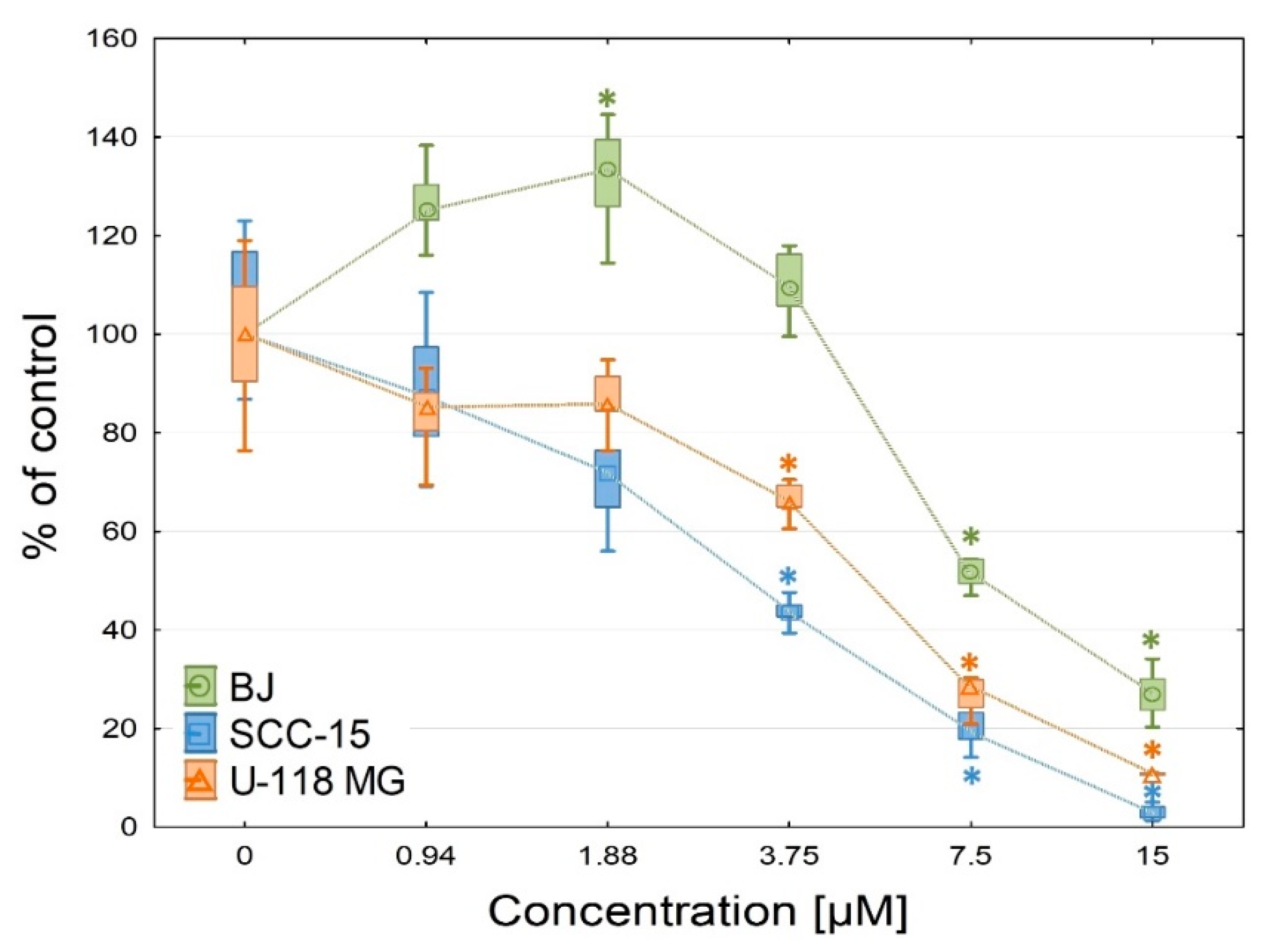
2.2.3. Anti-Proliferation
3. Materials and Methods
3.1. Materials
3.2. Syntheses
3.2.1. Synthesis of PAMAM G0 Dendrimer Substituted with 8 Glycidol Molecules, G0gl
3.2.2. Synthesis of PAMAM G3 Dendrimer Substituted with 64 Glycidol Molecules (G3gl): The Megamer Macromolecular Core
3.2.3. Synthesis of N-(4-Nitrophenoxycarbonyl) Nimesulid: N-(4-Nitrophenoxycarbonyl), N-(4-Nitro-2-Phenoxyphenyl) Methanesulfonamide, 1
3.2.4. Synthesis of PAMAM G0-Bis-Carbonylnimesulide (G02N) and Single Fluorescein-Labeled PAMAM G0 (G0F)
3.2.5. Synthesis of megamers G3gl-12G02N and G3gl-4G0F
3.3. Cell Cultures
3.3.1. Cytotoxicity Neutral Red Assay (NR)
3.3.2. Cellular Accumulation of Megamer
3.3.3. Confocal Microscopy
3.3.4. Proliferation Assay
3.4. Methods
3.4.1. Spectroscopy
3.4.2. Atomic Force Microscopy Studies
3.4.3. Molecular Weight Estimation with Gel Permeation Chromatography
3.4.4. Dynamic Light Scattering Measurements
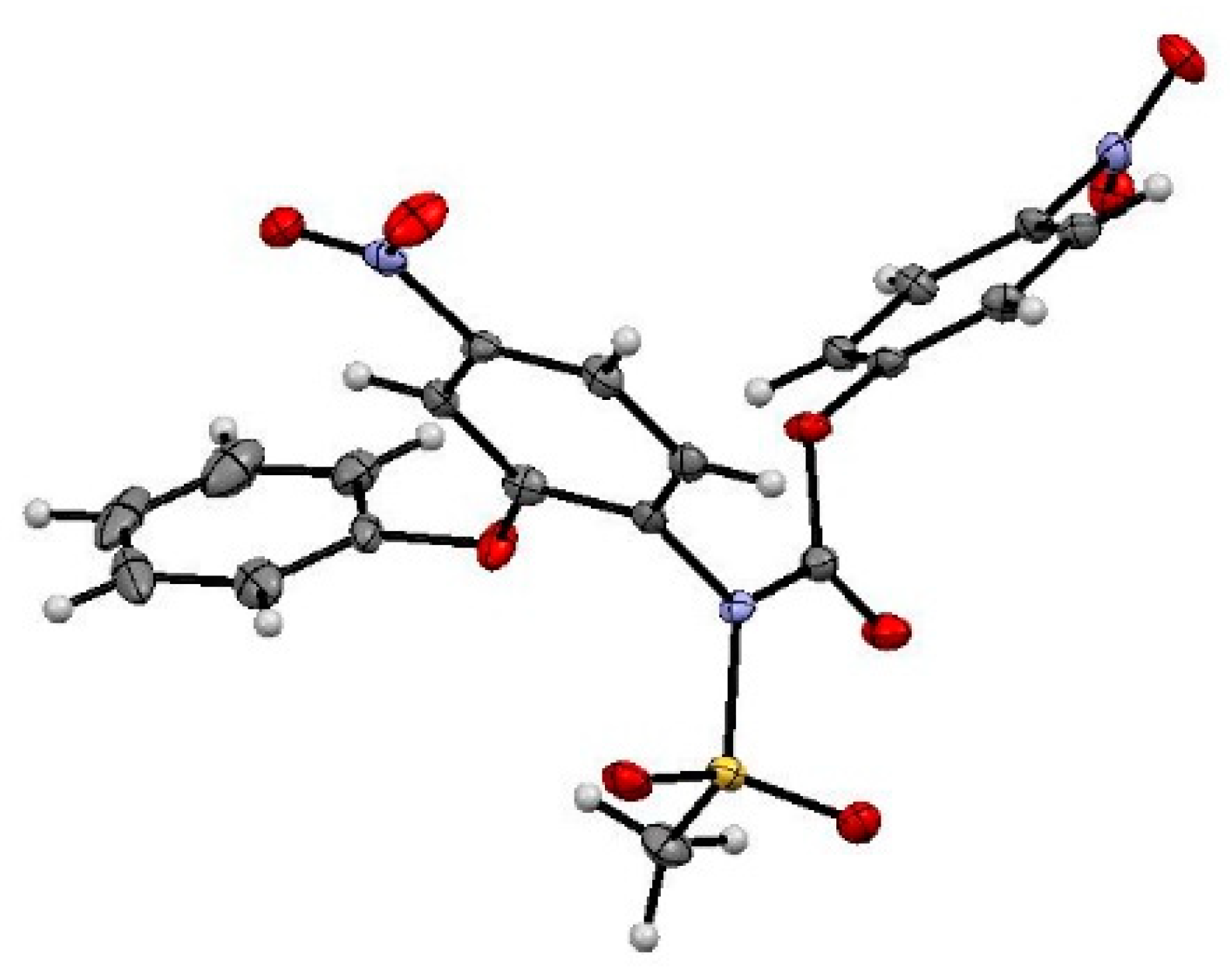
3.4.5. Crystallographic Measurements
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Bond Lengths [Å] | Torsion Angles [°] |
|---|---|
| S1A O2A 1.424(2) | O2A S1A O1A 120.34(14) |
| S1A O1A 1.428(2) | O2A S1A N1A 103.73(14) |
| S1A N1A 1.691(3) | O1A S1A N1A 108.47(14) |
| S1A C1A 1.757(3) | O2A S1A C1A 109.51(16) |
| N1A C14A 1.379(4) | O1A S1A C1A 108.95(16) |
| N1A C2A 1.456(4) | N1A S1A C1A 104.64(15) |
| C2A C7A 1.364(4) | C14A N1A C2A 121.7(3) |
| C2A C3A 1.399(4) | C14A N1A S1A 119.7(2) |
| O3A C3A 1.373(4) | C2A N1A S1A 118.3(2) |
| O3A C8A 1.411(4) | C7A C2A C3A 121.0(3) |
| C3A C4A 1.377(4) | C7A C2A N1A 120.8(3) |
| O4A N5A 1.238(3) | C3A C2A N1A 118.2(3) |
| C4A C5A 1.377(4) | C3A O3A C8A 118.5(2) |
| O5A N5A 1.218(3) | O3A C3A C4A 124.3(3) |
| N5A C5A 1.483(4) | O3A C3A C2A 116.1(3) |
| C5A C6A 1.381(4) | C4A C3A C2A 119.6(3) |
| O6A C14A 1.359(4) | C5A C4A C3A 118.1(3) |
| O6A C15A 1.410(4) | O5A N5A O4A 124.1(3) |
| C6A C7A 1.392(4) | O5A N5A C5A 118.3(3) |
| O7A C14A 1.194(4) | O4A N5A C5A 117.6(3) |
| C8A C9A 1.365(5) | C4A C5A C6A 123.6(3) |
| C8A C13A 1.384(5) | C4A C5A N5A 116.8(3) |
| C9A C10A 1.391(5) | C6A C5A N5A 119.5(3) |
| C10A C11A 1.389(6) | C14A O6A C15A 117.2(2) |
| C11A C12A 1.372(6) | C5A C6A C7A 117.2(3) |
| C12A C13A 1.369(5) | C2A C7A C6A 120.5(3) |
| C15A C16A 1.373(4) | C9A C8A C13A 122.0(3) |
| C15A C20A 1.382(4) | C9A C8A O3A 121.5(3) |
| C16A C17A 1.397(4) | C13A C8A O3A 116.4(3) |
| C17A C18A 1.377(4) | C8A C9A C10A 119.0(4) |
| O18A N18A 1.233(3) | C11A C10A C9A 119.4(4) |
| N18A O19A 1.229(3) | C12A C11A C10A 120.3(4) |
| N18A C18A 1.459(4) | C13A C12A C11A 120.7(4) |
| C18A C19A 1.395(4) | C12A C13A C8A 118.6(4) |
| C19A C20A 1.390(4) | O7A C14A O6A 125.6(3) |
| S1 O2 1.424(2) | O7A C14A N1A 126.0(3) |
| S1 O1 1.427(2) | O6A C14A N1A 108.4(3) |
| S1 N1 1.687(3) | C16A C15A C20A 123.1(3) |
| S1 C1 1.748(4) | C16A C15A O6A 118.0(3) |
| N1 C14 1.393(4) | C20A C15A O6A 118.7(3) |
| N1 C2 1.448(4) | C15A C16A C17A 118.7(3) |
| C2 C7 1.378(4) | C18A C17A C16A 118.1(3) |
| C2 C3 1.394(4) | O19A N18A O18A 123.7(3) |
| O3 C3 1.368(4) | O19A N18A C18A 118.3(3) |
| O3 C8 1.412(4) | O18A N18A C18A 118.0(3) |
| C3 C4 1.384(4) | C17A C18A C19A 123.6(3) |
| C4 C5 1.380(5) | C17A C18A N18A 118.6(3) |
| O4 N5 1.231(4) | C19A C18A N18A 117.8(3) |
| O5 N5 1.224(3) | C20A C19A C18A 117.5(3) |
| N5 C5 1.480(4) | C15A C20A C19A 119.0(3) |
| C5 C6 1.385(4) | O2 S1 O1 119.91(14) |
| O6 C14 1.364(4) | O2 S1 N1 103.87(14) |
| O6 C15 1.409(4) | O1 S1 N1 108.49(13) |
| C6 C7 1.391(4) | O2 S1 C1 109.37(17) |
| O7 C14 1.189(4) | O1 S1 C1 108.88(16) |
| C8 C9 1.375(5) | N1 S1 C1 105.29(15) |
| C8 C13 1.380(5) | C14 N1 C2 122.4(3) |
| C9 C10 1.381(5) | C14 N1 S1 117.9(2) |
| C10 C11 1.379(5) | C2 N1 S1 118.8(2) |
| C11 C12 1.369(5) | C7 C2 C3 121.0(3) |
| C12 C13 1.375(5) | C7 C2 N1 120.3(3) |
| C15 C20 1.369(4) | C3 C2 N1 118.6(3) |
| C15 C16 1.387(4) | C3 O3 C8 117.8(2) |
| C16 C17 1.388(5) | O3 C3 C4 123.9(3) |
| C17 C18 1.385(4) | O3 C3 C2 116.0(3) |
| O18 N18 1.228(4) | C4 C3 C2 120.1(3) |
| N18 O19 1.233(3) | C5 C4 C3 117.3(3) |
| N18 C18 1.468(4) | O5 N5 O4 123.8(3) |
| C18 C19 1.381(4) | O5 N5 C5 118.5(3) |
| C19 C20 1.384(5) | O4 N5 C5 117.8(3) |
| Cl1 C100 1.737(4) | C4 C5 C6 124.2(3) |
| Cl2 C100 1.765(4) | C4 C5 N5 116.6(3) |
| Cl3 C100 1.754(4) | C6 C5 N5 119.1(3) |
| C14 O6 C15 115.5(2) | |
| C5 C6 C7 117.2(3) | |
| C2 C7 C6 120.1(3) | |
| C9 C8 C13 121.8(3) | |
| C9 C8 O3 121.0(3) | |
| C13 C8 O3 117.2(3) | |
| C8 C9 C10 118.6(3) | |
| C11 C10 C9 120.2(4) | |
| C12 C11 C10 120.1(3) | |
| C11 C12 C13 120.8(3) | |
| C12 C13 C8 118.5(3) | |
| O7 C14 O6 126.0(3) | |
| O7 C14 N1 125.6(3) | |
| O6 C14 N1 108.4(3) | |
| C20 C15 C16 122.7(3) | |
| C20 C15 O6 119.5(3) | |
| C16 C15 O6 117.8(3) | |
| C15 C16 C17 118.5(3) | |
| C18 C17 C16 118.1(3) | |
| O18 N18 O19 123.8(3) | |
| O18 N18 C18 118.4(3) | |
| O19 N18 C18 117.8(3) | |
| C19 C18 C17 123.2(3) | |
| C19 C18 N18 119.0(3) | |
| C17 C18 N18 117.7(3) | |
| C18 C19 C20 117.9(3) | |
| C15 C20 C19 119.4(3) | |
| Cl1 C100 Cl3 111.1(2) | |
| Cl1 C100 Cl2 110.9(2) | |
| Cl3 C100 Cl2 110.6(2) |
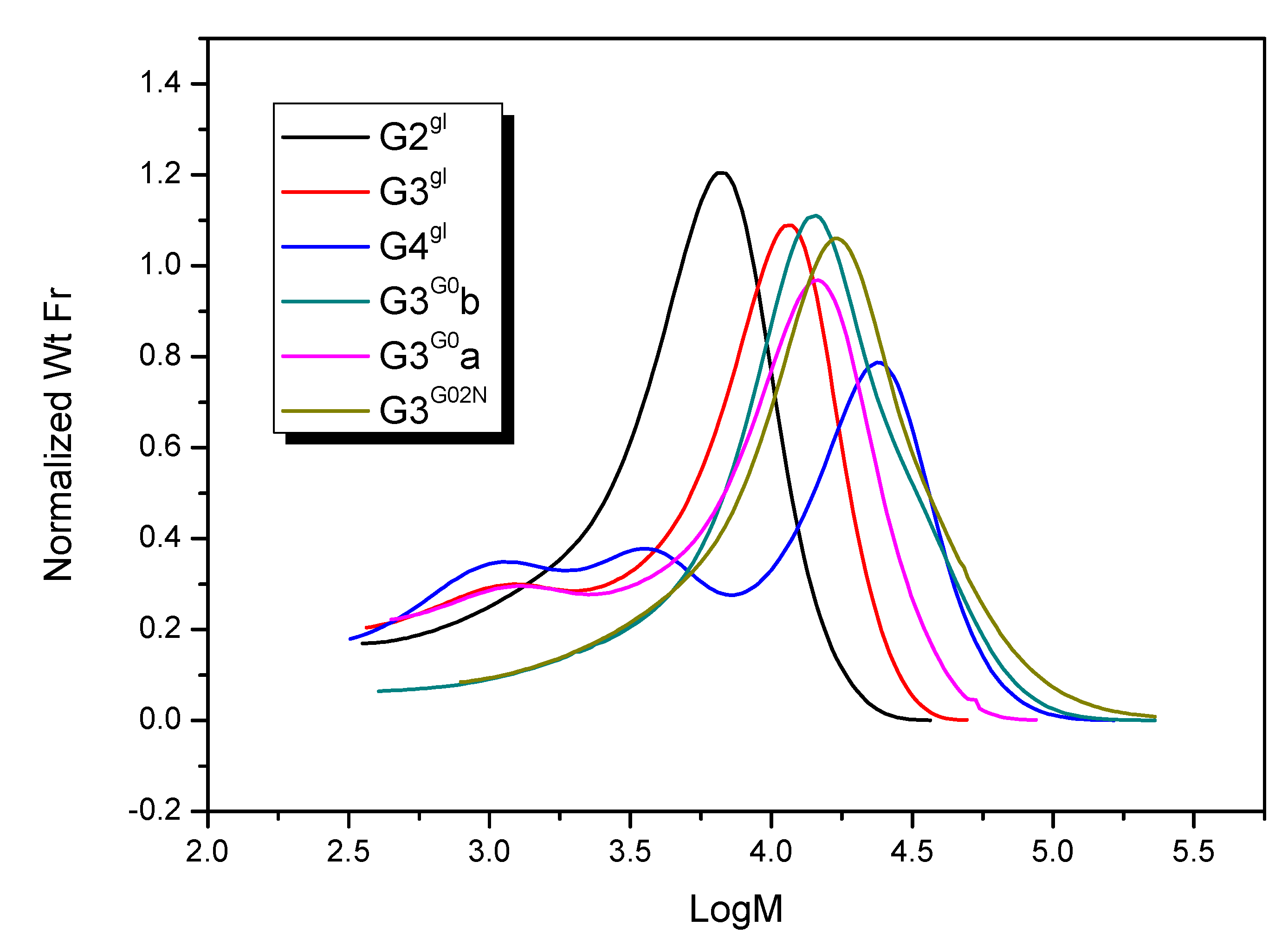
| Species | Mn | Mw | Mz | Mp | Mw/Mn |
|---|---|---|---|---|---|
| G3G0b | 11 860 | 18 980 | 30 930 | 14 370 | 1.60 |
| G3G0a | 11 010 | 15 050 | 20 220 | 14 920 | 1.37 |
| G3G02N | 13 740 | 22 400 | 37 230 | 17 310 | 1.63 |
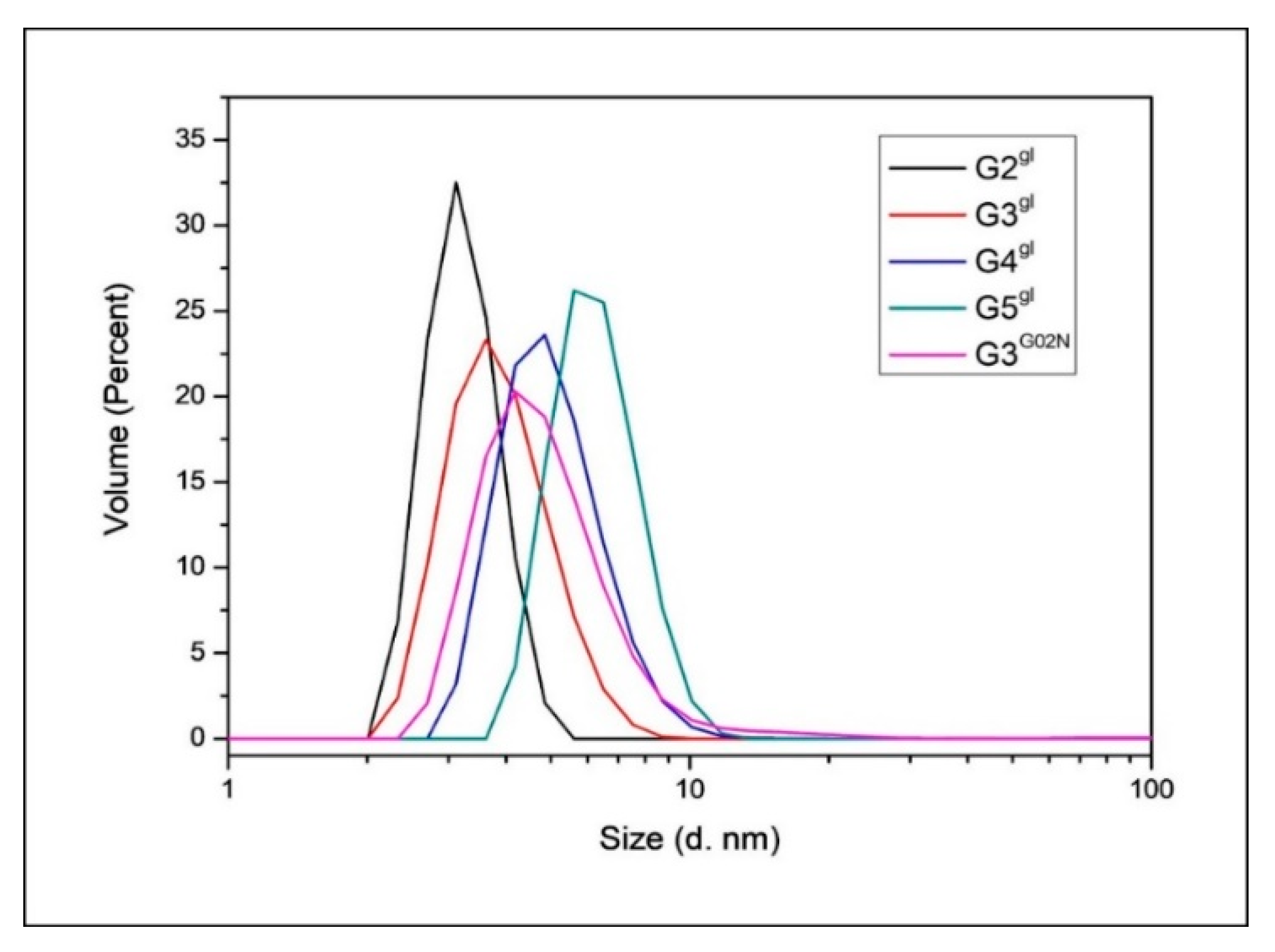
| Species | Diameter [nm] |
|---|---|
| G2gl | 3.24 ± 0.11 |
| G3gl | 3.94 ± 0.11 |
| G4gl | 5.12 ± 0.12 |
| G5gl | 6.35 ± 0.14 |
| G3G0N | 5.07 ± 0.16 |
References
- Dannhardt, G.; Kiefer, W. Cyclooxygenase inhibitors—Current status and future prospects. Eur. J. Med. Chem. 2001, 36, 109–126. [Google Scholar] [CrossRef]
- Subbaramaiah, K.; Dannenberg, A.J. Cyclooxygenase 2: A molecular target for cancer prevention and treatment. Trends Pharmcol. Sci. 2003, 24, 96–102. [Google Scholar] [CrossRef]
- Uram, Ł.; Filipowicz, A.; Misiorek, M.; Pieńkowska, N.; Markowicz, J.; Wałajtys-Rode, E.; Wołowiec, S. Biotinylated PAMAM G3 dendrimer conjugated with celecoxib and/or Fmoc-l-Leucine and its cytotoxicity for normal and cancer human cell lines. Eur. J. Pharm. Sci. 2018, 124, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Uram, Ł.; Filipowicz-Rachwał, A.; Misiorek, M.; Winiarz, A.; Wałajtys-Rode, E.; Wołowiec, S. Synthesis and Different Effects of Biotinylated PAMAM G3 Dendrimer Substituted with Nimesulide in Human Normal Fibroblasts and Squamous Carcinoma Cells. Biomolecules 2019, 9, 437. [Google Scholar] [CrossRef]
- Wang, W.; Xiong, W.; Zhu, Y.; Xu, H.; Yang, X. Protective effect of PEGylation against poly(amidoamine) dendrimer-induced hemolysis of human red blood cells. J. Biomed. Mater. Res. Part. B Appl. Biomater. 2010, 93, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Uram, Ł.; Szuster, M.; Filipowicz, A.; Zaręba, M.; Wałajtys-Rode, E.; Wołowiec, S. Cellular uptake of glucoheptoamidated poly(amidoamine) PAMAM G3 dendrimer with amide-conjugated biotin, a potential carrier of anticancer drugs. Bioorg. Med. Chem. 2017, 25, 706–713. [Google Scholar] [CrossRef]
- Tomalia, D.A.; Uppuluri, S.; Swanson, D.R.; Li, J. Dendrimers as reactive modules for the synthesis of new structure-controlled, higher-complexity megamers. Pure Appl. Chem. 2000, 72, 2343–2358. [Google Scholar] [CrossRef] [Green Version]
- Magalhães, T.M.; Guerra, R.C.; da Silva San Gil, R.A.; Val, A.P.; Simão, R.A.; Soares, B.G.; de Carvalho Mendes, T.; dos Santos Pyrrho, A.; de Sousa, V.P.; Rodrigues-Furtado, V.L. PAMAM dendrimer hydrogel film—Biocompatible material to an efficient dermal delivery of drugs. J. Nanopart. Res. 2017, 19, 277. [Google Scholar]
- Khopade, A.J.; Möhwald, H. Statistical Megamer Morphologies and Materials from PAMAM Dendrimers. Macromol. Rapid Commun. 2005, 26, 445–449. [Google Scholar] [CrossRef]
- van Dongen, M.A.; Vaidyanathan, S.; Banaszak Holl, M.M. PAMAM Dendrimers as Quantized Building Blocks for Novel Nanostructures. Soft Matter 2013, 9. [Google Scholar] [CrossRef]
- Atapour, M.H.; Mojarad, M.; Raoofian, R.; Baghebani, F.; Louie, O.; Massoudi, A.; Soukhtanloo, M.; Hooshang, V. PAMAM Megamer (G2-G2) as a versatile tool in gene delivery. Clin. Biochem. 2011, 13, S281–S282. [Google Scholar] [CrossRef]
- Filipowicz, A.; Wołowiec, S. Bioconjugates of PAMAM dendrimers with trans-retinal, pyridoxal, and pyridoxal phosphate. Int. J. Nanomed. 2012, 7, 4819–4828. [Google Scholar] [Green Version]
- Uram, Ł.; Szuster, M.; Filipowicz, A.; Gargasz, K.; Wołowiec, S.; Wałajtys-Rode, E. Different patterns of nuclear and mitochondrial penetration by the G3 PAMAM dendrimer and its biotin-pyridoxal bioconjugate BC-PAMAM in normal and cancer cells in vitro. Int. J. Nanomed. 2015, 10, 5647–5661. [Google Scholar] [CrossRef] [PubMed]
- Czarnik, J.; Kwaśniak, K.; Tutaj, K.; Filiks, I.; Stompor, M.; Tabarkiewicz, J.; Wołowiec, S.; Uram, Ł. Glucoheptoamidated polyamidoamine PAMAM G3 dendrimer as a vehicle for succinate linked doxorubicin; enhanced toxicity of DOX against grade IV glioblastoma U-118 MG cells. J. Drug Deliv. Sci. Technol. 2019. under review. [Google Scholar]
- Lubczak, J.; Kania, E.; Myśliwiec, B. The Kinetics and Mechanism of the Reaction between Barbituric Acid and Glycidol, Part I: The Products Analysis. Int. J. Chem. Kinet. 2017, 49, 259–266. [Google Scholar] [CrossRef]
- Zhou, Z.; D’Emanuele, A.; Lennon, K.; Attwood, D. Synthesis and Micellization of Linear−Dendritic Copolymers and Their Solubilization Ability for Poorly Water-Soluble Drugs. Macromolecules 2009, 42, 7936–7944. [Google Scholar] [CrossRef]
- Dupont, L.; Pirotte, B.; Masereel, B.; Delarge, J.; Geczy, J. Nimesulide. Acta Crystallogr. Sect. C 1995, 51, 507–509. [Google Scholar] [CrossRef]
- Renard, J.-F.O.; Julémont, F.; de Leval, X.; Pirotte, B. The use of nimesulide and its analogues in cancer chemoprevention. Anticancer Agents Med. Chem. 2006, 6, 233–237. [Google Scholar] [CrossRef]
- Zhong, B.; Cai, X.; Chennamaneni, S.; Yi, X.; Liu, L.; Pink, J.J.; Dowlati, A.; Xu, Y.; Zhou, A.; Su, B. From COX-2 inhibitor nimesulide to potent anti-cancer agent: Synthesis, in vitro, in vivo and pharmacokinetic evaluation. Eur. J. Med. Chem. 2012, 47, 432–444. [Google Scholar] [CrossRef]
- Najlah, M.; Freeman, S.; Attwood, D.; D’Emanuele, A. In vitro evaluation of dendrimer prodrugs for oral drug delivery. Int. J. Pharm. 2007, 336, 183–190. [Google Scholar] [CrossRef]
- Zeng, Y.; Kurokawa, Y.; Win-Shwe, T.-T.; Zeng, Q.; Hirano, S.; Zhang, Z.; Sone, H. Effects of PAMAM dendrimers with various surface functional groups and multiple generations on cytotoxicity and neuronal differentiation using human neural progenitor cells. J. Toxicol. Sci. 2016, 41, 351–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uram, Ł.; Szuster, M.; Gargasz, K.; Filipowicz, A.; Wałajtys-Rode, E.; Wołowiec, S. In vitro cytotoxicity of the ternary PAMAM G3-pyridoxal-biotin bioconjugate. Int. J. Nanomed. 2013, 8, 4707–4720. [Google Scholar]
- van der Pauw, M.T.; Van den Bos, T.; Everts, V.; Beertsen, W. Phagocytosis of fibronectin and collagens type I, III, and V by human gingival and periodontal ligament fibroblasts in vitro. J. Periodontol. 2001, 72, 1340–1347. [Google Scholar] [CrossRef] [PubMed]
- Bronger, H.; König, J.; Kopplow, K.; Steiner, H.-H.; Ahmadi, R.; Herold-Mende, C.; Keppler, D.; Nies, A.T. ABCC drug efflux pumps and organic anion uptake transporters in human gliomas and the blood-tumor barrier. Cancer Res. 2005, 65, 11419–11428. [Google Scholar] [CrossRef] [PubMed]
- Lai, P.S.; Shieh, M.-J.; Pai, C.L.; Wang, C.Y.; Lou, P.P. Studies on the Intracellular Trafficking of PAMAM Dendrimer. In Proceedings of the NSTI Nanotechnology Conference and Trade Show, Anaheim Marriott & Convention Center, Anaheim, CA, USA, 8–12 May 2005; pp. 232–235. [Google Scholar]
- Zhang, J.; Liu, D.; Zhang, M.; Sun, Y.; Zhang, X.; Guan, G.; Zhao, X.; Qiao, M.; Chen, D.; Hu, H. The cellular uptake mechanism, intracellular transportation, and exocytosis of polyamidoamine dendrimers in multidrug-resistant breast cancer cells. Int. J. Nanomed. 2016, 11, 3677–3690. [Google Scholar] [CrossRef] [PubMed]
- Periasamy, J.; Muthuswami, M.; Ramesh, V.; Muthusamy, T.; Jain, A.; Karthikeyan, C.; Trivedi, P.; Kumar, R.S.; Gunasekaran, P.; Rha, S.; et al. Nimesulide and celecoxib inhibits multiple oncogenic pathways in gastric cancer cells. J. Cancer Sci. Ther. 2013, 5, 126–136. [Google Scholar] [CrossRef]
- Hida, T.; Kozaki, K.; Muramatsu, H.; Masuda, A.; Shimizu, S.; Mitsudomi, T.; Sugiura, T.; Ogawa, M.; Takahashi, T. Cyclooxygenase-2 inhibitor induces apoptosis and enhances cytotoxicity of various anticancer agents in non-small cell lung cancer cell lines. Clin. Cancer Res. 2000, 6, 2006–2011. [Google Scholar]
- Khodaie, F.; Khazaei-Poul, Y.; Moini-Zanjani, T. Anti-Proliferative Effects of Piroxicam and Nimesulide on A431 Human Squamous Carcinoma Cell Line. Int. J. Cancer Manag. 2017, 10. [Google Scholar] [CrossRef]
- Parimi, S.; Barnes, T.J.; Callen, D.F.; Prestidge, C.A. Mechanistic Insight into Cell Growth, Internalization, and Cytotoxicity of PAMAM Dendrimers. Biomacromolecules 2010, 11, 382–389. [Google Scholar] [CrossRef]
- Tomalia, D.A.; Huang, B.; Swanson, D.R.; Brothers, H.M.; Klimash, J.W. Structure control within poly(amidoamine) dendrimers: Size, shape and regio-chemical mimicry of globular proteins. Tetrahedron 2003, 59, 3799–3813. [Google Scholar] [CrossRef]
- Uram, Ł.; Nizioł, J.; Maj, P.; Sobich, J.; Rode, W.; Ruman, T. N(4)-[B-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan) methyl]-2′-deoxycytidine as a potential boron delivery agent with respect to glioblastoma. Biomed. Pharm. 2017, 95, 749–755. [Google Scholar] [CrossRef] [PubMed]
- Sekuła, J.; Nizioł, J.; Rode, W.; Ruman, T. Gold nanoparticle-enhanced target (AuNPET) as universal solution for laser desorption/ionization mass spectrometry analysis and imaging of low molecular weight compounds. Anal. Chim. Acta 2015, 875, 61–72. [Google Scholar] [CrossRef] [PubMed]
- CrysAlisPro, Agilent Technologies, Version 1.171.36.21t, Rigaku Oxford Diffraction (Poland): Wrocław, Poland, 2001.
- Sheldrick, G.M. A short history of SHELX. Acta Cryst. AFound. Cryst. 2008, 64, 112–122. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaręba, M.; Sareło, P.; Kopaczyńska, M.; Białońska, A.; Uram, Ł.; Walczak, M.; Aebisher, D.; Wołowiec, S. Mixed-Generation PAMAM G3-G0 Megamer as a Drug Delivery System for Nimesulide: Antitumor Activity of the Conjugate Against Human Squamous Carcinoma and Glioblastoma Cells. Int. J. Mol. Sci. 2019, 20, 4998. https://doi.org/10.3390/ijms20204998
Zaręba M, Sareło P, Kopaczyńska M, Białońska A, Uram Ł, Walczak M, Aebisher D, Wołowiec S. Mixed-Generation PAMAM G3-G0 Megamer as a Drug Delivery System for Nimesulide: Antitumor Activity of the Conjugate Against Human Squamous Carcinoma and Glioblastoma Cells. International Journal of Molecular Sciences. 2019; 20(20):4998. https://doi.org/10.3390/ijms20204998
Chicago/Turabian StyleZaręba, Magdalena, Przemysław Sareło, Marta Kopaczyńska, Agata Białońska, Łukasz Uram, Małgorzata Walczak, David Aebisher, and Stanisław Wołowiec. 2019. "Mixed-Generation PAMAM G3-G0 Megamer as a Drug Delivery System for Nimesulide: Antitumor Activity of the Conjugate Against Human Squamous Carcinoma and Glioblastoma Cells" International Journal of Molecular Sciences 20, no. 20: 4998. https://doi.org/10.3390/ijms20204998
APA StyleZaręba, M., Sareło, P., Kopaczyńska, M., Białońska, A., Uram, Ł., Walczak, M., Aebisher, D., & Wołowiec, S. (2019). Mixed-Generation PAMAM G3-G0 Megamer as a Drug Delivery System for Nimesulide: Antitumor Activity of the Conjugate Against Human Squamous Carcinoma and Glioblastoma Cells. International Journal of Molecular Sciences, 20(20), 4998. https://doi.org/10.3390/ijms20204998