Long Non-Coding RNAs and Related Molecular Pathways in the Pathogenesis of Epilepsy
Abstract
1. Introduction
2. Expression Profiling Studies Firstly Suggested the Role of lncRNA in Epilepsy
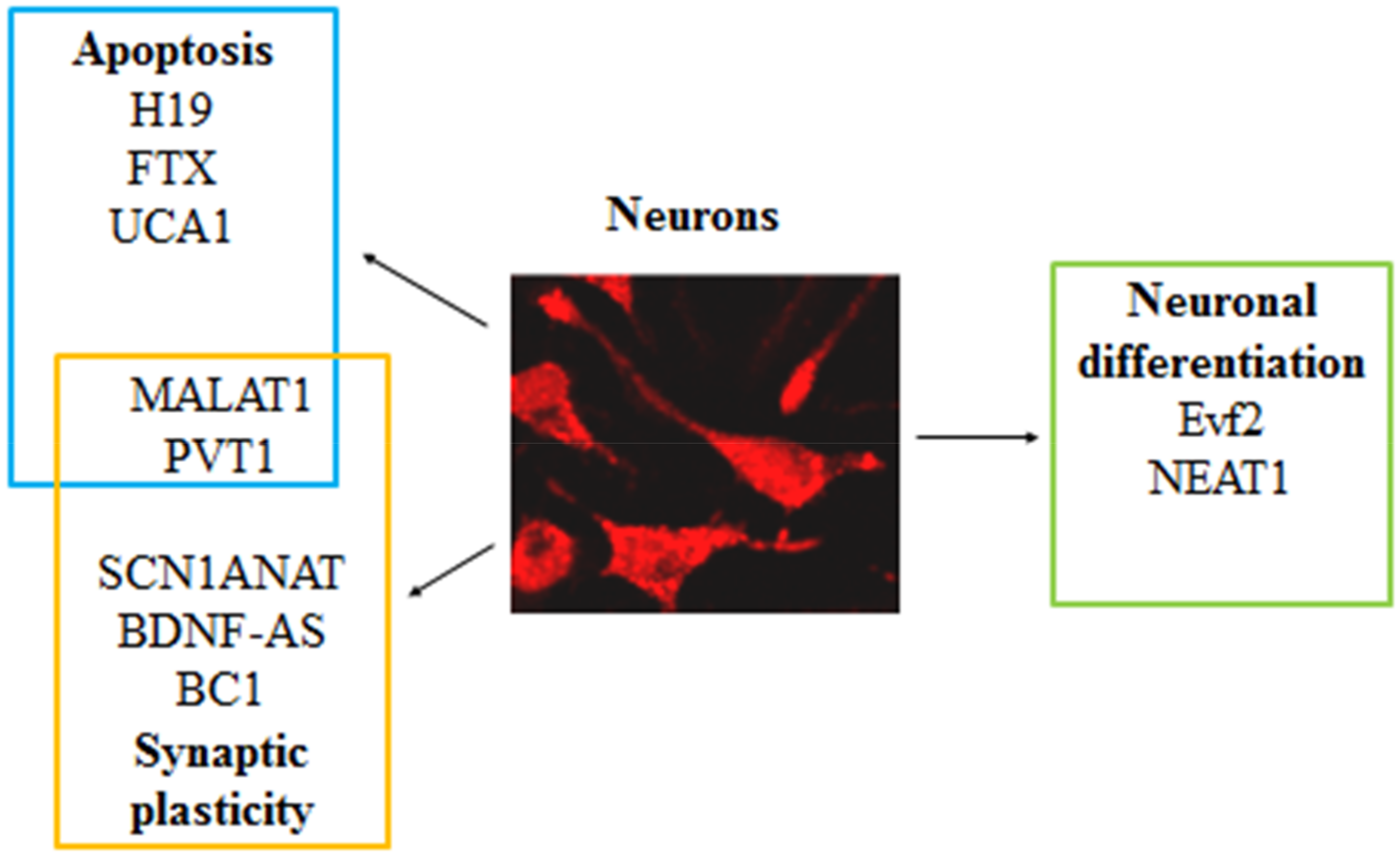
3. LncRNAs Associated with Epilepsy and Involved in Synaptic Plasticity
3.1. Brain Cytoplasmic 1 RNA (BC1)
3.2. Brain-Derived Neurotrophic Factor Antisense RNA (BDNF-AS)
3.3. SCN1ANAT: Natural Antisense Transcript (NAT) of Sodium Voltage-Gated Channel α-1 Subunit (SCN1A)
4. LncRNAs Associated with Epilepsy and Involved in Neuron Apoptosis
4.1. H19
4.2. FTX
4.3. Urothelial Cancer Associated 1 (UCA1)
5. LncRNAs Associated with Epilepsy and Involved in Both Synaptic Plasticity and Apoptosis
5.1. Metastasis-Associated Lung Adenocarcinoma Transcript 1 (MALAT1)
5.2. Plasmacytoma Variant Translocation 1 (PVT1)
6. LncRNAs Associated with Epilepsy and Involved in Neuronal Differentiation
6.1. Evf2
6.2. Nuclear-Enriched Abundant Transcript 1 (NEAT1)
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Qureshi, I.A.; Mattick, J.S.; Mehler, M.F. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010, 1338, 20–35. [Google Scholar] [CrossRef] [PubMed]
- Ulitsky, I.; Bartel, D.P. LincRNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Meng, X.; Jiang, N.; Jin, X.; Zhou, C.; Xu, D.; Gong, Z. Insights into the Noncoding RNA-encoded Peptides. Protein Pept. Lett. 2018, 25, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, C. Coding or Noncoding, the Converging Concepts of RNAs. Front. Genet. 2019, 10, 496. [Google Scholar] [CrossRef] [PubMed]
- Rackham, O.; Shearwood, A.M.; Mercer, T.R.; Davies, S.M.; Mattick, J.S.; Filipovska, A. Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA 2011, 17, 2085–2093. [Google Scholar] [CrossRef] [PubMed]
- Van Heesch, S.; van Iterson, M.; Jacobi, J.; Boymans, S.; Essers, P.B.; de Bruijn, E.; Hao, W.; MacInnes, A.W.; Cuppen, E.; Simonis, M. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 2014, 15, R6. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.W.; Wang, Y.; Chen, L.L. Cellular functions of long noncoding RNAs. Nat. Cell. Biol. 2019, 21, 542–551. [Google Scholar] [CrossRef]
- Cai, Y.; Wan, J. Competing Endogenous RNA Regulations in Neurodegenerative Disorders: Current Challenges and Emerging Insights. Front. Mol. Neurosci. 2018, 11, 370. [Google Scholar] [CrossRef]
- Fernandes, J.C.R.; Acuña, S.M.; Aoki, J.I.; Floeter-Winter, L.M.; Muxel, S.M. Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease. Noncoding RNA 2019, 5, 17. [Google Scholar] [CrossRef]
- Hu, G.; Niu, F.; Humburg, B.A.; Liao, K.; Bendi, S.; Callen, S.; Fox, H.S.; Buch, S. Molecular mechanisms of long noncoding RNAs and their role in disease pathogenesis. Oncotarget 2018, 9, 18648–18663. [Google Scholar] [CrossRef] [PubMed]
- Vance, K.W.; Ponting, C.P. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014, 30, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.Y.; Lin, L.; Soh, B.S.; Stanton, L.W. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. 2013, 29, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Zuo, X.; Deng, H.; Liu, X.; Liu, L.; Ji, A. Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res. Bull. 2013, 97, 69–80. [Google Scholar] [CrossRef]
- Wan, P.; Su, W.; Zhuo, Y. The Role of Long Noncoding RNAs in Neurodegenerative Diseases. Mol. Neurobiol. 2017, 54, 2012–2021. [Google Scholar] [CrossRef] [PubMed]
- Cortini, F.; Roma, F.; Villa, C. Emerging roles of long non-coding RNAs in the pathogenesis of Alzheimer’s disease. Ageing Res. Rev. 2019, 50, 16–26. [Google Scholar] [CrossRef]
- De Boer, H.M.; Mula, M.; Sander, J.W. The global burden and stigma of epilepsy. Epilepsy Behav. 2008, 12, 540–546. [Google Scholar] [CrossRef]
- Gong, X.W.; Li, J.B.; Lu, Q.C.; Liang, P.J.; Zhang, P.M. Effective connectivity of hippocampal neural network and its alteration in Mg2+-free epilepsy model. PLoS ONE 2014, 9, e92961. [Google Scholar] [CrossRef]
- Pitkänen, A.; Lukasiuk, K. Mechanisms of epileptogenesis and potential treatment targets. Lancet Neurol. 2011, 10, 173–186. [Google Scholar] [CrossRef]
- Steinlein, O.K. Genetics and epilepsy. Dialogues Clin. Neurosci. 2008, 10, 29–38. [Google Scholar]
- Henshall, D.C.; Kobow, K. Epigenetics and Epilepsy. Cold Spring Harb. Perspect. Med. 2015, 5, a022731. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.Y.; Moon, J.; Lee, S.T.; Jung, K.H.; Park, D.K.; Yoo, J.S.; Sunwoo, J.S.; Byun, J.I.; Lim, J.A.; Kim, T.J.; et al. Dysregulation of long non-coding RNAs in mouse models of localization-related epilepsy. Biochem. Biophys. Res. Commun. 2015, 462, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Cao, Y.; Long, H.; Luo, Z.; Li, S.; Deng, N.; Wang, J.; Lu, X.; Wang, T.; Ning, S.; et al. Genome-Wide DNA Methylation Patterns Analysis of Noncoding RNAs in Temporal Lobe Epilepsy patients. Mol. Neurobiol. 2018, 55, 793–803. [Google Scholar] [CrossRef]
- Jang, Y.; Moon, J.; Lee, S.T.; Jun, J.S.; Kim, T.J.; Lim, J.A.; Park, B.S.; Yu, J.S.; Park, D.K.; Yang, A.R.; et al. Dysregulated long non-coding RNAs in the temporal lobe epilepsy mouse model. Seizure 2018, 58, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Y.; Li, S.; Lu, J.; Chen, J.; Wang, Y.; Li, Y.; Xu, J.; Li, X. Genome-wide methylome analysis reveals epigenetically dysregulated ncRNAs in human breast cancer. Sci. Rep. 2015, 5, 8790. [Google Scholar] [CrossRef]
- Sosunov, A.A.; Wu, X.; McGovern, R.A.; Coughlin, D.G.; Mikell, C.B.; Goodman, R.R.; McKhann, G.M., 2nd. The mTOR pathway is activated in glial cells in mesial temporal sclerosis. Epilepsia 2012, 53, 78–86. [Google Scholar] [CrossRef]
- Goldberg, E.M.; Coulter, D.A. Mechanisms of epileptogenesis: A convergence on neural circuit dysfunction. Nat. Rev. Neurosci. 2013, 14, 337–349. [Google Scholar] [CrossRef]
- Leal, G.; Comprido, D.; Duarte, C.B. BDNF-induced local protein synthesis and synaptic plasticity. Neuropharmacology 2014, 76 Pt C, 639–656. [Google Scholar] [CrossRef]
- Maag, J.L.; Panja, D.; Sporild, I.; Patil, S.; Kaczorowski, D.C.; Bramham, C.R.; Dinger, M.E.; Wibrand, K. Dynamic expression of long noncoding RNAs and repeat elements in synaptic plasticity. Front. Neurosci. 2015, 9, 351. [Google Scholar] [CrossRef]
- Muddashetty, R.; Khanam, T.; Kondrashov, A.; Bundman, M.; Iacoangeli, A.; Kremerskothen, J.; Duning, K.; Barnekow, A.; Huttenhofer, A.; Tiedge, H.; et al. Poly(A)-binding protein is associated with neuronal BC1 and BC200 ribonucleoprotein particles. J. Mol. Biol. 2002, 321, 433–445. [Google Scholar] [CrossRef]
- Tiedge, H.; Fremeau, R.T., Jr.; Weinstock, P.H.; Arancio, O.; Brosius, J. Dendritic location of neural BC1 RNA. Proc. Natl. Acad. Sci. USA 1991, 88, 2093–2097. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Pestova, T.V.; Hellen, C.U.; Tiedge, H. Translational control by a small RNA: Dendritic BC1 RNA targets the eukaryotic initiation factor4A helicase mechanism. Mol. Cell. Biol. 2008, 28, 3008–3019. [Google Scholar] [CrossRef]
- Zalfa, F.; Adinolfi, S.; Napoli, I.; Kühn-Hölsken, E.; Urlaub, H.; Achsel, T.; Pastore, A.; Bagni, C. Fragile X mental retardation protein (FMRP) binds specifically to the brain cytoplasmic RNAs BC1/BC200 via a novel RNA-binding motif. J. Biol. Chem. 2005, 280, 33403–33410. [Google Scholar] [CrossRef]
- Zhong, J.; Chuang, S.C.; Bianchi, R.; Zhao, W.; Lee, H.; Fenton, A.A.; Wong, R.K.; Tiedge, H. BC1 regulation of metabotropic glutamate receptor-mediated neuronal excitability. J. Neurosci. 2009, 29, 9977–9986. [Google Scholar] [CrossRef]
- Gitaí, D.L.; Fachin, A.L.; Mello, S.S.; Elias, C.F.; Bittencourt, J.C.; Leite, J.P.; Passos, G.A.; Garcia-Cairasco, N.; Paçó-Larson, M.L. The non-coding RNA BC1 is down-regulated in the hippocampus of Wistar Audiogenic Rat (WAR) strain after audiogenic kindling. Brain Res. 2011, 1367, 114–121. [Google Scholar] [CrossRef]
- Zeng, X.; Zong, W.; Gao, Q.; Chen, S.; Chen, L.; Zeng, G.; Huang, W.; Li, Z.; Zeng, C.; Xie, Y.; et al. The Expression Alteration of BC1 RNA and its Interaction with Eukaryotic Translation Initiation Factor eIF4A Post-Status Epilepticus. Neurochem. Res. 2018, 43, 1328–1338. [Google Scholar] [CrossRef]
- Iughetti, L.; Lucaccioni, L.; Fugetto, F.; Predieri, B.; Berardi, A.; Ferrari, F. Brain-derived neurotrophic factor and epilepsy: A systematic review. Neuropeptides 2018, 72, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Modarresi, F.; Faghihi, M.A.; Lopez-Toledano, M.A.; Fatemi, R.P.; Magistri, M.; Brothers, S.P.; van der Brug, M.P.; Wahlestedt, C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 2012, 30, 453–459. [Google Scholar] [CrossRef]
- Lipovich, L.; Dachet, F.; Cai, J.; Bagla, S.; Balan, K.; Jia, H.; Loeb, J.A. Activity-dependent human brain coding/noncoding gene regulatory networks. Genetics 2012, 192, 1133–1148. [Google Scholar] [CrossRef]
- Escayg, A.; Goldin, A.L. Sodium channel SCN1A and epilepsy: Mutations and mechanisms. Epilepsia 2010, 51, 1650–1658. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, J.; Yuan, T.Y.; Tsai, M.S.; Lu, C.Y.; Lin, Y.C.; Lee, M.L.; Lin, S.W.; Chang, F.C.; Liu Pimentel, H.; Olive, C.; et al. Upregulation of Haploinsufficient Gene Expression in the Brain by Targeting a Long Non-coding RNA Improves Seizure Phenotype in a Model of Dravet Syndrome. EBioMedicine 2016, 9, 257–277. [Google Scholar] [CrossRef] [PubMed]
- Dutra, M.R.H.; Feliciano, R.D.S.; Jacinto, K.R.; Gouveia, T.L.F.; Brigidio, E.; Morris, M.; Naffah-Mazzacoratti, M.D.G.; Silva, J.A., Jr. Protective role of UCP2 in oxidative stress and apoptosis during the silent phase of an experimental model of epilepsy induced by pilocarpine. Oxid. Med. Cell. Longev. 2018, 2018, 6736721. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Gao, Y.; Yan, N.; Liu, C.; Zhang, J.G.; Xing, W.M.; Kong, D.M.; Meng, F.G. High-frequency stimulation of the hippocampus protects against seizure activity and hippocampal neuronal apoptosis induced by kainic acid administration in macaques. Neuroscience 2014, 256, 370–378. [Google Scholar] [CrossRef]
- Jiang, X.; Yan, Y.; Hu, M.; Chen, X.; Wang, Y.; Dai, Y.; Wu, D.; Wang, Y.; Zhuang, Z.; Xia, H. Increased level of H19 long noncoding RNA promotes invasion, angiogenesis, and stemness of glioblastoma cells. J. Neurosurg. 2016, 124, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Han, C.L.; Liu, Y.P.; Zhao, X.M.; Wang, K.L.; Chen, N.; Hu, W.; Zhang, J.G.; Ge, M.; Meng, F.G. Whole-transcriptome screening reveals the regulatory targets and functions of long non-coding RNA H19 in epileptic rats. Biophys. Res. Commun. 2017, 489, 262–269. [Google Scholar] [CrossRef]
- Han, C.L.; Ge, M.; Liu, Y.P.; Zhao, X.M.; Wang, K.L.; Chen, N.; Hu, W.; Zhang, J.G.; Li, L.; Meng, F.G. Long non-coding RNA H19 contributes to apoptosis of hippocampal neurons by inhibiting let-7b in a rat model of temporal lobe epilepsy. Cell. Death Dis. 2018, 9, 617. [Google Scholar] [CrossRef] [PubMed]
- Han, C.L.; Ge, M.; Liu, Y.P.; Zhao, X.M.; Wang, K.L.; Chen, N.; Meng, W.J.; Hu, W.; Zhang, J.G.; Li, L.; et al. LncRNA H19 contributes to hippocampal glial cell activation via JAK/STAT signaling in a rat model of temporal lobe epilepsy. J. Neuroinflammation 2018, 15, 103. [Google Scholar] [CrossRef]
- Chureau, C.; Chantalat, S.; Romito, A.; Galvani, A.; Duret, L.; Avner, P.; Rougeulle, C. Ftx is a non-coding RNA which affects Xist expression and chromatin structure within the X-inactivation center Region. Hum. Mol. Genet. 2011, 20, 705–718. [Google Scholar] [CrossRef]
- Zhang, W.; Bi, Y.; Li, J.; Peng, F.; Li, H.; Li, C.; Wang, L.; Ren, F.; Xie, C.; Wang, P.; et al. Long noncoding RNA FTX is upregulated in gliomas and promotes proliferation and invasion of glioma cells by negatively regulating miR-342-3p. Lab. Investig. 2017, 97, 447–457. [Google Scholar] [CrossRef]
- Liu, F.; Yuan, J.H.; Huang, J.F.; Yang, F.; Wang, T.T.; Ma, J.Z.; Zhang, L.; Zhou, C.C.; Wang, F.; Yu, J.; et al. Long noncoding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR-374a. Oncogene 2016, 35, 5422–5434. [Google Scholar] [CrossRef] [PubMed]
- Long, B.; Li, N.; Xu, X.X.; Li, X.X.; Xu, X.J.; Guo, D.; Zhang, D.; Wu, Z.H.; Zhang, S.Y. Long noncoding RNA FTX regulates cardiomyocyte apoptosis by targeting miR-29b-1-5p and Bcl2l2. Biochem. Biophys. Res. Commun. 2018, 495, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Giri, V.; Cui, Y.; Yin, M.; Xian, Z.; Li, J. LncRNA FTX inhibits hippocampal neuron apoptosis by regulating miR-21-5p/SOX7 axis in a rat model of temporal lobe epilepsy. Biochem. Biophys Res. Commun. 2019, 512, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Yi, D.; Liu, Y.; Wang, M.; Zhu, Y.; Shi, H. Long noncoding RNA UCA1 regulates neural Stem cell differentiation by controlling miR1/Hes1 expression. Am. J. Transl. Res. 2017, 9, 3696–3704. [Google Scholar] [PubMed]
- Miller-Delaney, S.F.; Bryan, K.; Das, S.; McKiernan, R.C.; Bray, I.M.; Reynolds, J.P.; Gwinn, R.; Stallings, R.L.; Henshall, D.C. Differential DNA methylation profiles of coding and non-coding genes define hippocampal sclerosis in human temporal lobe epilepsy. Brain 2015, 138, 616–631. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.K.; Yan, H.; Wang, K.; Wang, J. Dynamic regulation effect of long non-coding RNA-UCA1 on NF-kB in hippocampus of epilepsy rats. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 3113–3119. [Google Scholar]
- Wang, W.; Wu, Y.; Zhang, G.; Fang, H.; Wang, H.; Zang, H.; Xie, T.; Wang, W. Activation of Nrf2-ARE signal pathway protects the brain from damage induced by epileptic seizure. Brain Res. 2014, 1544, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Geng, J.F.; Liu, X.; Zhao, H.B.; Fan, W.F.; Geng, J.J.; Liu, X.Z. LncRNA UCA1 inhibits epilepsy and seizure-induced brain injury by regulating miR-495/Nrf2-ARE signal pathway. Int. J. Biochem. Cell. Biol. 2018, 99, 133–139. [Google Scholar] [CrossRef]
- Zhang, X.; Hamblin, M.H.; Yin, K.J. The long noncoding RNA Malat1: Its physiological and pathophysiological functions. RNA Biol. 2017, 14, 1705–1714. [Google Scholar] [CrossRef]
- Bernard, D.; Prasanth, K.V.; Tripathi, V.; Colasse, S.; Nakamura, T.; Xuan, Z.; Zhang, M.Q.; Sedel, F.; Jourdren, L.; Coulpier, F.; et al. A long nuclear-retained non-coding RNA regulates Synaptogenesis by modulating gene expression. EMBO J. 2010, 29, 3082–3093. [Google Scholar] [CrossRef]
- Grant, S.G. Synaptopathies: Diseases of the synaptome. Curr. Opin. Neurobiol. 2012, 22, 522–529. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Yi, X. Down-regulation of Long Noncoding RNA MALAT1 Protects Hippocampal Neurons Against Excessive Autophagy and Apoptosis via the PI3K/Akt Signaling Pathway in Rats with Epilepsy. J. Mol. Neurosci. 2018, 65, 234–245. [Google Scholar] [CrossRef] [PubMed]
- Xie, N.; Cao, L.; Zhao, X.; Jiang, H.; Chi, Z. Diazoxide preconditioning against seizure-induced oxidative injury is via the PI3K/Akt pathway in epileptic rat. Neurosci. Lett. 2011, 495, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Luo, P.; Wang, Q.; Ye, Y.; Wang, B. LncRNA PVT1 in cancer: A review and meta-analysis. Clin. Chim. Acta 2017, 474, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Ding, Y.; Li, M.; Zhou, C.; Lin, W. Silencing lncRNA PVT1 inhibits activation of astrocytes and increases BDNF expression in hippocampus tissues of rats with epilepsy by downregulating the Wnt signaling pathway. J. Cell. Physiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Qureshi, I.A.; Gokhan, S.; Dinger, M.E.; Li, G.; Mattick, J.S.; Mehler, M.F. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Roberts, T.C.; Morris, K.V.; Wood, M.J. The role of long non-coding RNAs in neurodevelopment, brain function and neurological disease. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2014, 369. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Bi, C.; Clark, B.S.; Mady, R.; Shah, P.; Kohtz, J.D. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006, 20, 1474–1480. [Google Scholar] [CrossRef]
- Bond, A.M.; Vangompel, M.J.; Sametsky, E.A.; Clark, M.F.; Savage, J.C.; Disterhoft, J.F.; Kohtz, J.D. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 2009, 12, 1020–1027. [Google Scholar] [CrossRef]
- Cajigas, I.; Chakraborty, A.; Swyter, K.R.; Luo, H.; Bastidas, M.; Nigro, M.; Morris, E.R.; Chen, S.; VanGompel, M.J.W.; Leib, D.; et al. The Evf2 Ultraconserved Enhancer lncRNA Functionally and Spatially Organizes Megabase Distant Genes in the Developing Forebrain. Mol. Cell. 2018, 71, 956–972. [Google Scholar] [CrossRef]
- An, H.; Williams, N.G.; Shelkovnikoava, T.A. NEAT1 and paraspeckles in neurodegenerative diseases: A missing lnc found? Noncoding RNA Res. 2018, 3, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Hempelmann, A.; Taylor, K.P.; Heils, A.; Lorenz, S.; Prud’homme, J.F.; Nabbout, R.; Dulac, O.; Rudolf, G.; Zara, F.; Bianchi, A.; et al. Exploration of the genetic architecture of idiopathic generalized epilepsies. Epilepsia 2006, 47, 1682–1690. [Google Scholar] [CrossRef] [PubMed]
- Barry, G.; Briggs, J.A.; Hwang, D.W.; Nayler, S.P.; Fortuna, P.R.; Jonkhout, N.; Dachet, F.; Maag, J.L.; Mestdagh, P.; Singh, E.M.; et al. The long non-coding RNA NEAT1 is responsive to neuronal activity and is associated with hyperexcitability states. Sci. Rep. 2017, 7, 40127. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Ward, A.J.; Chun, S.; Bennett, C.F.; Beaudet, A.L.; Rigo, F. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature 2015, 518, 409–412. [Google Scholar] [CrossRef]
- Adams, B.D.; Parsons, C.; Walker, L.; Zhang, W.C.; Slack, F.J. Targeting noncoding RNAs in Disease. J. Clin. Investig. 2017, 127, 761–771. [Google Scholar] [CrossRef] [PubMed]
- Miller, T.M.; Pestronk, A.; David, W.; Rothstein, J.; Simpson, E.; Appel, S.H.; Andres, P.L.; Mahoney, K.; Allred, P.; Alexander, K.; et al. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: A phase 1, randomised, first-in-man study. Lancet Neurol. 2013, 12, 435–442. [Google Scholar] [CrossRef]
- Chiriboga, C.A.; Swoboda, K.J.; Darras, B.T.; Iannaccone, S.T.; Montes, J.; De Vivo, D.C.; Norris, D.A.; Bennett, C.F.; Bishop, K.M. Results from a phase 1 study of nusinersen (ISIS-SMN(Rx)) in children with spinal muscular atrophy. Neurology 2016, 86, 890–897. [Google Scholar] [CrossRef]
- Pan, J.-J.; Xie, X.-J.; Li, X.; Chen, W. Long Non-coding RNAs and Drug Resistance. Asian Pac. J. Cancer Prev. 2015, 16, 8067–8073. [Google Scholar] [CrossRef]
- Zhou, X.; Yin, C.; Dang, Y.; Ye, F.; Zhang, G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci. Rep. 2015, 5, 11516. [Google Scholar] [CrossRef]
- Martignano, F.; Rossi, L.; Maugeri, A.; Gallà, V.; Conteduca, V.; De Giorgi, U.; Casadio, V.; Schepisi, G. Urinary RNA-based biomarkers for prostate cancer detection. Clin. Chim. Acta 2017, 473, 96–105. [Google Scholar] [CrossRef]
- Akers, J.C.; Gonda, D.; Kim, R.; Carter, B.S.; Chen, C.C. Biogenesis of extracellular vesicles (EV): Exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J. Neurooncol. 2013, 113, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sarfi, M.; Abbastabar, M.; Khalili, E. Long noncoding RNAs biomarker-based cancer assessment. J. Cell. Physiol. 2019, 2019. [Google Scholar] [CrossRef] [PubMed]
- Kogo, R.; Shimamura, T.; Mimori, K.; Kawahara, K.; Imoto, S.; Sudo, T.; Tanaka, F.; Shibata, K.; Suzuki, A.; Komune, S.; et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011, 71, 6320–6326. [Google Scholar] [CrossRef] [PubMed]
| LncRNA | Model/Tissue, Species | Effects/Findings | References |
|---|---|---|---|
| BC1 | BC1-null model, mouse | lowered seizure threshold | [35] |
| WAR model, rat | decreased levels | [36] | |
| PILO model, rat | altered levels at different time points after SE | [37] | |
| BDNF-AS | resected neocortex from TLE patients, human | decreased levels correlating with upregulated BDNF | [40] |
| SCN1ANAT | DS model, mouse | improved seizure phenotype through its specifically blocking | [42] |
| H19 | KA model, rat | involved in a broad spectrum of epileptogenic processes | [46] |
| resected hippocampus from TLE patients, human | upregulated in the latent period of TLE and contributed to apoptosis by inhibiting let-7b | [47] | |
| KA model, rat | |||
| resected hippocampus from TLE patients, human | involved in microglia activation modulating JAK/STAT signaling | [48] | |
| KA model, rat | |||
| FTX | PILO model, rat | ameliorated seizure activity by inhibiting apoptosis | [53] |
| UCA1 | resected hippocampus from TLE patients, human | abnormally methylated | [55] |
| PILO model, rat | increased expression positively correlating with the nuclear transcription factor NF-kB | [56] | |
| PILO model, rat | suppressed epilepsy by inhibiting apoptosis | [58] | |
| MALAT1 | PILO model, rat | inhibited apoptosis and autophagy by its downregulation | [62] |
| PVT1 | PILO model, rat | decreased neuronal loss and increased BDNF expression through its silencing | [65] |
| Evf2 | knock-out model, mouse | increased susceptibility to more severe and frequently seizures | [70] |
| NEAT1 | resected neocortex from TLE patients, human | upregulated in high activity regions | [40,73] |
| PILO and KA models, rat | transiently downregulated in response to acute activity | [73] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villa, C.; Lavitrano, M.; Combi, R. Long Non-Coding RNAs and Related Molecular Pathways in the Pathogenesis of Epilepsy. Int. J. Mol. Sci. 2019, 20, 4898. https://doi.org/10.3390/ijms20194898
Villa C, Lavitrano M, Combi R. Long Non-Coding RNAs and Related Molecular Pathways in the Pathogenesis of Epilepsy. International Journal of Molecular Sciences. 2019; 20(19):4898. https://doi.org/10.3390/ijms20194898
Chicago/Turabian StyleVilla, Chiara, Marialuisa Lavitrano, and Romina Combi. 2019. "Long Non-Coding RNAs and Related Molecular Pathways in the Pathogenesis of Epilepsy" International Journal of Molecular Sciences 20, no. 19: 4898. https://doi.org/10.3390/ijms20194898
APA StyleVilla, C., Lavitrano, M., & Combi, R. (2019). Long Non-Coding RNAs and Related Molecular Pathways in the Pathogenesis of Epilepsy. International Journal of Molecular Sciences, 20(19), 4898. https://doi.org/10.3390/ijms20194898







