Association of Polymorphisms in Genes Involved in One-Carbon Metabolism with MTHFR Methylation Levels
Abstract
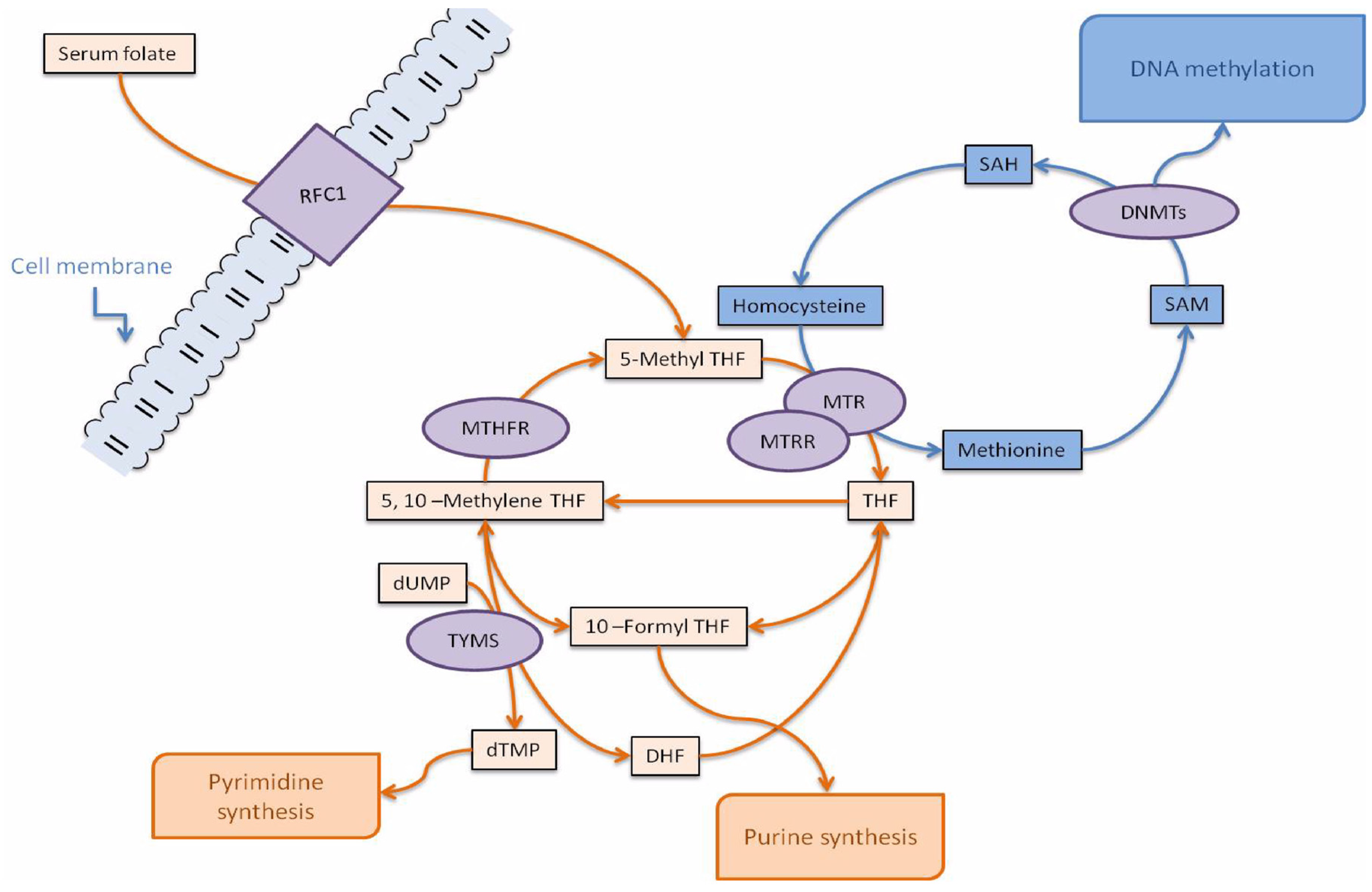
1. Introduction
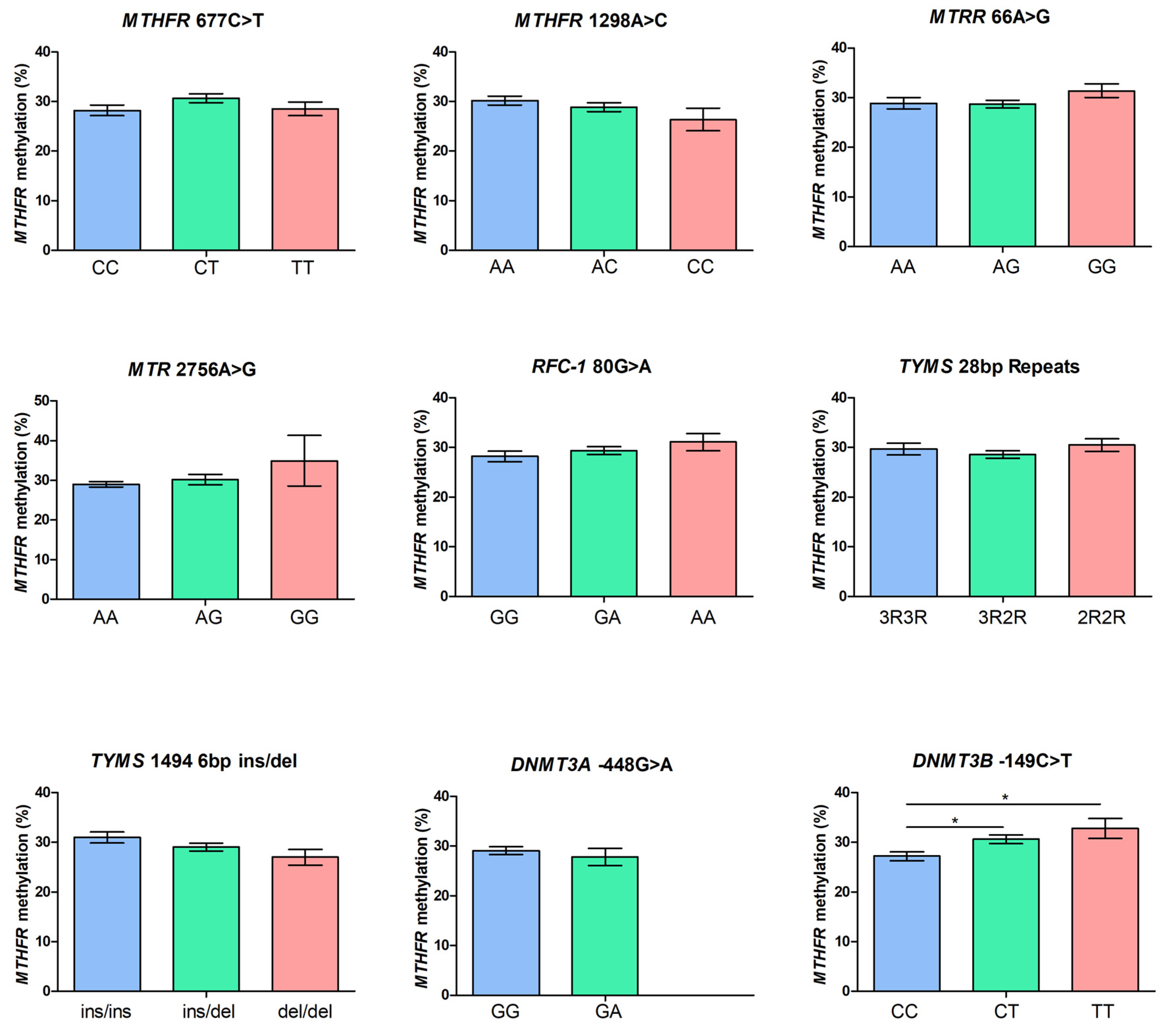
2. Results
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Analysis of MTHFR Methylation Levels
4.3. Analysis of Common Polymorphisms in One-Carbon Metabolism Genes
4.4. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| DNMT | DNA methyltransferase |
| HWE | Hardy-Weinberg equilibrium |
| MS-HRM | Methylation-sensitive high-resolution melting |
| MTHFR | Methylenetetrahydrofolate reductase |
| MTR | Methionine synthase |
| MTRR | Methionine synthase reductase |
| RFC1 | Reduced folate carrier 1 |
| SAM | S-adenosylmethionine |
| SD | Standard deviation |
| SEM | Mean standard error |
| THF | Tetrahydrofolate |
| TYMS | Thymidilate synthase |
| VPA | Valproic acid |
References
- Coppedè, F. The genetics of folate metabolism and maternal risk of birth of a child with Down syndrome and associated congenital heart defects. Front. Genet. 2015, 25, 6–223. [Google Scholar] [CrossRef] [PubMed]
- Huemer, M.; Diodato, D.; Martinelli, D.; Olivieri, G.; Blom, H.; Gleich, F.; Kölker, S.; Kožich, V.; Morris, A.A.; Seifert, B.; et al. Phenotype, treatment practice and outcome in the cobalamin-dependent remethylation disorders and MTHFR deficiency: Data from the E-HOD registry. J. Inherit. Metab. Dis. 2019, 42, 333–352. [Google Scholar] [CrossRef] [PubMed]
- Shane, B.; Pangilinan, F.; Mills, J.L.; Fan, R.; Gong, T.; Cropp, C.D.; Kim, Y.; Ueland, P.M.; Bailey-Wilson, J.E.; Wilson, A.F.; et al. The 677C→T variant of MTHFR is the major genetic modifier of biomarkers of folate status in a young, healthy Irish population. Am. J. Clin. Nutr. 2018, 108, 1334–1341. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, T.K.; Böttiger, A.K.; Henríquez, P.; Serra Majem, L. MTHFR polymorphisms and serum cobalamin affect plasma homocysteine concentrations differentially in females and males. Mol. Med. Rep. 2014, 10, 2706–2712. [Google Scholar] [CrossRef] [PubMed]
- Coppedè, F.; Tannorella, P.; Pezzini, I.; Migheli, F.; Ricci, G.; Caldarazzo lenco, E.; Piaceri, I.; Polini, A.; Nacmias, B.; Monzani, F.; et al. Folate, homocysteine, vitamin B12, and polymorphisms of genes participating in one-carbon metabolism in late-onset Alzheimer’s disease patients and healthy controls. Antioxid. Redox Signal. 2012, 17, 195–204. [Google Scholar] [CrossRef]
- Gupta, N.; Gupta, S.; Dama, M.; David, A.; Khanna, G.; Khanna, A.; Rajender, S. Strong association of 677 C>T substitution in the MTHFR gene with male infertility—A study on an indian population and a meta-analysis. PLoS ONE 2011, 6, e22277. [Google Scholar] [CrossRef]
- Zhang, Y.; He, X.; Xiong, X.; Chuan, J.; Zhong, L.; Chen, G.; Yu, D. The association between maternal methylenetetrahydrofolate reductase C677T and A1298C polymorphism and birth defects and adverse pregnancy outcomes. Prenat. Diagn. 2019, 39, 3–9. [Google Scholar] [CrossRef]
- Zhang, R.; Huo, C.; Wang, X.; Dang, B.; Mu, Y.; Wang, Y. Two Common MTHFR Gene Polymorphisms (C677T and A1298C) and Fetal Congenital Heart Disease Risk: An Updated Meta-Analysis with Trial Sequential Analysis. Cell. Physiol. Biochem. 2018, 45, 2483–2496. [Google Scholar] [CrossRef]
- Meneses-Sanchez, P.; Garcia-Hernandez, S.C.; Porchia, L.M.; Pérez-Fuentes, R.; Torres-Rasgado, E.; Del Angel Soto, A.; Gonzalez-Mejia, M.E. C677T and A1298C methylenetetrahydrofolate reductase polymorphisms and breast cancer susceptibility among Latinos: A meta-analysis. Breast Cancer 2019. [Google Scholar] [CrossRef]
- Shiao, S.P.K.; Lie, A.; Yu, C.H. Meta-analysis of homocysteine-related factors on the risk of colorectal cancer. Oncotarget 2018, 9, 25681–25697. [Google Scholar] [CrossRef]
- Song, Y.; Li, B.; Wang, C.; Wang, P.; Gao, X.; Liu, G. Association between 5,10-Methylenetetrahydrofolate Reductase C677T Gene Polymorphism and Risk of Ischemic Stroke: A Meta-analysis. J. Stroke Cerebrovasc. Dis. 2016, 25, 679–687. [Google Scholar] [CrossRef]
- Yuan, Y.; Shao, W.; Li, Y. Associations between C677T and A1298C polymorphisms of MTHFR and susceptibility to rheumatoid arthritis: A systematic review and meta-analysis. Rheumatol. Int. 2017, 37, 557–569. [Google Scholar] [CrossRef]
- Stoccoro, A.; Tannorella, P.; Salluzzo, M.G.; Ferri, R.; Romano, C.; Nacmias, B.; Siciliano, G.; Migliore, L.; Coppedè, F. The Methylenetetrahydrofolate Reductase C677T Polymorphism and Risk for Late-Onset Alzheimer’s disease: Further Evidence in an Italian Multicenter Study. J. Alzheimers Dis. 2017, 56, 1451–1457. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, L.; Guo, L.; Yu, Q.; Li, H.; Teng, J.; Xie, A. MTHFR C677T and A1298C polymorphisms may contribute to the risk of Parkinson’s disease: A meta-analysis of 19 studies. Neurosci. Lett. 2018, 662, 339–345. [Google Scholar] [CrossRef]
- Khazamipour, N.; Noruzinia, M.; Fatehmanesh, P.; Keyhanee, M.; Pujol, P. MTHFR promoter hypermethylation in testicular biopsies of patients with non-obstructive azoospermia: The role of epigenetics in male infertility. Hum. Reprod. 2009, 24, 2361–2364. [Google Scholar] [CrossRef]
- Wu, W.; Shen, O.; Qin, Y.; Niu, X.; Lu, C.; Xia, Y.; Song, L.; Wang, S.; Wang, X. Idiopathic male infertility is strongly associated with aberrant promoter methylation of methylenetetrahydrofolate reductase (MTHFR). PLoS ONE 2010, 5, e13884. [Google Scholar] [CrossRef]
- Rotondo, J.C.; Bosi, S.; Bazzan, E.; Di Domenico, M.; De Mattei, M.; Selvatici, R.; Patella, A.; Marci, R.; Tognon, M.; Martini, F. Methylenetetrahydrofolate reductase gene promoter hypermethylation in semen samples of infertile couples correlates with recurrent spontaneous abortion. Hum. Reprod. 2012, 27, 3632–3638. [Google Scholar] [CrossRef]
- Saraswathy, K.N.; Kaur, L.; Talwar, S.; Mishra, J.; Huidrom, S.; Sachdeva, M.P.; Puri, M. Methylenetetrahydrofolate Reductase Gene-specific Methylation and Recurrent Miscarriages: A Case-Control Study from North India. J. Hum. Reprod. Sci. 2018, 11, 142–147. [Google Scholar] [CrossRef]
- Ge, J.; Wang, J.; Zhang, F.; Diao, B.; Song, Z.F.; Shan, L.L.; Wang, W.; Cao, H.J.; Li, X.Q. Correlation between MTHFR gene methylation and pre-eclampsia, and its clinical significance. Genet. Mol. Res. 2015, 14, 8021–8028. [Google Scholar] [CrossRef]
- Coppedè, F.; Denaro, M.; Tannorella, P.; Migliore, L. Increased MTHFR promoter methylation in mothers of Down syndrome individuals. Mutat. Res. 2016, 787, 1–6. [Google Scholar] [CrossRef]
- Asim, A.; Agarwal, S.; Panigrahi, I.; Saiyed, N.; Bakshi, S. MTHFR promoter hypermethylation may lead to congenital heart defects in Down syndrome. Intractable Rare Dis. Res. 2017, 6, 295–298. [Google Scholar] [CrossRef]
- Santana Bezerra, H.; Severo de Assis, C.; Dos Santos Nunes, M.K.; Wanderley de Queiroga Evangelista, I.; Modesto Filho, J.; Alves Pegado Gomes, C.N.; Ferreira do Nascimento, R.A.; Pordeus Luna, R.C.; de Carvalho Costa, M.J.; de Oliveira, N.F.P.; et al. The MTHFR promoter hypermethylation pattern associated with the A1298C polymorphism influences lipid parameters and glycemic control in diabetic patients. Diabetol. Metab. Syndr. 2019, 11, 4. [Google Scholar] [CrossRef]
- Dos Santos Nunes, M.K.; Silva, A.S.; de Queiroga Evangelista, I.W.; Filho, J.M.; Gomes, C.N.A.P.; do Nascimento, R.A.F.; Luna, R.C.P.; de Carvalho Costa, M.J.; de Oliveira, N.F.P.; Persuhn, D.C. Hypermethylation in the promoter of the MTHFR gene is associated with diabetic complications and biochemical indicators. Diabetol. Metab. Syndr. 2017, 9, 84. [Google Scholar] [CrossRef]
- Vaissière, T.; Hung, R.J.; Zaridze, D.; Moukeria, A.; Cuenin, C.; Fasolo, V.; Ferro, G.; Paliwal, A.; Hainaut, P.; Brennan, P.; et al. Quantitative analysis of DNA methylation profiles in lung cancer identifies aberrant DNA methylation of specific genes and its association with gender and cancer risk factors. Cancer Res. 2009, 69, 243–252. [Google Scholar] [CrossRef]
- Botezatu, A.; Socolov, D.; Iancu, I.V.; Huica, I.; Plesa, A.; Ungureanu, C.; Anton, G. Methylenetetrahydrofolate reductase (MTHFR) polymorphisms and promoter methylation in cervical oncogenic lesions and cancer. J. Cell. Mol. Med. 2013, 17, 543–549. [Google Scholar] [CrossRef]
- Wei, L.K.; Sutherland, H.; Au, A.; Camilleri, E.; Haupt, L.M.; Gan, S.H.; Griffiths, L.R. A potential epigenetic marker mediating serum folate and vitamin B12 levels contributes to the risk of ischemic stroke. Biomed. Res. Int. 2015, 2015, 167976. [Google Scholar]
- Grossi, E.; Stoccoro, A.; Tannorella, P.; Migliore, L.; Coppedè, F. Artificial Neural Networks Link One-Carbon Metabolism to Gene-Promoter Methylation in Alzheimer’s Disease. J. Alzheimers Dis. 2016, 53, 1517–1522. [Google Scholar] [CrossRef]
- Pauwels, S.; Ghosh, M.; Duca, R.C.; Bekaert, B.; Freson, K.; Huybrechts, I.; Langie, S.A.S.; Koppen, G.; Devlieger, R.; Godderis, L. Maternal intake of methyl-group donors affects DNA methylation of metabolic genes in infants. Clin. Epigenet. 2017, 9, 16. [Google Scholar] [CrossRef]
- McKay, J.A.; Groom, A.; Potter, C.; Coneyworth, L.J.; Ford, D.; Mathers, J.C.; Relton, C.L. Genetic and non-genetic influences during pregnancy on infant global and site specific DNA methylation: Role for folate gene variants and vitamin B12. PLoS ONE 2012, 7, e33290. [Google Scholar] [CrossRef]
- Haggarty, P.; Hoad, G.; Horgan, G.W.; Campbell, D.M. DNA methyltransferase candidate polymorphisms, imprinting methylation, and birth outcome. PLoS ONE 2013, 8, e68896. [Google Scholar] [CrossRef]
- Coppedè, F.; Migheli, F.; Lopomo, A.; Failli, A.; Legitimo, A.; Consolini, R.; Fontanini, G.; Sensi, E.; Servadio, A.; Seccia, M.; et al. Gene promoter methylation in colorectal cancer and healthy adjacent mucosa specimens: Correlation with physiological and pathological characteristics, and with biomarkers of one-carbon metabolism. Epigenetics 2014, 9, 621–633. [Google Scholar] [CrossRef]
- Llanos, A.A.; Marian, C.; Brasky, T.M.; Dumitrescu, R.G.; Liu, Z.; Mason, J.B.; Makambi, K.H.; Spear, S.L.; Kallakury, B.V.; Freudenheim, J.L.; et al. Associations between genetic variation in one-carbon metabolism and LINE-1 DNA methylation in histologically normal breast tissues. Epigenetics 2015, 10, 727–735. [Google Scholar] [CrossRef]
- Song, M.A.; Brasky, T.M.; Marian, C.; Weng, D.Y.; Taslim, C.; Llanos, A.A.; Dumitrescu, R.G.; Liu, Z.; Mason, J.B.; Spear, S.L.; et al. Genetic variation in one-carbon metabolism in relation to genome-wide DNA methylation in breast tissue from heathy women. Carcinogenesis 2016, 37, 471–480. [Google Scholar] [CrossRef]
- Lopomo, A.; Ricciardi, R.; Maestri, M.; De Rosa, A.; Melfi, F.; Lucchi, M.; Mussi, A.; Coppedè, F.; Migliore, L. Gene-Specific Methylation Analysis in Thymomas of Patients with Myasthenia Gravis. Int. J. Mol. Sci. 2016, 17, e2121. [Google Scholar] [CrossRef]
- Tannorella, P.; Stoccoro, A.; Tognoni, G.; Petrozzi, L.; Salluzzo, M.G.; Ragalmuto, A.; Siciliano, G.; Haslberger, A.; Bosco, P.; Bonuccelli, U.; et al. Methylation analysis of multiple genes in blood DNA of Alzheimer’s disease and healthy individuals. Neurosci. Lett. 2015, 600, 143–147. [Google Scholar] [CrossRef]
- Tannorella, P.; Stoccoro, A.; Tognoni, G.; Bonuccelli, U.; Migliore, L.; Coppedè, F. Association study between the DNMT3A -448A>G polymorphism and risk of Alzheimer’s disease in Caucasians of Italian origin. Am. J. Neurodegener. Dis. 2016, 5, 85–93. [Google Scholar]
- Giusti, B.; Saracini, C.; Bolli, P.; Magi, A.; Sestini, I.; Sticchi, E.; Pratesi, G.; Pulli, R.; Pratesi, C.; Abbate, R. Genetic analysis of 56 polymorphisms in 17 genes involved in methionine metabolism in patients with abdominal aortic aneurysm. J. Med. Genet. 2008, 45, 721–730. [Google Scholar] [CrossRef]
- Shen, H.; Wang, L.; Spitz, M.R.; Hong, W.K.; Mao, L.; Wei, Q. A novel polymorphism in human cytosine DNA-methyltransferase-3B promoter is associated with an increased risk of lung cancer. Cancer Res. 2002, 62, 4992–4995. [Google Scholar]
- Xiao, Y.; Word, B.; Hammons, G.; Lyn-Cook, B. Transcriptional activity of DNMT3B in pancreatic cancer cells: Effects of −149 (C→T) promoter polymorphism. Biochem. Biophys. Res. Commun. 2011, 415, 220–223. [Google Scholar] [CrossRef]
- Gagliardi, M.; Strazzullo, M.; Matarazzo, M.R. DNMT3B Functions: Novel Insights from Human Disease. Front. Cell. Dev. Biol. 2018, 6, 140. [Google Scholar] [CrossRef]
- Lai, C.Y.; Huang, C.C.; Tsai, C.H.; Wang, J.Y.; Kerr, C.L.; Chen, Y.Y.; Cai, Y.W.; Wong, R.H. The DNA Methyltransferase 3B -149 Genetic Polymorphism Modulates Lung Cancer Risk from Smoking Asian. Pac. J. Cancer Prev. 2017, 18, 2717–2723. [Google Scholar]
- Duan, F.; Cui, S.; Song, C.; Dai, L.; Zhao, X.; Zhang, X. Systematic evaluation of cancer risk associated with DNMT3B polymorphisms. J. Cancer Res. Clin. Oncol. 2015, 141, 1205–1220. [Google Scholar] [CrossRef]
- Khoram-Abadi, K.M.; Forat-Yazdi, M.; Kheirandish, S.; Saeidi, N.; Zarezade, Z.; Mehrabi, N.; Neamatzadeh, H. DNMT3B -149 C>T and -579 G>T Polymorphisms and Risk of Gastric and Colorectal Cancer: A Meta-analysis. Asian Pac. J. Cancer Prev. 2016, 17, 3015–3020. [Google Scholar]
- Naghibalhossaini, F.; Mokarram, P.; Khalili, E.; Naghibalhossaini, S. DNMT3b -149C/T promoter variants and methylation of colorectal cancer-associated genes. Cancer Biomark. 2015, 15, 227–233. [Google Scholar] [CrossRef]
- Rey, R.; Chauvet-Gelinier, J.C.; Suaud-Chagny, M.F.; Ragot, S.; Bonin, B.; d’Amato, T.; Teyssier, J.R. Distinct Expression Pattern of Epigenetic Machinery Genes in Blood Leucocytes and Brain Cortex of Depressive Patients. Mol. Neurobiol. 2019, 56, 4697–4707. [Google Scholar] [CrossRef]
- Remuzgo-Martínez, S.; Genre, F.; López-Mejías, R.; Ubilla, B.; Mijares, V.; Pina, T.; Corrales, A.; Blanco, R.; Martín, J.; Llorca, J.; et al. Decreased expression of methylene tetrahydrofolate reductase (MTHFR) gene in patients with rheumatoid arthritis. Clin. Exp. Rheumatol. 2016, 34, 106–110. [Google Scholar]
- Coppedè, F.; Bosco, P.; Tannorella, P.; Romano, C.; Antonucci, I.; Stuppia, L.; Romano, C.; Migliore, L. DNMT3B promoter polymorphisms and maternal risk of birth of a child with Down syndrome. Hum. Reprod. 2013, 28, 545–550. [Google Scholar] [CrossRef]
- Jaiswal, S.K.; Sukla, K.K.; Kumari, N.; Lakhotia, A.R.; Kumar, A.; Rai, A.K. Maternal risk for down syndrome and polymorphisms in the promoter region of the DNMT3B gene: A case-control study. Birth Defects Res. A Clin. Mol. Teratol. 2015, 103, 299–305. [Google Scholar] [CrossRef]
- Moura, C.M.; Bastos, P.R.; Ribeiro, J.S.V.; Ribeiro, M.G.; Amorim, M.R.; Costa-Lima, M.A. DNA (cytosine-5)-methyltransferase 3B (DNMT 3B) polymorphism and risk of Down syndrome offspring. Saudi J. Biol. Sci. 2018, 25, 101–104. [Google Scholar] [CrossRef]
- Pesmatzoglou, M.; Lourou, M.; Goulielmos, G.N.; Stiakaki, E. DNA methyltransferase 3B gene promoter and interleukin-1 receptor antagonist polymorphisms in childhood immune thrombocytopenia. Clin. Dev. Immunol. 2012, 2012, 352059. [Google Scholar] [CrossRef]
- Gouda, H.M.; Kamel, N.M.; Meshaal, S.S. Association of DNA Methyltransferase 3B Promotor Polymorphism with Childhood Chronic Immune Thrombocytopenia. Lab. Med. 2016, 47, 312–317. [Google Scholar] [CrossRef]
- Cai, T.T.; Zhang, J.; Wang, X.; Song, R.H.; Qin, Q.; Muhali, F.S.; Zhou, J.Z.; Xu, J.; Zhang, J.A. Gene-gene and gene-sex epistatic interactions of DNMT1; DNMT3A and DNMT3B in autoimmune thyroid disease. Endocr. J. 2016, 63, 643–653. [Google Scholar] [CrossRef]
- Coppedè, F.; Zitarosa, M.T.; Migheli, F.; Lo Gerfo, A.; Bagnoli, S.; Dardano, A.; Nacmias, B.; Mancuso, M.; Monzani, F.; Siciliano, G.; et al. DNMT3B promoter polymorphisms and risk of late onset Alzheimer’s disease. Curr. Alzheimer Res. 2012, 9, 550–554. [Google Scholar] [CrossRef]
- Pezzi, J.C.; Ens, C.M.; Borba, E.M.; Schumacher-Schuh, A.F.; de Andrade, F.M.; Chaves, M.L.; Fiegenbaum, M.; Camozzato, A.L. DNA methyltransferase haplotype is associated with Alzheimer’s disease. Neurosci. Lett. 2014, 579, 70–74. [Google Scholar] [CrossRef]
- Pezzi, J.C.; de Bem, C.M.; da Rocha, T.J.; Schumacher-Schuh, A.F.; Chaves, M.L.; Rieder, C.R.; Hutz, M.H.; Fiegenbaum, M.; Camozzato, A.L. Association between DNA methyltransferase gene polymorphism and Parkinson’s disease. Neurosci. Lett. 2017, 639, 146–150. [Google Scholar] [CrossRef]
- Pan, H.; Shen, J.Y.; Du, J.J.; Cui, S.S.; Liu, J.; Lin, Y.Q.; He, Y.X.; Fu, Y.; Gao, C.; Li, G.; et al. Lack of Association Between DNMT3B Polymorphisms and Sporadic Parkinson’s Disease in a Han Chinese Population. Neurosci. Bull. 2018, 34, 867–869. [Google Scholar] [CrossRef]
- Erichsen, L.; Ghanjati, F.; Beermann, A.; Poyet, C.; Hermanns, T.; Schulz, W.A.; Seifert, H.H.; Wild, P.J.; Buser, L.; Kröning, A.; et al. Aberrant methylated key genes of methyl group metabolism within the molecular etiology of urothelial carcinogenesis. Sci. Rep. 2018, 8, 3477. [Google Scholar] [CrossRef]
- Tang, Q.; Chen, Y.; Wu, W.; Ding, H.; Xia, Y.; Chen, D.; Wang, X. Idiopathic male infertility and polymorphisms in the DNA methyltransferase genes involved in epigenetic marking. Sci. Rep. 2017, 7, 11219. [Google Scholar] [CrossRef]
- Liu, Y.; Zheng, H.; Guo, P.; Feng, S.; Zhou, X.; Ye, D.; Chen, X.; Chen, S. DNA methyltransferase 3A promoter polymorphism is associated with the risk of human spontaneous abortion after assisted reproduction techniques and natural conception. J. Assist. Reprod. Genet. 2017, 34, 245–252. [Google Scholar] [CrossRef]
- Liu, C.H.; Tao, T.; Jiang, L.; Xu, B.; Zhang, L.; Lu, K.; Zhang, X.W.; Chen, S.Q.; Liu, D.C.; Chen, M. DNMT3A-448A>G polymorphism and cancer risk: A meta-analysis. Genet. Mol. Res. 2015, 14, 3640–3649. [Google Scholar] [CrossRef]
- Ni, G.; Qin, J.; Chen, Z.; Li, H.; Zhou, J.; Huang, M.; Zhou, L. Associations between genetic variation in one-carbon metabolism and leukocyte DNA methylation in valproate-treated patients with epilepsy. Clin. Nutr. 2018, 37, 308–312. [Google Scholar] [CrossRef]
- Sharma, T.K.; Vardey, S.K.; Sitaraman, S. Evaluate the Effect of Valproate Monotherapy on the Serum Homocysteine; Folate and Vitamin B12 Levels in Epileptic Children. Clin. Lab. 2015, 61, 933–940. [Google Scholar] [CrossRef]
- Ni, G.; Qin, J.; Li, H.; Chen, Z.; Zhou, Y.; Fang, Z.; Chen, Y.; Zhou, J.; Huang, M.; Zhou, L. Effects of antiepileptic drug monotherapy on one-carbon metabolism and DNA methylation in patients with epilepsy. PLoS ONE 2015, 10, e0125656. [Google Scholar] [CrossRef]
- Migheli, F.; Stoccoro, A.; Coppedè, F.; Wan Omar, W.A.; Failli, A.; Consolini, R.; Seccia, M.; Spisni, R.; Miccoli, P.; Mathers, J.C.; et al. Comparison study of MS-HRM and pyrosequencing techniques for quantification of APC and CDKN2A gene methylation. PLoS ONE 2013, 8, e52501. [Google Scholar] [CrossRef]
- Coppedè, F.; Grossi, E.; Buscema, M.; Migliore, L. Application of artificial neural networks to investigate one-carbon metabolism in Alzheimer’s disease and healthy matched individuals. PLoS ONE 2013, 8, e74012. [Google Scholar] [CrossRef]
| Total Subjects | Age (Mean ± SD) | Gender | MTHFR Methylation (Mean ± SD) |
|---|---|---|---|
| 206 | 71.4 ± 15.4 | M: 67 | 29.3 ± 9.3% |
| F: 139 |
| Polymorphism | Genotypes: N° of Subjects (%) |
|---|---|
| MTHFR 677C>T | CC: 72 (35.0%), CT: 91 (44.2%), TT: 43 (20.8%) |
| MTHFR 1298A>C | AA: 95 (46.1%), AC: 95 (46.1%), CC: 16 (7.8%) |
| MTRR 66A>G | AA: 61 (29.6%), AG: 105 (51.0%), GG: 40 (19.4%) |
| MTR 2756A>G | AA: 157 (76.2%), AG: 47 (22.8%), GG: 2 (1.0%) |
| RFC-1 80G>A | GG: 62 (30%), GA: 113 (54.9%), AA: 31 (15.1%) |
| TYMS 28bp Repeats | 3R3R: 52 (25.2%), 3R2R: 108 (52.4%), 2R2R: 46 (22.4%) |
| TYMS 1494 6bp ins/del | ins/ins:64 (31.1%), ins/del: 108 (52.4%), del/del: 34 (16.5%) |
| DNMT3A -448G>A | GG: 170 (82.3%), GA: 36 (17.7%), AA: 0 (0.0%) |
| DNMT3B -149C>T | CC: 90 (43.7%), CT: 96 (46.6%), TT: 20 (9.7%) |
| Primer Sequences | Ta | Amplicon Lenght | Region | CpG Sites |
|---|---|---|---|---|
| F: 5′-TTTTAATTTTTGTTTGGAGGGTAGT-3′ R: 5′-AAAAAAACCACTTATCACCAAATTC-3′ | 54 °C | 155 bp | From +30 to +184 | 7 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coppedè, F.; Stoccoro, A.; Tannorella, P.; Gallo, R.; Nicolì, V.; Migliore, L. Association of Polymorphisms in Genes Involved in One-Carbon Metabolism with MTHFR Methylation Levels. Int. J. Mol. Sci. 2019, 20, 3754. https://doi.org/10.3390/ijms20153754
Coppedè F, Stoccoro A, Tannorella P, Gallo R, Nicolì V, Migliore L. Association of Polymorphisms in Genes Involved in One-Carbon Metabolism with MTHFR Methylation Levels. International Journal of Molecular Sciences. 2019; 20(15):3754. https://doi.org/10.3390/ijms20153754
Chicago/Turabian StyleCoppedè, Fabio, Andrea Stoccoro, Pierpaola Tannorella, Roberta Gallo, Vanessa Nicolì, and Lucia Migliore. 2019. "Association of Polymorphisms in Genes Involved in One-Carbon Metabolism with MTHFR Methylation Levels" International Journal of Molecular Sciences 20, no. 15: 3754. https://doi.org/10.3390/ijms20153754
APA StyleCoppedè, F., Stoccoro, A., Tannorella, P., Gallo, R., Nicolì, V., & Migliore, L. (2019). Association of Polymorphisms in Genes Involved in One-Carbon Metabolism with MTHFR Methylation Levels. International Journal of Molecular Sciences, 20(15), 3754. https://doi.org/10.3390/ijms20153754






