Novel Phototransformable Fluorescent Protein SAASoti with Unique Photochemical Properties
Abstract
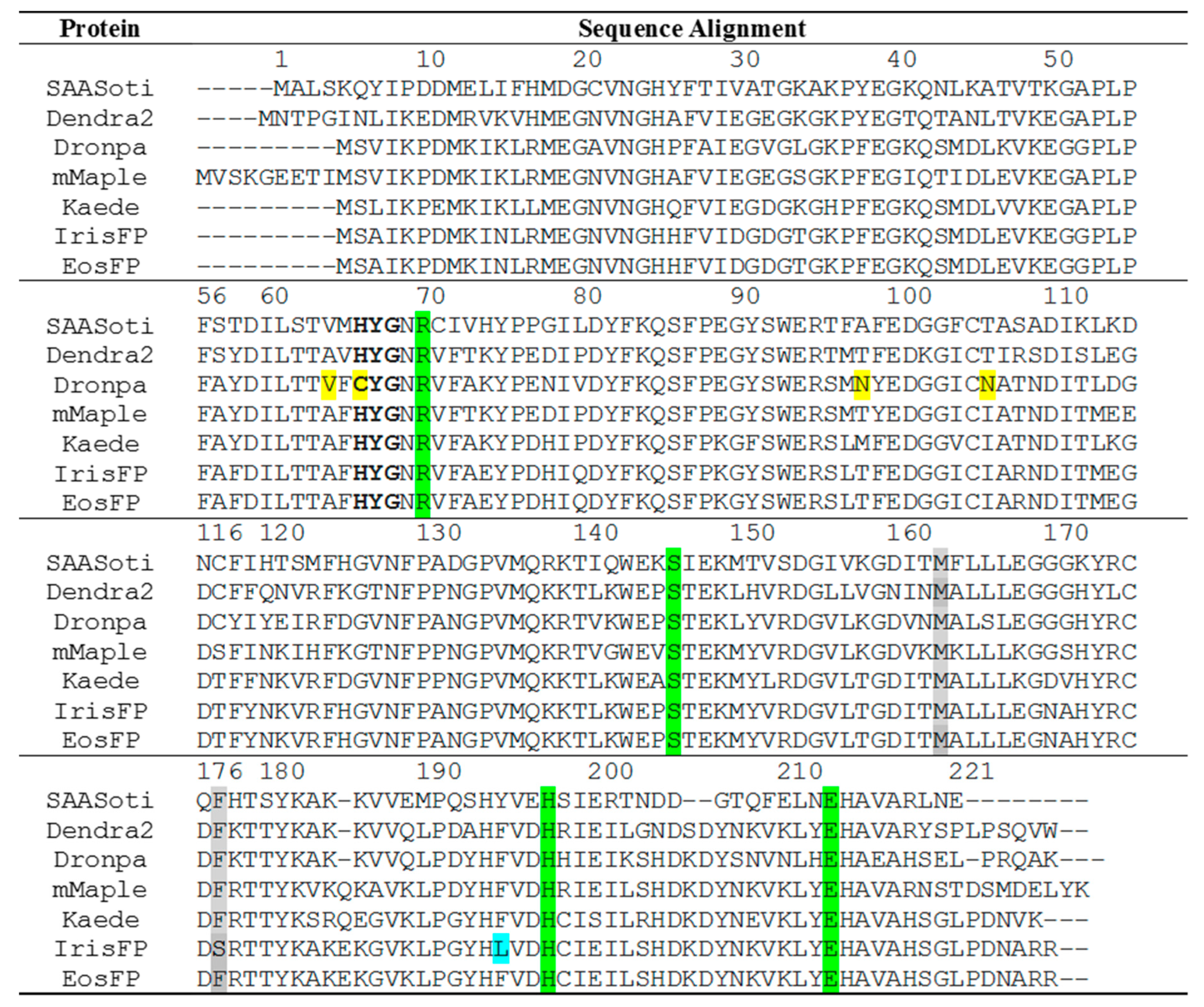
1. Introduction
2. Results
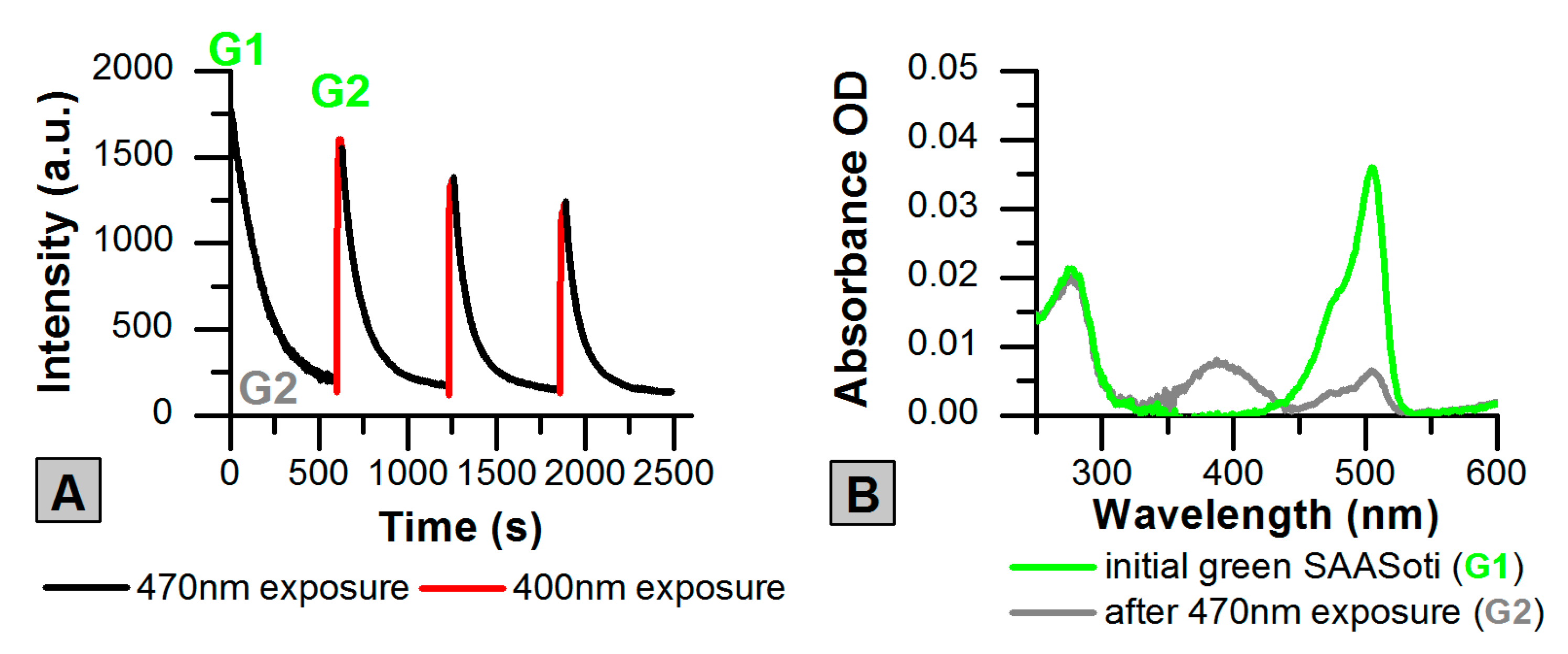
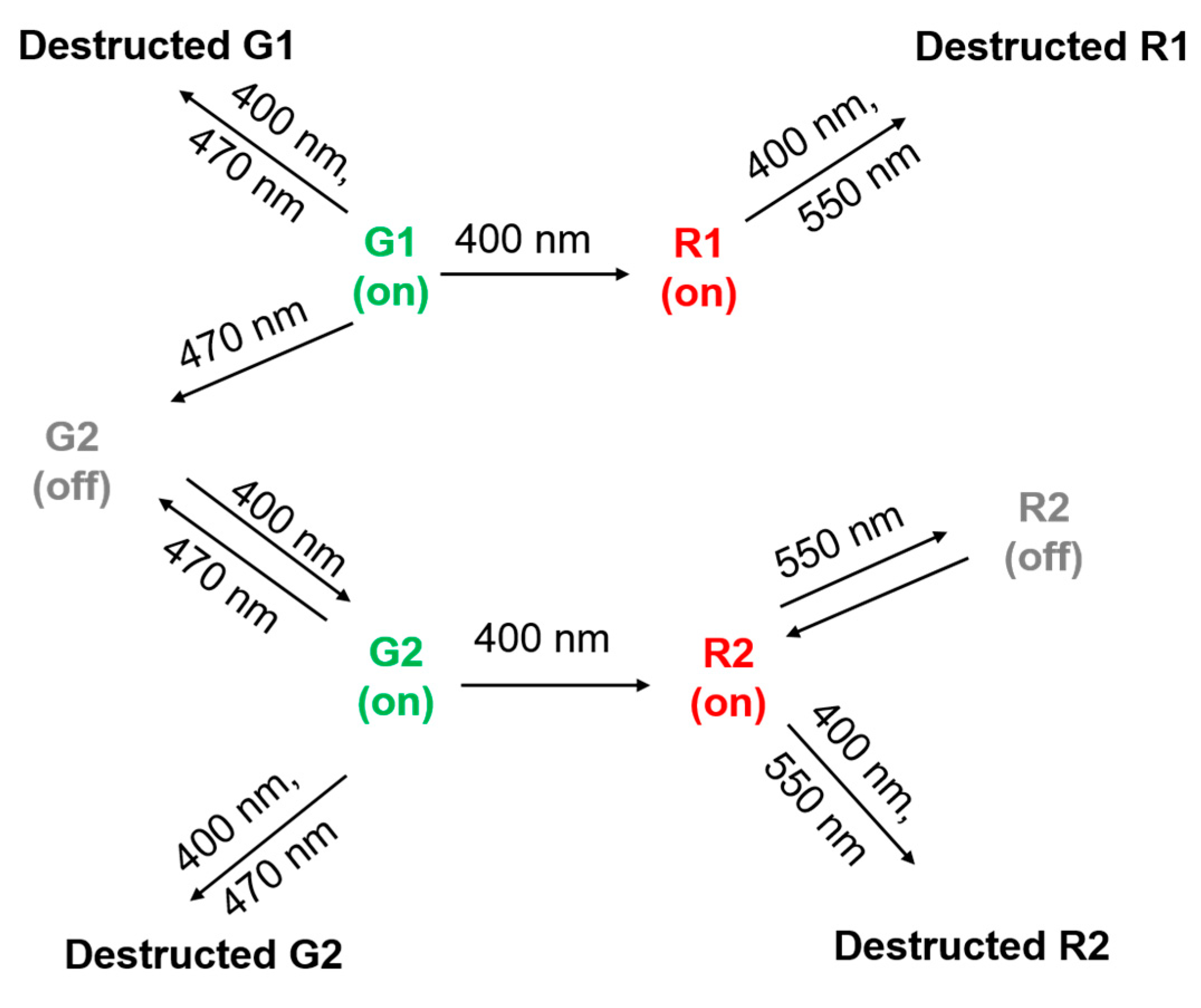
2.1. Photoswitching of the Green Form of SAASoti
2.2. Green-to-Red Photoconversion of SAASoti
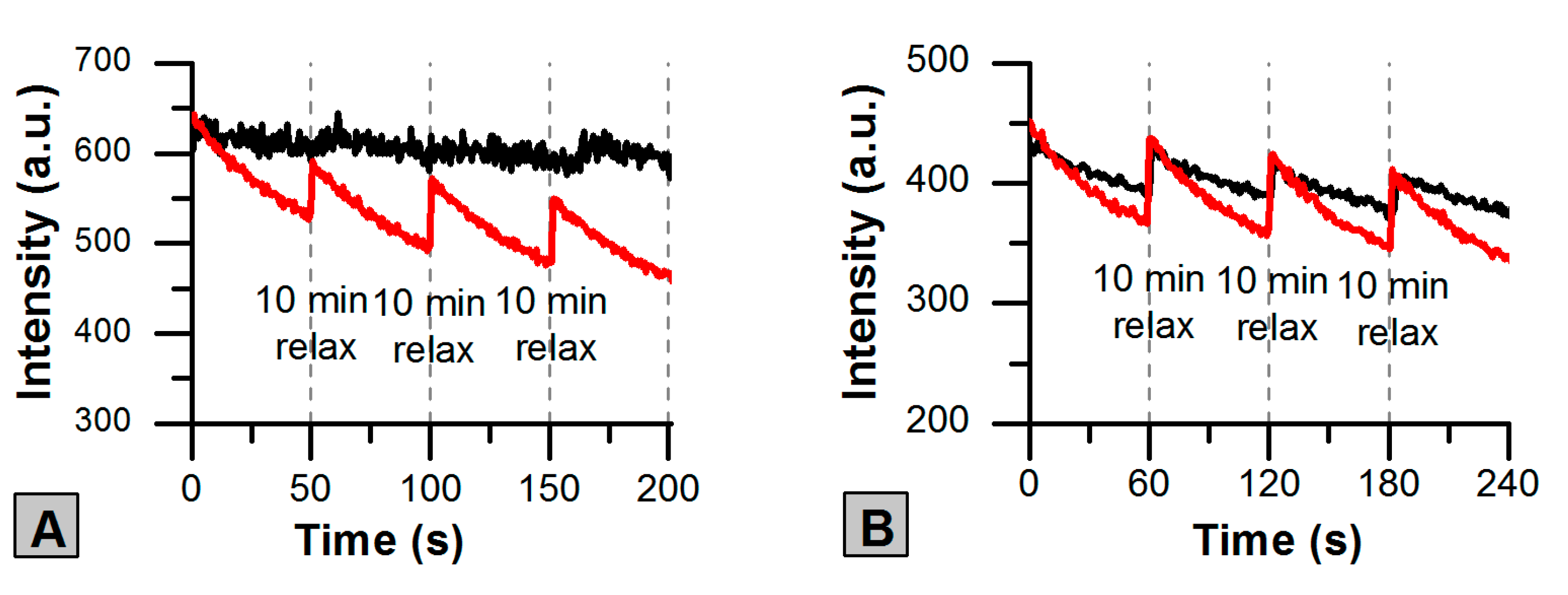
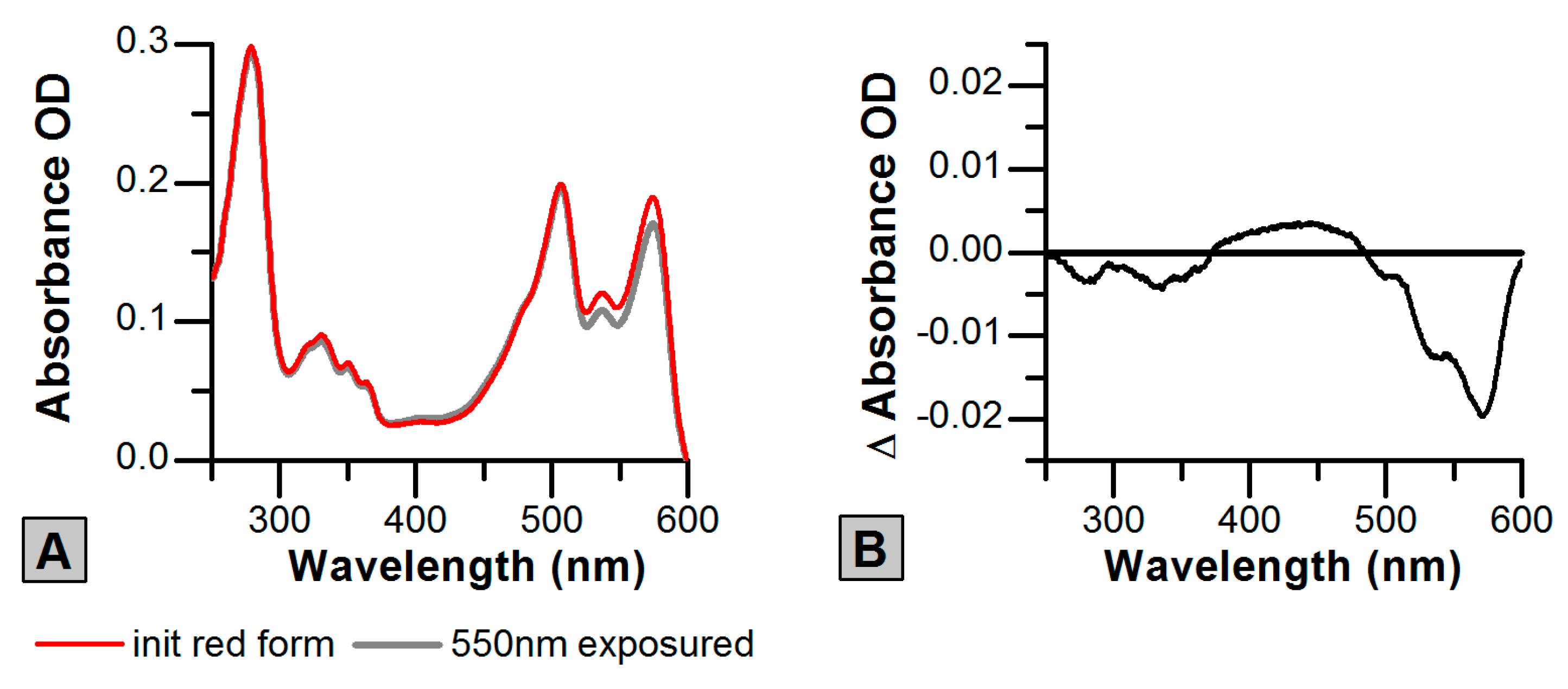
2.3. Photoswitching of the Red SAASoti Form
3. Discussion
4. Materials and Methods
4.1. Protein Expression and Purification
4.2. Absorbance and Fluorescence Measurements
4.3. Mass Spectrometry
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Betzig, E.; Patterson, G.H.; Sougrat, R.; Lindwasser, O.W.; Olenych, S.; Bonifacino, J.S.; Davidson, M.W.; Lippincott-Schwartz, J.; Hess, H.F. Imaging Intracellular Fluorescent Proteins at Nanometer Resolution. Science 2006, 313, 1642–1645. [Google Scholar] [CrossRef] [PubMed]
- Hess, S.T.; Girirajan, T.P.K.; Mason, M.D. Ultra-High Resolution Imaging by Fluorescence Photoactivation Localization Microscopy. Biophys. J. 2006, 91, 4258–4272. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Gurskaya, N.; Merzlyak, E.; Staroverov, D.; Mudrik, N.; Samarkina, O.; Vinokurov, L.; Lukyanov, S.; Lukyanov, K. Method for real-time monitoring of protein degradation at the single cell level. BioTechniques 2007, 42, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, J.; Böhme, S.; Oswald, F.; Hedde, P.N.; Krause, M.; Wiedenmann, J.; Nienhaus, G.U. A photoactivatable marker protein for pulse-chase imaging with superresolution. Nat. Methods 2010, 7, 627–630. [Google Scholar] [CrossRef] [PubMed]
- Patterson, G.H.; Lippincott-Schwartz, J. A Photoactivatable GFP for Selective Photolabeling of Proteins and Cells. Science 2002, 297, 1873–1877. [Google Scholar] [CrossRef] [PubMed]
- Subach, F.V.; Patterson, G.H.; Renz, M.; Lippincott-Schwartz, J.; Verkhusha, V.V. Bright Monomeric Photoactivatable Red Fluorescent Protein for Two-Color Super-Resolution sptPALM of Live Cells. J. Am. Chem. Soc. 2010, 132, 6481–6491. [Google Scholar] [CrossRef] [PubMed]
- Subach, F.V.; Patterson, G.H.; Manley, S.; Gillette, J.M.; Lippincott-Schwartz, J.; Verkhusha, V.V. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat. Methods 2009, 6, 153–159. [Google Scholar] [CrossRef]
- Wiedenmann, J.; Ivanchenko, S.; Oswald, F.; Schmitt, F.; Röcker, C.; Salih, A.; Spindler, K.-D.; Nienhaus, G.U. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc. Natl. Acad. Sci. USA 2004, 101, 15905–15910. [Google Scholar] [CrossRef]
- Ando, R.; Hama, H.; Yamamoto-Hino, M.; Mizuno, H.; Miyawaki, A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc. Natl. Acad. Sci. USA 2002, 99, 12651–12656. [Google Scholar] [CrossRef]
- McEvoy, A.L.; Hoi, H.; Bates, M.; Platonova, E.; Cranfill, P.J.; Baird, M.A.; Davidson, M.W.; Ewers, H.; Liphardt, J.; Campbell, R.E. mMaple: A Photoconvertible Fluorescent Protein for Use in Multiple Imaging Modalities. PLoS ONE 2012, 7, e51314. [Google Scholar] [CrossRef]
- Gurskaya, N.G.; Verkhusha, V.V.; Shcheglov, A.S.; Staroverov, D.B.; Chepurnykh, T.V.; Fradkov, A.F.; Lukyanov, S.; Lukyanov, K.A. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat. Biotechnol. 2006, 24, 461–465. [Google Scholar] [CrossRef] [PubMed]
- Subach, O.M.; Patterson, G.H.; Ting, L.-M.; Wang, Y.; Condeelis, J.S.; Verkhusha, V.V. A photoswitchable orange-to-far-red fluorescent protein, PSmOrange. Nat. Methods 2011, 8, 771–777. [Google Scholar] [CrossRef] [PubMed]
- Dedecker, P.; Hotta, J.-I.; Ando, R.; Miyawaki, A.; Engelborghs, Y.; Hofkens, J. Fast and Reversible Photoswitching of the Fluorescent Protein Dronpa as Evidenced by Fluorescence Correlation Spectroscopy. Biophys. J. 2006, 91, L45–L47. [Google Scholar] [CrossRef][Green Version]
- Ai, H.; Campbell, R. Teal fluorescent proteins: Characterization of a reversibly photoswitchable variant. Proc. SPIE 2008, 6868, 68680D. [Google Scholar]
- Grotjohann, T.; Testa, I.; Leutenegger, M.; Bock, H.; Urban, N.T.; Lavoie-Cardinal, F.; Willig, K.I.; Eggeling, C.; Jakobs, S.; Hell, S.W. Diffraction-unlimited all-optical imaging and writing with a photochromic GFP. Nature 2011, 478, 204–208. [Google Scholar] [CrossRef]
- Brakemann, T.; Stiel, A.C.; Weber, G.; Andresen, M.; Testa, I.; Grotjohann, T.; Leutenegger, M.; Plessmann, U.; Urlaub, H.; Eggeling, C.; et al. A reversibly photoswitchable GFP-like protein with fluorescence excitation decoupled from switching. Nat. Biotechnol. 2011, 29, 942–947. [Google Scholar] [CrossRef]
- Gunewardene, M.S.; Subach, F.V.; Gould, T.J.; Penoncello, G.P.; Gudheti, M.V.; Verkhusha, V.V.; Hess, S.T. Superresolution Imaging of Multiple Fluorescent Proteins with Highly Overlapping Emission Spectra in Living Cells. Biophys. J. 2011, 101, 1522–1528. [Google Scholar] [CrossRef]
- Lukyanov, K.A. Natural Animal Coloration Can be Determined by a Nonfluorescent Green Fluorescent Protein Homolog. J. Boil. Chem. 2000, 275, 25879–25882. [Google Scholar] [CrossRef]
- Adam, V.; Lelimousin, M.; Boehme, S.; Desfonds, G.; Nienhaus, K.; Field, M.J.; Wiedenmann, J.; McSweeney, S.; Nienhaus, G.U.; Bourgeois, D. Structural characterization of IrisFP, an optical highlighter undergoing multiple photo-induced transformations. Proc. Natl. Acad. Sci. USA 2008, 105, 18343–18348. [Google Scholar] [CrossRef]
- Adam, V.; Moeyaert, B.; David, C.C.; Mizuno, H.; Lelimousin, M.; Dedecker, P.; Ando, R.; Miyawaki, A.; Michiels, J.; Engelborghs, Y.; et al. Rational Design of Photoconvertible and Biphotochromic Fluorescent Proteins for Advanced Microscopy Applications. Chem. Boil. 2011, 18, 1241–1251. [Google Scholar] [CrossRef]
- Moeyaert, B.; Bich, N.N.; De Zitter, E.; Rocha, S.; Clays, K.; Mizuno, H.; Van Meervelt, L.; Hofkens, J.; Dedecker, P. Green-to-Red Photoconvertible Dronpa Mutant for Multimodal Super-resolution Fluorescence Microscopy. ACS Nano 2014, 8, 1664–1673. [Google Scholar] [CrossRef] [PubMed]
- Lapshin, G.; Salih, A.; Kolosov, P.; Golovkina, M.; Zavorotnyi, Y.; Ivashina, T.; Vinokurov, L.; Bagratashvili, V.; Savitsky, A. Fluorescence color diversity of great barrier reef corals. J. Innov. Opt. Heal. Sci. 2015, 8, 1550028. [Google Scholar] [CrossRef]
- Solovyev, I.D.; Gavshina, A.V.; Katti, A.S.; Chizhik, A.I.; Vinokurov, L.M.; Lapshin, G.D.; Ivashina, T.V.; Khrenova, M.G.; Kireev, I.I.; Gregor, I.; et al. Monomerization of the photoconvertible fluorescent protein SAASoti by rational mutagenesis of single amino acids. Sci. Rep. 2018, 8, 15542. [Google Scholar] [CrossRef] [PubMed]
- Solovyev, I.; Gavshina, A.; Savitsky, A. Reversible photobleaching of photoconvertible SAASoti-FP. J. Biomed. Photon. Eng. 2017, 3. [Google Scholar] [CrossRef]
- Voliani, V.; Bizzarri, R.; Nifosì, R.; Abbruzzetti, S.; Grandi, E.; Viappiani, C.; Beltram, F. Cis−Trans Photoisomerization of Fluorescent-Protein Chromophores. J. Phys. Chem. B 2008, 112, 10714–10722. [Google Scholar] [CrossRef]
- Klementieva, N.V.; Lukyanov, K.A.; Markina, N.M.; Lukyanov, S.A.; Zagaynova, E.V.; Mishin, A.S. Green-to-red primed conversion of Dendra2 using blue and red lasers. Chem. Commun. 2016, 52, 13144–13146. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.; Adam, V.; Byrdin, M.; Ridard, J.; Kieffer-Jaquinod, S.; Morlot, C.; Arcizet, D.; Demachy, I.; Bourgeois, D. Structural Evidence for a Two-Regime Photobleaching Mechanism in a Reversibly Switchable Fluorescent Protein. J. Am. Chem. Soc. 2013, 135, 15841–15850. [Google Scholar] [CrossRef]
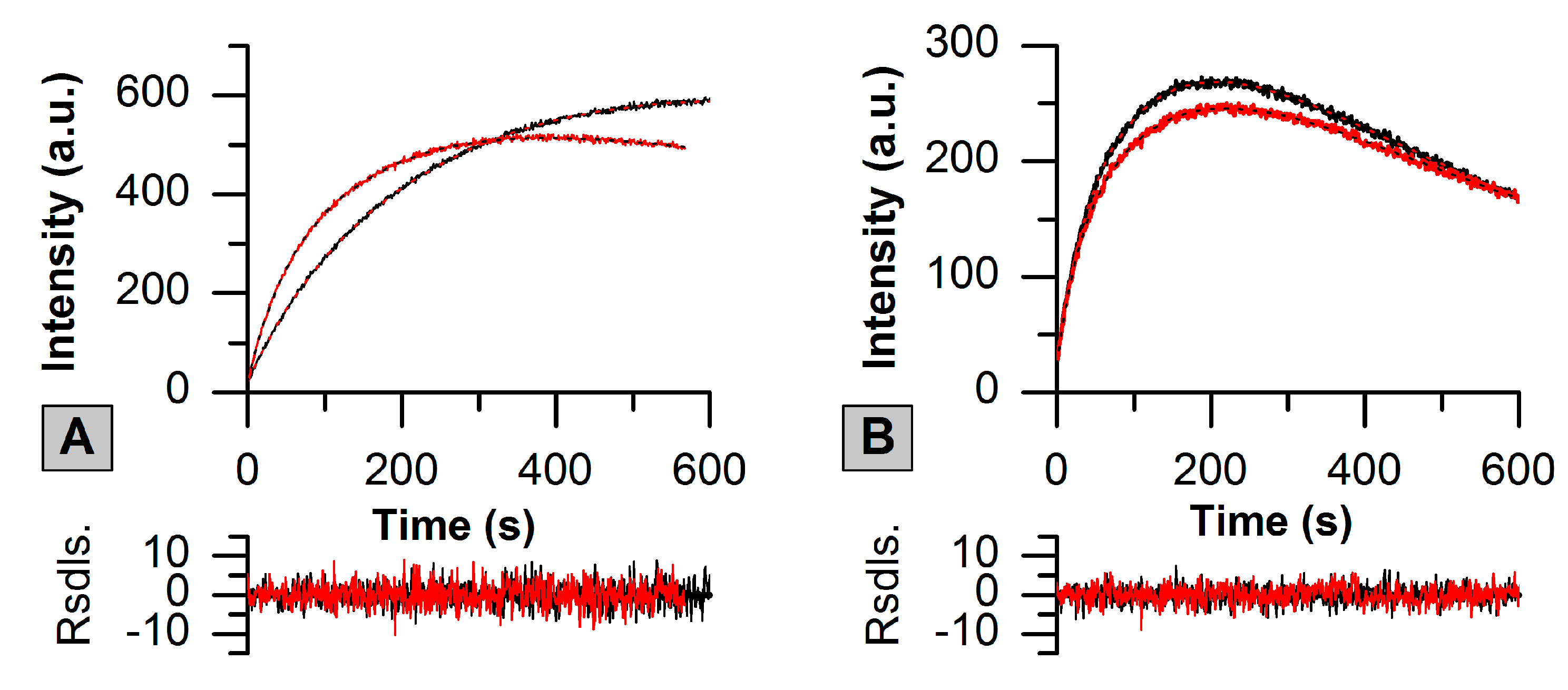
| Conversion | k1 × 103 (s−1) | k2 × 104 (s−1) | k3 × 104 (s−1) | A1 (a.u.) | A2 (a.u.) | A3 (a.u.) |
|---|---|---|---|---|---|---|
| G1 | 13 ± 1 | 24 ± 3 | 8 ± 1 | −110 ± 20 | −1280 ± 240 | 1380 ± 260 |
| G2 | 28 ± 2 | 59 ± 3 | 6 ± 1 | −150 ± 10 | −540 ± 10 | 680 ± 20 |
| ‘old’ G1 | 36 ± 2 | 68 ± 3 | 23 ± 1 | −88 ± 5 | −490 ± 20 | 570 ± 30 |
| ‘old’ G2 | 42 ± 2 | 69 ± 2 | 19 ± 5 | −81 ± 4 | −381 ± 12 | 456 ± 16 |
| Switching | k1 × 102 (s−1) | k2 × 104 (s−1) | A1 (a.u.) | A2 (a.u.) |
|---|---|---|---|---|
| R1 | 5 ± 2 | 12 ± 2 | 6 ± 1 | 352 ± 2 |
| R2 | 24 ± 1 | 6 ± 1 | 140 ± 6 | 470 ± 7 |
| ‘old’ R1 | 13 ± 1 | 3 ± 1 | 52 ± 1 | 326 ±1 |
| ‘old’ R2 | 17 ± 1 | 3 ± 1 | 99 ± 1 | 277 ± 1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Solovyev, I.D.; Gavshina, A.V.; Savitsky, A.P. Novel Phototransformable Fluorescent Protein SAASoti with Unique Photochemical Properties. Int. J. Mol. Sci. 2019, 20, 3399. https://doi.org/10.3390/ijms20143399
Solovyev ID, Gavshina AV, Savitsky AP. Novel Phototransformable Fluorescent Protein SAASoti with Unique Photochemical Properties. International Journal of Molecular Sciences. 2019; 20(14):3399. https://doi.org/10.3390/ijms20143399
Chicago/Turabian StyleSolovyev, Ilya D., Alexandra V. Gavshina, and Alexander P. Savitsky. 2019. "Novel Phototransformable Fluorescent Protein SAASoti with Unique Photochemical Properties" International Journal of Molecular Sciences 20, no. 14: 3399. https://doi.org/10.3390/ijms20143399
APA StyleSolovyev, I. D., Gavshina, A. V., & Savitsky, A. P. (2019). Novel Phototransformable Fluorescent Protein SAASoti with Unique Photochemical Properties. International Journal of Molecular Sciences, 20(14), 3399. https://doi.org/10.3390/ijms20143399





