DNA Supercoiling, Topoisomerases, and Cohesin: Partners in Regulating Chromatin Architecture?
Abstract
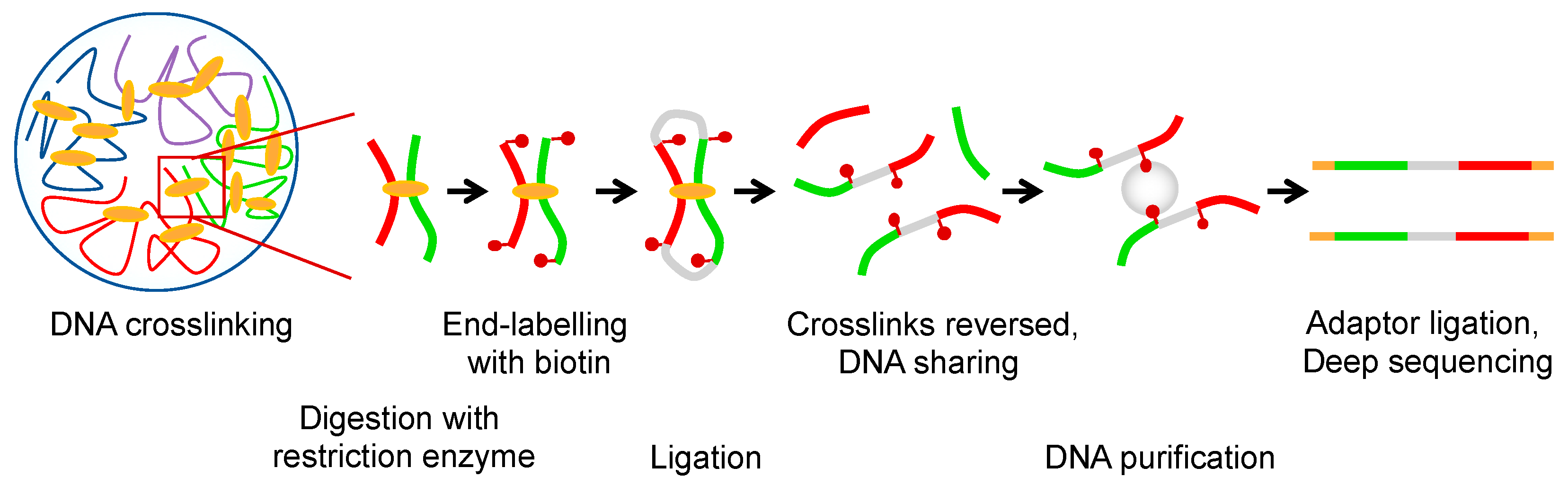
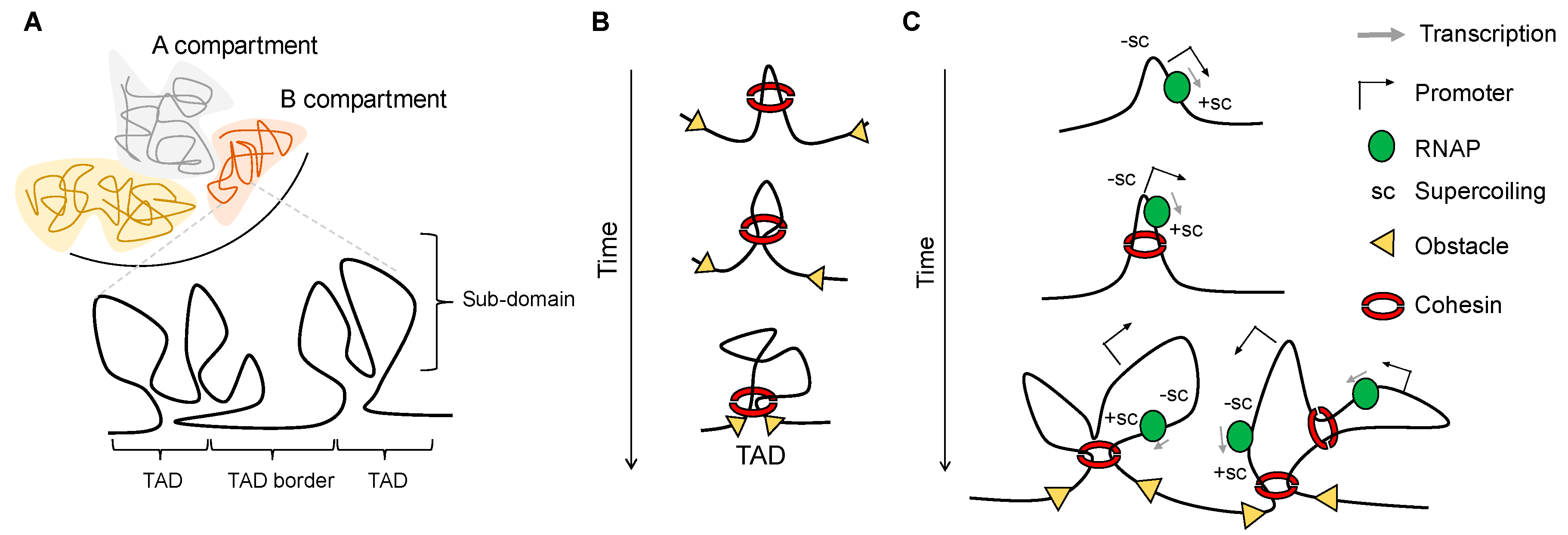
1. Hierarchy and Principles of Genome Organization
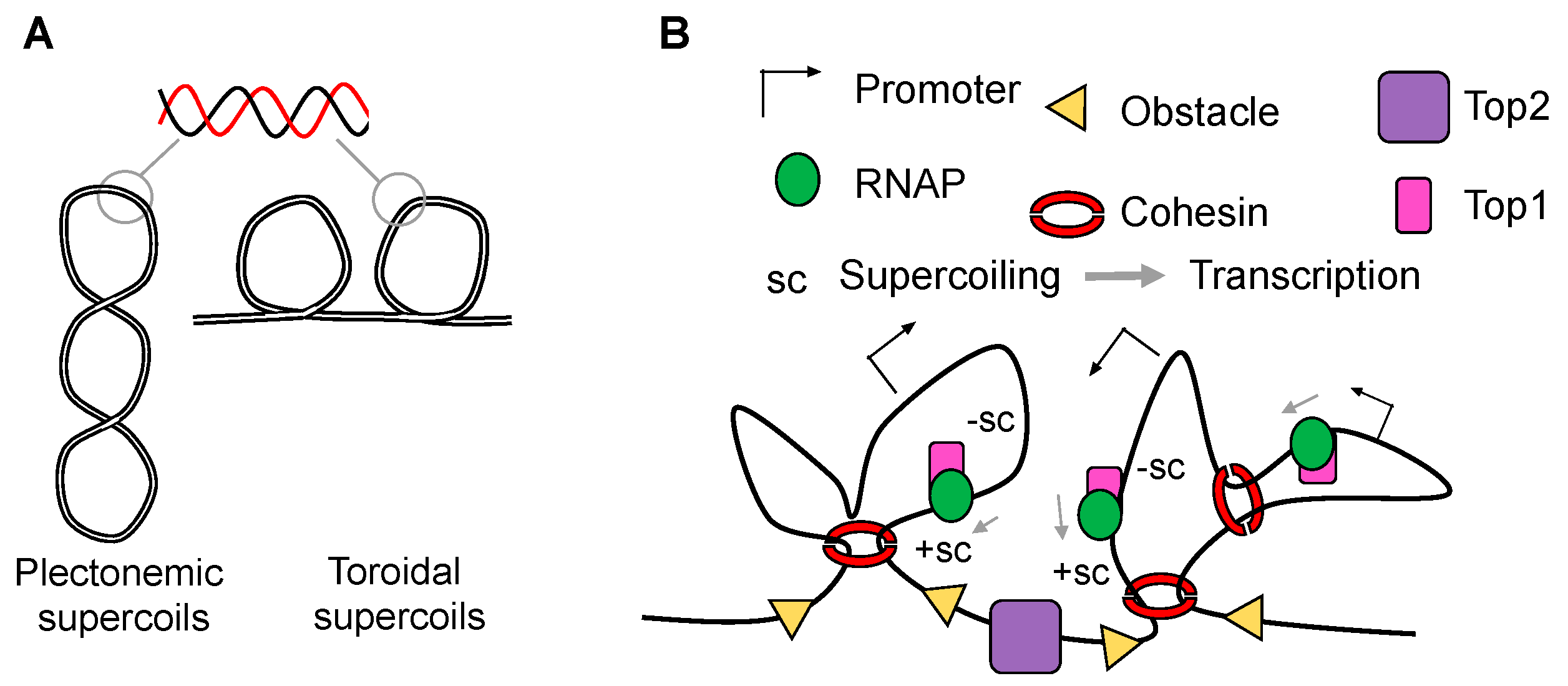
2. Transcription-Generated Supercoiling as a Driving Force for Domain Organization
3. Independent Layers of Chromatin Organization
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Fraser, J.; Williamson, I.; Bickmore, W.A.; Dostie, J. An Overview of Genome Organization and How We Got There: From FISH to Hi-C. Microbiol. Mol. Biol. Rev. 2015, 79, 347–372. [Google Scholar] [CrossRef] [PubMed]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Apostolou, E.; Thanos, D. Virus infection induces NF-κb-dependent interchromosomal associations mediating monoallelic ifn-beta gene expression. Cell 2008, 134, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Cecchini, K.R.; Kim, T.H. Conserved, developmentally regulated mechanism couples chromosomal looping and heterochromatin barrier activity at the homeobox gene a locus. Proc. Natl. Acad. Sci. USA 2011, 108, 7391–7396. [Google Scholar] [CrossRef] [PubMed]
- Rowley, M.J.; Corces, V.G. The three-dimensional genome: Principles and roles of long-distance interactions. Curr. Opin. Cell Biol. 2016, 40, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Dekker, J.; Mirny, L. The 3D genome as moderator of chromosomal communication. Cell 2016, 164, 1110–1121. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Diaz, E.; Corces, V.G. Architectural proteins: Regulators of 3D genome organization in cell fate. Trends Cell Biol. 2014, 24, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Abdullaev, Z.K.; Smith, A.D.; Ching, K.A.; Loukinov, D.I.; Green, R.D.; Zhang, M.Q.; Lobanenkov, V.V.; Ren, B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 2007, 128, 1231–1245. [Google Scholar] [CrossRef] [PubMed]
- Ohlsson, R.; Renkawitz, R.; Lobanenkov, V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001, 17, 520–527. [Google Scholar] [CrossRef]
- Gruber, S.; Haering, C.H.; Nasmyth, K. Chromosomal cohesin forms a ring. Cell 2003, 112, 765–777. [Google Scholar] [CrossRef]
- Haering, C.H.; Lowe, J.; Hochwagen, A.; Nasmyth, K. Molecular architecture of smc proteins and the yeast cohesin complex. Mol. Cell 2002, 9, 773–788. [Google Scholar] [CrossRef]
- Guacci, V.; Koshland, D.; Strunnikov, A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of mcd1 in S. Cerevisiae. Cell 1997, 91, 47–57. [Google Scholar] [CrossRef]
- Michaelis, C.; Ciosk, R.; Nasmyth, K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell 1997, 91, 35–45. [Google Scholar] [CrossRef]
- Hadjur, S.; Williams, L.M.; Ryan, N.K.; Cobb, B.S.; Sexton, T.; Fraser, P.; Fisher, A.G.; Merkenschlager, M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 2009, 460, 410–413. [Google Scholar] [CrossRef] [PubMed]
- Kagey, M.H.; Newman, J.J.; Bilodeau, S.; Zhan, Y.; Orlando, D.A.; van Berkum, N.L.; Ebmeier, C.C.; Goossens, J.; Rahl, P.B.; Levine, S.S.; et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 2010, 467, 430–435. [Google Scholar] [CrossRef] [PubMed]
- Nativio, R.; Wendt, K.S.; Ito, Y.; Huddleston, J.E.; Uribe-Lewis, S.; Woodfine, K.; Krueger, C.; Reik, W.; Peters, J.M.; Murrell, A. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009, 5, e1000739. [Google Scholar] [CrossRef] [PubMed]
- Uuskula-Reimand, L.; Hou, H.; Samavarchi-Tehrani, P.; Rudan, M.V.; Liang, M.; Medina-Rivera, A.; Mohammed, H.; Schmidt, D.; Schwalie, P.; Young, E.J.; et al. Topoisomerase ii β interacts with cohesin and CTCF at topological domain borders. Genome Biol. 2016, 17, 182. [Google Scholar] [PubMed]
- Zuin, J.; Dixon, J.R.; van der Reijden, M.I.; Ye, Z.; Kolovos, P.; Brouwer, R.W.; van de Corput, M.P.; van de Werken, H.J.; Knoch, T.A.; van, I.W.F.; et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl. Acad. Sci. USA 2014, 111, 996–1001. [Google Scholar] [CrossRef] [PubMed]
- Nasmyth, K. Disseminating the genome: Joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 2001, 35, 673–745. [Google Scholar] [CrossRef] [PubMed]
- Alipour, E.; Marko, J.F. Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res. 2012, 40, 11202–11212. [Google Scholar] [CrossRef] [PubMed]
- Fudenberg, G.; Imakaev, M.; Lu, C.; Goloborodko, A.; Abdennur, N.; Mirny, L.A. Formation of chromosomal domains by loop extrusion. Cell Rep. 2016, 15, 2038–2049. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.H.; Weiner, A.; Lajoie, B.; Dekker, J.; Friedman, N.; Rando, O.J. Mapping nucleosome resolution chromosome folding in yeast by micro-C. Cell 2015, 162, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.S.; Huntley, M.H.; Durand, N.C.; Stamenova, E.K.; Bochkov, I.D.; Robinson, J.T.; Sanborn, A.L.; Machol, I.; Omer, A.D.; Lander, E.S.; et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014, 159, 1665–1680. [Google Scholar] [CrossRef] [PubMed]
- Terakawa, T.; Bisht, S.; Eeftens, J.M.; Dekker, C.; Haering, C.H.; Greene, E.C. The condensin complex is a mechanochemical motor that translocates along DNA. Science 2017, 358, 672–676. [Google Scholar] [CrossRef] [PubMed]
- Ganji, M.; Shaltiel, I.A.; Bisht, S.; Kim, E.; Kalichava, A.; Haering, C.H.; Dekker, C. Real-time imaging of DNA loop extrusion by condensing. Science 2018, eaar7831. [Google Scholar] [CrossRef] [PubMed]
- Mizuguchi, T.; Fudenberg, G.; Mehta, S.; Belton, J.M.; Taneja, N.; Folco, H.D.; FitzGerald, P.; Dekker, J.; Mirny, L.; Barrowman, J.; et al. Cohesin-dependent globules and heterochromatin shape 3D genome architecture in S. pombe. Nature 2014, 516, 432–435. [Google Scholar] [CrossRef] [PubMed]
- Busslinger, G.A.; Stocsits, R.R.; van der Lelij, P.; Axelsson, E.; Tedeschi, A.; Galjart, N.; Peters, J.M. Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 2017, 544, 503–507. [Google Scholar] [CrossRef] [PubMed]
- Baranello, L.; Levens, D.; Gupta, A.; Kouzine, F. The importance of being supercoiled: How DNA mechanics regulate dynamic processes. Biochim. Biophys. Acta 2012, 1819, 632–638. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.F.; Wang, J.C. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA 1987, 84, 7024–7027. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.C. Cellular roles of DNA topoisomerases: A molecular perspective. Nat. Rev. Mol. Cell Biol. 2002, 3, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Droge, P. Differential control of transcription-induced and overall DNA supercoiling by eukaryotic topoisomerases in vitro. EMBO J. 1996, 15, 581–589. [Google Scholar] [PubMed]
- Koster, D.A.; Croquette, V.; Dekker, C.; Shuman, S.; Dekker, N.H. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase ib. Nature 2005, 434, 671–674. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Kouzine, F.; Nie, Z.; Chung, H.J.; Elisha-Feil, Z.; Weber, A.; Zhao, K.; Levens, D. The FUSE/FBP/FIR/TFIIH system is a molecular machine programming a pulse of c-myc expression. EMBO J. 2006, 25, 2119–2130. [Google Scholar] [CrossRef] [PubMed]
- Teves, S.S.; Henikoff, S. Transcription-generated torsional stress destabilizes nucleosomes. Nat. Struct. Mol. Biol. 2014, 21, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Kegel, A.; Betts-Lindroos, H.; Kanno, T.; Jeppsson, K.; Strom, L.; Katou, Y.; Itoh, T.; Shirahige, K.; Sjogren, C. Chromosome length influences replication-induced topological stress. Nature 2011, 471, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Decorsiere, A.; Mueller, H.; van Breugel, P.C.; Abdul, F.; Gerossier, L.; Beran, R.K.; Livingston, C.M.; Niu, C.; Fletcher, S.P.; Hantz, O.; et al. Hepatitis b virus x protein identifies the Smc5/6 complex as a host restriction factor. Nature 2016, 531, 386–389. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Bai, L.; Wang, M.D. Transcription under torsion. Science 2013, 340, 1580–1583. [Google Scholar] [CrossRef] [PubMed]
- Kouzine, F.; Sanford, S.; Elisha-Feil, Z.; Levens, D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat. Struct. Mol. Biol. 2008, 15, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Jupe, E.R.; Sinden, R.R.; Cartwright, I.L. Stably maintained microdomain of localized unrestrained supercoiling at a drosophila heat shock gene locus. EMBO J. 1993, 12, 1067–1075. [Google Scholar] [PubMed]
- Joshi, R.S.; Pina, B.; Roca, J. Positional dependence of transcriptional inhibition by DNA torsional stress in yeast chromosomes. EMBO J. 2010, 29, 740–748. [Google Scholar] [CrossRef] [PubMed]
- Kouzine, F.; Gupta, A.; Baranello, L.; Wojtowicz, D.; Ben-Aissa, K.; Liu, J.; Przytycka, T.M.; Levens, D. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat. Struct. Mol. Biol. 2013, 20, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Naughton, C.; Avlonitis, N.; Corless, S.; Prendergast, J.G.; Mati, I.K.; Eijk, P.P.; Cockroft, S.L.; Bradley, M.; Ylstra, B.; Gilbert, N. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat. Struct. Mol. Biol. 2013, 20, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Kouzine, F.; Levens, D.; Baranello, L. DNA topology and transcription. Nucleus 2014, 5, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Baranello, L.; Wojtowicz, D.; Cui, K.; Devaiah, B.N.; Chung, H.J.; Chan-Salis, K.Y.; Guha, R.; Wilson, K.; Zhang, X.; Zhang, H.; et al. RNA polymerase ii regulates topoisomerase 1 activity to favor efficient transcription. Cell 2016, 165, 357–371. [Google Scholar] [CrossRef] [PubMed]
- Sinden, R.R.; Carlson, J.O.; Pettijohn, D.E. Torsional tension in the DNA double helix measured with trimethylpsoralen in living E. coli cells: Analogous measurements in insect and human cells. Cell 1980, 21, 773–783. [Google Scholar] [CrossRef]
- Villeponteau, B.; Lundell, M.; Martinson, H. Torsional stress promotes the DNAase I sensitivity of active genes. Cell 1984, 39, 469–478. [Google Scholar] [CrossRef]
- Bancaud, A.; Conde e Silva, N.; Barbi, M.; Wagner, G.; Allemand, J.F.; Mozziconacci, J.; Lavelle, C.; Croquette, V.; Victor, J.M.; Prunell, A.; et al. Structural plasticity of single chromatin fibers revealed by torsional manipulation. Nat. Struct. Mol. Biol. 2006, 13, 444–450. [Google Scholar] [CrossRef] [PubMed]
- Lavelle, C. DNA torsional stress propagates through chromatin fiber and participates in transcriptional regulation. Nat. Struct Mol. Biol 2008, 15, 123–125. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, F.; Dorier, J.; Burnier, Y.; Stasiak, A. Models that include supercoiling of topological domains reproduce several known features of interphase chromosomes. Nucleic Acids Res. 2014, 42, 2848–2855. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, F.; Dorier, J.; Stasiak, A. Effects of supercoiling on enhancer-promoter contacts. Nucleic Acids Res. 2014, 42, 10425–10432. [Google Scholar] [CrossRef] [PubMed]
- Vernimmen, D.; Bickmore, W.A. The hierarchy of transcriptional activation: From enhancer to promoter. Trends Genet. 2015, 31, 696–708. [Google Scholar] [CrossRef] [PubMed]
- Cozzarelli, N.R.; Wang, J.C. DNA Topology and Its Biological Effects; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1990; 480p. [Google Scholar]
- Puc, J.; Kozbial, P.; Li, W.; Tan, Y.; Liu, Z.; Suter, T.; Ohgi, K.A.; Zhang, J.; Aggarwal, A.K.; Rosenfeld, M.G. Ligand-dependent enhancer activation regulated by topoisomerase-I activity. Cell 2015, 160, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Bansal, K.; Yoshida, H.; Benoist, C.; Mathis, D. The transcriptional regulator Aire binds to and activates super-enhancers. Nat. Immunol. 2017, 18, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Phillips-Cremins, J.E.; Sauria, M.E.; Sanyal, A.; Gerasimova, T.I.; Lajoie, B.R.; Bell, J.S.; Ong, C.T.; Hookway, T.A.; Guo, C.; Sun, Y.; et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 2013, 153, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Arner, E.; Daub, C.O.; Vitting-Seerup, K.; Andersson, R.; Lilje, B.; Drablos, F.; Lennartsson, A.; Ronnerblad, M.; Hrydziuszko, O.; Vitezic, M.; et al. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science 2015, 347, 1010–1014. [Google Scholar] [CrossRef] [PubMed]
- Roca, J.; Wang, J.C. The probabilities of supercoil removal and decatenation by yeast DNA topoisomerase ii. Genes Cells 1996, 1, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Baranello, L.; Kouzine, F.; Levens, D. DNA topoisomerases beyond the standard role. Transcription 2013, 4, 232–237. [Google Scholar] [CrossRef] [PubMed]
- Canela, A.; Maman, Y.; Jung, S.; Wong, N.; Callen, E.; Day, A.; Kieffer-Kwon, K.R.; Pekowska, A.; Zhang, H.; Rao, S.S.P.; et al. Genome organization drives chromosome fragility. Cell 2017, 170, 507–521 e518. [Google Scholar] [CrossRef] [PubMed]
- Miller, E.L.; Hargreaves, D.C.; Kadoch, C.; Chang, C.Y.; Calarco, J.P.; Hodges, C.; Buenrostro, J.D.; Cui, K.; Greenleaf, W.J.; Zhao, K.; et al. Top2 synergizes with Baf chromatin remodeling for both resolution and formation of facultative heterochromatin. Nat. Struct. Mol. Biol. 2017, 24, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, F.; Racko, D.; Dorier, J.; Burnier, Y.; Stasiak, A. Transcription-induced supercoiling explains formation of self-interacting chromatin domains in S. pombe. Nucleic Acids Res. 2017, 45, 9850–9859. [Google Scholar] [CrossRef] [PubMed]
- Racko, D.; Benedetti, F.; Dorier, J.; Stasiak, A. Transcription-induced supercoiling as the driving force of chromatin loop extrusion during formation of tads in interphase chromosomes. Nucleic Acids Res. 2017, 46, 1648–1660. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, X.; Diaz-Ingelmo, O.; Martinez-Garcia, B.; Roca, J. Chromatin regulates DNA torsional energy via topoisomerase ii-mediated relaxation of positive supercoils. EMBO J. 2014, 33, 1492–1501. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Le, T.B.; Imakaev, M.V.; Mirny, L.A.; Laub, M.T. High-resolution mapping of the spatiasl organization of a bacterial chromosome. Science 2013, 342, 731–734. [Google Scholar] [CrossRef] [PubMed]
- Mumbach, M.R.; Rubin, A.J.; Flynn, R.A.; Dai, C.; Khavari, P.A.; Greenleaf, W.J.; Chang, H.Y. Hichip: Efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 2016, 13, 919–922. [Google Scholar] [CrossRef] [PubMed]
- Rowley, M.J.; Nichols, M.H.; Lyu, X.; Ando-Kuri, M.; Rivera, I.S.M.; Hermetz, K.; Wang, P.; Ruan, Y.; Corces, V.G. Evolutionarily conserved principles predict 3D chromatin organization. Mol. Cell 2017, 67, 837–852.e837. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Sun, Q.; Czajkowsky, D.M.; Shao, Z. Sub-kb Hi-C in D. melanogaster reveals conserved characteristics of tads between insect and mammalian cells. Nat. Commun. 2018, 9, 188. [Google Scholar] [CrossRef] [PubMed]
- Schwarzer, W.; Abdennur, N.; Goloborodko, A.; Pekowska, A.; Fudenberg, G.; Loe-Mie, Y.; Fonseca, N.A.; Huber, W.; C, H.H.; Mirny, L.; et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature 2017, 551, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.S.P.; Huang, S.C.; Glenn St Hilaire, B.; Engreitz, J.M.; Perez, E.M.; Kieffer-Kwon, K.R.; Sanborn, A.L.; Johnstone, S.E.; Bascom, G.D.; Bochkov, I.D.; et al. Cohesin loss eliminates all loop domains. Cell 2017, 171, 305–320.e324. [Google Scholar] [CrossRef] [PubMed]
- Hnisz, D.; Shrinivas, K.; Young, R.A.; Chakraborty, A.K.; Sharp, P.A. A phase separation model for transcriptional control. Cell 2017, 169, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Strom, A.R.; Emelyanov, A.V.; Mir, M.; Fyodorov, D.V.; Darzacq, X.; Karpen, G.H. Phase separation drives heterochromatin domain formation. Nature 2017, 547, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Whyte, W.A.; Orlando, D.A.; Hnisz, D.; Abraham, B.J.; Lin, C.Y.; Kagey, M.H.; Rahl, P.B.; Lee, T.I.; Young, R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013, 153, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Finan, K.; Cook, P.R.; Marenduzzo, D. Non-specific (entropic) forces as major determinants of the structure of mammalian chromosomes. Chromosome Res. 2011, 19, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Weber, S.C.; Brangwynne, C.P. Inverse size scaling of the nucleolus by a concentration-dependent phase transition. Curr. Biol. 2015, 25, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Kouzine, F.; Wojtowicz, D.; Baranello, L.; Yamane, A.; Nelson, S.; Resch, W.; Kieffer-Kwon, K.R.; Benham, C.J.; Casellas, R.; Przytycka, T.M.; et al. Permanganate/s1 nuclease footprinting reveals non-b DNA structures with regulatory potential across a mammalian genome. Cell Syst. 2017, 4, 344–356. [Google Scholar] [CrossRef] [PubMed]
- Tomonaga, T.; Michelotti, G.A.; Libutti, D.; Uy, A.; Sauer, B.; Levens, D. Unrestraining genetic processes with a protein-DNA hinge. Mol. Cell 1998, 1, 759–764. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Björkegren, C.; Baranello, L. DNA Supercoiling, Topoisomerases, and Cohesin: Partners in Regulating Chromatin Architecture? Int. J. Mol. Sci. 2018, 19, 884. https://doi.org/10.3390/ijms19030884
Björkegren C, Baranello L. DNA Supercoiling, Topoisomerases, and Cohesin: Partners in Regulating Chromatin Architecture? International Journal of Molecular Sciences. 2018; 19(3):884. https://doi.org/10.3390/ijms19030884
Chicago/Turabian StyleBjörkegren, Camilla, and Laura Baranello. 2018. "DNA Supercoiling, Topoisomerases, and Cohesin: Partners in Regulating Chromatin Architecture?" International Journal of Molecular Sciences 19, no. 3: 884. https://doi.org/10.3390/ijms19030884
APA StyleBjörkegren, C., & Baranello, L. (2018). DNA Supercoiling, Topoisomerases, and Cohesin: Partners in Regulating Chromatin Architecture? International Journal of Molecular Sciences, 19(3), 884. https://doi.org/10.3390/ijms19030884




