Transcriptome Analysis of Long Non-Coding RNA in the Bovine Mammary Gland Following Dietary Supplementation with Linseed Oil and Safflower Oil
Abstract
1. Introduction
2. Results
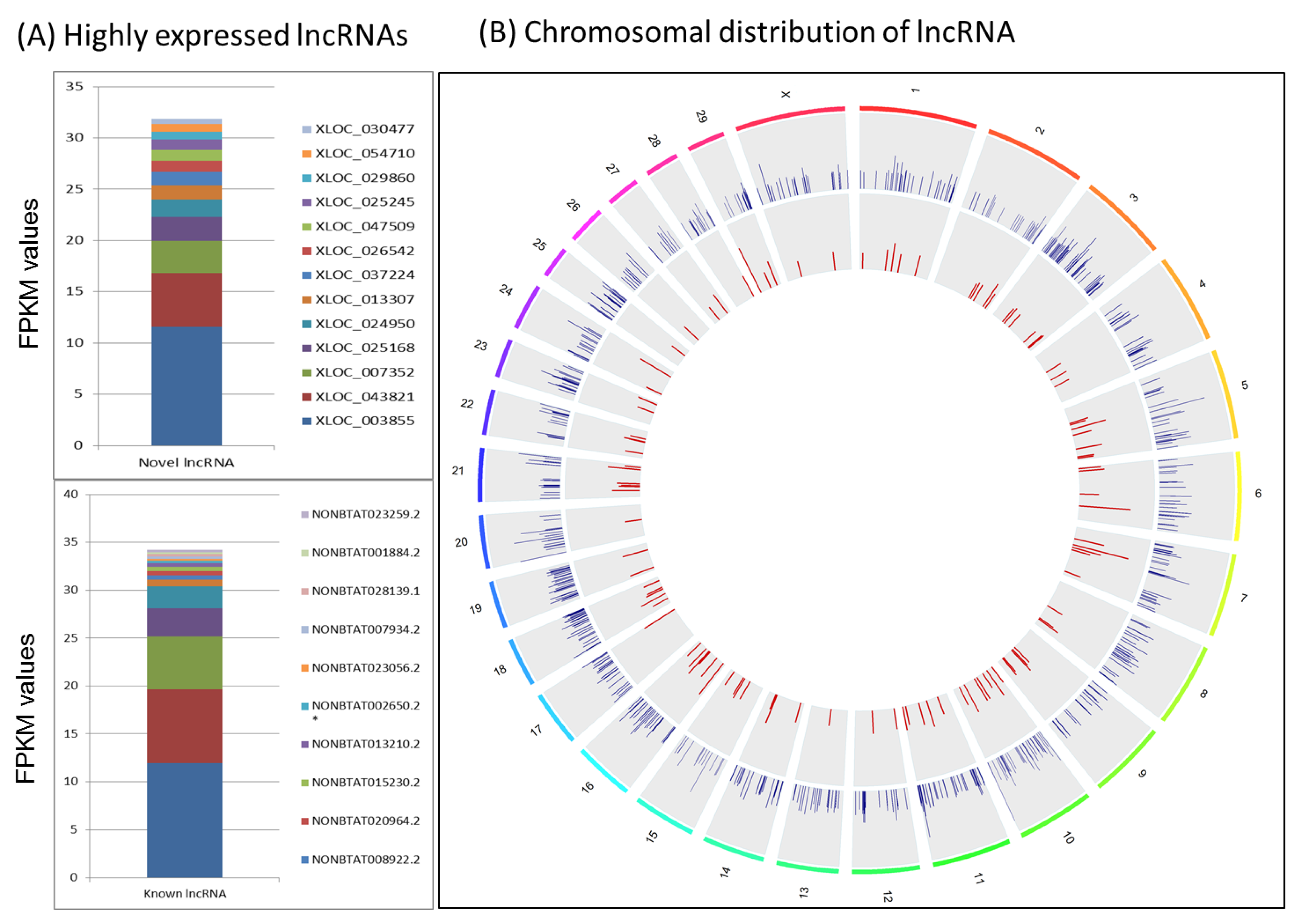
2.1. Expressed LncRNAs in the Bovine Mammary Gland
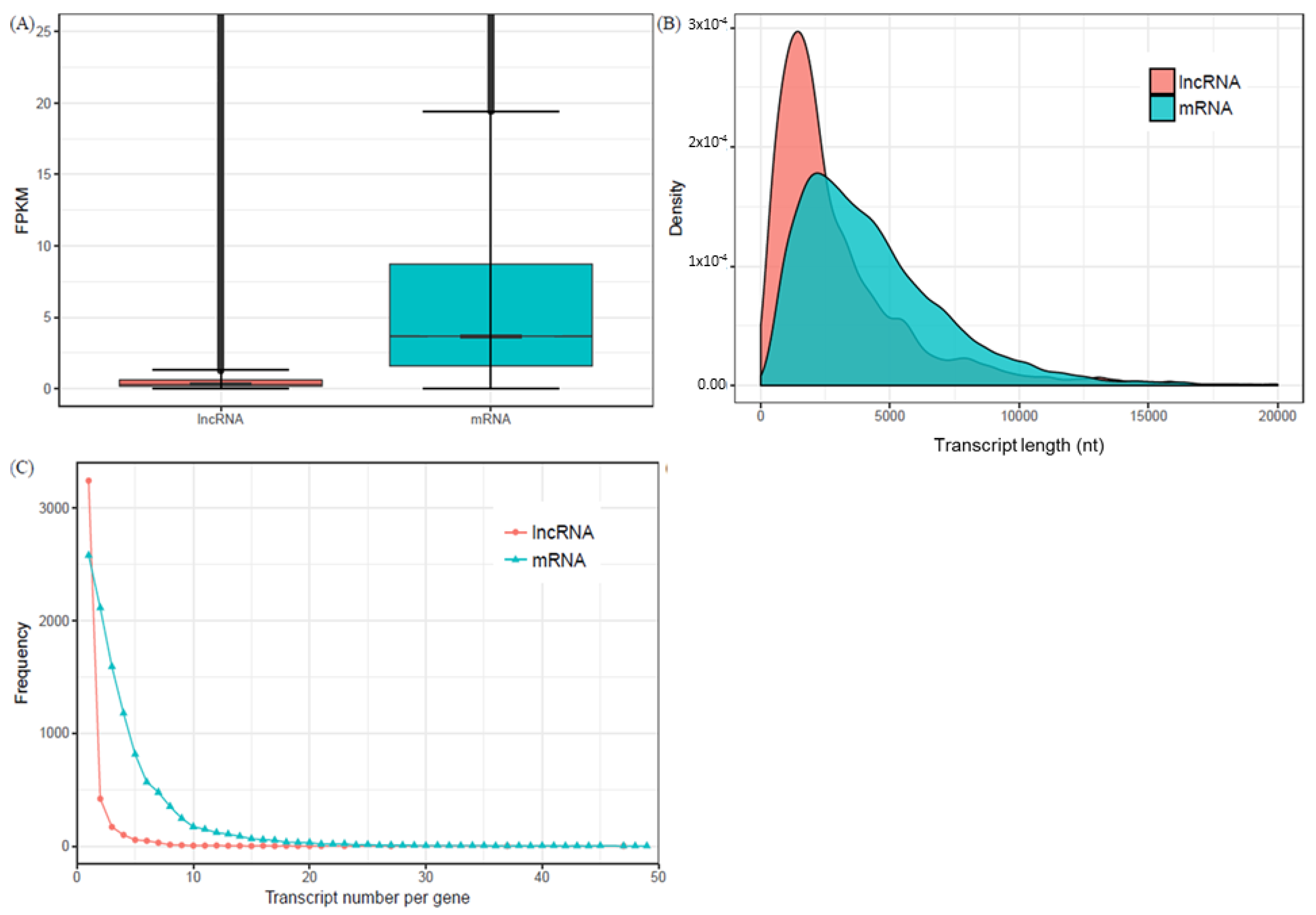
2.2. Characteristics of Expressed LncRNAs
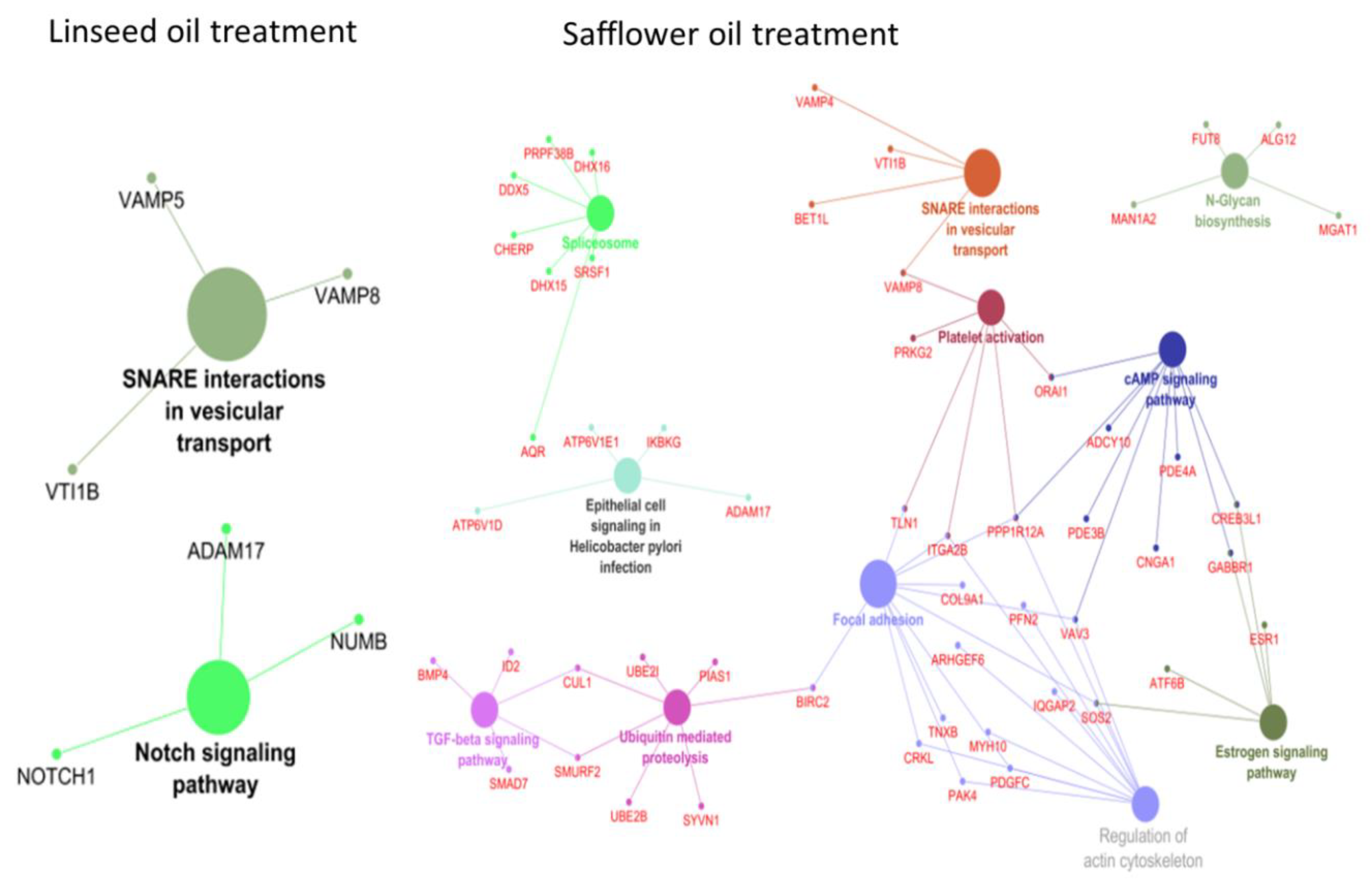
2.3. Function Enrichment via Potential cis Target Genes of lncRNAs
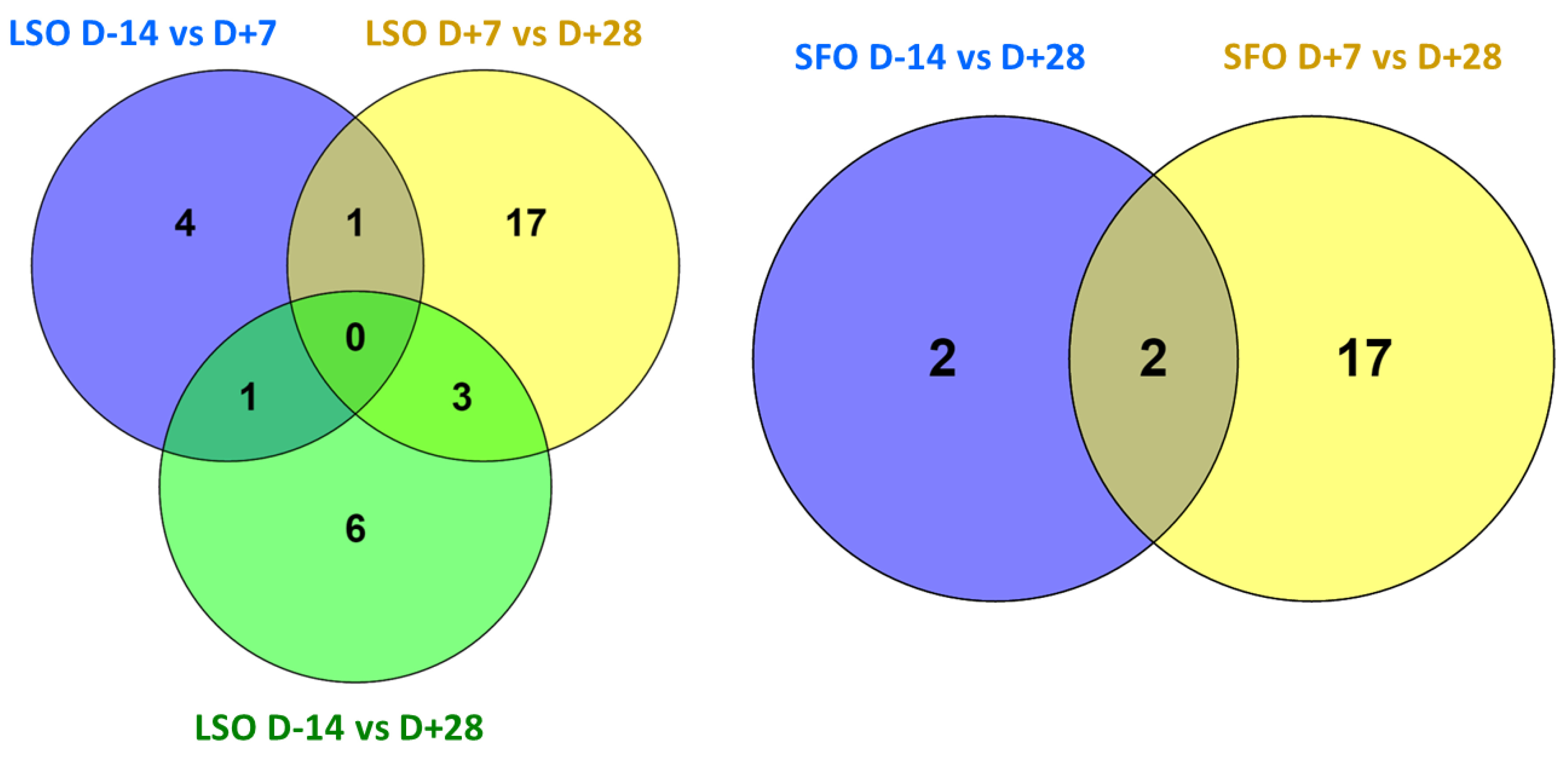
2.4. Effects of Diets Rich in Unsaturated Fatty Acids on lncRNA Expression
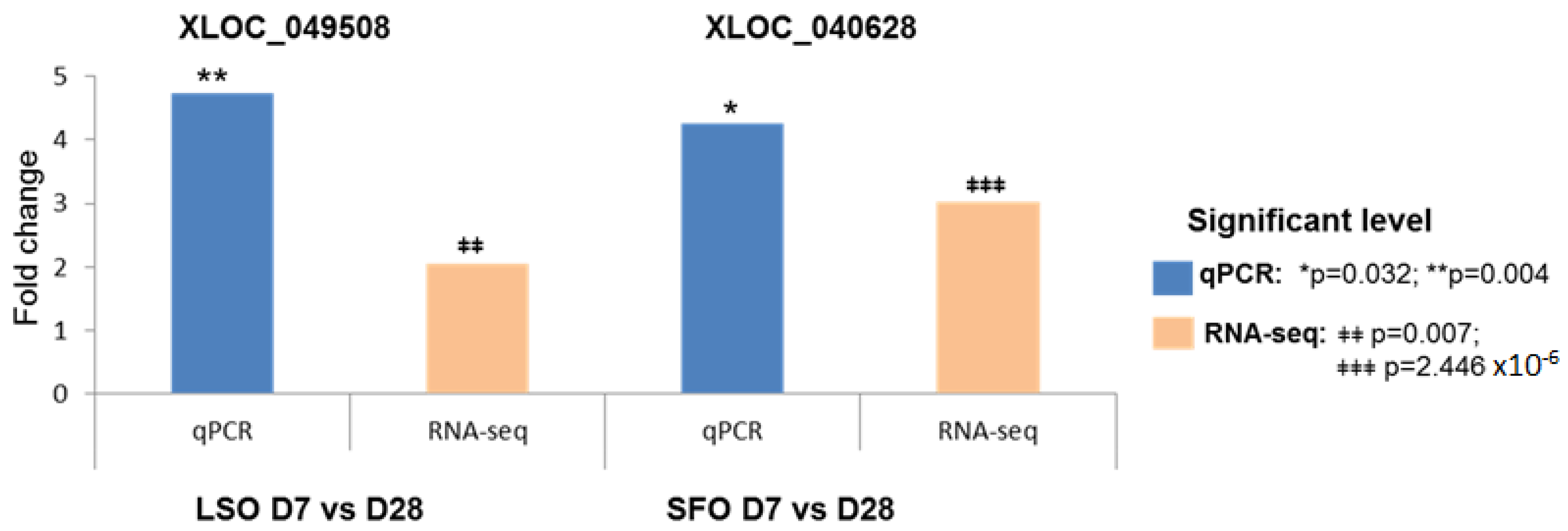
2.5. Reversed Transcribed PCR (RT-PCR) Verification of the Detection of lncRNA and Real Time Quantitative PCR (qPCR) Verification of the Expression Level of lncRNA
3. Discussion
4. Materials and Methods
4.1. Experimental Animals and Tissue Sampling
4.2. RNA Isolation and Sequencing
4.3. RNA-Sequence Read Alignment and Identification of lncRNA
4.4. Gene Ontology and Pathways Enrichment for lncRNA cis Target Genes
4.5. LncRNA Expression and Differential Gene Expression Analysis
4.6. Reversed Transcribed (RT)-PCR
4.7. Real-Time qPCR Verification of lncRNA Expression
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Availability of supporting data
Abbreviations
| ANGPTL4 | Angiopoietin like 4 |
| BCL6 | B-cell CLL/lymphoma 6 |
| bp | Base pair |
| BTA | Bovine chromosome |
| CLA | Conjugated linoleic acid |
| CNCI | Coding-Non-Coding Index |
| CPAT | Coding Potential Assessment Tool |
| DE | Differentially expressed |
| DM | Dry matter |
| ERK | Extracellular signal–regulated kinase |
| EST | Expressed sequence tag |
| FPKM | Fragments per kilo base of transcript per million mapped reads |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| GO | Geno ontology |
| HHIPL2 | HHIP like 2 |
| kb | Kilo base pairs |
| KCNF1 | Potassium voltage-gated channel modifier subfamily F member 1 |
| KEGG | Kyoto encyclopedia of genes and genomes |
| LncRNA | Long non-coding RNA |
| LSO | Linseed oil |
| MALAT1 | Metastasis associated lung adenocarcinoma transcript 1 |
| Mb | Mega base pairs |
| miRNA | Micro RNA |
| MMD | Macrophage differentiation associated |
| mRNA | Messenger RNA |
| Mt | Mitochondria |
| ncRNA | Non-coding RNA |
| nt | Nucleotides |
| NXPE2 | Neurexophilin and PC-esterase domain family member 2 |
| PINC | Pregnancy-induced non coding RNA |
| QTL | Quantitative trait loci |
| rRNA | ribosomal RNA |
| RPS15 | Ribosomal protein S15 |
| RT-PCR | Reverse transcription polymerase chain reaction |
| SFO | Safflower oil |
| snoRNA | Small nucleolar RNA |
| snRNA | Small nuclear RNA |
| STARD13 | StAR related lipid transfer domain containing 13 |
| TNF-α | Tumor necrosis factor-alpha |
| tRNA | Transfer RNA |
| XIST | X inactive specific transcript |
| Zfas1 | ZNFX1 antisense RNA 1 |
| ZNFX1 | Zinc finger NFX1-type containing 1 |
References
- Kretz, M.; Siprashvili, Z.; Chu, C.; Webster, D.; Zehnder, A.; Qu, K.; Lee, C.; Flockhart, R.; Groff, A.; Chow, J.; et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 2013, 493, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Satpathy, A.T.; Chang, H.Y. Long noncoding RNA in hematopoiesis and immunity. Immunity 2015, 42, 792–804. [Google Scholar] [CrossRef] [PubMed]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Caballero, J.; Gilbert, I.; Fournier, E.; Gagne, D.; Scantland, S.; Macaulay, A.; Robert, C. Exploring the function of long non-coding RNA in the development of bovine early embryos. Reprod. Fertil. Dev. 2014, 27, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Devaux, Y.; Zangrando, J.; Schroen, B.; Creemers, E.E.; Pedrazzini, T.; Chang, C.-P.; Dorn, G.W., II; Thum, T.; Heymans, S. Long noncoding RNAs in cardiac development and ageing. Nat. Rev. Cardiol. 2015, 12, 415–425. [Google Scholar] [PubMed]
- Hansji, H.; Leung, E.Y.; Baguley, B.C.; Finlay, G.J.; Askarian-Amiri, M.E. Keeping abreast with long non-coding RNAs in mammary gland development and breast cancer. Front. Genet. 2014, 5, 379. [Google Scholar] [CrossRef] [PubMed]
- McHugh, C.A.; Chen, C.-K.; Chow, A.; Surka, C.F.; Tran, C.; McDonel, P.; Pandya-Jones, A.; Blanco, M.; Burghard, C.; Moradian, A.; et al. The Xist lncRNA interacts directly with sharp to silence transcription through HDAC3. Nature 2015, 521, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Flynn, R.A.; Chang, H.Y. Long noncoding RNAs in cell fate programming and reprogramming. Cell. Stem Cell 2014, 14, 752–761. [Google Scholar] [CrossRef] [PubMed]
- Melissari, M.T.; Grote, P. Roles for long non-coding RNAs in physiology and disease. Pflügers Arch. 2016, 468, 945–958. [Google Scholar] [CrossRef] [PubMed]
- Ulitsky, I.; Bartel, D.P. LincRNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Geisler, S.; Coller, J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell. Biol. 2013, 14, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Wutz, A.; Jaenisch, R. A shift from reversible to irreversible x inactivation is triggered during es cell differentiation. Mol. Cell 2000, 5, 695–705. [Google Scholar] [CrossRef]
- Wutz, A.; Rasmussen, T.P.; Jaenisch, R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 2002, 30, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA hotair reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Kogo, R.; Shimamura, T.; Mimori, K.; Kawahara, K.; Imoto, S.; Sudo, T.; Tanaka, F.; Shibata, K.; Suzuki, A.; Komune, S.; et al. Long noncoding RNA hotair regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011, 71, 6320–6326. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Liu, X.; Qu, S.; Song, E.; Zou, H.; Gong, C. Long non-coding RNA hotair is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci. 2013, 104, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Gutschner, T.; Hammerle, M.; Eissmann, M.; Hsu, J.; Kim, Y.; Hung, G.; Revenko, A.; Arun, G.; Stentrup, M.; Gross, M.; et al. The noncoding RNA malat1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013, 73, 1180–1189. [Google Scholar] [CrossRef] [PubMed]
- Billerey, C.; Boussaha, M.; Esquerré, D.; Rebours, E.; Djari, A.; Meersseman, C.; Klopp, C.; Gautheret, D.; Rocha, D. Identification of large intergenic non-coding RNAs in bovine muscle using next-generation transcriptomic sequencing. BMC Genom. 2014, 15, 499. [Google Scholar] [CrossRef] [PubMed]
- Weikard, R.; Hadlich, F.; Kuehn, C. Identification of novel transcripts and noncoding RNAs in bovine skin by deep next generation sequencing. BMC Genom. 2013, 14, 789. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Long, N.; Khatib, H. Genome-wide identification and initial characterization of bovine long non-coding RNAs from EST data. Anim. Genet. 2012, 43, 674–682. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.; Adelson, D.L. Bovine ncRNAs are abundant, primarily intergenic, conserved and associated with regulatory genes. PLoS ONE 2012, 7, e42638. [Google Scholar] [CrossRef] [PubMed]
- Koufariotis, L.T.; Chen, Y.-P.P.; Chamberlain, A.; Vander Jagt, C.; Hayes, B.J. A catalogue of novel bovine long noncoding RNA across 18 tissues. PLoS ONE 2015, 10, e0141225. [Google Scholar] [CrossRef] [PubMed]
- Tong, C.; Chen, Q.; Zhao, L.; Ma, J.; Ibeagha-Awemu, E.M.; Zhao, X. Identification and characterization of long intergenic noncoding RNAs in bovine mammary glands. BMC Genom. 2017, 18, 468. [Google Scholar] [CrossRef] [PubMed]
- Ibeagha-Awemu, E.; Do, D.; Dudemaine, P.-L.; Fomenky, B.; Bissonnette, N. Integration of lncRNA and mRNA transcriptome analyses reveals genes and pathways potentially involved in calf intestinal growth and development during the early weeks of life. Genes 2018, 9, 142. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, G.K.; Milevskiy, M.J.G.; Wilson, W.; Shewan, A.M.; Brown, M.A. Non-coding RNAs in mammary gland development and disease. In Non-coding RNA and the Reproductive System; Wilhelm, D., Bernard, P., Eds.; Springer: Dordrecht, The Netherlands, 2016; pp. 121–153. [Google Scholar]
- Ginger, M.R.; Gonzalez-Rimbau, M.F.; Gay, J.P.; Rosen, J.M. Persistent changes in gene expression induced by estrogen and progesterone in the rat mammary gland. Mol. Endocrinol. 2001, 15, 1993–2009. [Google Scholar] [CrossRef] [PubMed]
- Ginger, M.R.; Shore, A.N.; Contreras, A.; Rijnkels, M.; Miller, J.; Gonzalez-Rimbau, M.F.; Rosen, J.M. A noncoding RNA is a potential marker of cell fate during mammary gland development. Proc. Natl. Acad. Sci. USA 2006, 103, 5781–5786. [Google Scholar] [CrossRef] [PubMed]
- Askarian-Amiri, M.E.; Crawford, J.; French, J.D.; Smart, C.E.; Smith, M.A.; Clark, M.B.; Ru, K.; Mercer, T.R.; Thompson, E.R.; Lakhani, S.R.; et al. Snord-host RNA ZFAS1 is a regulator of mammary development and a potential marker for breast cancer. RNA 2011, 17, 878–891. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Ibeagha-Awemu, E.M.; Liang, G.; Beaudoin, F.; Zhao, X.; Guan, L.L. Transcriptome microRNA profiling of bovine mammary epithelial cells challenged with escherichia coli or staphylococcus aureus bacteria reveals pathogen directed microRNA expression profiles. BMC Genom. 2014, 15, 181. [Google Scholar] [CrossRef] [PubMed]
- Le Guillou, S.; Marthey, S.; Laloë, D.; Laubier, J.; Mobuchon, L.; Leroux, C.; Le Provost, F. Characterisation and comparison of lactating mouse and bovine mammary gland mirnomes. PLoS ONE 2014, 9, e91938. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Beaudoin, F.; Ammah, A.; Bissonnette, N.; Benchaar, C.; Zhao, X.; Lei, C.; Ibeagha-Awemu, E. Deep sequencing shows microRNA involvement in bovine mammary gland adaptation to diets supplemented with linseed oil or safflower oil. BMC Genom. 2015, 16, 884. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Dudemaine, P.-L.; Zhao, X.; Lei, C.; Ibeagha-Awemu, E.M. Comparative analysis of the mirnome of bovine milk fat, whey and cells. PLoS ONE 2016, 11, e0154129. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhang, C.-L.; Liao, X.-X.; Chen, D.; Wang, W.-Q.; Zhu, Y.-H.; Geng, X.-H.; Ji, D.-J.; Mao, Y.-J.; Gong, Y.-C.; et al. Transcriptome microRNA profiling of bovine mammary glands infected with Staphylococcus aureus. Int. J. Mol. Sci. 2015, 16, 4997–5013. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, H.; Jin, X.; Lo, L.; Liu, J. Expression profiles of microRNAs from lactating and non-lactating bovine mammary glands and identification of miRNA related to lactation. BMC Genom. 2012, 13, 731. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Moisá, S.; Khan, M.J.; Wang, J.; Bu, D.; Loor, J.J. MicroRNA expression patterns in the bovine mammary gland are affected by stage of lactation. J. Dairy Sci. 2012, 95, 6529–6535. [Google Scholar] [CrossRef] [PubMed]
- Wicik, Z.; Gajewska, M.; Majewska, A.; Walkiewicz, D.; Osińska, E.; Motyl, T. Characterization of microRNA profile in mammary tissue of dairy and beef breed heifers. J. Anim. Breed. Gen. 2016, 133, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Matboli, M.; Shafei, A.; Ali, M.; Kamal, K.M.; Noah, M.; Lewis, P.; Habashy, A.; Ehab, M.; Gaber, A.I.; Abdelzaher, H.M. Emerging role of nutrition and the non-coding landscape in type 2 diabetes mellitus: A review of literature. Gene 2018. [Google Scholar] [CrossRef] [PubMed]
- Ruan, X. Long non-coding RNA central of glucose homeostasis. J. Cell. Biochem. 2016, 117, 1061–1065. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.-Y.; Lin, J.D. Long noncoding RNAs: A new regulatory code in metabolic control. Trends Biochem. Sci. 2015, 40, 586–596. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, P.; Yang, W.; Ruan, X.; Kiesewetter, K.; Zhu, J.; Cao, H. Integrative transcriptome analyses of metabolic responses in mice define pivotal lncRNA metabolic regulators. Cell. Metabolism 2016, 24, 627–639. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Xin, L.; Zhu, W.; Li, L.; Li, C.; Wang, Y.; Mu, Y.; Yang, S.; Li, K. Characterization of long non-coding RNA transcriptome in high-energy diet induced nonalcoholic steatohepatitis minipigs. Sci. Rep. 2016, 6, 30709. [Google Scholar] [CrossRef] [PubMed]
- Weikard, R.; Hadlich, F.; Hammon, H.M.; Frieten, D.; Gerbert, C.; Koch, C.; Dusel, G.; Kuehn, C. Long noncoding RNAs are associated with metabolic and cellular processes in the jejunum mucosa of pre-weaning calves in response to different diets. Oncotarget 2018, 9, 21052–21069. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Huo, X.-S.; Yuan, S.-X.; Zhang, L.; Zhou, W.-P.; Wang, F.; Sun, S.-H. Repression of the long noncoding RNA-let by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol. Cell 2013, 49, 1083–1096. [Google Scholar] [CrossRef] [PubMed]
- Haug, A.; Høstmark, A.T.; Harstad, O.M. Bovine milk in human nutrition–a review. Lipids Health Dis. 2007, 6, 25. [Google Scholar] [CrossRef] [PubMed]
- Lock, A.L.; Bauman, D.E. Modifying milk fat composition of dairy cows to enhance fatty acids beneficial to human health. Lipids 2004, 39, 1197–1206. [Google Scholar] [CrossRef] [PubMed]
- Loor, J.; Ferlay, A.; Ollier, A.; Doreau, M.; Chilliard, Y. Relationship among trans and conjugated fatty acids and bovine milk fat yield due to dietary concentrate and linseed oil. J. Dairy Sci. 2005, 88, 726–740. [Google Scholar] [CrossRef]
- Kelly, M.L.; Berry, J.R.; Dwyer, D.A.; Griinari, J.; Chouinard, P.Y.; Van Amburgh, M.E.; Bauman, D.E. Dietary fatty acid sources affect conjugated linoleic acid concentrations in milk from lactating dairy cows. J. Nutr. 1998, 128, 881–885. [Google Scholar] [CrossRef] [PubMed]
- Glasser, F.; Ferlay, A.; Chilliard, Y. Oilseed lipid supplements and fatty acid composition of cow milk: A meta-analysis. J. Dairy Sci. 2008, 91, 4687–4703. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.; Griinari, J.; Kennelly, J. Effect of safflower oil, flaxseed oil, monensin, and vitamin e on concentration of conjugated linoleic acid in bovine milk fat. J. Dairy Sci. 2006, 89, 733–748. [Google Scholar] [CrossRef]
- Ammah, A.A.; Benchaar, C.; Bissonnette, N.; Gévry, N.; Ibeagha-Awemu, E.M. Treatment and post-treatment effects of dietary supplementation with safflower oil and linseed oil on milk components and blood metabolites of canadian holstein cows. J. Appl. Anim. Res. 2018, 46, 898–906. [Google Scholar] [CrossRef]
- Ammah, A.; Do, D.; Bissonnette, N.; Gévry, N.; Ibeagha-Awemu, E. Co-expression network analysis identifies miRNA–mRNA networks potentially regulating milk traits and blood metabolites. Int. J. Mol. Sci. 2018, 19, 2500. [Google Scholar] [CrossRef] [PubMed]
- Ibeagha-Awemu, E.M.; Li, R.; Ammah, A.A.; Dudemaine, P.-L.; Bissonnette, N.; Benchaar, C.; Zhao, X. Transcriptome adaptation of the bovine mammary gland to diets rich in unsaturated fatty acids shows greater impact of linseed oil over safflower oil on gene expression and metabolic pathways. BMC Genom. 2016, 17, 104. [Google Scholar] [CrossRef] [PubMed]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The gencode v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Donaghey, J.; Carey, B.W.; Garber, M.; Grenier, J.K.; Munson, G.; Young, G.; Lucas, A.B.; Ach, R.; Bruhn, L.; et al. LincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011, 477, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Kotake, Y.; Nakagawa, T.; Kitagawa, K.; Suzuki, S.; Liu, N.; Kitagawa, M.; Xiong, Y. Long non-coding RNA anril is required for the prc2 recruitment to and silencing of p15(ink4b) tumor suppressor gene. Oncogene 2011, 30, 1956–1962. [Google Scholar] [CrossRef] [PubMed]
- Maamar, H.; Cabili, M.N.; Rinn, J.; Raj, A. Linc-hoxa1 is a noncoding RNA that represses hoxa1 transcription in cis. Genes Dev. 2013, 27, 1260–1271. [Google Scholar] [CrossRef] [PubMed]
- Ponjavic, J.; Oliver, P.L.; Lunter, G.; Ponting, C.P. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009, 5, e1000617. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human hox loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Yang, Y.W.; Liu, B.; Sanyal, A.; Corces-Zimmerman, R.; Chen, Y.; Lajoie, B.R.; Protacio, A.; Flynn, R.A.; Gupta, R.A.; et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011, 472, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Ebisuya, M.; Yamamoto, T.; Nakajima, M.; Nishida, E. Ripples from neighbouring transcription. Nat. Cell Biol. 2008, 10, 1106–1113. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, S.; Aiello, D.; Atianand, M.K.; Ricci, E.P.; Gandhi, P.; Hall, L.L.; Byron, M.; Monks, B.; Henry-Bezy, M.; Lawrence, J.B. A long noncoding RNA mediates both activation and repression of immune response genes. Science 2013, 341, 789–792. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, K.A.; Caffrey, D.R. Long noncoding RNAs in innate and adaptive immunity. Curr. Opin. Immunol. 2014, 26, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Li, X.; Qi, S.; Li, X.; Zhou, K.; Qing, S.; Zhang, Y.; Gao, M.-Q. LncRNA h19 is involved in tgf-β1-induced epithelial to mesenchymal transition in bovine epithelial cells through pi3k/akt signaling pathway. PeerJ 2017, 5, e3950. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Li, L.; Tang, Y.; Fu, Q.; Liu, S.; Hu, S.; Qiao, J.; Chen, C.; Ni, W. Analyses of long non-coding RNAs and mRNA profiling through RNA sequencing of mdbk cells at different stages of bovine viral diarrhea virus infection. Res. Vet. Sci. 2017, 115, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Flintoft, L. Non-coding RNA: Structure and function for lncRNAs. Nat. Rev. Genet. 2013, 14, 598. [Google Scholar] [CrossRef] [PubMed]
- Callahan, R.; Raafat, A. Notch signaling in mammary gland tumorigenesis. J. Mammary Gland Biol. Neoplasia 2001, 6, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Callahan, R.; Egan, S.E. Notch signaling in mammary development and oncogenesis. J. Mammary Gland Biol. Neoplasia 2004, 9, 145–163. [Google Scholar] [CrossRef] [PubMed]
- Do, D.; Dudemaine, P.-L.; Li, R.; Ibeagha-Awemu, E. Co-expression network and pathway analyses reveal important modules of miRNAs regulating milk yield and component traits. Int. J. Mol. Sci. 2017, 18, 1560. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Kim, H.J.; Kim, S.W.; Park, S.-A.; Chun, K.-H.; Cho, N.H.; Song, Y.S.; Kim, Y.T. The long non-coding RNA hotair increases tumour growth and invasion in cervical cancer by targeting the notch pathway. Oncotarget 2016, 7, 44558. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Reinisch, K.; Ferro-Novick, S. Coats, tethers, rabs, and snares work together to mediate the intracellular destination of a transport vesicle. Dev. Cell 2007, 12, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.R. Transport vesicle docking: Snares and associates. Annu. Rev. Cell Dev. Biol. 1996, 12, 441–461. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.A.; Scheller, R.H. Snare-mediated membrane fusion. Nat. Rev. Mol. Cell Biol. 2001, 2, 98. [Google Scholar] [CrossRef] [PubMed]
- Do, D.N.; Bissonnette, N.; Lacasse, P.; Miglior, F.; Sargolzaei, M.; Zhao, X.; Ibeagha-Awemu, E. Genome-wide association analysis and pathways enrichment for lactation persistency in canadian holstein cattle. J. Dairy Sci. 2017, 100, 1955–1970. [Google Scholar] [CrossRef] [PubMed]
- Do, D.N.; Li, R.; Dudemaine, P.-L.; Ibeagha-Awemu, E.M. MicroRNA roles in signalling during lactation: An insight from differential expression, time course and pathway analyses of deep sequence data. Sci. Rep. 2017, 7, 44605. [Google Scholar] [CrossRef] [PubMed]
- Benchaar, C.; Romero-Pérez, G.A.; Chouinard, P.Y.; Hassanat, F.; Eugene, M.; Petit, H.V.; Côrtes, C. Supplementation of increasing amounts of linseed oil to dairy cows fed total mixed rations: Effects on digestion, ruminal fermentation characteristics, protozoal populations, and milk fatty acid composition. J. Dairy Sci. 2012, 95, 4578–4590. [Google Scholar] [CrossRef] [PubMed]
- Palmquist, D.L.; Lock, A.L.; Shingfield, K.J.; Bauman, D.E. Biosynthesis of conjugated linoleic acid in ruminants and humans. Adv. Food Nutr. Res. 2005, 50, 179–217. [Google Scholar] [PubMed]
- Jacobs, A.A.A.; van Baal, J.; Smits, M.A.; Taweel, H.Z.H.; Hendriks, W.H.; van Vuuren, A.M.; Dijkstra, J. Effects of feeding rapeseed oil, soybean oil, or linseed oil on stearoyl-coa desaturase expression in the mammary gland of dairy cows. J. Dairy Sci. 2011, 94, 874–887. [Google Scholar] [CrossRef] [PubMed]
- Soccio, R.E.; Breslow, J.L. Star-related lipid transfer (start) proteins: Mediators of intracellular lipid metabolism. J. Biol. Chem. 2003, 278, 22183–22186. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Zhang, R.; He, Y.; Zou, M.; Guo, L.; Xi, T. MicroRNA-125b induces metastasis by targeting stard13 in mcf-7 and mda-mb-231 breast cancer cells. PLoS ONE 2012, 7, e35435. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zheng, J.; Yin, D.D.; Xiang, J.; He, F.; Wang, Y.C.; Liang, L.; Qin, H.Y.; Liu, L.; Liang, Y.M.; et al. Monocyte to macrophage differentiation-associated (mmd) positively regulates erk and akt activation and tnf-alpha and no production in macrophages. Mol. Biol. Rep. 2012, 39, 5643–5650. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. Cluego: A cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Farr, V.C.; Stelwagen, K.; Cate, L.R.; Molenaar, A.J.; McFadden, T.B.; Davis, S.R. An improved method for the routine biopsy of bovine mammary tissue. J. Dairy Sci. 1996, 79, 543–549. [Google Scholar] [CrossRef]
- Zimin, A.V.; Delcher, A.L.; Florea, L.; Kelley, D.R.; Schatz, M.C.; Puiu, D.; Hanrahan, F.; Pertea, G.; Van Tassell, C.P.; Sonstegard, T.S. A whole-genome assembly of the domestic cow, bos taurus. Genome Biol. 2009, 10, R42. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. Tophat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with tophat and cufflinks. Nat. Protocols 2012, 7, 562. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.-P.; Li, W. Cpat: Coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, H.; Fang, S.; Kang, Y.; Wu, W.; Hao, Y.; Li, Z.; Bu, D.; Sun, N.; Zhang, M.Q.; et al. Noncode 2016: An informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016, 44, D203–D208. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than blast. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Bu, D.; Yu, K.; Sun, S.; Xie, C.; Skogerbø, G.; Miao, R.; Xiao, H.; Liao, Q.; Luo, H.; Zhao, G. Noncode v3.0: Integrative annotation of long noncoding RNAs. Nucleic Acids Res. 2011, 40, D210–D215. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. Bedtools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with deseq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2−δδct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
| GOID | GO Term | Ontology Source | p_Value | p_FDR |
|---|---|---|---|---|
| GO:1904375 | Regulation of protein localization to cell periphery | GO_BP | 8.40 × 10−5 | 3.60 × 10−4 |
| GO:1903729 | Regulation of plasma membrane organization | GO_BP | 1.10 × 10−4 | 3.80 × 10−4 |
| GO:1903076 | Regulation of protein localization to plasma membrane | GO_BP | 7.20 × 10−5 | 4.60 × 10−4 |
| GO:0060412 | Ventricular septum morphogenesis | GO_BP | 2.00 × 10−4 | 5.40 × 10−4 |
| GO:0048546 | Digestive tract morphogenesis | GO_BP | 4.40 × 10−4 | 8.30 × 10−4 |
| GO:0030858 | Positive regulation of epithelial cell differentiation | GO_BP | 6.90 × 10−4 | 1.00 × 10−3 |
| GO:0060411 | Cardiac septum morphogenesis | GO_BP | 1.00 × 10−3 | 1.30 × 10−3 |
| GO:0003281 | Ventricular septum development | GO_BP | 1.00 × 10−3 | 1.30 × 10−3 |
| GO:0055006 | Cardiac cell development | GO_BP | 1.20 × 10−3 | 1.40 × 10−3 |
| GO:0048663 | Neuron fate commitment | GO_BP | 1.30 × 10−3 | 1.40 × 10−3 |
| GO:0048708 | Astrocyte differentiation | GO_BP | 1.40 × 10−3 | 1.40 × 10−3 |
| GO:0002040 | Sprouting angiogenesis | GO_BP | 1.40 × 10−3 | 1.40 × 10−3 |
| GO:0055038 | Recycling endosome membrane | GO_CC | 4.30 × 10−5 | 5.50 × 10−4 |
| GO:0031201 | SNARE complex | GO_CC | 6.60 × 10−4 | 1.00 × 10−3 |
| GO:0005484 | SNAP receptor activity | GO_MF | 2.20 × 10−4 | 4.90 × 10−4 |
| GOID | GO Term | Ontology Source | p_Value | p_FDR |
|---|---|---|---|---|
| GO:0048294 | Negative regulation of isotype switching to IgE isotypes | GO_BP | 1.50 × 10−5 | 2.60 × 10−3 |
| GO:0045910 | Negative regulation of DNA recombination | GO_BP | 1.10 × 10−5 | 4.20 × 10−3 |
| GO:0045829 | Negative regulation of isotype switching | GO_BP | 5.90 × 10−5 | 7.00 × 10−3 |
| GO:0010633 | Negative regulation of epithelial cell migration | GO_BP | 8.60 × 10−5 | 7.70 × 10−3 |
| GO:0006396 | RNA processing | GO_BP | 1.00 × 10−4 | 7.70 × 10−3 |
| GO:0002262 | Myeloid cell homeostasis | GO_BP | 2.20 × 10−4 | 1.10 × 10−2 |
| GO:0000018 | Regulation of DNA recombination | GO_BP | 3.20 × 10−4 | 1.10 × 10−2 |
| GO:0030218 | Erythrocyte differentiation | GO_BP | 3.00 × 10−4 | 1.10 × 10−2 |
| GO:1902679 | Negative regulation of RNA biosynthetic process | GO_BP | 2.10 × 10−4 | 1.20 × 10−2 |
| GO:0048289 | Isotype switching to IgE isotypes | GO_BP | 4.90 × 10−4 | 1.50 × 10−2 |
| GO:0048293 | Regulation of isotype switching to IgE isotypes | GO_BP | 4.90 × 10−4 | 1.50 × 10−2 |
| GO:0045646 | Regulation of erythrocyte differentiation | GO_BP | 6.20 × 10−4 | 1.70 × 10−2 |
| GO:0034101 | Erythrocyte homeostasis | GO_BP | 6.20 × 10−4 | 1.80 × 10−2 |
| GO:0002638 | Negative regulation of immunoglobulin production | GO_BP | 7.70 × 10−4 | 1.80 × 10−2 |
| GO:0045654 | Positive regulation of megakaryocyte differentiation | GO_BP | 7.70 × 10−4 | 1.80 × 10−2 |
| GO:0045892 | Negative regulation of transcription, DNA-templated | GO_BP | 7.60 × 10−4 | 1.90 × 10−2 |
| GO:0060840 | Artery development | GO_BP | 9.60 × 10−4 | 2.00 × 10−2 |
| GO:0016071 | mRNA metabolic process | GO_BP | 1.20 × 10−3 | 2.40 × 10−2 |
| GO:1903706 | Regulation of hemopoiesis | GO_BP | 1.60 × 10−3 | 2.70 × 10−2 |
| GO:0010720 | Positive regulation of cell development | GO_BP | 1.80 × 10−3 | 2.80 × 10−2 |
| GO:0060602 | Branch elongation of an epithelium | GO_BP | 1.90 × 10−3 | 2.80 × 10−2 |
| GO:0002829 | Negative regulation of type 2 immune response | GO_BP | 2.10 × 10−3 | 3.00 × 10−2 |
| GO:0003158 | Endothelium development | GO_BP | 2.30 × 10−3 | 3.20 × 10−2 |
| GO:0010632 | Regulation of epithelial cell migration | GO_BP | 2.50 × 10−3 | 3.20 × 10−2 |
| GO:0006260 | DNA replication | GO_BP | 3.70 × 10−3 | 3.40 × 10−2 |
| GO:0002064 | Epithelial cell development | GO_BP | 2.70 × 10−3 | 3.40 × 10−2 |
| GO:0060442 | Branching involved in prostate gland morphogenesis | GO_BP | 2.80 × 10−3 | 3.40 × 10−2 |
| GO:0045623 | Negative regulation of T-helper cell differentiation | GO_BP | 2.80 × 10−3 | 3.40 × 10−2 |
| GO:0045648 | Positive regulation of erythrocyte differentiation | GO_BP | 3.60 × 10−3 | 3.40 × 10−2 |
| GO:0035561 | Regulation of chromatin binding | GO_BP | 3.10 × 10−3 | 3.60 × 10−2 |
| GO:1903708 | Positive regulation of hemopoiesis | GO_BP | 3.40 × 10−3 | 3.60 × 10−2 |
| GO:0045620 | Negative regulation of lymphocyte differentiation | GO_BP | 3.20 × 10−3 | 3.70 × 10−2 |
| GO:0070076 | Histone lysine demethylation | GO_BP | 4.70 × 10−3 | 3.80 × 10−2 |
| GO:1903573 | Negative regulation of response to endoplasmic reticulum stress | GO_BP | 4.90 × 10−3 | 3.80 × 10−2 |
| GO:1902105 | Regulation of leukocyte differentiation | GO_BP | 4.40 × 10−3 | 3.80 × 10−2 |
| GO:0030968 | Endoplasmic reticulum unfolded protein response | GO_BP | 5.60 × 10−3 | 4.20 × 10−2 |
| GO:0016577 | Histone demethylation | GO_BP | 6.10 × 10−3 | 4.30 × 10−2 |
| GO:1902106 | Negative regulation of leukocyte differentiation | GO_BP | 6.20 × 10−3 | 4.30 × 10−2 |
| GO:0016447 | Somatic recombination of immunoglobulin gene segments | GO_BP | 5.90 × 10−3 | 4.30 × 10−2 |
| GO:0006349 | Regulation of gene expression by genetic imprinting | GO_BP | 6.60 × 10−3 | 4.50 × 10−2 |
| GO:0002467 | Germinal center formation | GO_BP | 6.60 × 10−3 | 4.50 × 10−2 |
| GO:0045064 | T-helper 2 cell differentiation | GO_BP | 6.60 × 10−3 | 4.50 × 10−2 |
| GO:0045652 | Regulation of megakaryocyte differentiation | GO_BP | 6.60 × 10−3 | 4.50 × 10−2 |
| GO:0001568 | Blood vessel development | GO_BP | 7.20 × 10−3 | 4.70 × 10−2 |
| GO:0034620 | Cellular response to unfolded protein | GO_BP | 7.10 × 10−3 | 4.70 × 10−2 |
| GO:0006482 | Protein demethylation | GO_BP | 7.70 × 10−3 | 4.80 × 10−2 |
| GO:0050869 | Negative regulation of B cell activation | GO_BP | 7.70 × 10−3 | 4.80 × 10−2 |
| GO:2000241 | Regulation of reproductive process | GO_BP | 8.20 × 10−3 | 4.90 × 10−2 |
| GO:0048872 | Homeostasis of number of cells | GO_BP | 7.70 × 10−3 | 4.90 × 10−2 |
| GO:0031252 | Cell leading edge | GO_CC | 8.30 × 10−4 | 1.80 × 10−2 |
| GO:0031256 | Leading edge membrane | GO_CC | 1.50 × 10−3 | 2.50 × 10−2 |
| GO:0001726 | Ruffle | GO_CC | 1.40 × 10−3 | 2.60 × 10−2 |
| GO:0042581 | Specific granule | GO_CC | 3.80 × 10−3 | 3.30 × 10−2 |
| GO:0032039 | Integrator complex | GO_CC | 3.50 × 10−3 | 3.50 × 10−2 |
| GO:0031253 | Cell projection membrane | GO_CC | 3.40 × 10−3 | 3.70 × 10−2 |
| GO:0098858 | Actin-based cell projection | GO_CC | 4.40 × 10−3 | 3.80 × 10−2 |
| GO:0005902 | Microvillus | GO_CC | 5.80 × 10−3 | 4.30 × 10−2 |
| GO:0055037 | Recycling endosome | GO_CC | 6.60 × 10−3 | 4.40 × 10−2 |
| GO:0051731 | Polynucleotide 5′-hydroxyl-kinase activity | GO_MF | 2.80 × 10−4 | 1.20 × 10−2 |
| GO:0008134 | Transcription factor binding | GO_MF | 1.40 × 10−3 | 2.60 × 10−2 |
| GO:0051020 | GTPase binding | GO_MF | 3.60 × 10−3 | 3.40 × 10−2 |
| GO:0019787 | Ubiquitin-like protein transferase activity | GO_MF | 3.50 × 10−3 | 3.60 × 10−2 |
| GO:0030374 | Ligand-dependent nuclear receptor transcription coactivator activity | GO_MF | 3.30 × 10−3 | 3.70 × 10−2 |
| GO:0005089 | Rho guanyl-nucleotide exchange factor activity | GO_MF | 4.80 × 10−3 | 3.80 × 10−2 |
| GO:0060589 | Nucleoside-triphosphatase regulator activity | GO_MF | 4.70 × 10−3 | 3.90 × 10−2 |
| GO:0035591 | Signaling adaptor activity | GO_MF | 5.80 × 10−3 | 4.30 × 10−2 |
| GO:0031267 | Small GTPase binding | GO_MF | 7.80 × 10−3 | 4.80 × 10−2 |
| Periods of Comparison | Known lncRNA Notation | Chr | Chr Start..End | Nearest Gene | FC | log2FC | p-Value | Padj |
|---|---|---|---|---|---|---|---|---|
| D-14 vs. D+7 | ||||||||
| XLOC_044813 | NONBTAT030934.1 | 6 | 17675271..17680447 | - | - | −1.459 | 2.247 × 10−7 | 0.0010 |
| NONBTAG014563.2 | 2.749 | |||||||
| XLOC_032807 | New | 26 | 33011966..33019072 | - | 2.310 | 1.208 | 7.345 × 10−7 | 0.0017 |
| XLOC_041145 | NONBTAT020143.2 | 4 | 95417314..95613931 | - | - | −1.131 | 3.393 × 10−6 | 0.0051 |
| NONBTAG013424.2 | 2.190 | |||||||
| XLOC_021427 | New | 19 | 31640546..31641078 | - | −2.378 | −1.250 | 8.409 × 10−6 | 0.0095 |
| XLOC_021420 | New | 19 | 31604279..31663126 | - | −2.258 | −1.175 | 3.207 × 10−5 | 0.0289 |
| XLOC_004564 | NONBTAT002269.2 | 10 | 49527965..49605076 | RORA (ENSBTAT00000021144) | 1.720 | 0.782 | 0.0001 | 0.0769 |
| NONBTAG013424.2 | ||||||||
| D+7 vs. D+28 | ||||||||
| XLOC_050004 | New | 8 | 64796589..64820744 | - | - | −1.111 | 4.666 × 10−8 | 0.0001 |
| 2.160 | ||||||||
| XLOC_004564 | NONBTAT002269.2 | 10 | 49527965..49605076 | RORA (ENSBTAT00000021144) | - | −0.886 | 1.084 × 10−5 | 0.0065 |
| NONBTAG001608.2 | 1.848 | |||||||
| XLOC_018587 | New | 18 | 11048799..11055932 | CRISPLD2 (ENSBTAT00000028221) | - | −0.814 | 8.899 × 10−6 | 0.0065 |
| 1.758 | ||||||||
| XLOC_049508 | New | 8 | 22729811..22734496 | ENSBTAG00000047195 | - | −1.045 | 8.973 × 10−6 | 0.0065 |
| (ENSBTAT00000048617) | 2.063 | |||||||
| XLOC_026857 | New | 21 | 31732594..31860453 | FBXO22 (ENSBTAT00000003665) | - | −0.557 | 2.408 × 10−5 | 0.0116 |
| 1.471 | ||||||||
| XLOC_024438 | New | 2 | 134829905..134832425 | - | - | −0.865 | 7.330 × 10−5 | 0.0196 |
| 1.821 | ||||||||
| XLOC_039327 | New | 3 | 114773040..114828419 | - | - | −0.762 | 6.304 × 10−5 | 0.0196 |
| 1.696 | ||||||||
| XLOC_049790 | NONBTAT031343.1 | 8 | 42264400..42279290 | - | 1.873 | 0.905 | 6.640 × 10−5 | 0.0196 |
| NONBTAG022051.1 | ||||||||
| XLOC_049791 | New | 8 | 42273319..42275306 | - | 2.172 | 1.119 | 6.569 × 10−5 | 0.0196 |
| XLOC_049767 | New | 8 | 42141722..42245895 | - | 1.911 | 0.934 | 8.906 × 10−5 | 0.0214 |
| XLOC_049792 | NONBTAT031344.1 | 8 | 42279395..42321282 | - | 2.032 | 1.023 | 0.0002 | 0.0410 |
| NONBTAG016235.2 | ||||||||
| XLOC_011302 | New | 14 | 84116708..84118510 | - | 2.035 | 1.025 | 0.0002 | 0.0466 |
| XLOC_030043 | New | 23 | 36290019..36292016 | - | 1.691 | 0.758 | 0.0003 | 0.0467 |
| XLOC_004276 | New | 10 | 26691672..26693523 | - | 1.657 | 0.729 | 0.0003 | 0.0590 |
| XLOC_007663 | New | 12 | 27541269..28034473 | STARD13 (ENSBTAT00000029081) | - | −0.411 | 0.0004 | 0.0592 |
| 1.330 | ||||||||
| XLOC_005960 | New | 11 | 86699810..86710266 | - | - | −0.868 | 0.0006 | 0.0825 |
| 1.825 | ||||||||
| XLOC_014482 | New | 16 | 52617661..52619004 | - | - | −0.708 | 0.0006 | 0.0825 |
| 1.634 | ||||||||
| XLOC_050157 | New | 8 | 77681842..77683706 | - | - | −0.956 | 0.0006 | 0.0825 |
| 1.940 | ||||||||
| XLOC_051249 | New | 8 | 84443959..84447116 | - | - | −0.751 | 0.0007 | 0.0825 |
| 1.683 | ||||||||
| XLOC_040832 | New | 4 | 66327795..66329807 | - | - | −0.922 | 0.0008 | 0.0913 |
| 1.895 | ||||||||
| XLOC_044269 | New | 5 | 100899101..100938587 | - | - | −0.504 | 0.0009 | 0.0985 |
| 1.418 | ||||||||
| D-14 vs. D+28 | ||||||||
| XLOC_032807 | New | 26 | 33011966..33019072 | - | 2.178 | 1.123 | 3.826 × 10−6 | 0.0023 |
| XLOC_040082 | New | 4 | 93460873..93469656 | HIG2 (ENSBTAT00000045181) | 2.310 | 1.208 | 2.246 × 10−6 | 0.0023 |
| XLOC_044264 | New | 5 | 100888960..100892632 | - | - | −1.056 | 4.000 × 10−6 | 0.0023 |
| 2.079 | ||||||||
| XLOC_044269 | New | 5 | 100899101..100938587 | - | - | −0.613 | 4.937 × 10−5 | 0.0152 |
| 1.529 | ||||||||
| XLOC_045228 | New | 6 | 87225278..87228405 | - | - | −0.924 | 4.182 × 10−5 | 0.0152 |
| 1.897 | ||||||||
| XLOC_053316 | New | Mt | 2..360 | - | - | −0.978 | 5.330 × 10−5 | 0.0152 |
| 1.970 | ||||||||
| XLOC_049790 | NONBTAT031343.1 | 8 | 42264400..42279290 | - | 1.813 | 0.858 | 0.0001 | 0.0349 |
| NONBTAG022051.1 | ||||||||
| XLOC_054333 | New | X | 123683291..124283250 | ENSBTAG00000048092 | 1.580 | 0.660 | 0.0004 | 0.0847 |
| (ENSBTAT00000030016) | ||||||||
| XLOC_002555 | New | 1 | 153149175..153164789 | - | - | −0.766 | 0.0006 | 0.0973 |
| 1.701 | ||||||||
| XLOC_049791 | New | 8 | 42273319..42275306 | - | 1.957 | 0.969 | 0.0005 | 0.0973 |
| Periods of Comparison | LncRNA Type | Chr | Chr Start..End | Nearest Gene | FC | log2FC | p-Value | Padj |
|---|---|---|---|---|---|---|---|---|
| D-14 vs. D+28 | ||||||||
| XLOC_053295 | NONBTAT026075.2 | Mt | 1453..3023 | ENSBTAG00000043570 | 1.683 | 0.751 | 2.491 × 10−6 | 0.0107 |
| NONBTAG017440.2 | (ENSBTAT00000060540) | |||||||
| XLOC_014422 | New | 16 | 50833181..50845563 | ARHGEF16 (ENSBTAT00000027769) | - | −1.014 | 7.395 × 10−6 | 0.0159 |
| 2.020 | ||||||||
| XLOC_033615 | New | 27 | 24828725..24847714 | - | 1.709 | 0.773 | 2.105 × 10−5 | 0.0302 |
| XLOC_049508 | New | 8 | 22729811..22734496 | ENSBTAG00000047195 | - | −0.895 | 4.575 × 10−5 | 0.0492 |
| (ENSBTAT00000048617) | 1.860 | |||||||
| D+7 vs. D+28 | ||||||||
| XLOC_040628 | New | 4 | 36035624..36063375 | - | - | −1.601 | 9.914 × 10−10 | 2.446 × 10−6 |
| 3.034 | ||||||||
| XLOC_039658 | New | 4 | 36060986..36063955 | - | - | −1.005 | 3.385 × 10−5 | 0.0417 |
| 2.007 | ||||||||
| XLOC_001923 | New | 1 | 80169821..80179076 | - | - | −0.603 | 0.0002 | 0.0828 |
| 1.519 | ||||||||
| XLOC_005093 | New | 10 | 100872512..100938144 | - | - | −0.505 | 0.00030 | 0.0828 |
| 1.419 | ||||||||
| XLOC_012186 | New | 15 | 25534554..25535977 | - | - | −0.973 | 0.0003 | 0.0828 |
| 1.963 | ||||||||
| XLOC_014185 | New | 16 | 26739288..26747603 | TAF1A (ENSBTAT00000017928) | - | −0.610 | 0.0005 | 0.0828 |
| 1.526 | ||||||||
| XLOC_016131 | New | 17 | 64537633..64544131 | - | - | −0.721 | 0.0005 | 0.0828 |
| 1.648 | ||||||||
| XLOC_020830 | NONBTAT028906.1 | 19 | 5793670..5835852 | MMD (ENSBTAT00000000244) | 1.396 | 0.481 | 0.0004 | 0.0828 |
| NONBTAG019735.1 | ||||||||
| XLOC_034163 | New | 27 | 41333596..41335357 | - | - | −0.944 | 0.0003 | 0.0828 |
| 1.924 | ||||||||
| XLOC_040082 | New | 4 | 93460873..93469656 | HIG2 (ENSBTAT00000045181) | 1.806 | 0.853 | 0.0005 | 0.0828 |
| XLOC_042624 | New | 5 | 82039036..82042254 | - | - | −0.887 | 0.0002 | 0.0828 |
| 1.849 | ||||||||
| XLOC_044264 | New | 5 | 100888960..100892632 | - | - | −0.834 | 0.0004 | 0.0828 |
| 1.783 | ||||||||
| XLOC_049508 | New | 8 | 22729811..22734496 | ENSBTAG00000047195 | - | −0.788 | 0.0003 | 0.0828 |
| (ENSBTAT00000048617) | 1.727 | |||||||
| XLOC_052993 | New | 9 | 95309003..95312277 | - | - | −0.952 | 0.0003 | 0.0828 |
| 1.935 | ||||||||
| XLOC_053295 | NONBTAT026075.2 | Mt | 1453..3023 | ENSBTAG00000043570 | 1.476 | 0.562 | 0.0004 | 0.0828 |
| NONBTAG017440.2 | (ENSBTAT00000060540) | |||||||
| XLOC_015543 | New | 16 | 67180319..67190790 | - | - | −0.429 | 0.0006 | 0.0878 |
| 1.346 | ||||||||
| XLOC_047068 | New | 7 | 27353120..27714937 | CTXN3 (ENSBTAT00000044060) | - | −0.748 | 0.0006 | 0.0878 |
| 1.679 | ||||||||
| XLOC_002568 | New | 1 | 153860742..153955586 | ENSBTAG00000044519 | - | −0.648 | 0.0007 | 0.0971 |
| (ENSBTAT00000061952) | 1.567 | |||||||
| XLOC_043291 | New | 5 | 6768479..6777191 | - | - | −0.778 | 0.0007 | 0.0971 |
| 1.715 |
| Treatment | LncRNA | Gene | Correlation | p-Value | Chromosome | Gene.Start | Gene.End |
|---|---|---|---|---|---|---|---|
| Linseed oil | XLOC_005960 | KCNF1 | 0.821379 | 2.93 × 10−5 | 11 | 86759338 | 86761647 |
| Linseed oil | XLOC_007663 | STARD13 | 0.894839 | 5.38 × 10−7 | 12 | 27866074 | 28033238 |
| Safflower oil | XLOC_001923 | BCL6 | 0.797951 | 7.25 × 10−5 | 1 | 80179482 | 80202388 |
| Safflower oil | XLOC_012186 | NXPE2 | 0.822692 | 2.77 × 10−5 | 15 | 25515542 | 25524857 |
| Safflower oil | XLOC_014185 | HHIPL2 | 0.739728 | 0.00045 | 16 | 26713225 | 26741364 |
| Safflower oil | XLOC_020830 | MMD | 0.940505 | 6.54 × 10−9 | 19 | 5819716 | 5840124 |
© 2018 by her Majesty the Queen in Right of Canada as represented by the Minister of Agriculture and Agri-Food. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ibeagha-Awemu, E.M.; Li, R.; Dudemaine, P.-L.; Do, D.N.; Bissonnette, N. Transcriptome Analysis of Long Non-Coding RNA in the Bovine Mammary Gland Following Dietary Supplementation with Linseed Oil and Safflower Oil. Int. J. Mol. Sci. 2018, 19, 3610. https://doi.org/10.3390/ijms19113610
Ibeagha-Awemu EM, Li R, Dudemaine P-L, Do DN, Bissonnette N. Transcriptome Analysis of Long Non-Coding RNA in the Bovine Mammary Gland Following Dietary Supplementation with Linseed Oil and Safflower Oil. International Journal of Molecular Sciences. 2018; 19(11):3610. https://doi.org/10.3390/ijms19113610
Chicago/Turabian StyleIbeagha-Awemu, Eveline M., Ran Li, Pier-Luc Dudemaine, Duy N. Do, and Nathalie Bissonnette. 2018. "Transcriptome Analysis of Long Non-Coding RNA in the Bovine Mammary Gland Following Dietary Supplementation with Linseed Oil and Safflower Oil" International Journal of Molecular Sciences 19, no. 11: 3610. https://doi.org/10.3390/ijms19113610
APA StyleIbeagha-Awemu, E. M., Li, R., Dudemaine, P.-L., Do, D. N., & Bissonnette, N. (2018). Transcriptome Analysis of Long Non-Coding RNA in the Bovine Mammary Gland Following Dietary Supplementation with Linseed Oil and Safflower Oil. International Journal of Molecular Sciences, 19(11), 3610. https://doi.org/10.3390/ijms19113610






