Inhibition of LINE-1 Retrotransposition by Capsaicin
Abstract
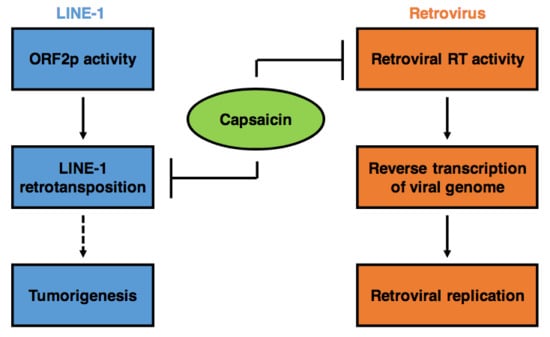
1. Introduction
2. Results
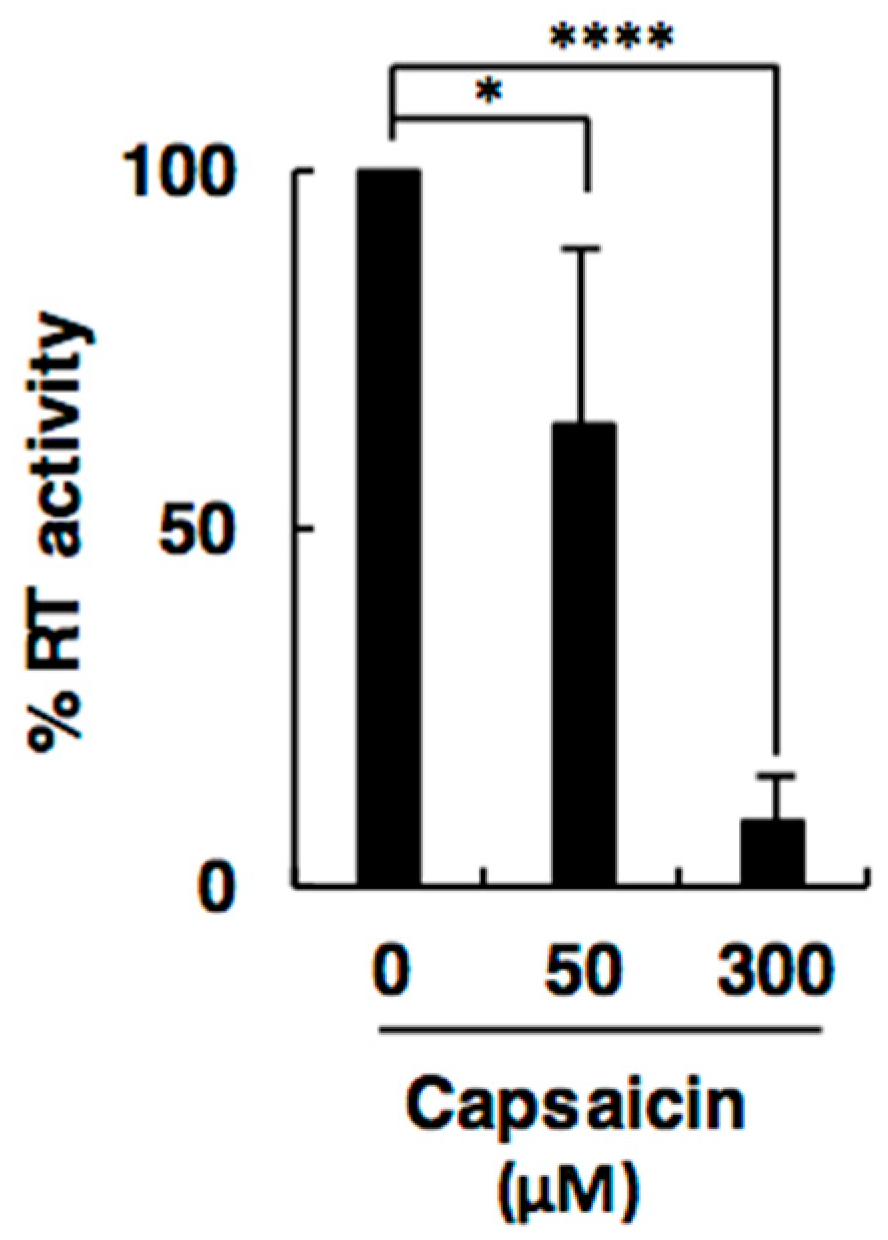
2.1. Capsaicin Inhibits Activity of Retroviral RT
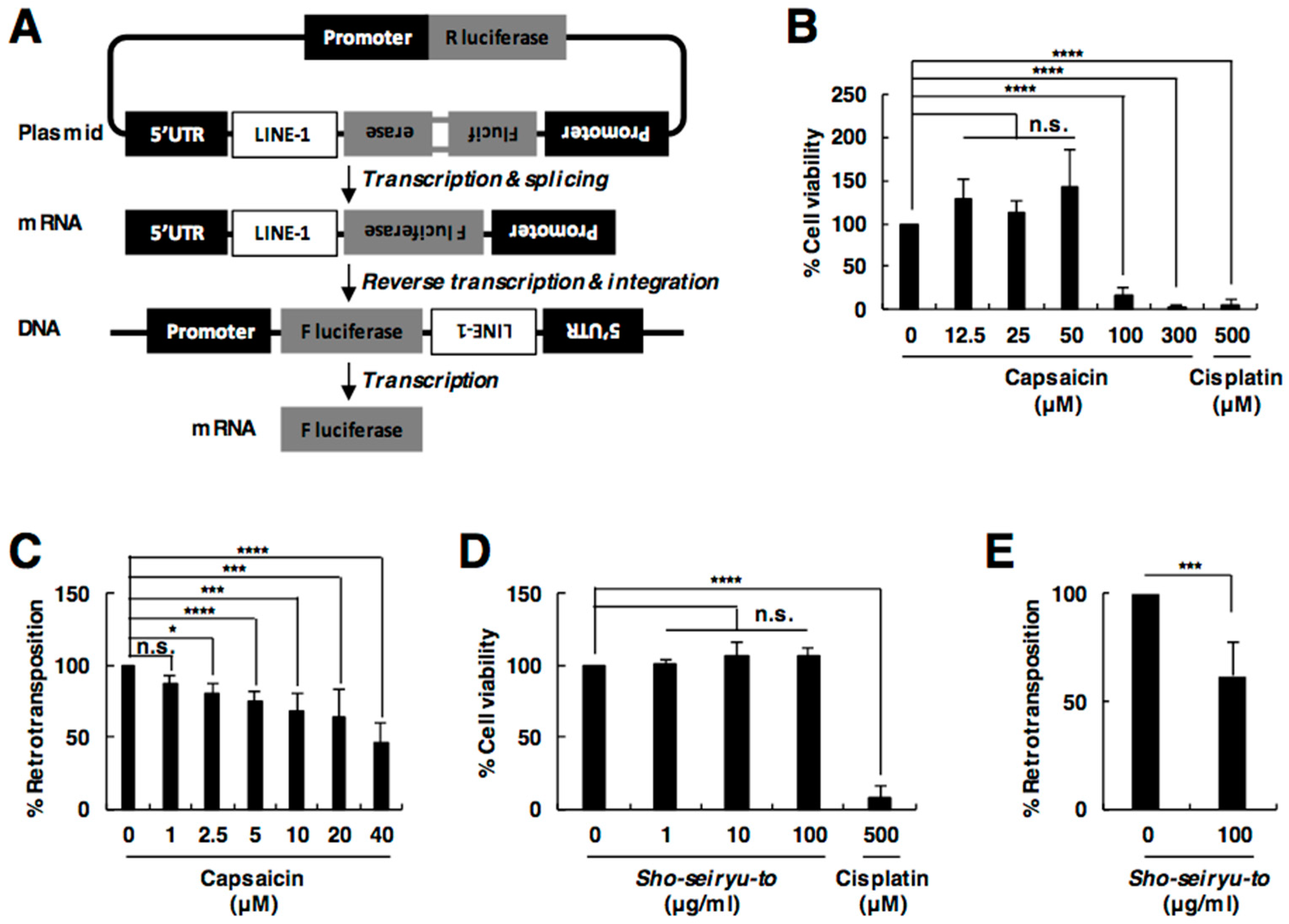
2.2. Capsaicin Suppresses L1 Retrotransposition
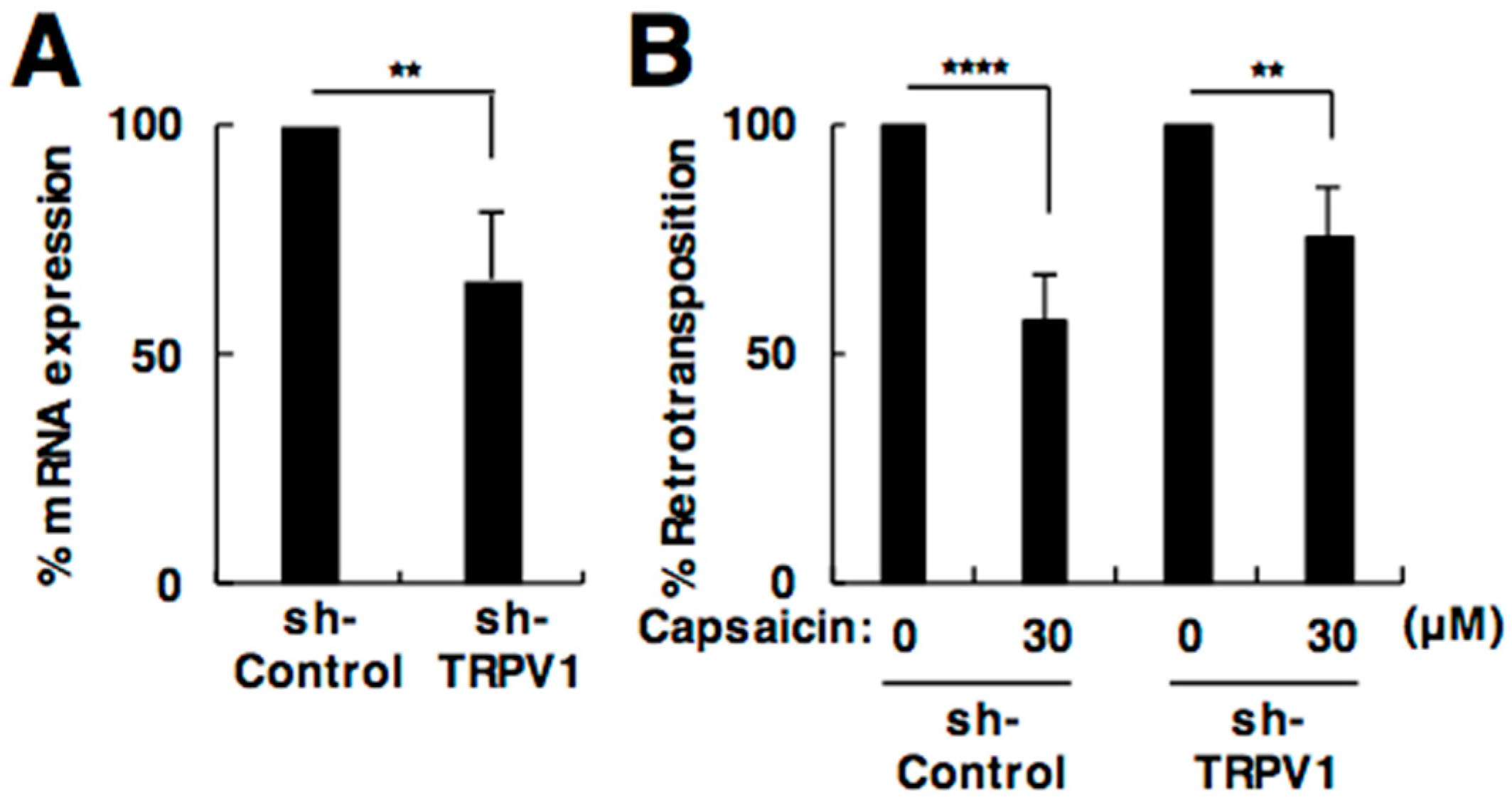
2.3. The Effect of Capsaicin on L1 Is Independent of TRPV1
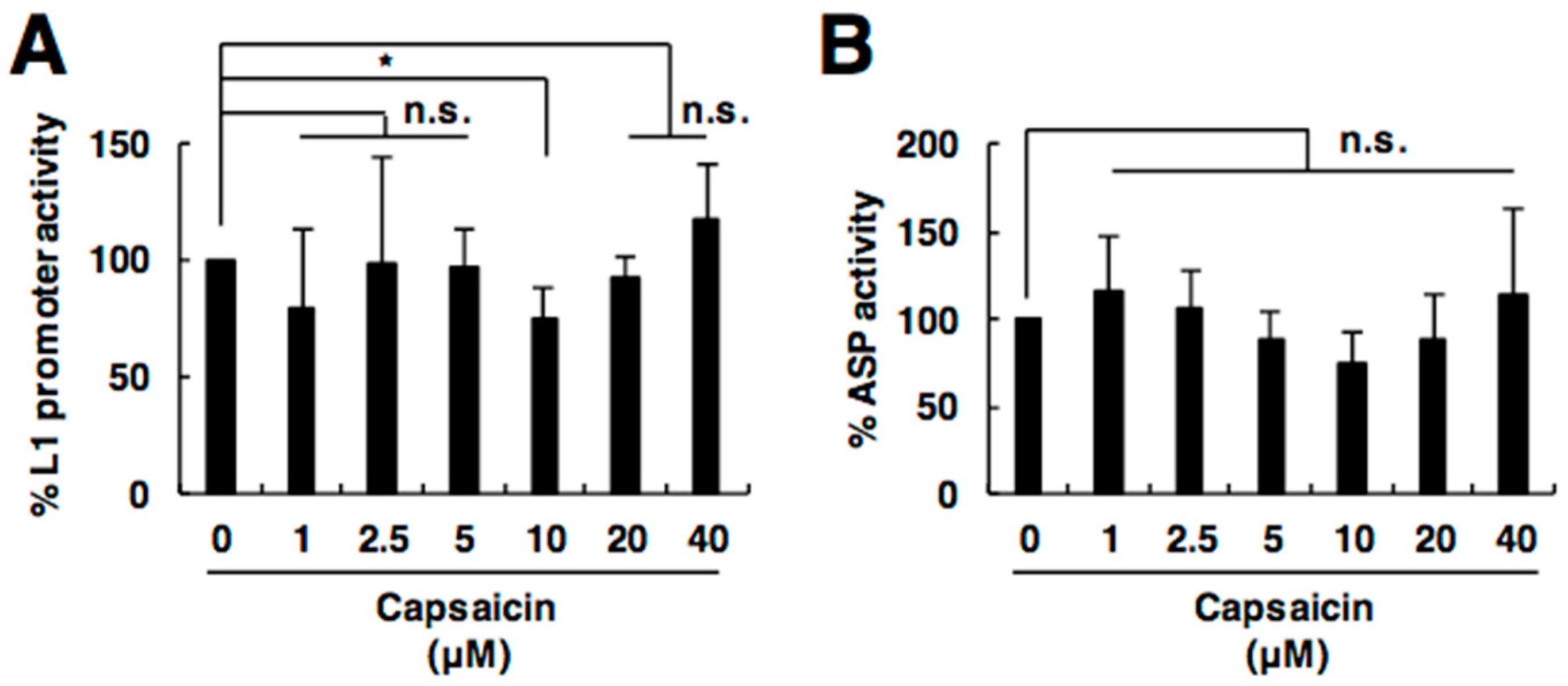
2.4. Capsaicin Does Not Affect L1 Promoter or Antisense Promoter Activity
3. Discussion
4. Materials and Methods
4.1. Cells
4.2. Chemicals
4.3. Plasmids
4.4. In Vitro RT Assay
4.5. L1 Retrotransposition Assay
4.6. Cell Viability Assay
4.7. L1 Promoter and ASP Assays
4.8. Real-Time RT-PCR
4.9. Primers
4.10. Statistics
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Khazina, E.; Truffault, V.; Büttner, R.; Schmidt, S.; Coles, M.; Weichenrieder, O. Trimeric structure and flexibility of the L1ORF1 protein in human L1 retrotransposition. Nat. Struct. Mol. Biol. 2011, 18, 1006–1014. [Google Scholar] [CrossRef] [PubMed]
- Beck, C.R.; Collier, P.; Macfarlane, C.; Malig, M.; Kidd, J.M.; Eichler, E.E.; Badge, R.M.; Moran, J. V LINE-1 retrotransposition activity in human genomes. Cell 2010, 141, 1159–1170. [Google Scholar] [CrossRef] [PubMed]
- Brouha, B.; Schustak, J.; Badge, R.M.; Lutz-Prigge, S.; Farley, A.H.; Moran, J.V.; Kazazian, H.H. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. USA 2003, 100, 5280–5285. [Google Scholar] [CrossRef] [PubMed]
- Burns, K.H.; Boeke, J.D. Human Transposon Tectonics. Cell 2012, 149, 740–752. [Google Scholar] [CrossRef] [PubMed]
- Levin, H.L.; Moran, J.V. Dynamic interactions between transposable elements and their hosts. Nat. Rev. Genet. 2011, 12, 615–627. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Iskow, R.; Yang, L.; Gokcumen, O.; Haseley, P.; Luquette, L.J.; Lohr, J.G.; Harris, C.C.; Ding, L.; Wilson, R.K.; et al. Landscape of somatic retrotransposition in human cancers. Science 2012, 337, 967–971. [Google Scholar] [CrossRef] [PubMed]
- Shukla, R.; Upton, K.R.; Muñoz-Lopez, M.; Gerhardt, D.J.; Fisher, M.E.; Nguyen, T.; Brennan, P.M.; Baillie, J.K.; Collino, A.; Ghisletti, S.; et al. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell 2013, 153, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Honda, T. Links between Human LINE-1 Retrotransposons and Hepatitis Virus-Related Hepatocellular Carcinoma. Front. Chem. 2016, 4, 21. [Google Scholar] [CrossRef] [PubMed]
- Honda, T. Potential Links between Hepadnavirus and Bornavirus Sequences in the Host Genome and Cancer. Front. Microbiol. 2017, 8, 2537. [Google Scholar] [CrossRef] [PubMed]
- Bundo, M.; Toyoshima, M.; Okada, Y.; Akamatsu, W.; Ueda, J.; Nemoto-Miyauchi, T.; Sunaga, F.; Toritsuka, M.; Ikawa, D.; Kakita, A.; et al. Increased l1 retrotransposition in the neuronal genome in schizophrenia. Neuron 2014, 81, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Kazazian, H.H.; Wong, C.; Youssoufian, H.; Scott, A.F.; Phillips, D.G.; Antonarakis, S.E. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 1988, 332, 164–166. [Google Scholar] [CrossRef] [PubMed]
- Holmes, S.E.; Dombroski, B.A.; Krebs, C.M.; Boehm, C.D.; Kazazian, H.H. A new retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric insertion. Nat. Genet. 1994, 7, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Terasaki, N.; Goodier, J.L.; Cheung, L.E.; Wang, Y.J.; Kajikawa, M.; Kazazian, H.H.; Okada, N. In vitro screening for compounds that enhance human L1 mobilization. PLoS ONE 2013, 8, e74629. [Google Scholar] [CrossRef] [PubMed]
- Giorgi, G.; Marcantonio, P.; Del Re, B. LINE-1 retrotransposition in human neuroblastoma cells is affected by oxidative stress. Cell Tissue Res. 2011, 346, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.R.; Nolan, N.A.; Miles, S.L.; Brown, K.C.; Akers, A.T.; Colclough, K.W.; Seidler, J.W.; Rimoldi, J.M.; Valentovic, M.A.; Dasgupta, P. Anti-cancer Activity of Natural and Synthetic Capsaicin Analogs. J. Pharmacol. Exp. Ther. 2017, 364, 462–474. [Google Scholar] [CrossRef] [PubMed]
- Basith, S.; Cui, M.; Hong, S.; Choi, S. Harnessing the Therapeutic Potential of Capsaicin and Its Analogues in Pain and Other Diseases. Molecules 2016, 21, 966. [Google Scholar] [CrossRef] [PubMed]
- Julius, D.; Caterina, M.J.; Schumacher, M.A.; Tominaga, M.; Rosen, T.A.; Levine, J.D. The capsaicin receptor: A heat-activated ion channel in the pain pathway. Nature 1997, 389, 816–824. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Huang, Q.; Boeke, J.D. Effect of reverse transcriptase inhibitors on LINE-1 and Ty1 reverse transcriptase activities and on LINE-1 retrotransposition. BMC Biochem. 2011, 12, 18. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Rosser, J.M.; Thompson, T.L.; Boeke, J.D.; An, W. Characterization of L1 retrotransposition with high-throughput dual-luciferase assays. Nucleic Acids Res. 2011, 39, e16. [Google Scholar] [CrossRef] [PubMed]
- Dasari, S.; Bernard Tchounwou, P. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 2014, 740, 364–378. [Google Scholar] [CrossRef] [PubMed]
- Denli, A.M.; Narvaiza, I.; Kerman, B.E.; Pena, M.; Benner, C.; Marchetto, M.C.N.; Diedrich, J.K.; Aslanian, A.; Ma, J.; Moresco, J.J.; et al. Primate-Specific ORF0 Contributes to Retrotransposon-Mediated Diversity. Cell 2015, 163, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Kassiotis, G.; Stoye, J.P. Immune responses to endogenous retroelements: Taking the bad with the good. Nat. Rev. Immunol. 2016, 16, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Chapa-Oliver, A.M.; Mejía-Teniente, L. Capsaicin: From Plants to a Cancer-Suppressing Agent. Molecules 2016, 21, 931. [Google Scholar] [CrossRef] [PubMed]
- Clark, R.; Lee, S.-H. Anticancer Properties of Capsaicin Against Human Cancer. Anticancer Res. 2016, 36, 837–843. [Google Scholar] [PubMed]
- Caprodossi, S.; Amantini, C.; Nabissi, M.; Morelli, M.B.; Farfariello, V.; Santoni, M.; Gismondi, A.; Santoni, G. Capsaicin promotes a more aggressive gene expression phenotype and invasiveness in null-TRPV1 urothelial cancer cells. Carcinogenesis 2011, 32, 686–694. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Bhattacharjee, S.; Mandal, D.P. Induction of Apoptosis by Eugenol and Capsaicin in Human Gastric Cancer AGS Cells—Elucidating the Role of p53. Asian Pac. J. Cancer Prev. 2015, 16, 6753–6759. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Yang, Z.; Wang, Y.; Zhu, G.; Wang, X. Capsaicin induces cycle arrest by inhibiting cyclin-dependent-kinase in bladder carcinoma cells. Int. J. Urol. 2012, 19, 662–668. [Google Scholar] [CrossRef] [PubMed]
- Speek, M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol. Cell. Biol. 2001, 21, 1973–1985. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nishikawa, Y.; Nakayama, R.; Obika, S.; Ohsaki, E.; Ueda, K.; Honda, T. Inhibition of LINE-1 Retrotransposition by Capsaicin. Int. J. Mol. Sci. 2018, 19, 3243. https://doi.org/10.3390/ijms19103243
Nishikawa Y, Nakayama R, Obika S, Ohsaki E, Ueda K, Honda T. Inhibition of LINE-1 Retrotransposition by Capsaicin. International Journal of Molecular Sciences. 2018; 19(10):3243. https://doi.org/10.3390/ijms19103243
Chicago/Turabian StyleNishikawa, Yuki, Ryota Nakayama, Shunsuke Obika, Eriko Ohsaki, Keiji Ueda, and Tomoyuki Honda. 2018. "Inhibition of LINE-1 Retrotransposition by Capsaicin" International Journal of Molecular Sciences 19, no. 10: 3243. https://doi.org/10.3390/ijms19103243
APA StyleNishikawa, Y., Nakayama, R., Obika, S., Ohsaki, E., Ueda, K., & Honda, T. (2018). Inhibition of LINE-1 Retrotransposition by Capsaicin. International Journal of Molecular Sciences, 19(10), 3243. https://doi.org/10.3390/ijms19103243