Nach Is a Novel Subgroup at an Early Evolutionary Stage of the CNC-bZIP Subfamily Transcription Factors from the Marine Bacteria to Humans
Abstract
1. Introduction
2. Results
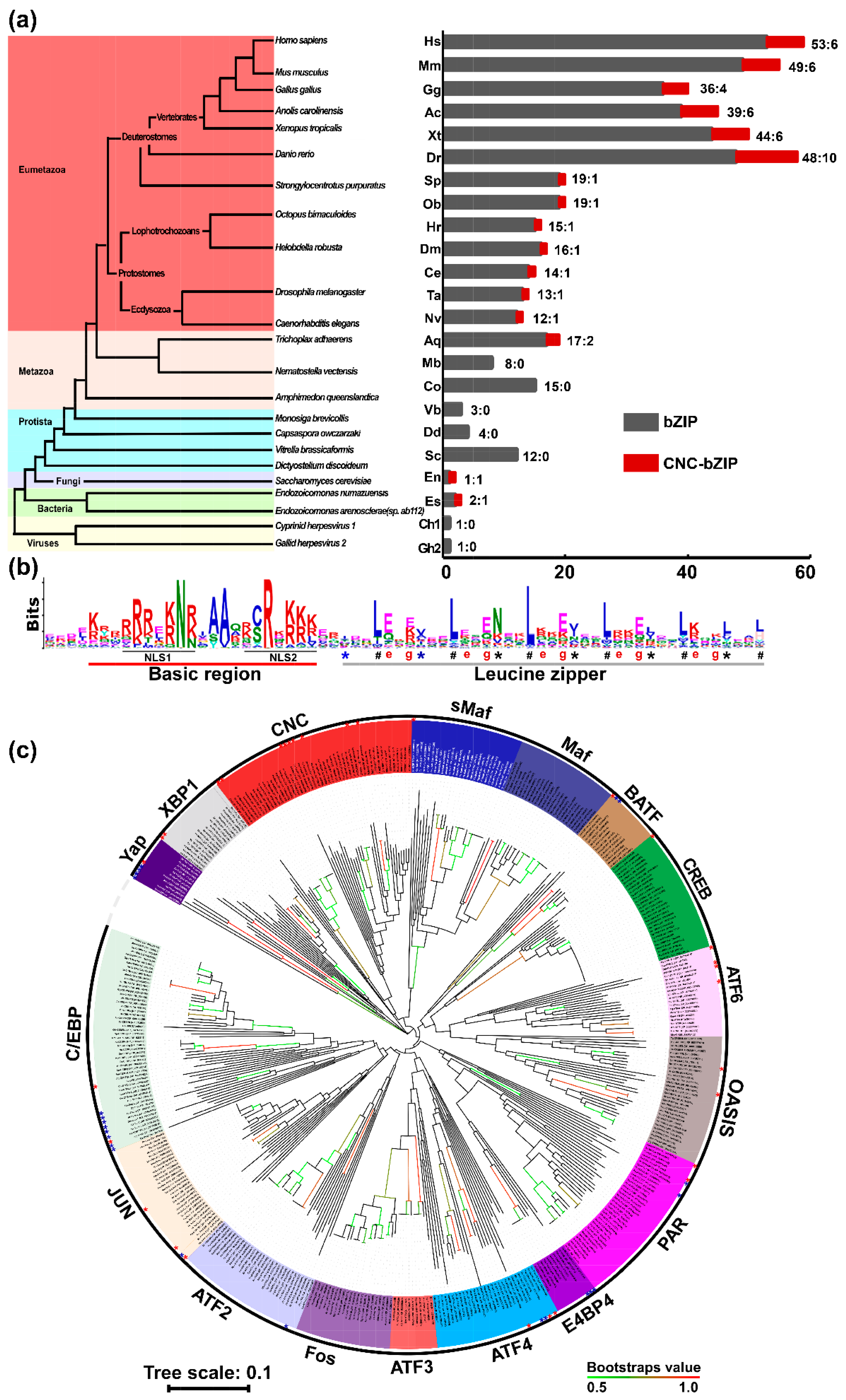
2.1. Species Distribution and Phylogenetic Analysis of bZIP Transcription Factors
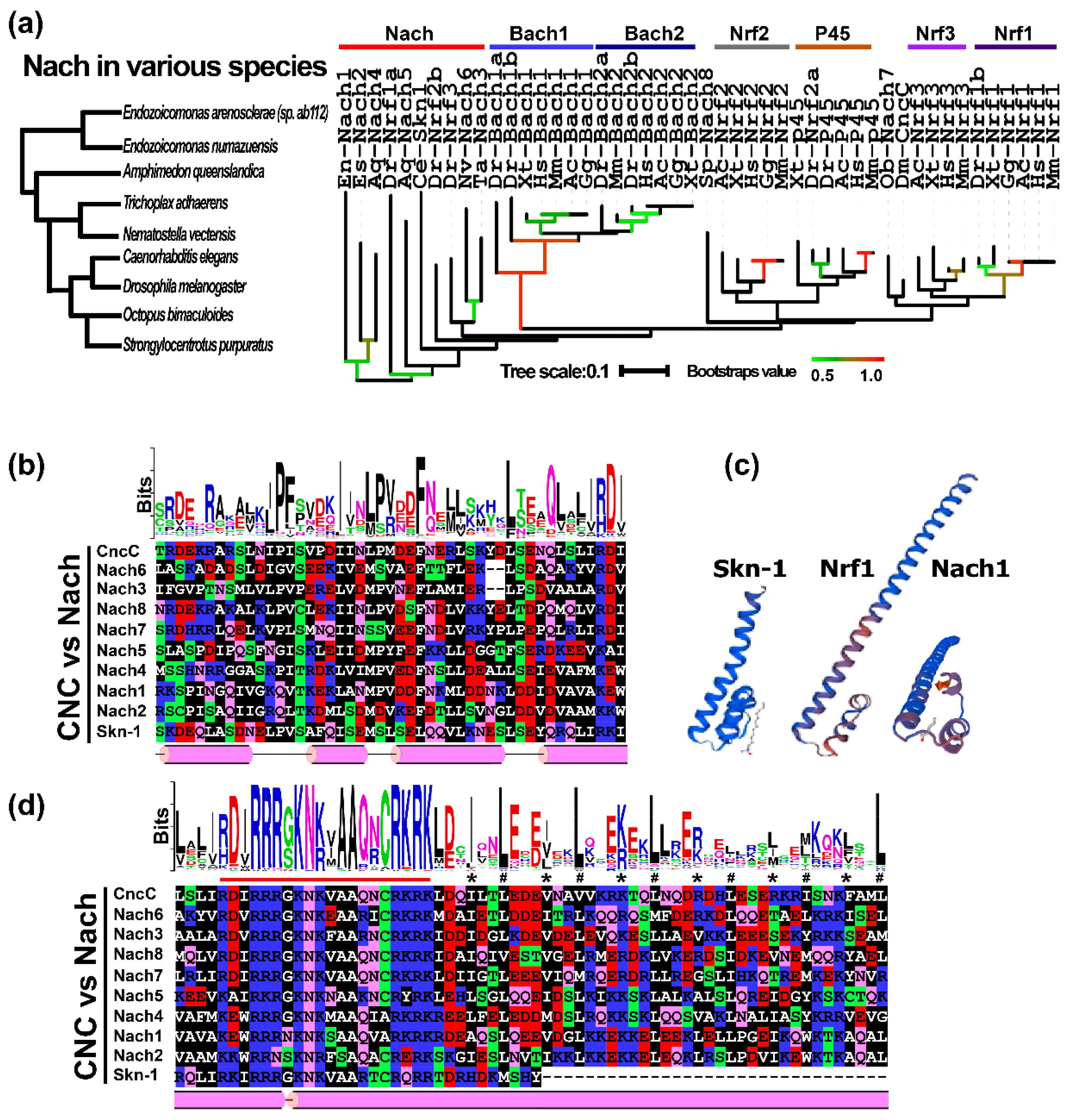
2.2. A Novel Evolutionary Branch of the CNC-bZIP Subfamily from Ancestral Nach Proteins
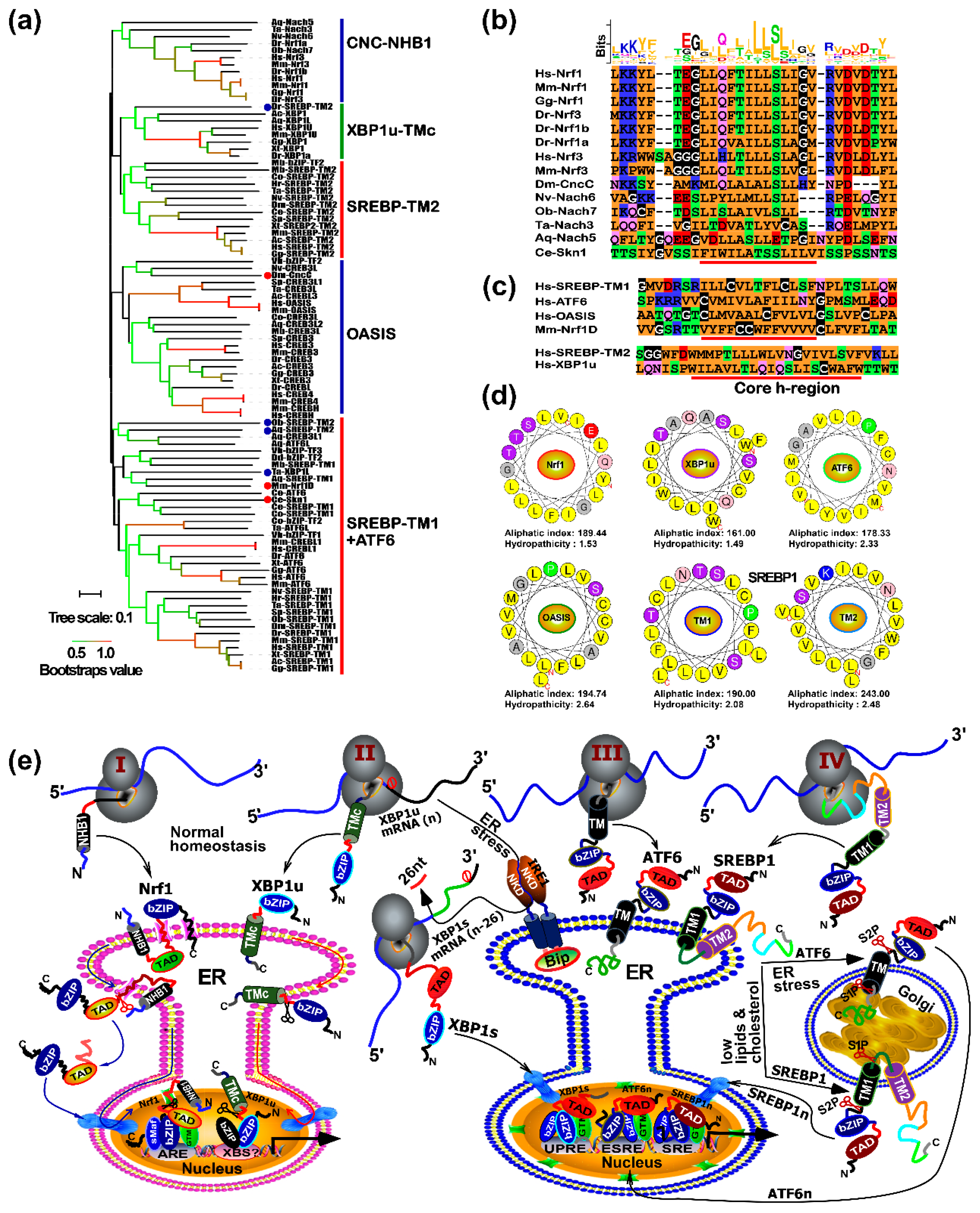
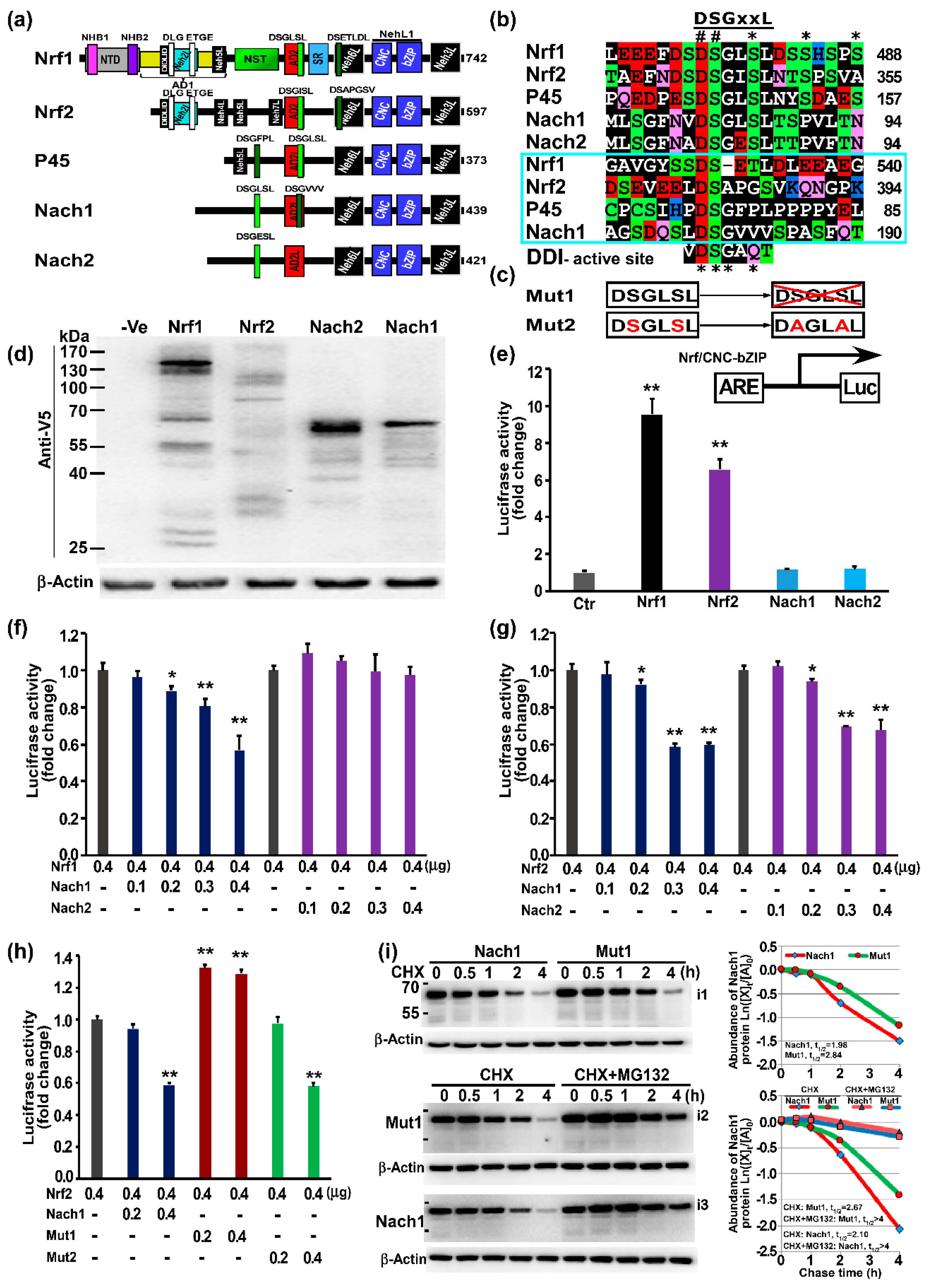
2.3. Distinct Subgroups of the Membrane-Bound bZIP Transcription Factors
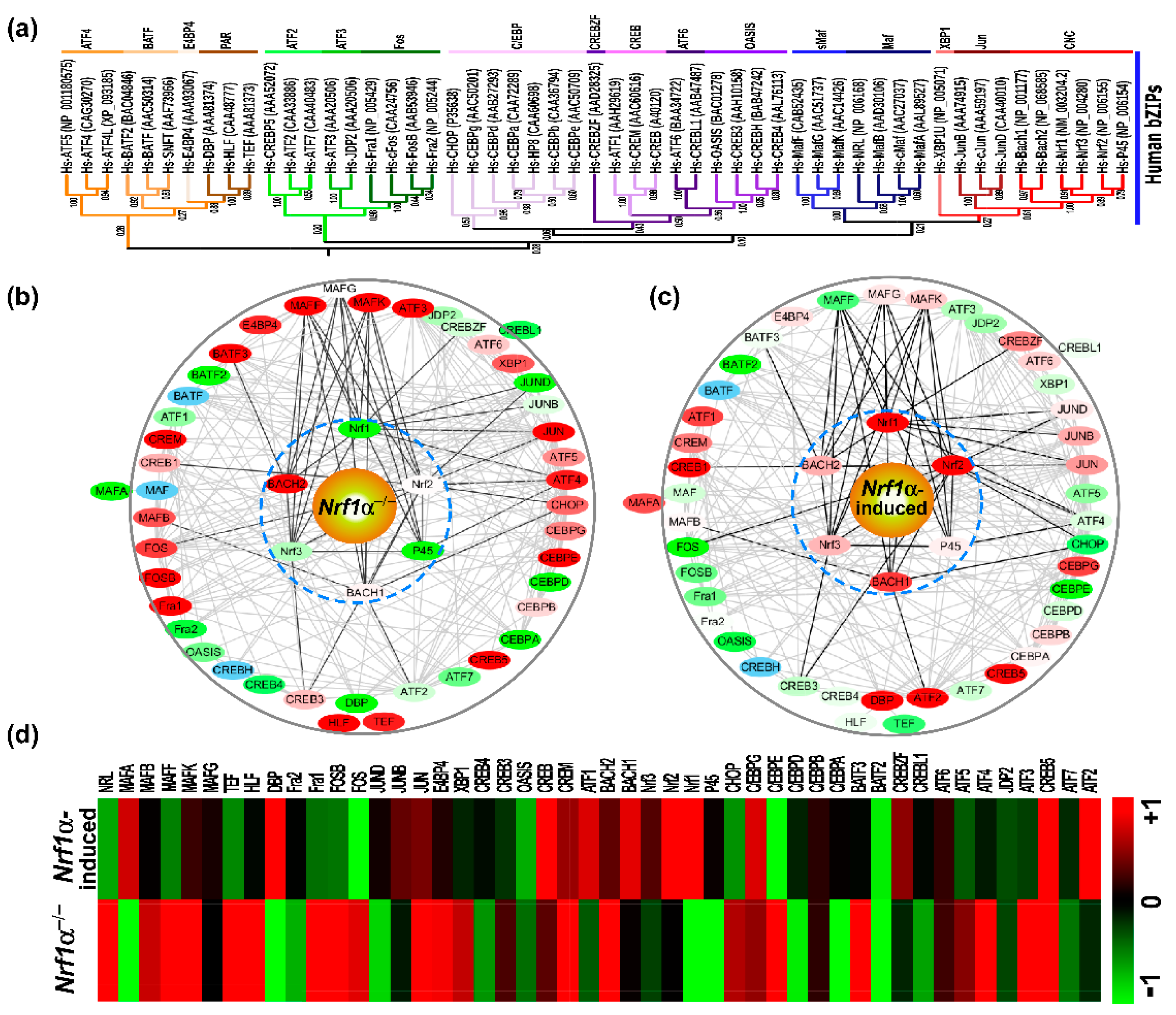
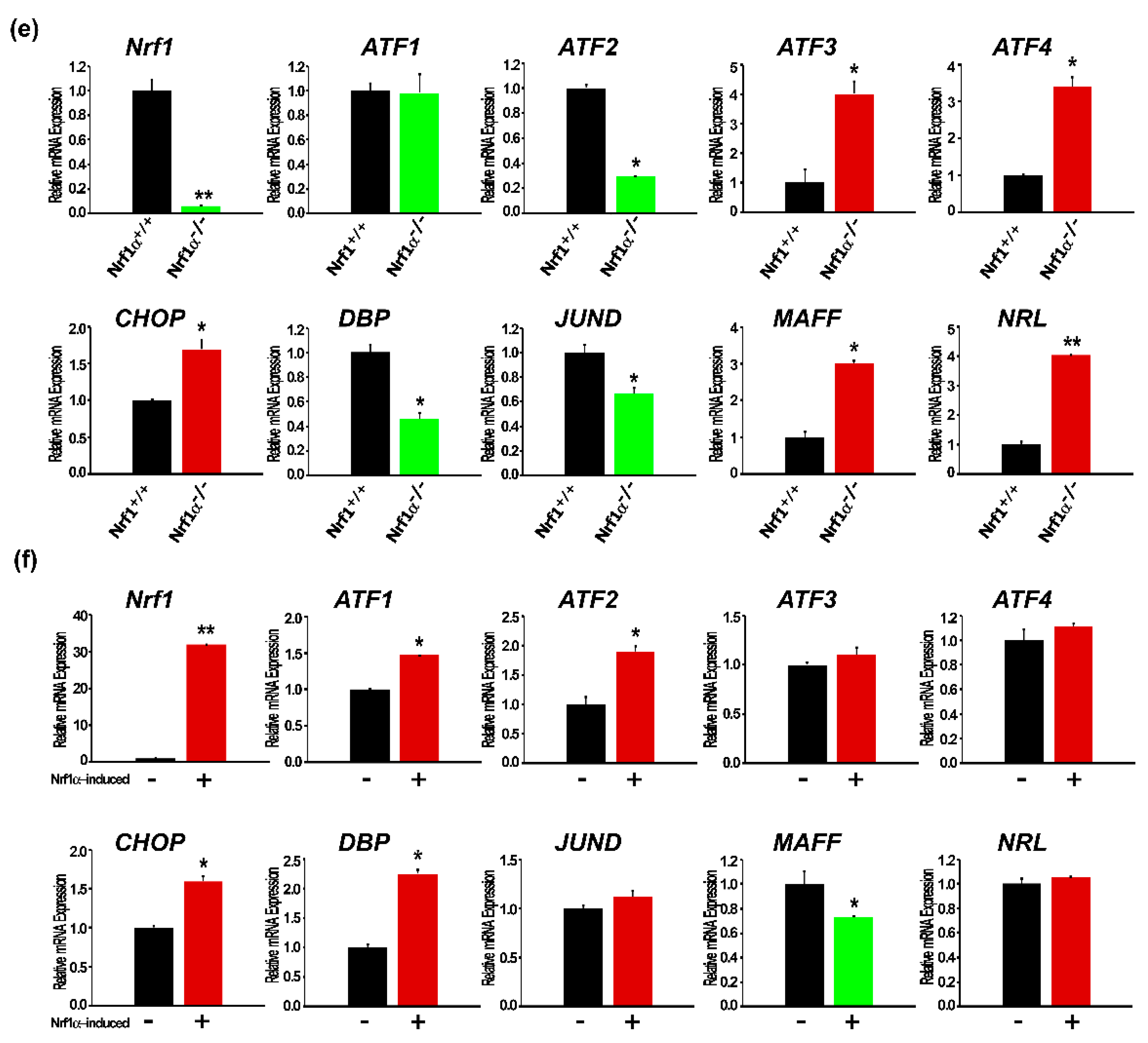
2.4. Expressive Differentiations of Human bZIP Factors within Their Endogenous Interaction Networks
2.5. Nach1 Shares Conserved Domains with Other CNC-bZIP Factors at Differently Regulating Target Genes
3. Discussion
3.1. A Phylogenetic Web of the bZIP Transcription Factors
3.2. Nach Is Buded at the Early Evolutionary Stage of the CNC-bZIP Transcription Factors
3.3. Distinct Functions of Nach1 and Nrf1 in Regulating Target Genes
4. Materials and Methods
4.1. Identification of bZIP Proteins
4.2. Phylogenic Analysis of Structural Domains
4.3. Bioinformatic Analysis of TM-Containing Transcription Factors
4.4. Interaction Network and Transcriptomic Analysis
4.5. Experimental Cell Lines
4.6. Validation of Gene Expression by qRT-PCR
4.7. The Pulse-Chase Experiments Followed by Western Blotting
4.8. ARE-Driven Reporter Gene Assays
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van de Peer, Y.; Chapelle, S.; De Wachter, R. A quantitative map of nucleotide substitution rates in bacterial rRNA. Nucleic Acids Res. 1996, 24, 3381–3391. [Google Scholar] [CrossRef] [PubMed]
- Iwama, H.; Gojobori, T. Highly conserved upstream sequences for transcription factor genes and implications for the regulatory network. Proc. Natl. Acad. Sci. USA 2004, 101, 17156–17161. [Google Scholar] [CrossRef] [PubMed]
- Ellegren, H.; Sheldon, B.C. Genetic basis of fitness differences in natural populations. Nature 2008, 452, 169–175. [Google Scholar] [CrossRef]
- Vaquerizas, J.M.; Kummerfeld, S.K.; Teichmann, S.A.; Luscombe, N.M. A census of human transcription factors: Function, expression and evolution. Nat. Rev. Genet. 2009, 10, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Hughes, T.R. Introduction to “a handbook of transcription factors”. In A Handbook of Transcription Factors; Sub-Cellular Biochemistry; Springer: Dordrecht, The Netherlands, 2011; Volume 52, pp. 1–6. [Google Scholar]
- Shlyueva, D.; Stampfel, G.; Stark, A. Transcriptional enhancers: From properties to genome-wide predictions. Nat. Rev. Genet. 2014, 15, 272–286. [Google Scholar] [CrossRef] [PubMed]
- Miller, M. The importance of being flexible: The case of basic region leucine zipper transcriptional regulators. Curr. Protein Pept. Sci. 2009, 10, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Davudian, S.; Mansoori, B.; Shajari, N.; Mohammadi, A.; Baradaran, B. BACH1, the master regulator gene: A novel candidate target for cancer therapy. Gene 2016, 588, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Liu, Q.; Liang, G.; Wang, L.; Luo, S.; Tang, Q.; Zhao, H.; Su, Y.; Yung, S.; Chan, T.M.; et al. E4BP4 overexpression: A protective mechanism in CD4+ T cells from SLE patients. J. Autoimmun. 2013, 41, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, K.; Kurosaki, T.; Roychoudhuri, R. BACH transcription factors in innate and adaptive immunity. Nat. Rev. Immunol. 2017, 17, 437–450. [Google Scholar] [CrossRef] [PubMed]
- Kaser, A.; Lee, A.H.; Franke, A.; Glickman, J.N.; Zeissig, S.; Tilg, H.; Nieuwenhuis, E.E.; Higgins, D.E.; Schreiber, S.; Glimcher, L.H.; et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 2008, 134, 743–756. [Google Scholar] [CrossRef] [PubMed]
- Hurst, H.C. Transcription factors 1: bZIP proteins. Protein Profile 1994, 1, 123–168. [Google Scholar] [PubMed]
- Mitchell, P.J.; Tjian, R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science 1989, 245, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Ptashne, M.; Gann, A. Transcriptional activation by recruitment. Nature 1997, 386, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F.; Group, B. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Nijhawan, A.; Jain, M.; Tyagi, A.K.; Khurana, J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008, 146, 333–350. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Guo, R.; Guo, C.; Hou, H.; Wang, X.; Gao, H. Evolutionary and eExpression analyses of the apple basic leucine zipper tTranscription factor family. Front. Plant Sci. 2016, 7, 376. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.; Chen, J.; Wang, Y.; Chen, Y.; Chen, S.; Lin, Y.; Pan, S.; Zhong, X.; Xie, D. Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 2012, 19, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Sebe-Pedros, A.; de Mendoza, A.; Lang, B.F.; Degnan, B.M.; Ruiz-Trillo, I. Unexpected repertoire of metazoan transcription factors in the unicellular holozoan Capsaspora owczarzaki. Mol. Biol. Evol. 2010, 28, 1241–1254. [Google Scholar] [CrossRef] [PubMed]
- Amoutzias, G.; Veron, A.; Weiner, J.; Robinson-Rechavi, M.; Bornberg-Bauer, E.; Oliver, S.; Robertson, D. One billion years of bZIP transcription factor evolution: Conservation and change in dimerization and DNA-binding site specificity. Mol. Biol. Evol. 2006, 24, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Li, J.; Glass, N.L. Exploring the bZIP transcription factor regulatory network in Neurospora crassa. Microbiology 2011, 157, 747–759. [Google Scholar] [CrossRef] [PubMed]
- Castro, P.H.; Lilay, G.H.; Munoz-Merida, A.; Schjoerring, J.K.; Azevedo, H.; Assuncao, A.G.L. Phylogenetic analysis of F-bZIP transcription factors indicates conservation of the zinc deficiency response across land plants. Sci. Rep. 2017, 7, 3806. [Google Scholar] [CrossRef] [PubMed]
- Jindrich, K.; Degnan, B.M. The diversification of the basic leucine zipper family in eukaryotes correlates with the evolution of multicellularity. BMC Evol. Biol. 2016, 16, 28. [Google Scholar] [CrossRef] [PubMed]
- Bowerman, B.; Eaton, B.A.; Priess, J.R. Skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell 1992, 68, 1061–1075. [Google Scholar] [CrossRef]
- Mohler, J.; Vani, K.; Leung, S.; Epstein, A. Segmentally restricted, cephalic expression of a leucine zipper gene during Drosophila embryogenesis. Mech. Dev. 1991, 34, 3–9. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiang, Y. Molecular and cellular basis for the unique functioning of Nrf1, an indispensable transcription factor for maintaining cell homoeostasis and organ integrity. Biochem. J. 2016, 473, 961–1000. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Crouch, D.H.; Yamamoto, M.; Hayes, J.D. Negative regulation of the Nrf1 transcription factor by its N-terminal domain is independent of Keap1: Nrf1, but not Nrf2, is targeted to the endoplasmic reticulum. Biochem. J. 2006, 399, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lucocq, J.M.; Yamamoto, M.; Hayes, J.D. The NHB1 (N-terminal homology box 1) sequence in transcription factor Nrf1 is required to anchor it to the endoplasmic reticulum and also to enable its asparagine-glycosylation. Biochem. J. 2007, 408, 161–172. [Google Scholar] [CrossRef] [PubMed]
- von Heijne, G. Membrane-protein topology. Nat. Rev. Mol. Cell Biol. 2006, 7, 909–918. [Google Scholar] [CrossRef] [PubMed]
- Vitrac, H.; Bogdanov, M.; Dowhan, W. In vitro reconstitution of lipid-dependent dual topology and postassembly topological switching of a membrane protein. Proc. Natl. Acad. Sci. USA 2013, 110, 9338–9343. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ren, Y.; Li, S.; Hayes, J. Transcription factor Nrf1 is topologically repartitioned across membranes to enable target gene transactivation through its acidic glucose-responsive domains. PLoS ONE 2014, 9, e93456. [Google Scholar] [CrossRef] [PubMed]
- Tirosh, B.; Iwakoshi, N.N.; Glimcher, L.H.; Ploegh, H.L. Rapid turnover of unspliced Xbp-1 as a factor that modulates the unfolded protein response. J. Biol. Chem. 2006, 281, 5852–5860. [Google Scholar] [CrossRef] [PubMed]
- Yanagitani, K.; Imagawa, Y.; Iwawaki, T.; Hosoda, A.; Saito, M.; Kimata, Y.; Kohno, K. Cotranslational targeting of XBP1 protein to the membrane promotes cytoplasmic splicing of its own mRNA. Mol. Cell 2009, 34, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Yanagitani, K.; Kimata, Y.; Kadokura, H.; Kohno, K. Translational pausing ensures membrane targeting and cytoplasmic splicing of XBP1u mRNA. Science 2011, 331, 586–589. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Rawson, R.B.; Komuro, R.; Chen, X.; Dave, U.P.; Prywes, R.; Brown, M.S.; Goldstein, J.L. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 2000, 6, 1355–1364. [Google Scholar] [CrossRef]
- Rawson, R.B. Regulated intramembrane proteolysis: From the endoplasmic reticulum to the nucleus. Essays Biochem. 2002, 38, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sato, R.; Brown, M.S.; Hua, X.; Goldstein, J.L. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell 1994, 77, 53–62. [Google Scholar] [CrossRef]
- Ren, Y.; Qiu, L.; Lu, F.; Ru, X.; Li, S.; Xiang, Y.; Yu, S.; Zhang, Y. TALENs-directed knockout of the full-length transcription factor Nrf1alpha that represses malignant behaviour of human hepatocellular carcinoma (HepG2) cells. Sci. Rep. 2016, 6, 23775. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hayes, J.D. The membrane-topogenic vectorial behaviour of Nrf1 controls its post-translational modification and transactivation activity. Sci. Rep. 2013, 3, 2006. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, S.; Xiang, Y.; Qiu, L.; Zhao, H.; Hayes, J.D. The selective post-translational processing of transcription factor Nrf1 yields distinct isoforms that dictate its ability to differentially regulate gene expression. Sci. Rep. 2015, 5, 12983. [Google Scholar] [CrossRef] [PubMed]
- Ao, P. Laws in Darwinian evolutionary theory. Phys. Life Rev. 2006, 2, 117–156. [Google Scholar] [CrossRef]
- Reinke, A.; Baek, J.; Ashenberg, O.; Keating, A.E. Networks of bZIP protein-protein interactions diversified over a billion years of evolution. Science 2013, 340, 730–734. [Google Scholar] [CrossRef] [PubMed]
- Moitra, J.; Szilak, L.D.; Vinson, C. Leucine is the most stabilizing aliphatic amino acid in the d position of a dimeric leucine zipper coiled coil. Biochemistry 1997, 36, 12567–12573. [Google Scholar] [CrossRef] [PubMed]
- Newman, J.R.; Keating, A.E. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science 2003, 300, 2097–2101. [Google Scholar] [CrossRef] [PubMed]
- Deppmann, C.D.; Acharya, A.; Rishi, V.; Wobbes, B.; Smeekens, S.; Taparowsky, E.J.; Vinson, C. Dimerization specificity of all 67 B-ZIP motifs in Arabidopsis thaliana: A comparison to Homo sapiens B-ZIP motifs. Nucleic Acids Res. 2004, 32, 3435–3445. [Google Scholar] [CrossRef] [PubMed]
- Harbury, P.B.; Zhang, T.; Kim, P.S.; Alber, T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 1993, 262, 1401–1407. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, L., Jr.; Woolfson, D.N.; Alber, T. Buried polar residues and structural specificity in the GCN4 leucine zipper. Nat. Struct. Biol. 1996, 3, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Nishijima, M.; Adachi, K.; Katsuta, A.; Shizuri, Y.; Yamasato, K. Endozoicomonas numazuensis sp. nov., a gammaproteobacterium isolated from marine sponges, and emended description of the genus Endozoicomonas Kurahashi and Yokota 2007. Int. J. Syst. Evol. Microbiol. 2013, 63, 709–714. [Google Scholar] [CrossRef] [PubMed]
- Gaudray, G.; Gachon, F.; Basbous, J.; Biard-Piechaczyk, M.; Devaux, C.; Mesnard, J.M. The complementary strand of the human T-cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down-regulates viral transcription. J. Virol. 2002, 76, 12813–12822. [Google Scholar] [CrossRef] [PubMed]
- Basbous, J.; Arpin, C.; Gaudray, G.; Piechaczyk, M.; Devaux, C.; Mesnard, J.M. The HBZ factor of human T-cell leukemia virus type I dimerizes with transcription factors JunB and c-Jun and modulates their transcriptional activity. J. Biol. Chem. 2003, 278, 43620–43627. [Google Scholar] [CrossRef] [PubMed]
- Moreira, D.; López-García, P. Ten reasons to exclude viruses from the tree of life. Nat. Rev. Microbiol. 2009, 7, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Aswad, A.; Katzourakis, A. Convergent capture of retroviral superantigens by mammalian herpesviruses. Nat. Commun. 2015, 6, 8299. [Google Scholar] [CrossRef] [PubMed]
- Cavalier-Smith, T. Obcells as proto-organisms: Membrane heredity, lithophosphorylation, and the origins of the genetic code, the first cells, and photosynthesis. J. Mol. Evol. 2001, 53, 555–595. [Google Scholar] [CrossRef] [PubMed]
- Fournier, G.P.; Andam, C.P.; Gogarten, J.P. Ancient horizontal gene transfer and the last common ancestors. BMC Evol. Biol. 2015, 15, 70. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Nei, M. Origin and evolution of influenza virus hemagglutinin genes. Mol. Biol. Evol. 2002, 19, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Fu, Y.; Jiang, D.; Li, G.; Xie, J.; Cheng, J.; Peng, Y.; Ghabrial, S.A.; Yi, X. Widespread horizontal gene transfer from double-stranded RNA viruses to eukaryotic nuclear genomes. J. Virol. 2010, 84, 11876–11887. [Google Scholar] [CrossRef] [PubMed]
- Crisp, A.; Boschetti, C.; Perry, M.; Tunnacliffe, A.; Micklem, G. Expression of multiple horizontally acquired genes is a hallmark of both vertebrate and invertebrate genomes. Genome Biol. 2015, 16, 50. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, A.J. bZIP proteins of human gammaherpesviruses. J. Gen. Virol. 2003, 84, 1941–1949. [Google Scholar] [CrossRef] [PubMed]
- Soucy, S.M.; Huang, J.; Gogarten, J.P. Horizontal gene transfer: Building the web of life. Nat. Rev. Genet. 2015, 16, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Sykiotis, G.P.; Bohmann, D. Stress-activated cap’n’collar transcription factors in aging and human disease. Sci. Signal. 2010, 3, re3. [Google Scholar] [CrossRef] [PubMed]
- Blank, V. Small Maf proteins in mammalian gene control: Mere dimerization partners or dynamic transcriptional regulators? J. Mol. Biol. 2008, 376, 913–925. [Google Scholar] [CrossRef] [PubMed]
- Jeyapaul, J.; Jaiswal, A.K. Nrf2 and c-Jun regulation of antioxidant response element (ARE)-mediated expression and induction of gamma-glutamylcysteine synthetase heavy subunit gene. Biochem. Pharmacol. 2000, 59, 1433–1439. [Google Scholar] [CrossRef]
- Giudice, A.; Montella, M. Activation of the Nrf2-ARE signaling pathway: A promising strategy in cancer prevention. Bioessays 2006, 28, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Lombard, J.; López-García, P.; Moreira, D. The early evolution of lipid membranes and the three domains of life. Nat. Rev. Microbiol. 2012, 10, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.S.; Goldstein, J.L. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc. Natl. Acad. Sci. USA 1999, 96, 11041–11048. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Chen, X.; Hendershot, L.; Prywes, R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell 2002, 3, 99–111. [Google Scholar] [CrossRef]
- Nadanaka, S.; Yoshida, H.; Sato, R.; Mori, K. Analysis of ATF6 activation in Site-2 protease-deficient Chinese hamster ovary cells. Cell Struct. Funct. 2006, 31, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Slusky, J.S.; Berger, B.W.; Walters, R.S.; Vilaire, G.; Litvinov, R.I.; Lear, J.D.; Caputo, G.A.; Bennett, J.S.; Degrado, W.F. Computational design of peptides that target transmembrane helices. Science 2007, 315, 1817–1822. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Matsui, T.; Yamamoto, A.; Okada, T.; Mori, K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 2001, 107, 881–891. [Google Scholar] [CrossRef]
- Pinney, J.W.; Amoutzias, G.D.; Rattray, M.; Robertson, D.L. Reconstruction of ancestral protein interaction networks for the bZIP transcription factors. Proc. Natl. Acad. Sci. USA 2007, 104, 20449–20453. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Martínez, J.A.; Reinke, A.W.; Bhimsaria, D.; Keating, A.E.; Ansari, A.Z. Combinatorial bZIP dimers display complex DNA-binding specificity landscapes. Elife Sci. 2017, 6, e19272. [Google Scholar] [CrossRef] [PubMed]
- Yuan, R.; Zhu, X.; Wang, G.; Li, S.; Ao, P. Cancer as robust intrinsic state shaped by evolution: A key issues review. Rep. Prog. Phys. 2017, 80, 042701. [Google Scholar] [CrossRef] [PubMed]
- McMahon, M.; Thomas, N.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J. Biol. Chem. 2004, 279, 31556–31567. [Google Scholar] [CrossRef] [PubMed]
- Chowdhry, S.; Zhang, Y.; McMahon, M.; Sutherland, C.; Cuadrado, A.; Hayes, J.D. Nrf2 is controlled by two distinct β-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene 2013, 32, 3765–3781. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, Y.; Morita, T.; Kim, M.; Iemura, S.; Natsume, T.; Yamamoto, M.; Kobayashi, A. Dual regulation of the transcriptional activity of Nrf1 by beta-TrCP- and Hrd1-dependent degradation mechanisms. Mol. Cell. Biol. 2011, 31, 4500–4512. [Google Scholar] [CrossRef] [PubMed]
- Gasiorek, J.J.; Blank, V. Regulation and function of the NFE2 transcription factor in hematopoietic and non-hematopoietic cells. Cell. Mol. Life Sci. 2015, 72, 2323–2335. [Google Scholar] [CrossRef] [PubMed]
- Chevillard, G.; Blank, V. NFE2L3 (NRF3): The Cinderella of the Cap’n’Collar transcription factors. Cell. Mol. Life Sci. 2011, 68, 3337–3348. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Kobayashi, A.; Yamamoto, M.; Hayes, J.D. The Nrf3 transcription factor is a membrane-bound glycoprotein targeted to the endoplasmic reticulum through its N-terminal homology box 1 sequence. J. Biol. Chem. 2009, 284, 3195–3210. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Letunic, I.; Okuda, S.; Kanehisa, M.; Bork, P. iPath2.0: Interactive pathway explorer. Nucleic Acids Res. 2011, 39, W412–W415. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qiu, L.; Li, S.; Xiang, Y.; Chen, J.; Ren, Y. The C-terminal domain of Nrf1 negatively regulates the full-length CNC-bZIP factor and its shorter isoform LCR-F1/Nrf1β; both are also inhibited by the small dominant-negative Nrf1γ/δ isoforms that down-regulate ARE-battery gene expression. PLoS ONE 2014, 9, e109159. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.-P.; Wang, M.; Xiang, Y.; Qiu, L.; Hu, S.; Zhang, Z.; Mattjus, P.; Zhu, X.; Zhang, Y. Nach Is a Novel Subgroup at an Early Evolutionary Stage of the CNC-bZIP Subfamily Transcription Factors from the Marine Bacteria to Humans. Int. J. Mol. Sci. 2018, 19, 2927. https://doi.org/10.3390/ijms19102927
Zhu Y-P, Wang M, Xiang Y, Qiu L, Hu S, Zhang Z, Mattjus P, Zhu X, Zhang Y. Nach Is a Novel Subgroup at an Early Evolutionary Stage of the CNC-bZIP Subfamily Transcription Factors from the Marine Bacteria to Humans. International Journal of Molecular Sciences. 2018; 19(10):2927. https://doi.org/10.3390/ijms19102927
Chicago/Turabian StyleZhu, Yu-Ping, Meng Wang, Yuancai Xiang, Lu Qiu, Shaofan Hu, Zhengwen Zhang, Peter Mattjus, Xiaomei Zhu, and Yiguo Zhang. 2018. "Nach Is a Novel Subgroup at an Early Evolutionary Stage of the CNC-bZIP Subfamily Transcription Factors from the Marine Bacteria to Humans" International Journal of Molecular Sciences 19, no. 10: 2927. https://doi.org/10.3390/ijms19102927
APA StyleZhu, Y.-P., Wang, M., Xiang, Y., Qiu, L., Hu, S., Zhang, Z., Mattjus, P., Zhu, X., & Zhang, Y. (2018). Nach Is a Novel Subgroup at an Early Evolutionary Stage of the CNC-bZIP Subfamily Transcription Factors from the Marine Bacteria to Humans. International Journal of Molecular Sciences, 19(10), 2927. https://doi.org/10.3390/ijms19102927





